Abstract
MicroRNA (miR)-31 serves a key role in various biological processes, including tumor development, angiogenesis and inflammation. Whether miR-31 is involved in the pathological processes of arteriosclerosis obliterans (ASO) remains to be elucidated, as does the mechanism of miR-31 regulation of arterial smooth muscle cells (ASMCs). In the present study, miR-31 expression was detected by reverse transcription-quantitative polymerase chain reaction and in situ hybridization, and was significantly upregulated in human ASO arterial walls compared with normal arterial walls (P<0.001). In addition, miR-31 proliferation was detected by Cell Counting Kit-8 and EdU assays; proliferation was significantly promoted in platelet-derived growth factor (PDGF)-BB-induced human ASMCs (HASMCs) (P<0.001). miR-31 migration was detected by transwell and wound closure assays, and was revealed to be promoted in PDGF-BB-induced HASMCs (P<0.001). Lastly, HASMCs were transfected with miR-31 mimics and inhibitors, and negative controls. A dual-luciferase reporter assay was performed to verify that mitofusin-2 (MFN2) was a direct target of miR-31 and that MFN2 expression was significantly downregulated by miR-31 at a post-transcriptional level in HASMCs as detected by western blotting (P<0.01). These findings suggest that miR-31 is able to promote the proliferation and migration of HASMCs, at least in part, by targeting MFN2. The results of the present study provide novel insight into the underlying mechanisms and roles of miR-31/MFN2 in the pathology of ASO, which may offer a potential therapeutic target for the treatment of ASO.
Keywords: microRNA-31, arteriosclerosis obliterans, arterial smooth muscle cell, proliferation, migration, mitofusin-2
Introduction
Atherosclerosis is a chronic, progressive systemic disease affecting arteries, particularly those in the brain, heart, and lower extremities. Arteriosclerosis obliterans (ASO) of the lower extremities is one of the most significant health-related issues worldwide and may have devastating consequences, including intermittent claudication, resting pain and gangrene (1–3). Notable progress has been made in endovascular surgical techniques to treat ASO; however, restenosis following angioplasty remains a primary limiting factor for the long-term success of artery reconstruction (4). Vascular smooth muscle cells (VSMCs) comprise the main component of blood vessel walls, and excessive proliferation and migration of VSMCs is a critical event in the pathogenesis of atherosclerosis and vascular stenosis (5,6). It is unclear whether the molecular mechanism underlying the proliferation and migration of human arterial smooth muscle cells (HASMCs) is associated with ASO.
MicroRNAs (miRNAs or miRs) have previously been shown to serve important roles in cardiovascular systems (7–9). miRNAs comprise a class of non-coding, short (18–22 nucleotides), endogenous RNAs that negatively regulate their targets through binding of the 3′-untransslated region (3′-UTR) at a post-translational level. Aberrant miRNA expression serves a key role in the development of ASO (10). There have been notable breakthroughs in studies of gene expression regulation in recent decades, and so miRNAs have been investigated for their functions in VSMC differentiation, proliferation, migration and apoptosis (11,12). A previous study by the present authors demonstrated that miR-21 promoted HASMC proliferation and migration, and inhibited apoptosis in ASO of the lower extremities (11). Another study revealed that miR-663 regulated the human VSMC phenotypic switch and vascular neointimal formation by targeting JunB/myosin light chain 9 expression, which suggests that targeting miR-663 or its targets may provide a promising approach for the treatment of proliferative vascular diseases (12).
Although miR-31 is involved in various biological processes, including tumor development (13–16), angiogenesis (17,18) and inflammation (19), the role of miR-31 in ASO, particularly the regulatory mechanism, remains elusive. The present study aimed to elucidate the potential roles of miR-31 in HASMCs within ASO processes, and to identify the underlying molecular mechanism.
Materials and methods
Source of artery specimens
ASO artery specimens were obtained from 6 patients with ASO (4 males and 2 females) with an average age of 65.3 years who had undergone major amputations, and normal artery specimens were acquired from 6 healthy organ donors (3 males and 3 females) with an average age of 55.1 years. Specimens were obtained between June 2013 and December 2014. ASO artery specimens were used for reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analyses, in situ hybridization (ISH), immunofluorescence, and 4′,6-diamidino-2-phenylindole (DAPI) and haematoxylin and eosin (H&E) staining. Artery specimens were harvested in a sterile state and some specimens were snap-frozen in liquid nitrogen immediately after arteriectomy and stored at −80°C for RNA extraction, while other specimens were fixed with paraformaldehyde (4%) at 4°C overnight and embedded in paraffin for further analysis. The present study was approved by the research ethics committee of the First Affiliated Hospital, Sun Yat-Sen University (Guangzhou, China) and prior informed consent was provided by the patients.
RT-qPCR analysis
The separation of three layers of artery samples was performed using a light microscope with anatomical lens (magnification, ×5), micro scissors and micro forceps (both Shanghai Medical Instruments (Group) Ltd., Corp. Surgical Instruments Factory, Shanghai, China). The normal tissues or untreated cells were used as control. Total RNA was extracted from arterial specimens or cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., USA), according to the manufacturer's protocol. RNA was reverse transcribed using a miRNA RT kit (Takara Biotechnology Co., Ltd., Dalian, China) at 37°C for 60 min, then 85°C for 5 sec. The cDNA was used template with the SYBR primeScript miRNA Real-Time PCR kit (Takara Biotechnology Co., Ltd.) for qPCR analysis, according to the manufacturer's protocol with the following primers: miR-31 5′-CTACGTTCTGGCATAGCTGAAA-3′, U6 5′-ACGCAAATTCGTGAAGCGTT-3′. The volume of the total PCR reaction was 25 µl and the contained the following: ddH2O (10.5 µl), miRNA Primer Mix (1 µl), cDNA (1 µl), 2X SYBR® Premix Ex Taq™ II (12.5 µl). PCR was performed using a Roche Lightcycler 480 Real-Time PCR System (Roche Diagnostics, Basel, Switzerland) under the following conditions: 95°C for 10 sec, followed by 40 cycles at 95°C for 5 sec and 60°C for 20 sec. U6 was used as a reference gene. Relative expression was analyzed using the 2−∆∆Cq method (20). All experiments were independent and performed at least in triplicate.
ISH, immunofluorescence, and DAPI and H&E staining
ISH was performed using a 5′-Digoxigenin (DIG)- and 3′-DIG-labeled miRCURY LNA Detection Probe (Exiqon A/S, Vedbaek, Denmark) to human miR-31 (5′-AGCTATGCCAGCATCTTGCCT-3′) in 4-µm-thick sections of tissues. The tissues were fixed with 4% paraformaldehyde at 4°C overnight, and embedded in paraffin then stored at room temperature for further analysis. Briefly, the sections were deparaffinized and rehydrated in graded dilutions of ethanol (100, 95, 75 and 60%, digested with 40 µg/ml proteinase K (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 10 min, rinsed in 0.2% glycine/PBS for 5 min, fixed with 4% paraformaldehyde at room temperature for 10 min, and acetylated for 10 min. Sections were subsequently prehybridized in a hybridization buffer [50X Denhardts's solution, 60% formamide, 300 µg/ml transfer RNA, 20X Saline Sodium Citrate (SSC) Buffer and 1 M dithiothreitol (Invitrogen; Thermo Fisher Scientific, Inc.)] at 49°C for 30 min and then hybridized with the miR-31 probe (1:500) at 49.5°C overnight. Sections were subsequently washed with 2X SSC for 5 min at 49.5°C and then blocked for 1 h at room temperature using 2% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and incubated with anti-DIG antibodies (1:200; cat. no. 11093274910; Roche Diagnostics) at 37°C for 2.5 h. Sections were incubated with tyramide signal amplification buffer (1:50; Focofish, Inc., Guangzhou, China) at room temperature for 20 min in the dark, washed with PBS and covered with a cover slip. Immunofluorescence was performed to determine SM-α-actin localization using anti-SM-α-actin primary antibodies (1:500; cat. no. ab5694; Abcam, Cambridge, MA, USA) at 37°C for 2 h (11) and Alexa Fluor™ 488-conjugated secondary antibodies (1:1,000; cat. no. A12379; Thermo Fisher Scientific, Inc.) at room temperature for 30 min.
DAPI staining was used as a nuclear counterstain in fluorescence microscope. Following the fluorescence staining of SM-α-actin, DAPI staining was performed using a SlowFade™ Diamond Antifade Mountant with DAPI reagent (cat. no. 36968; Invitrogen; Thermo Fisher Scientific, Inc.) in of tissue sections at room temperature overnight in the dark. Images of the sections were captured by an inverted fluorescence microscope (magnification, ×100; Axio Observer Z1, Carl Zeiss AG, Oberkochen, Germany) and analyzed by calculating the integrated optical density (IOD) value using Image Pro Plus software (version 6.0, Media Cybernetics, Inc., Rockville, MD, USA).
H&E staining was used to demonstrate arterial structures. The tissue sections were deparaffinized and rehydrated in graded dilutions of ethanol (100, 100, 95 and 75%) for 5 min each. The slides were the incubated with hematoxylin solution for 20 sec, then washed the slides with H2O for 1 min. The slides were then stained with 1% eosin solution for 3 min and dehydrated with graded dilutions of ethanol (75, 95, 100 and 100%) for 1 min each. Alcohol was extracted with two changes of xylene and covered with a cover slip. All of the above steps were performed at room temperature. Images of H&E sections were observed by a light microscope (magnification, ×100).
Western blotting
The HASMCs harvested from the femoral arterial walls of healthy organ donors were lysed at 0°C for 15 min in a lysis buffer (cat. no. 9806S; Cell Signaling Technology, Inc., Danvers, MA, USA) supplemented with a protease inhibitor cocktail (cat. no. 04693132001; Roche Diagnostics) and centrifuged in 12,500 × g at 4°C for 15 min. The proteins (20 µg/lane) were separated by 10% SDS-PAGE and transferred onto a polyvinylidene difluoride membrane (EMD Millipore, Billerica, MA, USA). The membranes were blocked with 5% non-fat milk in TBS at 37°C for 2 h, incubated with a rabbit anti-mitofusin-2 (MFN2) monoclonal antibody (1:1,000; cat. no. ab50843; Abcam) at 4°C overnight and washed with TBST at room temperature, three times for 10 min. The signals were detected using Luminol reagent (Pierce; Thermo Fisher Scientific, Inc.), imaged using a GE ImageQuant Las 4000 mini phosphorimager (GE Healthcare Life Sciences, Chalfont, UK) with ImageJ (version 1.48; National Institutes of Health, Bethesda, MD, USA) and presented as the density ratio vs. GAPDH (1:1,000; cat. no. 2118S; Cell Signaling Technology, Inc.). Densitometric analyses were taken as an average of three experiments.
Cell culture
The primary HASMCs were obtained from the femoral arterial walls of amputation from patients who suffered serious trauma, and the cells were prepared by the explant method, previously established in our lab (11). HASMCs from healthy donors were used in all subsequent experiments and the ASO cell model was established by platelet-derived growth factor-BB (PDGF-BB) treatment. The HASMCs were identified via staining with anti-SM-α-actin antibodies at 37°C for 2 h (1:1,000; cat. no. ab5694; Abcam), and passages from 4 to 9 were utilized in the present study. The cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS at 37°C in a humidified atmosphere containing 5% CO2, then the cells were added to a cell cryopreservation solution (10% dimethylsulphoxide and 90% FBS) and stored in liquid nitrogen.
HASMC transfection
Cells (2–3×103 cells/well) were seeded into wells and transfection was performed using Lipo RNA iMax (Invitrogen; Thermo Fisher Scientific, Inc.) 24 h later. For miR-31 function analysis, miR-31 mimics (50 nmol/l), miR-31 inhibitor (100 nmol/l) and negative control oligos (50 nmol/l; all Guangzhou RiboBio Co., Ltd., Guangzhou, China) were transfected into the cells, following the manufacturer's protocol.
Measurement of HASMC proliferation
Cell Counting Kit-8 (CCK-8) and EdU assays were performed to measure cell proliferation. HASMCs were seeded into 96-well plates (2–3×103 cells/well). Cells were incubated at 37°C in serum-free DMEM with or without 20 ng/ml PDGF-BB (R&D Systems, Inc., Minneapolis, MN, USA) for 24 h following transfection. For the CCK-8 (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) assay, 10 µl CCK-8 solution was added to each well, and the absorbance was measured at 450 nm following 3 h of incubation at 37°C.
For the EdU (Guangzhou RiboBio Co., Ltd.) assay, the cells were incubated at 37°C in EdU solution (50 nmol/l) for 2 h, and fixed at room temperature in 4% formaldehyde for 30 min. The cells were subsequently exposed to 1,000 µl Cell-Light™ EdU Apollo®488 In Vitro Imaging kit (100T) (938 µl dH2O, 50 µl reaction buffer, 10 µl catalyst solution, 3 µl fluorescent dye solution, 9 mg buffer additive) for 30 min, followed by nuclear staining at room temperature with Hoechst 33,342 for 30 min (all Guangzhou RiboBio Co., Ltd.). An inverted fluorescence microscope was used to capture and analyze the images (magnification, ×100; Axio Observer Z1).
Measurement of HASMC migration
Transwell and wound closure assays were used to assess cell migration. For the Transwell assay (Corning Incorporated, Corning, NY, USA), cells were resuspended following transfection and added to the upper chambers with 200 µl serum-free DMEM (5×105 cells/ml), and the lower chamber was filled with 500 µl serum-free DMEM with or without PDGF-BB (20 ng/ml). After 16 h, the migrated cells on the lower face of the chamber membrane were fixed with 4% formaldehyde and stained with 0.1% crystal violet.
For the wound closure assay, the HASMCs were seeded into 12-well plates following transfection (10,000 cells/well), and a straight scratch wound was created using a sterilized 200 µl disposable pipette tip in each well. The scratch wounds were visualized every hour at 37°C using a live cell imaging system (magnification, ×100; Axio Observer Z1). Image Pro Plus software (version 6.0) was used to measure the widths of the scratch wounds.
Luciferase reporter assay
The MFN2 mRNA 3′-UTR, containing putative or mutated binding sites for miR-31, were amplified by PCR using the aforementioned protocol and cloned into the pmiR-RB-Report vector (Guangzhou RiboBio Co., Ltd.). The HEK 293T cells (American Type Culture Collection, Manassas, VA, USA) were co-transfected with miR-31 mimics (50 nmol/l) or negative control oligos (50 nmol/l) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Luciferase values were determined 48 h later using the Dual-Luciferase Reporter Assay System kit (Promega Corporation, Madison, WI, USA), according to the manufacturer's protocol.
Statistical analysis
The data are presented as the mean ± standard deviation. Student's t-test and one-way analysis of variance, with multiple comparisons using the Newman-Keuls test, were used for the statistical analysis. The data analysis was performed with SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
miR-31 is upregulated in ASO arteries of the lower extremities
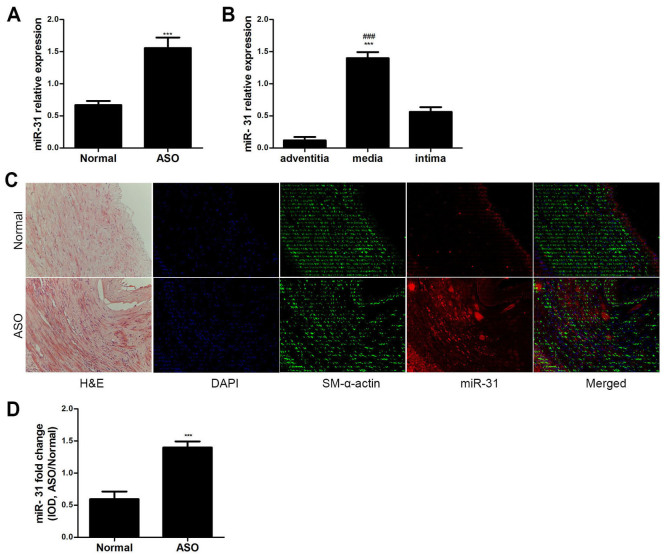
The miR-31 expression levels were analyzed in ASO and normal arterial walls from the lower extremities via RT-qPCR. miR-31 expression was significantly upregulated in the ASO arteries compared with the normal arteries (P<0.001; Fig. 1A). The expression level of miR-31 in the media was significantly higher compared with the adventitia and intima in ASO arteries (P<0.001; Fig. 1B). Furthermore, the ISH and immunofluorescence results demonstrated that miR-31 was primarily located in the artery media and neointima of ASO (Fig. 1C). The colocalization of miR-31 and SM-α-actin indicates that miR-31 is primarily expressed in ASO HASMCs. The IOD value also indicated that miR-31 expression was significantly increased in the ASO arteries compared with the normal arteries (P<0.001; Fig. 1D). These results suggest that miR-31 is upregulated in ASO HASMCs.
Figure 1.
Characteristics of miR-31 expression. (A) miR-31 expression in both ASO and normal arteries was determined by RT-qPCR (n=6). ***P<0.001 vs. normal group. (B) miR-31 expression in the three layers of the ASO artery wall was determined by RT-qPCR (n=6). ***P<0.001 vs. adventitia and ###P<0.001 vs. intima. (C) H&E staining revealed artery structures. Co-staining of miR-31 (red) and SM-α-actin (green) in artery sections. The ISH and immunofluorescence results showed that miR-31 was primarily localized in the arterial media and neointima. (D) IOD value of miR-31 staining in artery sections showed that miR-31 staining was significantly increased in ASO sections (n=6) compared with that in normal sections (n=6). Magnification, ×200. ***P<0.001 vs. normal group. miR, microRNA; ASO, arteriosclerosis obliterans; RT-qPCR; H&E, hematoxylin and eosin; SM, smooth muscle; IOD, integrated optical density; DAPI, 4′,6-diamidino-2-phenylindole; ISH, in situ hybridization.
Mir-31 promotes proliferation in PDGF-BB-induced HASMCs
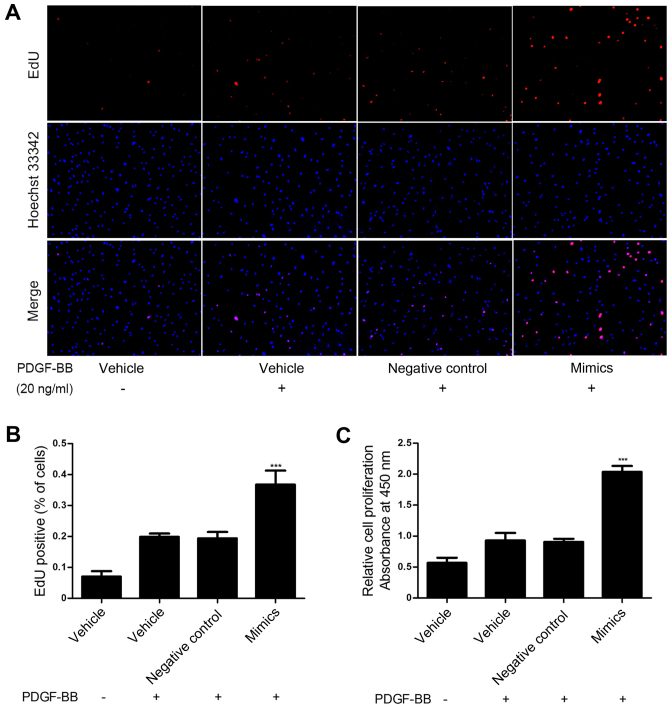
To investigate the role of miR-31 in HASMC proliferation induced by PDGF-BB, miR-31 mimics and negative control oligos were transfected into HASMCs and proliferation was assessed via an EdU and a CCK-8 assay. These assays demonstrated that upregulation of miR-31 significantly promoted HASMC proliferation compared with the control group (P<0.001; Fig. 2). These results suggest that miR-31 overexpression promotes HASMC proliferation in vitro.
Figure 2.
miR-31 promoted HASMC proliferation. HASMCs were transfected with miR-31 mimics (50 nmol/l) or negative control oligos (50 nmol/l). miR-31 mimic significantly increased PDGF-BB-induced (20 ng/ml) HASMC proliferation, as determined by (A and B) EdU assay and (C) Cell Counting Kit-8 assay. Magnification, ×100. ***P<0.001 vs. negative control. miR, microRNA; HASMC, human arterial smooth muscle cell; PDGF, platelet-derived growth factor.
Mir-31 promotes migration in HASMCs following PDGF-BB treatment
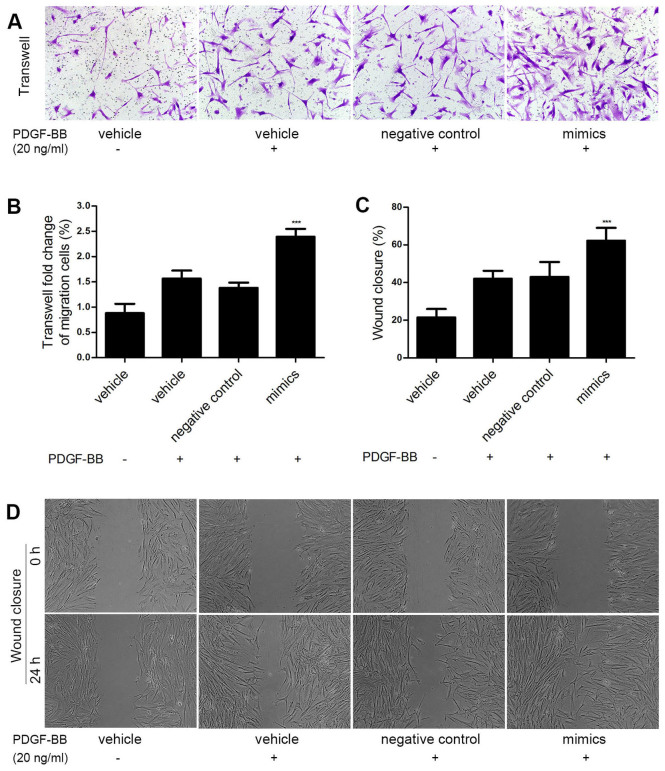
To explore whether miR-31 promoted HASMC migration induced by PDGF-BB, miR-31 mimics and negative control oligos were transfected into HASMCs to overexpress miR-31. Cell migration was measured via Transwell assay. As shown in Fig. 3A and B, the miR-31 mimics significantly enhanced PDGF BB-induced HASMC migration compared with the control group (P<0.001). Similar results were obtained in the wound closure assay (P<0.001; Fig. 3C and D). These results demonstrate that miR-31 overexpression promotes HASMC migration in vitro.
Figure 3.
miR-31 promoted HASMC migration. HASMCs were transfected with miR-31 mimics (50 nmol/l) or negative control oligos (50 nmol/l). miR-31 mimic markedly increased PDGF-BB-induced (20 ng/ml) HASMC migration, determined by the (A and B) Transwell assay and (C and D) wound closure assay. Magnification, ×100 (Transwell) or ×50 (wound closure). ***P<0.001 vs. negative control. miR, microRNA; HASMC, human arterial smooth muscle cell; PDGF, platelet-derived growth factor.
MiR-31 targets the 3′-UTR of MFN2 and downregulates its expression
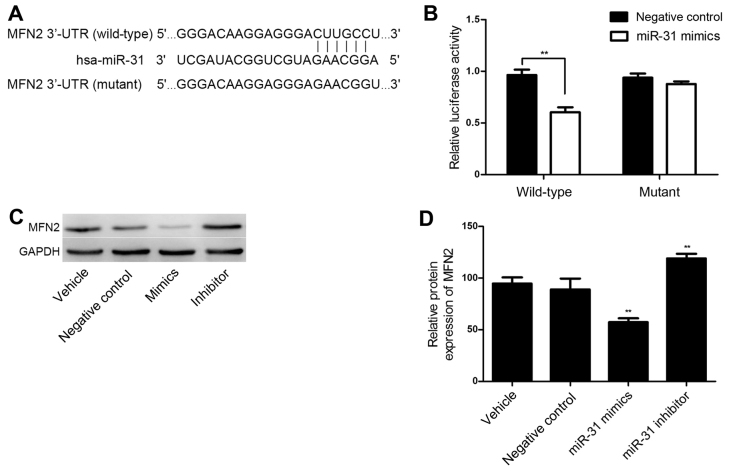
TargetScan online software was utilized for the bioinformatics prediction and MFN2 was identified as a potential target of miR-31. The putative seed sequences for miR-31 at the 3′-UTR of MFN2 were conserved (Fig. 4A). A dual-luciferase reporter assay was performed to verify whether MFN2 was a direct target gene of miR-31. As shown in Fig. 4B, in the wild-type MFN2 3′UTR group, the miR-31 mimics significantly reduced Renilla/firefly luciferase activity compared with the results in the negative control group (P<0.01). However, in the mutant type MFN2 $3′UTR group, there was no significant difference in Renilla/firefly luciferase activity between the miR-31 mimics and the negative control group. These results indicate that miR-31 directly binds to the 3′-UTR of MFN2.
Figure 4.
MFN2 was identified as a target of miR-31 in HASMCs. (A) Target sequences of miR-31 in 3′-UTR of the wild-type and mutant MFN2. (B) A luciferase reporter assay was used to determine whether MFN2 was a direct target of miR-31. miR-31 mimics significantly reduced the luciferase activity in the wild-type MFN2 compared with the negative control, whereas there was no significant difference between miR-31 and negative control transfection in the mutant MFN2. (C and D) Western blotting was performed to determine the levels of MFN2 protein in HASMCs transfected with miR-31 mimics (50 nmol/l), inhibitor (100 nmol/l) and negative control oligos (50 nmol/l). **P<0.01 vs. negative control. MFN2, mitofusin-2; miR, microRNA; HASMC, human arterial smooth muscle cell; UTR, untranslated region.
MFN2 expression is negatively regulated by miR-31 at a transcriptional level in HASMCs
To further investigate whether MFN2 is a functional target gene of miR-31 in HASMCs, HASMCs were transfected with miR-31 negative control oligos, mimics and an inhibitor, and the MFN2 expression levels were determined via western blotting. As shown in Fig. 4C and D, the protein level of MFN2 was significantly downregulated following miR-31 overexpression (P<0.01). However, MFN2 was significantly upregulated in the HASMCs transfected with a miR-31 inhibitor (P<0.01; Fig. 4C and D). These observations demonstrate that miR-31 negatively regulates the expression of MFN2 at a post-transcriptional level in HASMCs.
Discussion
It has been well established that miRNAs function as key mediators in multiple physiological and pathological processes, predominantly via modulating the expression of target genes at a post-transcriptional level (21). In the present study, miR-31 expression was markedly upregulated in human ASO arterial walls compared with normal arterial walls. Furthermore, an ASO cell model was established to investigate whether miR-31 overexpression had an effect on HASMCs. To the best of our knowledge, the present study demonstrated for the first time that miR-31 promotes proliferation and migration in PDGF-BB-induced HASMCs, most likely by regulating the expression of MFN2.
ASO of the lower extremities is one of the most painful vascular diseases, with high morbidity and mortality; ASO seriously affects survival and quality of life (22). It is well established that aberrant proliferation and migration of VSMCs serves an important role in arteriosclerosis and vascular stenosis (23). Despite great progress in the understanding of VSMC biology (24), the molecular mechanisms underlying the contribution of HASMCs to the pathogenesis of ASO has yet to be elucidated. miRNAs have previously been reported to serve important roles in cardiovascular systems (7–9). miR-31 functions as an oncogene in the majority of cancers, including colorectal cancer (13), lung cancer (14), and esophageal cancer (15); however, it has been demonstrated to act as a tumor suppressor in glioblastoma and liver cancer (16,25). In the present study, miR-31 was significantly upregulated in ASO arterial walls compared with normal arterial walls as determined by RT-qPCR and ISH. miR-31 was located primarily in the media and neointima, which indicates that HASMCs may be the main effector cells of miR-31 in the ASO pathological process. The characteristics of miR-31 distribution in he present study are consistent with the observations of a previous study, which was performed in rat carotid arteries with neointimal lesions (26).
The biological functions of miR-31 in the HASMC physiological and pathological processes require further study. To investigate the detailed roles of miR-31 in HASMCs in ASO, CCK-8 and EdU assays were performed to investigate cell proliferation, wound closure and Transwell assays were used to detect cell migration. In the present study, miR-31 was demonstrated to promote proliferation and migration in PDGF-BB-induced HASMCs, which was consistent with the observation of a previous study by Liu et al (27), who reported that VSMC proliferation in rats was significantly inhibited by miR-31 knockdown. Cell proliferation was increased by the overexpression of Rattus norvegicus-miR-31, which demonstrates that miR-31 has a pro-proliferative effect on VSMCs in rats. Wang et al (28) previously reported that miR-31 was involved in the regulation of a human thoracic ASMC phenotype switch by targeting the cellular repressor of E1A-stimulated genes.
MFN2, also known as a hyperplasia suppressor gene, has a crucial role in regulating cell proliferation, migration, apoptosis and differentiation, which are involved in the pathophysiological processes of severe cardiovascular diseases, including atherosclerosis, restenosis after angioplasty, hypertension and myocardial infarction (29–31). A previous study demonstrated that MFN2 acted as an anti-proliferation gene, by preventing VSMC proliferation in cultured cells and a rat carotid artery balloon-injury model (31). Another study revealed that overexpression of MFN2 reduced VSMC proliferation and increased apoptosis in human and experimental pulmonary arterial hypertension model (32). In the present study, a dual-luciferase reporter assay was applied to confirm that MFN2 was a direct target gene of miR-31. In addition, via gain-of-function and loss-of-function approaches, transfection with miR-31 mimics reduced the protein expression of MFN2 in HASMCs; however, transfection with an miR-31 inhibitor resulted in increased MFN2 expression. These previous observations and the results of the present study suggest that miR-31 has a role in promoting proliferation and migration in HASMCs, which is, at least in part, dependent on directly targeting MFN2.
In conclusion, miR-31 was significantly increased in arterial walls of patients with ASO. Furthermore, miR-31 promoted the proliferation and migration of HASMCs, at least partially by directly targeting the expression of MFN2. The results of the present study may provide a novel insight into the mechanisms and significant role of miR-31/MFN2 in the pathological processes of ASO and offers a potential therapeutic target for the treatment of ASO.
Acknowledgments
The present study was supported by the National Natural Science Foundation of China (nos. 81270378, 81070258 and 81370368), Guangdong Province Industry-Academia-Research Program (no. 2011B090400117), Guangdong Department of Science & Medicine Center grant (no. 2011A080300002) Guangdong Province Medical Science and Technology Research Project (no. A2016424) and Guangdong Science and Technology Plan Projects (no. 2017A020215124).
References
- 1.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, et al. Heart disease and stroke statistics-2013 update: A report from the American Heart Association. Circulation. 2013;127:e6–e245. doi: 10.1161/CIR.0b013e318282ab8f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohnishi H, Sawayama Y, Furusyo N, Maeda S, Tokunaga S, Hayashi J. Risk factors for and the prevalence of peripheral arterial disease and its relationship to carotid atherosclerosis: The kyushu and okinawa population study (KOPS) J Atheroscler Thromb. 2010;17:751–758. doi: 10.5551/jat.3731. [DOI] [PubMed] [Google Scholar]
- 3.Diehm C, Allenberg JR, Pittrow D, Mahn M, Tepohl G, Haberl RL, Darius H, Burghaus I, Trampisch HJ, German Epidemiological Trial on Ankle Brachial Index Study Group Mortality and vascular morbidity in older adults with asymptomatic versus symptomatic peripheral artery disease. Circulation. 2009;120:2053–2061. doi: 10.1161/CIRCULATIONAHA.109.865600. [DOI] [PubMed] [Google Scholar]
- 4.Setacci C, Castelli P, Chiesa R, Grego F, Simoni GA, Stella A, Galzerano G, Sirignano P, De Donato G, Setacci F. Restenosis: A challenge for vascular surgeon. J Cardiovasc Surg (Torino) 2012;53:735–746. [PubMed] [Google Scholar]
- 5.Hao H, Gabbiani G, Bochaton-Piallat ML. Arterial smooth muscle cell heterogeneity: Implications for atherosclerosis and restenosis development. Arterioscler Thromb Vasc Biol. 2003;23:1510–1520. doi: 10.1161/01.ATV.0000090130.85752.ED. [DOI] [PubMed] [Google Scholar]
- 6.Kim J, Zhang L, Peppel K, Wu JH, Zidar DA, Brian L, DeWire SM, Exum ST, Lefkowitz RJ, Freedman NJ. Beta-arrestins regulate atherosclerosis and neointimal hyperplasia by controlling smooth muscle cell proliferation and migration. Circ Res. 2008;103:70–79. doi: 10.1161/CIRCRESAHA.108.172338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 8.Condorelli G, Latronico MV, Cavarretta E. microRNAs in cardiovascular diseases: Current knowledge and the road ahead. J Am Coll Cardiol. 2014;63:2177–2187. doi: 10.1016/j.jacc.2014.01.050. [DOI] [PubMed] [Google Scholar]
- 9.Reilly S, Liu X, Carnicer R, Rajakumar T, Sayeed R, Krasopoulos G, Verheule S, Fulga T, Schotten U, Casadei B. Evaluation of the role of miR-31-dependent reduction in dystrophin and nNOS on atrial-fibrillation-induced electrical remodelling in man. Lancet. 2015;385(Suppl 1):S82. doi: 10.1016/S0140-6736(15)60397-X. [DOI] [PubMed] [Google Scholar]
- 10.Fish JE, Cybulsky MI. ApoE attenuates atherosclerosis via miR-146a. Circ Res. 2015;117:3–6. doi: 10.1161/CIRCRESAHA.115.306733. [DOI] [PubMed] [Google Scholar]
- 11.Wang M, Li Wen, Chang GQ, Ye CS, Ou JS, Li XX, Liu Y, Cheang TY, Huang XL, Wang SM. MicroRNA-21 regulates vascular smooth muscle cell function via targeting tropomyosin 1 in arteriosclerosis obliterans of lower extremities. Arterioscler Thromb Vasc Biol. 2011;31:2044–2053. doi: 10.1161/ATVBAHA.111.229559. [DOI] [PubMed] [Google Scholar]
- 12.Li P, Zhu N, Yi B, Wang N, Chen M, You X, Zhao X, Solomides CC, Qin Y, Sun J. MicroRNA-663 regulates human vascular smooth muscle cell phenotypic switch and vascular neointimal formation. Circ Res. 2013;113:1117–1127. doi: 10.1161/CIRCRESAHA.113.301306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu CW, Ng SC, Dong Y, Tian L, Ng SS, Leung WW, Law WT, Yau TO, Chan FK, Sung JJ, Yu J. Identification of microRNA-135b in stool as a potential noninvasive biomarker for colorectal cancer and adenoma. Clin Cancer Res. 2014;20:2994–3002. doi: 10.1158/1078-0432.CCR-13-1750. [DOI] [PubMed] [Google Scholar]
- 14.Liu X, Sempere LF, Ouyang H, Memoli VA, Andrew AS, Luo Y, Demidenko E, Korc M, Shi W, Preis M. MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. J Clin Invest. 2010;120:1298–1309. doi: 10.1172/JCI39566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang T, Wang Q, Zhao D, Cui Y, Cao B, Guo L, Lu SH. The oncogenetic role of microRNA-31 as a potential biomarker in oesophageal squamous cell carcinoma. Clin Sci (Lond) 2011;121:437–447. doi: 10.1042/CS20110207. [DOI] [PubMed] [Google Scholar]
- 16.Kim HS, Lee KS, Bae HJ, Eun JW, Shen Q, Park SJ, Shin WC, Yang HD, Park M, Park WS, et al. MicroRNA-31 functions as a tumor suppressor by regulating cell cycle and epithelial-mesenchymal transition regulatory proteins in liver cancer. Oncotarget. 2015;6:8089–8102. doi: 10.18632/oncotarget.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu YH, Hu TF, Chen YC, Tsai YN, Tsai YH, Cheng CC, Wang HW. The manipulation of miRNA-gene regulatory networks by KSHV induces endothelial cell motility. Blood. 2011;118:2896–2905. doi: 10.1182/blood-2011-01-330589. [DOI] [PubMed] [Google Scholar]
- 18.Shen J, Yang X, Xie B, Chen Y, Swaim M, Hackett SF, Campochiaro PA. MicroRNAs regulate ocular neovascularization. Mol Ther. 2008;16:1208–1216. doi: 10.1038/mt.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan S, Xu Z, Lou F, Zhang L, Ke F, Bai J, Liu Z, Liu J, Wang H, Zhu H. NF-κB-induced microRNA-31 promotes epidermal hyperplasia by repressing protein phosphatase 6 in psoriasis. Nat Commun. 2015;6:7652. doi: 10.1038/ncomms8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 22.Diehm C, Allenberg JR, Pittrow D, Mahn M, Tepohl G, Haberl RL, Darius H, Burghaus I, Trampisch HJ, German Epidemiological Trial on Ankle Brachial Index Study Group Mortality and vascular morbidity in older adults with asymptomatic versus symptomatic peripheral artery disease. Circulation. 2009;120:2053–2061. doi: 10.1161/CIRCULATIONAHA.109.865600. [DOI] [PubMed] [Google Scholar]
- 23.Kim J, Zhang L, Peppel K, Wu JH, Zidar DA, Brian L, DeWire SM, Exum ST, Lefkowitz RJ, Freedman NJ. Beta-arrestins regulate atherosclerosis and neointimal hyperplasia by controlling smooth muscle cell proliferation and migration. Circ Res. 2008;103:70–79. doi: 10.1161/CIRCRESAHA.108.172338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hao H, Gabbiani G, Bochaton-Piallat ML. Arterial smooth muscle cell heterogeneity: Implications for atherosclerosis and restenosis development. Arterioscler Thromb Vasc Biol. 2003;23:1510–1520. doi: 10.1161/01.ATV.0000090130.85752.ED. [DOI] [PubMed] [Google Scholar]
- 25.Rajbhandari R, McFarland BC, Patel A, Gerigk M, Gray GK, Fehling SC, Bredel M, Berbari NF, Kim H, Marks MP, et al. Loss of tumor suppressive microRNA-31 enhances TRADD/NF-κB signaling in glioblastoma. Oncotarget. 2015;6:17805–17816. doi: 10.18632/oncotarget.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steg PG, Bhatt DL, Wilson PW, D'Agostino R, Sr, Ohman EM, Röther J, Liau CS, Hirsch AT, Mas JL, Ikeda Y, et al. REACH Registry Investigators One-year cardiovascular event rates in outpatients with atherothrombosis. JAMA. 2007;297:1197–1206. doi: 10.1001/jama.297.11.1197. [DOI] [PubMed] [Google Scholar]
- 27.Liu X, Cheng Y, Chen X, Yang J, Xu L, Zhang C. MicroRNA-31 regulated by the extracellular regulated kinase is involved in vascular smooth muscle cell growth via large tumor suppressor homolog 2. J Biol Chem. 2011;286:42371–42380. doi: 10.1074/jbc.M111.261065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Yan CH, Li Y, Xu K, Tian XX, Peng CF, Tao J, Sun MY, Han YL. MicroRNA-31 controls phenotypic modulation of human vascular smooth muscle cells by regulating its target gene cellular repressor of E1A-stimulated genes. Exp Cell Res. 2013;319:1165–1175. doi: 10.1016/j.yexcr.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 29.Chen KH, Guo X, Ma D, Guo Y, Li Q, Yang D, Li P, Qiu X, Wen S, Xiao RP, Tang J. Dysregulation of HSG triggers vascular proliferative disorders. Nat Cell Biol. 2004;6:872–883. doi: 10.1038/ncb1161. [DOI] [PubMed] [Google Scholar]
- 30.Ma L, Liu Y, Geng C, Qi X, Jiang J. Estrogen receptor β inhibits estradiol-induced proliferation and migration of MCF-7 cells through regulation of mitofusin 2. Int J Oncol. 2013;42:1993–2000. doi: 10.3892/ijo.2013.1903. [DOI] [PubMed] [Google Scholar]
- 31.Guo X, Chen KH, Guo Y, Liao H, Tang J, Xiao RP. Mitofusin 2 triggers vascular smooth muscle cell apoptosis via mitochondrial death pathway. Circ Res. 2007;101:1113–1122. doi: 10.1161/CIRCRESAHA.107.157644. [DOI] [PubMed] [Google Scholar]
- 32.Ryan JJ, Marsboom G, Fang YH, Toth PT, Morrow E, Luo N, Piao L, Hong Z, Ericson K, Zhang HJ, et al. PGC1α-mediated mitofusin-2 deficiency in female rats and humans with pulmonary arterial hypertension. Am J Respir Crit Care Med. 2013;187:865–878. doi: 10.1164/rccm.201209-1687OC. [DOI] [PMC free article] [PubMed] [Google Scholar]