Abstract
Please cite this paper as: Balish et al. (2012) Analytical detection of influenza A(H3N2)v and other A variant viruses from the USA by rapid influenza diagnostic tests. Influenza and Other Respiratory Viruses 7(4), 491–496.
Background The performance of rapid influenza diagnostic tests (RIDTs) that detect influenza viral nucleoprotein (NP) antigen has been reported to be variable. Recent human infections with variant influenza A viruses that are circulating in pigs prompted the investigation of the analytical reactivity of RIDTs with these variant viruses.
Objectives To determine analytical reactivity of seven FDA‐cleared RIDTs with influenza A variant viruses in comparison with the reactivity with recently circulating seasonal influenza A viruses.
Methods Tenfold serial dilutions of cell culture–grown seasonal and variant influenza A viruses were prepared and tested in duplicate with seven RIDTs.
Results All RIDTs evaluated in this study detected the seasonal influenza A(H3N2) virus, although detection limits varied among assays. All but one examined RIDT identified the influenza A(H1N1)pdm09 virus. However, only four of seven RIDTs detected all influenza A(H3N2)v, A(H1N2)v, and A(H1N1)v viruses. Reduced sensitivity of RIDTs to variant influenza viruses may be due to amino acid differences between the NP proteins of seasonal viruses and the NP proteins from viruses circulating in pigs.
Conclusions Clinicians should be aware of the limitations of RIDTs to detect influenza A variant viruses. Specimens from patients with influenza‐like illness in whom H3N2v is suspected should be sent to public health laboratories for additional diagnostic testing.
Keywords: H3N2v, influenza A variant, rapid influenza diagnostic test
Introduction
Rapid influenza diagnostic tests (RIDTs) detect Influenza A and B viral nucleoprotein (NP) from respiratory specimens. Specific antibodies are used to capture the NP on a membrane with test results generated in 10–15 minutes. Rapid influenza diagnostic tests have been shown to be highly specific; however, several reports have shown that RIDT analytical and clinical sensitivity varies. 1 , 2 , 3 Additionally, the type of specimen collected, time from illness onset to collection of respiratory specimen, and prevalence of influenza activity in the population have been shown to affect RIDT results. 4
The emergence of a variant virus in 2009 highlighted the need to evaluate the reactivity of RIDTs as the NP from influenza A(H1N1)pdm09 originated from swine rather than human viruses. 5 Reduced reactivity of RIDTs with influenza A(H1N1)pdm09 in comparison with seasonal influenza viruses 6 , 7 , 8 , 9 , 10 is of public health concern. Rapid influenza diagnostic tests were found to be capable of detecting influenza A(H1N1)pdm09 in respiratory specimens, but many infections could be missed, especially in specimens with low viral loads. 10
Between 1990 and 2010, 27 human cases of infection with variant influenza viruses that contain one or more gene segments derived from viruses of swine origin were identified in the United States. 11 These variant viruses were of different subtype [A(H1N1)v, A(H1N2)v, and A(H3N2)v] and genome constellations. In 2010, the increase in the number of human infections with H3N2v viruses prompted the preparation of a candidate vaccine virus, A/Minnesota/11/2010 (H3N2)v. In July 2011, the first human case of infection with a new reassortant influenza A(H3N2)v virus containing the matrix (M) gene from influenza A(H1N1)pdm09 virus was detected. 12 , 13 From July 2011 through August 9, 2012, a total of 166 human infections with H3N2v viruses have been reported. 13 , 14
Increasing numbers of human infections with variant viruses prompted the investigation of the analytical reactivity of FDA‐cleared RIDTs with these viruses. To determine whether RIDTs can detect influenza A(H3N2)v in addition to A(H1N1)v and A(H1N2)v viruses, seven FDA‐cleared RIDTs were evaluated analytically with variant viruses as well as with circulating human influenza A(H3N2) and A(H1N1)09pdm viruses. A public health message describing updated influenza A(H3N2)v virus case counts and an abbreviated data set was published recently in the Morbidity and Mortality Weekly Report. 14 This manuscript provides additional data and discussion describing reactivity and genetic comparisons with seasonal viruses and additional influenza A variant viruses.
Materials and methods
RIDTs
Seven FDA‐cleared RIDTs were evaluated (Table 1). Four of the RIDTs were Clinical Laboratory Improvement Amendment (CLIA)‐waived. CLIA‐waived tests can be performed in point‐of‐care settings such as physician offices. Test time for all RIDTs was between 10 and 15 minutes. Two of the RIDTs required a manufacturer‐specific analyzer to determine the results of the test. BinaxNOW®(BinaxNOW, Scarborough, ME, USA), Directigen™ (Directigen, Sparks, MD, USA), FluAlert (FluAlert, San Antonio, TX, USA), QuickVue® (QuickVue, San Diego, CA, USA), and Sofia (Sofia, San Diego, CA, USA) tests were performed according to the procedures in the kit inserts for nasal washes or aspirates. Xpect™ (Xpect, Lenexa, KS, USA) tests were performed according to their procedure for nasal washes and swab specimens transported in liquid media. For the Veritor™ (Veritor, Sparks, MD, USA), 100 microliters of diluted sample were added directly to the reagent tube. Positive and negative controls contained in each RIDT were run prior to testing the viruses in the study to verify performance of each lot, with the exception of FluAlert, which does not provide controls. Five tenfold serial dilutions in 0·85% physiological saline were prepared for each virus immediately before testing all seven RIDTs in duplicate. Only specimens that tested positive for both replicates were interpreted as positive. If the internal control in the test was invalid or the analyzer reported an invalid result, the test was repeated.
Table 1.
Rapid Influenza Diagnostic Tests (RIDTs) used in the study
| RIDT (Manufacturer) | Abbreviated name | CLIA Complexity | Approved specimens* | Analyzer for interpretation |
|---|---|---|---|---|
| BinaxNOW® Influenza A&B (Alere) | BinaxNOW® | Waived | NP swab /Nasal wash/aspirate/swab | No |
| Directigen™ EZ Flu A + B (Becton‐Dickinson) | Directigen™ EZ | Not waived | NP wash/aspirate/swab /Throat swab | No |
| SAS™ FluAlert A&B (SA Scientific) | FluAlert | Not waived | Nasal wash/aspirate | No |
| QuickVue® Influenza A + B Test (Quidel) | QuickVue® | Waived | NP swab /Nasal wash/aspirate/swab | No |
| Sofia Influenza A + B (Quidel) | Sofia | Waived | NP aspirate/swab/wash /Nasal wash | Required |
| BD Veritor™ System for Rapid Detection of Flu A + B (Becton Dickinson) | Veritor™ | Waived | NP swab/nasal swab | Required |
| XPECT™ Flu A&B (Remel) | Xpect™ | Not waived | Nasal wash/swab /Throat swab | No |
*Approved respiratory specimens according to manufacturer’s package insert. Test performance has only been demonstrated for these specimen types.
NP, nasopharyngeal; CLIA, Clinical Laboratory Improvement Amendment.
Viruses
Influenza A viruses used in the study are listed in Table 2. The viruses were grown in Madin–Darby Canine Kidney cells from ATCC collection (CCL‐34) as previously described. 15 The 50% tissue culture infectious dose (TCID50/ml) was measured to estimate the amount of infectious virus in a sample as described. 16
Table 2.
Lowest virus concentration (TCID50/ml) detected in evaluated RIDTs
| Subtype | Strain designation | TCID50/ml | RIDT | ||||||
|---|---|---|---|---|---|---|---|---|---|
| BinaxNOW® | Directigen™ | FluAlert | QuickVue® | Sofia | Veritor™ | Xpect™ | |||
| H3N2 | A/Georgia/01/2011 | 106·0 | 102·0 | 101·0 | 104·0 | 101·0 | 101·0 | 102·0 | 103·0 |
| H1N1pdm09 | A/California/04/2009 | 106·0 | 104·0 | 102·0 | U | 104·0 | 103·0 | 102·0 | 103·0 |
| H3N2v | A/Kansas/13/2009 | 104·5 | 103·5 | 10−0·5 | U | U | 102·5 | 100·5 | 100·5 |
| H3N2v | A/Pennsylvania/14/2010 | 104·5 | 102·5 | 10−0·5 | U | 102·5 | 102·5 | 100·5 | 101·5 |
| H3N2v | A/Minnesota/11/2010 | 104·5 | U | 101·5 | U | U | 101·5 | 101·5 | 102·5 |
| H3N2v | A/Indiana/08/2011 | 106·0 | 105·0 | 103·0 | U | U | 104·0 | 103·0 | 104·0 |
| H3N2v | A/Indiana/10/2011 | 104·0 | U | 101·0 | U | U | 102·0 | 100·0 | 102·0 |
| H3N2v | A/West Virginia/06/2011 | 106·0 | 104·0 | 103·0 | U | 104·0 | 102·0 | 102·0 | 104·0 |
| H3N2v | A/Iowa/07/2011 | 104·5 | 102·5 | 100·5 | 103·5 | 103·5 | 101·5 | 100·5 | 101·5 |
| H1N1v | A/Wisconsin/28/2011 | 106·0 | 105·0 | 103·0 | 105·0 | 105·0 | 103·0 | 101·0 | 102·0 |
| H1N2v | A/Michigan/09/2007 | 106·0 | 105·0 | 104·0 | U | U | 104·0 | 102·0 | 104·0 |
| H1N2v | A/Minnesota/19/2011 | 105·0 | 104·0 | 101·0 | U | U | 102·0 | 100·0 | 103·0 |
RIDT, rapid influenza diagnostic test.
TCID50/ml, infectious titer of stock virus; U, undetected at any concentration tested.
Phylogenetic analysis
The evolutionary history of the influenza A NP gene was inferred by using the maximum likelihood method based on the data‐specific model. 17 The tree with the highest log likelihood (−21797·5176) is shown. Initial tree(s) for the heuristic search were obtained automatically as follows. When the number of common sites was <100 or less than one‐fourth of the total number of sites, the maximum parsimony method was used; otherwise, BIONJ method with MCL distance matrix was used. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 89 nucleotide sequences, and GISAID accession numbers are listed in Table S1. Codon positions included were 1st + 2nd + 3rd + Noncoding. All positions containing gaps and missing data were eliminated. There were a total of 1357 positions in the final data set. Evolutionary analyses were conducted in MEGA5. 18
Results
Table 2 shows the lowest virus concentration (TCID50/ml) detected by each RIDT for each influenza virus. All RIDTs detected the seasonal influenza A/Georgia/01/2011 (H3N2) virus, although at different infectious virus titers varying from 101·0 TCID50/ml (Directigen™, QuickVue®, Sofia) to 104·0 TCID50/ml (FluAlert). All but one of the RIDTs (FluAlert) were able to detect the A/California/04/2009 (H1N1)pdm09 virus, however, at higher virus concentrations compared with A/Georgia/01/2011 (H3N2). The lowest concentrations of A/California/04/2009 (H1N1)pdm09 detected by RIDTs that provided positive results varied from 102·0 TCID50/ml (Directigen™, Veritor™) to 104·0 TCID50/ml (BinaxNOW®, QuickVue®).
Four of seven RIDTs in this study (Directigen™, Sofia, Veritor™, and Xpect™) detected all influenza A(H3N2)v viruses (Table 2). The lowest A(H3N2)v virus concentrations detected by these four RIDTs were similar to those for A/California/04/2009 (H1N1)pdm09. BinaxNOW® detected five of seven and QuickVue® detected three of seven A(H3N2)v viruses. FluAlert detected only one of seven A(H3N2)v viruses.
Three influenza A(H1) variant viruses also were evaluated with the seven RIDTs: two A(H1N2)v isolates (A/Michigan/09/2007, A/Minnesota/19/2011) and one A(H1N1)v virus (A/Wisconsin/28/2011) (Table 2). Five of seven RIDTs (BinaxNOW®, Directigen™, Sofia, Veritor™, and Xpect™) detected all influenza A(H1)v viruses. FluAlert and QuickVue® were unable to detect two of three influenza A(H1)v viruses in this evaluation.
As the NP of influenza viruses is the target for all evaluated RIDTs, we compared predicted amino acid sequences of the NP based on available genetic data for tested viruses (Table 3). This amino acid alignment revealed that the NP of A/California/04/2009 (H1N1)pdm09 and the A(H3N2)v, A(H1N2)v, and A(H1N1)v viruses differ in amino acid sequences from the NP of the seasonal A/Georgia/01/2011 (H3N2) virus by 10·3–10·7%. The pandemic A/California/04/2011 (H1N1)pdm09 virus and all variant viruses tested here are different from each other by approximately 2%. There is <0·2% difference in the amino acid composition of the NP among influenza A(H3N2)v, A(H1N2)v, and A(H1N1)v viruses tested in this study. In addition, Figure 1 shows the phylogenetic analysis of influenza A NP genes from various species. Figure 1 further demonstrates that although the NP genes in North American swine and human seasonal influenza viruses are derived from a common ancestor related to the A(H1N1) from 1918, separate species‐specific evolutionary paths have since occurred. North American swine NP is quite divergent from the NP of seasonal human influenza A viruses but more closely genetically related to NP from influenza A(H1N1)pdm09 viruses.
Table 3.
Nucleoprotein amino acid difference comparisons between seasonal A(H3N2), A(H1N1)pdm09, and variant influenza A viruses
| Subtype | Amino acid distance* | Strain designation | 16 | 18 | 21 | 31 | 34 | 38 | 52 | 53 | 61 | 65 | 105 | 109 | 119 | 131 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H3N2 | A/Georgia/01/2011 | D | D | N | K | D | R | H | E | L | K | M | V | I | S | |
| H1N1pdm09 | 0·102 | A/California/04/2009 | G | E | D | R | G | Y | D | I | R | I | V | A | ||
| H3N2v | 0·104 | A/Kansas/13/2009 | G | E | D | R | G | K | Y | I | R | I | V | A | ||
| H3N2v | 0·104 | A/Pennsylvania/14/2010 | G | E | D | R | G | K | Y | I | R | I | V | A | ||
| H3N2v | 0·105 | A/Minnesota/11/2010 | G | E | D | R | G | K | Y | I | R | I | V | A | ||
| H3N2v | 0·106 | A/Indiana/08/2011 | G | E | D | R | G | K | Y | I | R | I | V | A | ||
| H3N2v | 0·104 | A/Indiana/10/2011 | G | E | D | R | G | K | Y | I | R | I | V | A | ||
| H3N2v | 0·104 | A/West Virginia/06/2011 | G | E | D | R | G | K | Y | I | R | I | V | A | ||
| H3N2v | 0·104 | A/Iowa/07/2011 | S | E | D | R | G | K | Y | I | R | I | V | A | ||
| H1N1v | 0·106 | A/Wisconsin/28/2011 | G | E | D | R | G | K | Y | I | R | I | I | V | A | |
| H1N2v | 0·104 | A/Michigan/09/2007 | G | E | D | R | S | K | Y | I | R | I | V | A | ||
| H1N2v | 0·104 | A/Minnesota/19/2011 | G | E | D | R | G | K | Y | I | R | I | V | A | ||
| Subtype | Amino acid distance* | Strain designation | 186 | 189 | 190 | 197 | 214 | 217 | 239 | 280 | 283 | 286 | 289 | 293 | 306 | 312 |
| H3N2 | A/Georgia/01/2011 | I | M | V | V | K | S | V | A | P | S | Y | K | L | I | |
| H1N1pdm09 | 0·102 | A/California/04/2009 | V | I | A | I | R | V | M | V | L | A | H | R | V | |
| H3N2v | 0·104 | A/Kansas/13/2009 | V | I | A | I | R | I | M | V | L | A | H | R | V | |
| H3N2v | 0·104 | A/Pennsylvania/14/2010 | V | I | A | I | R | I | M | V | L | A | H | R | V | |
| H3N2v | 0·105 | A/Minnesota/11/2010 | V | I | A | I | R | I | M | V | L | A | H | R | V | |
| H3N2v | 0·106 | A/Indiana/08/2011 | V | I | A | I | R | I | M | V | L | A | H | R | S | V |
| H3N2v | 0·104 | A/Indiana/10/2011 | V | I | A | I | R | I | M | V | L | A | H | R | V | |
| H3N2v | 0·104 | A/West Virginia/06/2011 | V | I | A | I | R | I | M | V | L | A | H | R | V | |
| H3N2v | 0·104 | A/Iowa/07/2011 | V | I | A | I | R | I | M | V | L | A | H | R | V | |
| H1N1v | 0·106 | A/Wisconsin/28/2011 | V | I | A | I | R | I | M | V | L | A | H | R | V | |
| H1N2v | 0·104 | A/Michigan/09/2007 | V | I | A | I | R | I | M | V | L | A | H | R | V | |
| H1N2v | 0·104 | A/Minnesota/19/2011 | V | I | A | I | R | I | M | V | L | A | H | R | V | |
| Subtype | Amino acid distance* | Strain Designation | 313 | 316 | 343 | 344 | 350 | 353 | 371 | 372 | 373 | 375 | 377 | 384 | 400 | 406 |
| H3N2 | A/Georgia/01/2011 | Y | I | L | L | T | S | M | D | N | G | S | G | R | T | |
| H1N1pdm09 | 0·102 | A/California/04/2009 | V | M | V | S | K | I | V | E | T | D | N | R | K | I |
| H3N2v | 0·104 | A/Kansas/13/2009 | F | V | S | K | I | V | E | A | D | N | R | K | I | |
| H3N2v | 0·104 | A/Pennsylvania/14/2010 | F | V | S | K | L | V | E | A | D | N | R | K | I | |
| H3N2v | 0·105 | A/Minnesota/11/2010 | F | V | S | K | L | V | E | A | D | N | R | K | I | |
| H3N2v | 0·106 | A/Indiana/08/2011 | F | V | S | K | L | V | E | A | D | N | R | K | I | |
| H3N2v | 0·104 | A/Indiana/10/2011 | F | V | S | K | L | V | E | A | D | N | R | K | I | |
| H3N2v | 0·104 | A/West Virginia/06/2011 | F | V | S | K | L | V | E | A | D | N | R | K | I | |
| H3N2v | 0·104 | A/Iowa/07/2011 | F | V | S | K | L | V | E | A | D | N | R | K | I | |
| H1N1v | 0·106 | A/Wisconsin/28/2011 | F | V | S | K | L | V | E | A | D | N | R | K | I | |
| H1N2v | 0·104 | A/Michigan/09/2007 | F | V | S | K | I | V | E | A | D | N | R | K | I | |
| H1N2v | 0·104 | A/Minnesota/19/2011 | F | V | S | K | L | V | E | A | D | N | R | K | I | |
| Subtype | Amino acid distance* | Strain Designation | 422 | 423 | 425 | 426 | 430 | 433 | 442 | 444 | 450 | 455 | 456 | 459 | 473 | 498 |
| H3N2 | A/Georgia/01/2011 | K | S | I | M | T | T | A | I | G | E | V | R | N | N | |
| H1N1pdm09 | 0·102 | A/California/04/2009 | R | A | V | S | N | T | V | S | D | L | Q | S | ||
| H3N2v | 0·104 | A/Kansas/13/2009 | R | A | V | L | S | N | T | V | S | D | L | Q | S | S |
| H3N2v | 0·104 | A/Pennsylvania/14/2010 | R | A | V | L | S | N | T | V | S | D | L | Q | S | S |
| H3N2v | 0·105 | A/Minnesota/11/2010 | R | A | V | L | S | N | T | V | S | D | L | Q | S | S |
| H3N2v | 0·106 | A/Indiana/08/2011 | R | A | V | L | S | N | T | V | S | D | L | Q | S | S |
| H3N2v | 0·104 | A/Indiana/10/2011 | R | A | V | L | S | N | T | V | S | D | L | Q | S | S |
| H3N2v | 0·104 | A/West Virginia/06/2011 | R | A | V | L | S | N | T | V | S | D | L | Q | S | S |
| H3N2v | 0·104 | A/Iowa/07/2011 | R | A | V | L | S | N | T | V | S | D | L | Q | S | S |
| H1N1v | 0·106 | A/Wisconsin/28/2011 | R | A | V | L | S | N | T | V | S | D | L | Q | S | S |
| H1N2v | 0·104 | A/Michigan/09/2007 | R | A | V | L | S | N | T | V | S | D | L | Q | S | S |
| H1N2v | 0·104 | A/Minnesota/19/2011 | R | A | V | L | S | N | T | V | S | D | L | Q | S | S |
*Compared with A/Georgia/01/2011 H3N2. Gaps in the table indicate identity to amino acids of A/Georgia/01/2011 (H3N2).
Figure 1.
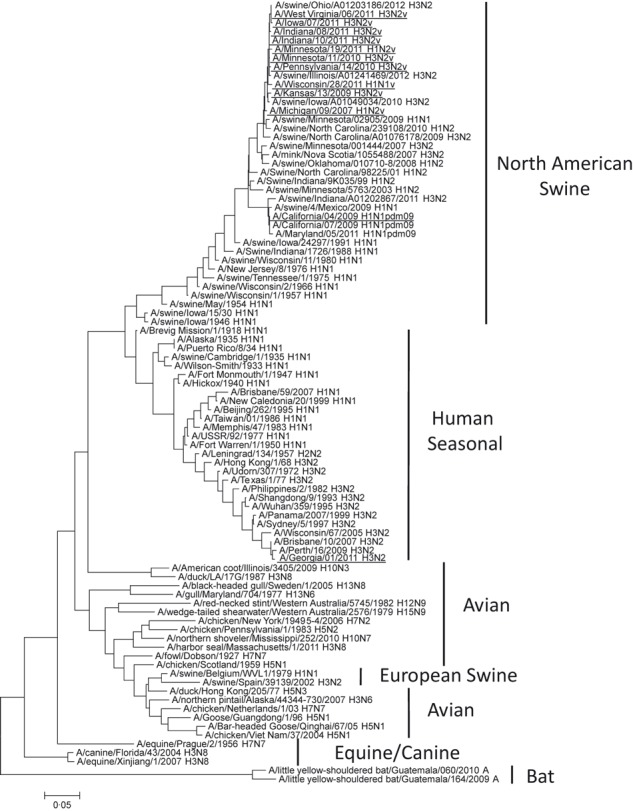
Phylogenetic analysis of influenza A nucleoprotein genes by maximum likelihood method. Viruses used in this study are underlined.
Discussion
As demonstrated by this evaluation, RIDTs may or may not detect the presence of NP derived from influenza viruses that are circulating in swine. Genetic sequence analysis indicates divergence of the NP of influenza A variant viruses and the A(H1N1)pdm09 virus compared with the NP of seasonal influenza A viruses, which may account for reduced reactivity with some RIDTs. It is important to note that all RIDTs have different proprietary antibodies to capture and detect NP, making it impossible to predict which RIDTs may detect NP that evolved from viruses that are not of human origin. As indicated in the manufacturer package inserts, influenza A viruses that have undergone minor amino acid changes in the target epitope region or regions may cause the antibodies to be less sensitive or not sensitive at all. Although the NP gene sequences are almost identical among variant viruses, several variant viruses were tested in this study because influenza virus preparations can be different in terms of amount of infectious/non‐infectious virus, protein concentration, etc. This may account for differences in reactivity of different viruses with RIDTs despite having similar infectious virus titers.
This first study to assess analytical reactivity of FDA‐cleared RIDTs with influenza A variant viruses has some limitations. Only seven RIDTs were evaluated when currently there are 15 FDA‐cleared RIDTs of which six are CLIA‐waived for use in point‐of‐care settings. 4 Cell culture virus isolates were used for this study. Limited volumes prevented a comparison of all seven RIDTs with clinical specimens. As in most analytical evaluations, influenza viruses in this study were characterized by their infectious virus titer but not by the NP concentration. Infectious virus titer can vary among preparations of the same virus, and the titers were not normalized for the purposes of this evaluation. A side‐by‐side performance evaluation of all cleared assays with multiple influenza virus preparations, dilutions, and replicates would provide a direct analytical comparison of all FDA‐cleared RIDTs.
This study was conducted to provide public health guidance for the use of RIDTs when a novel influenza virus infection is suspected. It is important for clinicians to minimize the occurrence of false RIDT results by strictly following the manufacturer’s instructions, collecting specimens soon after onset of influenza‐like illness symptoms (ideally within the first 72 hours), and confirming RIDT results by sending a specimen to a public health laboratory. 4 Qualified U.S. public health laboratories and global National Influenza Centers may utilize the CDC Flu rRT‐PCR Panel and a real‐time RT‐PCR assay, which allows for the preliminary identification of influenza variant viruses possessing the swine NP gene. 13 This evaluation reinforces the fact that a negative RIDT result should not be considered as conclusive evidence of a lack of infection with influenza virus and emphasizes that this may particularly be the case for influenza A(H3N2)v viruses.
Supporting information
Table S1. GISAD accession numbers for sequences in phylogenetic tree.
Supporting info item
Acknowledgements
We thank our collaborators from the following institutions: Indiana State Department of Health Laboratories, Iowa State Hygienic Laboratory, Kansas Department of Health and Environment, Maryland Department of Health and Mental Hygiene, Michigan Department of Community Health, Minnesota Department of Health, Pennsylvania Department of Health, West Virginia Office of Laboratory Services, Wisconsin State Laboratory of Hygiene.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
References
- 1. Uyeki TM. Influenza diagnosis and treatment in children: a review of studies on clinically useful tests and antiviral treatment for influenza. Pediatr Infect Dis J 2003; 22:164–177. [DOI] [PubMed] [Google Scholar]
- 2. Weinberg A, Walker ML. Evaluation of three immunoassay kits for rapid detection of influenza virus A and B. Clin Diagn Lab Immunol 2005; 12:367–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cruz AT, Cazacu AC, Greer JM, Demmler GJ. Rapid assays for the diagnosis of influenza A and B viruses in patients evaluated at a large tertiary care children’s hospital during two consecutive winter seasons. J Clin Virol 2008; 41:143–147. [DOI] [PubMed] [Google Scholar]
- 4. Centers for Disease Control and Prevention . Guidance for clinicians on the use of rapid influenza diagnostic tests, 2011; Available at http://www.cdc.gov/flu/professionals/diagnosis/clinician_guidance_ridt.htm (Accessed 17 April 2012).
- 5. Garten RJ, Davis CT, Russell CA et al. Antigenic and genetic characteristics of swine‐origin 2009 A(H1N1) influenza viruses circulating in humans. Science 2009; 325:197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Faix DJ, Sherman SS, Waterman SH. Rapid‐test sensitivity for novel swine‐origin influenza A (H1N1) virus in humans. N Engl J Med 2009; 361:728–729. [DOI] [PubMed] [Google Scholar]
- 7. Vasoo S, Stevens J, Singh K. Rapid antigen tests for diagnosis of pandemic (Swine) influenza A/H1N1. Clin Infect Dis 2009; 49:1090–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gao F, Loring C, Laviolette M, Bolton D, Daly ER, Bean C. Detection of 2009 pandemic influenza A(H1N1) virus Infection in different age groups by using rapid influenza diagnostic tests. Influenza Other Respir Viruses 2012; 6:e30–e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hara M, Morihara M, Takao S et al. Influenza viral load and rapid influenza diagnostic tests in children and adults. Diagn Microbiol Infect Dis 2012; 73:99–100. [DOI] [PubMed] [Google Scholar]
- 10. Centers for Disease Control and Prevention . Evaluation of rapid influenza diagnostic tests for detection of novel influenza A (H1N1) Virus – United States, 2009. MMWR Morb Mortal Wkly Rep 2009; 58:826–829. [PubMed] [Google Scholar]
- 11. Shu B, Garten R, Emery S et al. Genetic analysis and antigenic characterization of swine origin influenza viruses isolated from humans in the United States, 1990–2010. Virology 2012; 422:151–160. [DOI] [PubMed] [Google Scholar]
- 12. Centers for Disease Control and Prevention . Swine‐origin influenza A (H3N2) virus infection in two children – Indiana and Pennsylvania, July–August 2011. MMWR Morb Mortal Wkly Rep [Case Reports] 2011; 60:1213–1215. [PubMed] [Google Scholar]
- 13. Lindstrom S, Garten R, Balish A et al. Human infections with novel reassortant influenza A(H3N2)v viruses, United States, 2011. Emerg Infect Dis 2012; 18:834–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Centers for Disease Control and Prevention . Evaluation of Rapid Influenza Diagnostic Tests for Influenza A(H3N2)v Virus Infections and Updated Case Count – United States, 2012. MMWR Morb Mortal Wkly Rep 2012; 61:619–621. [PubMed] [Google Scholar]
- 15. Szretter KJ, Balish AL, Katz JM. Influenza: Propagation, Quantification and Storage. Current Protocols in Microbiology. John Wiley & Sons Inc., 2006. [DOI] [PubMed] [Google Scholar]
- 16. Klimov A, Balish A, Veguilla V et al. Influenza virus titration, antigenic characterization, and serological methods for antibody detection. Methods Mol Biol 2012; 865:25–51. [DOI] [PubMed] [Google Scholar]
- 17. Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford University Press, 2000. [Google Scholar]
- 18. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28:2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. GISAD accession numbers for sequences in phylogenetic tree.
Supporting info item


