Abstract
Please cite this paper as: Phungoen et al. (2011) Clinical factors predictive of PCR positive in pandemic H1N1 2009 influenza virus infection. Influenza and Other Respiratory Viruses 5(6), e558–e562.
Objective Pandemic H1N1 2009 influenza virus (H1N1) has been spreading globally. Clinical features might be predictive and may be different among countries. Even though the PCR test is a confirmatory test for this viral infection, it is expensive and limited in most Thai health care facilities. We studied predictive factors of PCR positive in H1N1 suspected patients.
Methods Consecutive patients who had influenza‐like illness less than seven days and had been tested for H1N1 by the real‐time PCR method between May and July 2009 were enrolled. Clinical data was collected and compared between those who had positive and negative PCR tests.
Results There were 6494 patients had flu‐like symptoms. Of those, 166 patients were done PCR test and 75 patients (45·18%) had positive PCR test. There were four predictors for positive PCR test including history of contact with confirmed H1N1 patients, headache, body temperature, and coryza with the adjusted odds ratio (95% confidence interval) of 2·84 (1·09–7·40), 6·25 (1·42–27·49), 1·69 (1·08–2·66), and 0·31 (0·12–0·79), respectively.
Conclusions Clinical factors can be both suggestive and protective factors for H1N1 infection. These factors may be helpful in clinical practice to assess the possibility of the H1N1 infection in people who are at risk; particularly in resource‐limited health care facilities.
Keywords: Clinical factors, H1N1, polymerase chain reaction, predictive
Introduction
Pandemic H1N1 2009 influenza has been spreading globally since June 11, 2009. Our hospital is a tertiary care hospital located in the northeastern part of Thailand. We have been monitored for the spread of this virus. The first polymerase chain reaction (PCR)‐confirmed patient with pandemic H1N1 2009 influenza was recorded on June 29, 2009, only 18 days after the pandemic announcement. Consequently, the public grew alert of the virus and even reached a state of panic in some instances. People wanted to be tested for the pandemic H1N1 2009 influenza virus infection, which may cause a large economic burden.
Clinical manifestations of pandemic H1N1 2009 influenza infection are different between countries. 1 , 2 Gastrointestinal symptoms such as nausea, vomiting, or diarrhea are prominent in the United States, 3 and headache is a dominant symptom in the U.K. 4 On the other hand, both gastrointestinal symptoms and headache are not common in a report from China. 1 Patients from Poland may have high numbers of neurological manifestations. 5
The PCR is used as a confirmatory test for pandemic H1N1 2009 influenza infection. The available PCR for pandemic H1N1 influenza is expensive in Thailand. During the outbreak, most suspected patients want to know if they are infected or not. If most suspected patients wish to be tested within a short time frame, this spike in test demand can cause a large economic burden in Thailand. The Ministry of Public Health set up the criteria for PCR test. However, the availability of the PCR test is limited. This study aims to examine the clinical features of the pandemic H1N1 influenza virus in Thailand and to find the clinical factors that are predictive for positive PCR tests in pandemic H1N1 2009 influenza suspected patients. Therefore, clinicians in resource‐limited facilities may be able to treat persons who are at risk properly.
Methods
We enrolled consecutive patients who had influenza‐like illness (ILI) for <7 days and had been tested for pandemic H1N1 2009 influenza at Srinagarind hospital, Khon Kaen University, between May and July 2009 (Figure 1). Influenza‐like illness was defined as fever (temperature of 37·8°C or greater) with cough or sore throat. The Ministry of Public Health in Thailand launched a clinical guideline for pandemic H1N1 2009 influenza infection in May 2009. The indications of PCR testing in suspicious cases are the high‐risk group, suspicion of pneumonia, or need for hospital admission.
Figure 1.
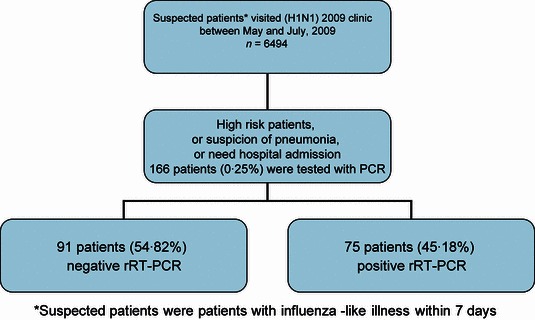
Flow chart of enrollment process.
High‐risk patients were patients with one of the following features: extreme age (<2 or more than 65 years), pregnancy, body mass index more than 25 kg/m2, chronic diseases such as asthma, chronic obstructive pulmonary disease, cardiovascular diseases, diabetes mellitus, thalassemia, or immunosuppressive conditions, such as HIV infection, cancer, and systemic lupus erythematosus.
Pneumonia is suspected if the patients have any one of the following symptoms or signs, including tachypnea (respiratory rate more than 60/minutes if age <2 months; 50/minutes if age 2–12 months; 40/minutes if age 1–5 years; 30/minutes if age 5–10 years; 24/minutes if age more than 10 years), dyspnea/chest pain, lung crepitation or consolidation signs upon physical examination, or oxygen saturation at room air being <95%.
Need for hospital admission is indicated if the patients need intravenous fluid therapy, electrolyte therapy, oxygen therapy, or close monitoring. If they are in the high‐risk group and are not able to follow‐up in the outpatient fashion, this is also an indication for hospital admission.
The PCR test for pandemic H1N1 2009 influenza infection was performed by using the commercial kit by Roche Applied Science, Germany (Real‐time ready Inf A/H1N1 Detection Set). The specimen for this kit is a nasopharyngeal swab. The study protocol was approved by the Khon Kaen University’s ethics committee for human research.
Clinical data were collected from the pandemic H1N1 2009 influenza report form. Descriptive statistics were used to compare clinical factors between those who had PCR positive and negative for pandemic H1N1 2009 influenza. All variables with P < 0·20 in the univariate analysis were included in subsequent multivariate logistic regression analyses. All variables with P > 0·20 in the multivariate model were excluded with the stepwise approach, whereas those with P < 0·10 were retained in the final model. Odds ratio (OR) was the risk of having PCR‐positive results compared to those with PCR‐negative results in patients with high‐risk pandemic H1N1 2009 influenza suspected. Analytical results were presented as crude ORs, adjusted ORs, and 95% confidence intervals (CIs). The goodness‐of‐fit of the final model was evaluated using Hosmer–Lemeshow statistics. 6 To evaluate the discriminatory power or accuracy of the model, c statistics or the area under the receiver that was operating characteristic curve was examined. 7 Data analyses were performed with sas software version 9.1 (Cary, NC, USA) and spss software version 17.0 (Chicago, IL, USA) on personal computer.
Results
There were 6494 patients with ILI who were attending the pandemic H1N1 2009 influenza clinic during the study period. Of those, 166 patients gave a nasopharyngeal swab for the pandemic H1N1 2009 influenza PCR test. There were 75 patients (45·18%) with positive PCR tests. Baseline characteristics, symptoms, signs, and laboratory results between those who had positive and negative PCR tests are shown in Table 1. The PCR‐positive group had more patients with a history of contact with the laboratory‐confirmed patients with pandemic H1N1 2009 influenza, body ache, headache, conjunctivitis, and higher BMI; however, the group had fewer patients with coryza and low platelet count.
Table 1.
Baseline characteristics and clinical factors of PCR negative and positive for pandemic H1N1 2009 influenza virus infection
| Variables | PCR negative n = 91 | PCR positive n = 75 | P value |
|---|---|---|---|
| Mean age (SD), years | 28·74 (21·10) | 24·71 (14·57) | 0·149 |
| Male, n (%) | 46 (50·55) | 30 (40·00) | 0·175 |
| Medical personnel, n | 17 (18·89) | 7 (9·33) | 0·083 |
| High risk for H1N1, n | 28 (31·11) | 17 (22·67) | 0·225 |
| Mean body weight (SD), kg | 57·01 (18·73) | 59·51 (13·56) | 0·424 |
| Mean height (SD), m | 1·62 (0·11) | 1·61 (0·11) | 0·701 |
| Mean BMI (SD), kg/m2 | 21·75 (6·65) | 23·23 (4·46) | 0·226 |
| History of contact with H1N1*, n | 26 (28·81) | 31 (41·33) | 0·094 |
| Chill, n | 10 (10·99) | 5 (6·67) | 0·334 |
| Cough, n | 82 (90·11) | 69 (92·00) | 0·673 |
| Sore throat, n | 39 (42·86) | 33 (44·00) | 0·882 |
| Conjunctivitis, n | 1 (1·10) | 6 (8·00) | 0·047 |
| Coryza, n | 47 (51·65) | 27 (36·00) | 0·044 |
| Body ache, n | 37 (40·66) | 40 (53·33) | 0·103 |
| Headache, n | 15 (16·48) | 21 (28·00) | 0·073 |
| Chest pain, n | 4 (4·40) | 2 (2·67) | 0·691 |
| Dyspnea, n | 11 (12·09) | 12 (16·00) | 0·468 |
| Stomachache, n | 10 (10·99) | 7 (9·33) | 0·726 |
| Diarrhea, n | 9 (9·89) | 6 (8·00) | 0·673 |
| Body temperature, °C | 38·50 (1·14) | 38·81 (1·00) | 0·076 |
| Pulse rate, beats/minutes | 103·09 (12·09) | 103·70 (12·21) | 0·750 |
| Respiratory rate, times/minutes | 26·08 (10·11) | 23·53 (6·82) | 0·061 |
| Duration of fever, days | 2·57 (1·52) | 2·63 (1·35) | 0·807 |
| Blood test | |||
| Mean total white blood count, cells/mm3 (SD) | 8857·59 (4770·56) | 8020·59 (4210·10) | 0·337 |
| Leukopenia, n | 7 (12·07) | 6 (11·76) | 0·961 |
| Leukocytosis, n | 16 (27·59) | 12 (23·53) | 0·629 |
| Band, % | 3·43 (4·33) | 3·21 (4·25) | 0·749 |
| PMN, % | 45·29 (33·05) | 51·27 (33·43) | 0·251 |
| Lymphocytes, % | 18·76 (18·41) | 22·50 (71·85) | 0·664 |
| Platelet count 100 000–144 000/μl, n | 12 (20·69) | 3 (6·00) | 0·028 |
| Platelet count <100 000/μl, n | 5 (8·62) | 3 (6·00) | 0·722 |
PCR, polymerase chain reaction.
Data presented in numbers, n (%) unless indicated. Total number in PCR‐negative and PCR‐positive group may not equal 91 and 75, respectively because of missing data. *indicating history of contact with laboratory‐confirmed patients with H1N1.
There were five factors remaining in the final model predictive for positive PCR test, including history of contact with patients with pandemic H1N1 2009 influenza, headache, body temperature, coryza, and platelet count between 100 000 and 144 000/μl. Only the first four factors were statistically significant. The adjusted odds ratio (95% CI) for history of contact with patients with pandemic H1N1 2009 influenza, headache, body temperature, and coryza were 2·84 (1·09–7·40), 6·25 (1·42–27·49), 1·69 (1·08–2·66), and 0·31 (0·12–0·79), respectively (Table 2). For the final model, the Hosmer–Lemeshow value and the c value were 12·96 (P = 0·11) and 0·78, respectively.
Table 2.
Results of multiple logistic regression analysis showing independent variables for polymerase chain reaction‐positive pandemic H1N1 2009 influenza infection and their adjusted odds ratio (OR) and 95% confidence interval (95% CI)
| Variables | Adjusted OR | 95% CI |
|---|---|---|
| History of contact with H1N1 | 2·84 | 1·09–7·40 |
| Headache | 6·25 | 1·42–27·49 |
| Body temperature | 1·69 | 1·08–2·66 |
| Coryza | 0·31 | 0·12–0·79 |
The body temperature of more than 38°C gave the sensitivity and specificity of having PCR‐positive results in suspected patients of 79·45% and 32·58%, respectively. Various cut‐off points by body temperature are shown in Table 3.
Table 3.
Sensitivities and specificities of various body temperature cut‐off points to predict the positive polymerase chain reaction results in patients with pandemic H1N1 2009 influenza virus suspected
| Cut‐off points (°C) | Sensitivity (%) | Specificity (%) |
|---|---|---|
| 38·05 | 79·42 | 32·58 |
| 39·05 | 42·46 | 66·29 |
| 39·95 | 12·33 | 88·76 |
Discussion
The predictors for pandemic H1N1 2009 influenza may be different across the world’s continents. As has been previously reported, gastrointestinal symptoms are prominent and suggesting for the pandemic H1N1 2009 influenza infection. 1 , 2 , 3 In the present study, clinical manifestations of the pandemic H1N1 2009 influenza virus infection are similar to the report from China, 1 but different from U.S. and U.K reports. In addition, there are both positive and negative clinical predictors for pandemic H1N1 2009 influenza infection. History of contact with confirmed patients of pandemic H1N1 influenza, headache, and high body temperature were positively associated with the positive PCR results; whereas coryza was a negative predictor.
The history of contact with confirmed patients of pandemic H1N1 2009 influenza increases the risk of being infected 2·84 times. This result indicated that the pandemic H1N1 2009 influenza infection is extremely contagious during the outbreak. Clinical factors suggestive for pandemic H1N1 2009 influenza infection are related to systemic responses such as headache, body ache, conjunctivitis, and high body temperature. After all symptoms were adjusted by the multiple logistic model, only headache and high body temperature are significantly related to positive PCR results (Table 2).
Having headache increases the risk of having pandemic H1N1 2009 influenza infection by 6·25 times, whereas every 1°C increase in body temperature increases risk by 69%. The pandemic H1N1 2009 influenza virus is a novel antigen and may cause more fever and headache. Coryza, on the other hand, is a protective factor for pandemic H1N1 2009 influenza infection and suggestive of upper respiratory tract infection caused by other viruses.
The cut‐off points of body temperature are another useful tool for predicting the possibility of having a pandemic H1N1 influenza virus infection. If suspected patients have body temperatures more than 38°C, the possibility of having the pandemic H1N1 influenza virus infection is 79·45% with the specificity of 32·58%. However, this prediction should be used with three other significant predictors: the history of contact with patients with pandemic H1N1 2009 influenza, headache, and coryza.
The results of the present study will apply only to those who are tested for pandemic H1N1 2009 influenza by the PCR test. Not all suspected patients were enrolled, only patients who were high risk or have severe conditions. The number of patients who had been tested for the pandemic H1N1 2009 influenza infection was quite low because of the strict criteria given by the guideline. The cost of a PCR test for pandemic H1N1 2009 influenza was expensive. Therefore, a large number of suspected patients in the outbreak received empirical treatment with oseltamivir. Goodness‐of‐fit statistics indicated a good fit to the model. The c statistic value or area under the ROC curve value of 0·78 showed good discrimination of a random pair of patients with negative and positive PCR results.
Conflict of interest
None declared. All authors are government employees.
Acknowledgement
This research was supported by the Office of the Higher Education Commission, the National Research Council of Thailand, and the National Research Project, Khon Kaen University, Thailand.
References
- 1. Cao B, Li XW, Mao Y et al. , National Influenza A Pandemic (H1N1) 2009 Clinical Investigation Group of China . Clinical features of the initial cases of 2009 pandemic influenza A (H1N1) virus infection in China. N Engl J Med 2009; 361:2507–2517. [DOI] [PubMed] [Google Scholar]
- 2. Centers for Disease Control and Prevention . Swine‐origin influenza A (H1N1) virus infections in a school – New York City, April 2009. MMWR Morb Mortal Wkly Rep 2009; 58:470–472. [PubMed] [Google Scholar]
- 3. Novel Swine‐Origin Influenza A (H1N1) Virus Investigation Team . Emergence of a novel swine‐origin influenza A (H1N1) virus in humans. N Engl J Med 2009; 360:2605–2615. [DOI] [PubMed] [Google Scholar]
- 4. Health Protection Agency and Health Protection Scotland new influenza A(H1N1) investigation teams . Epidemiology of new influenza A (H1N1) in the United Kingdom, April–May 2009. Eurosurveillance, 14(9):1–2. Available at http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19213 (accessed 30 May 2010). [DOI] [PubMed] [Google Scholar]
- 5. Cholewińska G, Higersberger J, Podlasin R et al. Clinical manifestations, diagnosis and treatment of swine flu (A/H1N1) infection among patients hospitalized in the Hospital of Infectious Diseases in Warsaw in 2009. Przegl Epidemiol 2010; 64:15–19. [PubMed] [Google Scholar]
- 6. Hosmer DW, Hosmer T, Le Cessie S, Lemeshow S. A comparison of goodness‐of‐fit tests for the logistic regression model. Stat Med 1997; 16:965–980. [DOI] [PubMed] [Google Scholar]
- 7. Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982; 143:29–36. [DOI] [PubMed] [Google Scholar]


