Abstract
Please cite this paper as: Pasricha et al. (2012) Comprehensive global amino acid sequence analysis of PB1F2 protein of influenza A H5N1 viruses and the Influenza A virus subtypes responsible for the 20th‐century pandemics. Influenza and Other Respiratory Viruses 7(4), 497–505.
Background PB1F2 is the 11th protein of influenza A virus translated from +1 alternate reading frame of PB1 gene. Since the discovery, varying sizes and functions of the PB1F2 protein of influenza A viruses have been reported. Selection of PB1 gene segment in the pandemics, variable size and pleiotropic effect of PB1F2 intrigued us to analyze amino acid sequences of this protein in various influenza A viruses.
Methods Amino acid sequences for PB1F2 protein of influenza A H5N1, H1N1, H2N2, and H3N2 subtypes were obtained from Influenza Research Database. Multiple sequence alignments of the PB1F2 protein sequences of the aforementioned subtypes were used to determine the size, variable and conserved domains and to perform mutational analysis.
Results Analysis showed that 96·4% of the H5N1 influenza viruses harbored full‐length PB1F2 protein. Except for the 2009 pandemic H1N1 virus, all the subtypes of the 20th‐century pandemic influenza viruses contained full‐length PB1F2 protein. Through the years, PB1F2 protein of the H1N1 and H3N2 viruses has undergone much variation. PB1F2 protein sequences of H5N1 viruses showed both human‐ and avian host‐specific conserved domains. Global database of PB1F2 protein revealed that N66S mutation was present only in 3·8% of the H5N1 strains. We found a novel mutation, N84S in the PB1F2 protein of 9·35% of the highly pathogenic avian influenza H5N1 influenza viruses.
Conclusions Varying sizes and mutations of the PB1F2 protein in different influenza A virus subtypes with pandemic potential were obtained. There was genetic divergence of the protein in various hosts which highlighted the host‐specific evolution of the virus. However, studies are required to correlate this sequence variability with the virulence and pathogenicity.
Keywords: Influenza A viruses, mutation, pandemics, PB1F2 protein, truncation
Introduction
Influenza is a viral disease where the virus continuously evolves causing emergence of newer epidemics and pandemics in human, which is of major health concern and economic burden worldwide. 1 Influenza A viruses are members of Orthomyxoviridae family, having eight segments of single‐stranded, negative‐sense RNA encoding for 12 proteins. 2 Highly mutable nature of influenza viruses is responsible for continuing evolution of viruses with prominent changes in the surface glycoprotein of influenza viruses. However, mutation in the internal gene segments particularly in PB1 is also of interest, because it was the only segment that was exchanged in the pandemic viruses of 1957 and 1968. 3 A novel PB1 gene was found in the 1998 swine reassortant viruses, implicating its role in pathogenesis of influenza. 4 Recent reconstruction of the 1918 virus has also confirmed that the viral polymerase is required for pathogenicity of the recombinant 1918 virus in mice. 5 Thus, the selection of PB1 gene in the previous pandemic strains is explainable because it has been shown to have potential to enhance the pathogenicity and virulence of the influenza A viruses. 6 , 7 In contrast to the pandemic strains, H5N1 viruses are extremely pathogenic, however, are not transmitted from human to human. There has been a widespread infection of poultry with H5N1 viruses in Asia, which increases the concern that this subtype may achieve human‐to‐human transmission and establish interspecies spread.
The PB1F2 is the 11th protein of influenza A virus that was identified and characterized almost 10 years ago by Chen et al. 7 It is encoded by the +1 alternate open reading frame of the PB1 gene and starts from the nucleotide position 120 surpassing three other initiation codons of the PB1 gene. PB1 gene has inefficient initiation of translation because it does not have consensus Kozak sequence, that is, it does not have a purine nucleotide in the −3 position of the ORF. The next initiation codon is surrounded by an exact Kozak sequence and serves for the synthesis of PB1F2. 8 A multifunctional role of PB1F2 protein has been elucidated, which includes a proapoptotic function in immune cells via mitochondrial pathway, 7 ability to cause increased pathogenesis in animal models 6 by causing dysregulation of cytokines 9 and inducing inflammation. 10 It has also been suggested that PB1F2 regulates the polymerase activity by colocalization with PB1 11 and causes enhanced secondary bacterial pneumonia. 10 However, there are ambiguities, regarding its proapoptotic role because there are reports suggesting that it is either strain or host/cell line specific. There are also unanswered questions regarding its antagonist or prognostic role toward the interferon response. 9 , 12 Furthermore, different truncated form of PB1F2 protein in different influenza A subtypes have been reported, 13 , 14 most recent being the H1N1 2009 pandemic having a C‐terminal‐truncated 11 amino acid PB1F2 protein. 15 Interestingly, the proapoptotic role of the protein is implicated to its C‐terminal end, 7 while the N‐terminal end is reported to regulate the polymerase activity. 8 The rate of synonymous and non‐synonymous substitutions in the PB1F2 gene is higher than that of PB1 gene; therefore, the amino acid sequence of PB1F2 in various subtypes is more diverse than that of PB1. 14 Thus, gaps in understanding the function of the protein and its variable length intrigued us to study this protein and determine size variability among different subtypes and identify conserved domains and putative pathogenic mutations in H5N1 viruses and also in viruses responsible for the 20th‐century pandemics.
Methods
Amino acid sequences for the PB1F2 protein were retrieved from National Institute of Allergy and Infectious Diseases (NIAID) influenza research database (IRD) online database (http://www.fludb.org). 16 All the PB1F2 sequences analyzed in the study were encoded by the alternative +1 open reading frame of the PB1 gene. All the sequences of PB1F2 protein available in the database for the H5N1, H1N1, H2N2, and H3N2 subtypes of influenza viruses available till November 2011 were included in the study. A total 919 amino acid sequences of the H5N1 subtype, 1530 sequences of the H1N1 subtype, 83 of the H2N2 subtype, and 2566 amino acid sequences of the H3N2 subtypes were analyzed in this study. ClustalX 2.1 (UCD, Dublin, Ireland) was used to align all the available sequences of the aforementioned subtypes studied and the phylogenetic trees were constructed using mega version 5.0 (Tempe, AZ, USA). The evolutionary history was inferred using the Neighbor‐Joining method, and the evolutionary distances were computed using the Poisson correction method. Tree topology was determined by bootstrap analysis with 500 replicates, and pairwise deletion was used for gaps or missing data treatment.
Results
PB1F2 sequences of a total of 919 influenza A H5N1 viruses isolated between 1959 and November 2011 were analyzed, of which 666 (72·5%) were isolated from avian species, 220 (24%) from human, 16 (1·7%) from environment, 13 (1·4%) from swine, and four (0·4%) from other hosts (Table 1). Ninety‐six percent of the avian H5N1 sequences (643 of 666) had a complete 90 amino acid PB1F2 fragment, while only 1·7% (11 of 666) had an N‐terminal‐truncated PB1F2 protein of 52 amino acid length. As a full‐length PB1F2 protein was present in 96·42% of the H5N1 strain, it implies that it has significant role in the life cycle of the virus. Sequence analysis of H5N1 PB1F2 protein of the avian isolates revealed that there were 10 conserved amino acids in 99·8% of the strains and they included 1M, 7T, 9W, 10T, 24G, 38L, 52H, 61W, 62L, and 72L (Figure 1B). Calculation of number of variants per amino acid position revealed that 10 amino acid positions showed variability above 10%, and they were two (35·8%), five (10·6), six (17·4%), 37 (15·6%), 40 (13·5%), 42 (10·6%), 50 (40·3%), 57 (11·5%), 74 (18·2%), and 89 (14·7%) (Figure 1B). Further analysis of the PB1F2 amino acid sequences of 666 avian strains showed that 271 strains were from chicken, 207 from ducks, 24, 19, 35, and 102 from geese, swan, openbill stork, and other avian species, respectively (Table 2). As chickens and ducks comprised most of the avian species in this study and also the fact they are the most studied, we further analyzed the conserved and variable regions in these two avian species (Figure 1C,D). Apart from the 10 amino acid positions that are conserved in all the avian species, another 14 amino acid positions were identified in 271 strains from chickens and another 15 in 207 strains from chicken. Thus, a total of 24 conserved amino acids were identified in PB1F2 protein sequences from chicken and 25 from the ducks.
Table 1.
PB1F2 variants present in IAV strains (H5N1, H1N1, H2N2, and H3N2) from various hosts
| S.No | Strains/Host | No. of analyzed strains | 101aa | 90aa | 87aa | N79aa* | N57aa** | C52aa*** | Varied size |
|---|---|---|---|---|---|---|---|---|---|
| 1 | H5N1 | 919 | – | 886 | 2 | 5 | 4 | 21 | 1 |
| Human | 220 | – | 211 | – | 3 | 2 | 4 | – | |
| Swine | 13 | – | 13 | – | – | – | – | – | |
| Avian | 666 | – | 643 | 2 | 2 | 1 | 17 | 1 | |
| Environment | 16 | – | 15 | – | – | 1 | – | – | |
| Others | 4 | – | 4 | – | – | – | – | – | |
| 2 | H1N1 | 1530 | – | 176 | 42 | – | 1080 | 221 | 11 |
| Human | 1155 | – | 32 | 41 | – | 1073 | 6 | 3 | |
| Swine | 261 | – | 43 | 1 | – | 7 | 202 | 8 | |
| Avian | 112 | – | 99 | – | – | – | 13 | – | |
| Environment | 1 | – | 1 | – | – | – | – | – | |
| Others | 1 | – | 1 | – | – | – | – | – | |
| 3 | H2N2 | 83 | 1 | 81 | – | – | 1 | – | – |
| Human | 54 | 1 | 52 | – | – | 1 | – | – | |
| Swine | – | – | – | – | – | – | – | – | |
| Avian | 22 | – | 22 | – | – | – | – | – | |
| Environment | 1 | – | 1 | – | – | – | – | – | |
| Others | 6 | – | 6 | – | – | – | – | – | |
| 4 | H3N2 | 2566 | 26 | 2105 | 45 | – | – | 337 | 53 |
| Human | 2419 | 25 | 1979 | 44 | – | – | 324 | 47 | |
| Swine | 76 | – | 63 | 1 | – | – | 7 | 5 | |
| Avian | 68 | 1 | 60 | – | – | – | 6 | 1 | |
| Environment | 2 | – | 2 | – | – | – | – | – | |
| Others | 1 | – | 1 | – | – | – | – | – |
* 79 amino acid PB1F2 fragment with C‐terminal end truncated.
** 57 amino acid fragment with C‐terminal end truncated.
*** 52 amino acid fragment with N‐terminal truncated.
Figure 1.
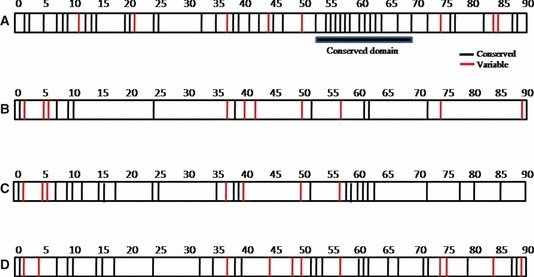
(A) Conserved and variable amino acid position of the PB1F2 protein analyzed from 220 H5N1 strains isolated from human. There were 41 amino acids positions that were conserved for all the strains, while for eight amino acid positions, the variability was >10%. A conserved domain spanning between amino acid positions 52–69 was identified, which is marked by black horizontal bar. (B) Conserved and variable amino acid positions of the PB1F2 protein analyzed from 666 H5N1 strains isolated from avian hosts. Unlike for human, there were only 10 amino acid positions that were completely conserved in the avian strains and 10 amino acid positions that showed variability >10%. (C) Conserved and variable amino acid positions of the PB1F2 protein analyzed from 279 H5N1 strains isolated from chickens. There were a total 24 amino acids that were conserved in 99·6% of the strains and seven amino acid positions that showed variability >10%. (D) Conserved and variable amino acid positions of the PB1F2 protein analyzed from 207 H5N1 strains isolated from ducks. There were a total 25 amino acids that were conserved 99·6% of the strains and 11 amino acid positions that showed variability >10%.
Table 2.
PB1F2 variants present in 666 avian strains belonging to H5N1 subtype
| Strains/Host | No. of analyzed strains | 90aa | 87aa | N79aa* | N57aa** | C52aa*** | Varied size |
|---|---|---|---|---|---|---|---|
| Total Avian | 666 | 643 | 2 | 2 | 1 | 17 | 1 |
| Chicken | 279 | 271 | – | – | 1 | 7 | – |
| Duck | 207 | 199 | 1 | – | – | 6 | 1 |
| Goose | 24 | 23 | – | – | – | 1 | – |
| Swan | 19 | 19 | – | – | – | – | – |
| Openbill stork | 35 | 35 | – | – | – | – | – |
| Other avian | 102 | 96 | 1 | 2 | – | 3 | – |
* 79 amino acid PB1F2 fragment with C‐terminal end truncated.
** 57 amino acid fragment with C‐terminal end truncated.
*** 52 amino acid fragment with N‐terminal truncated.
Among the 220 H5N1 strains isolated from human, 212 (96·3%) had the complete PB1F2 fragment, while four (1·8%) strains had a 52 amino acid N‐terminal‐truncated PB1F2 protein (henceforth designated as C‐52 in this study) and other four (1·8%) isolates analyzed had C‐terminal‐truncated PB1F2 designated as N57 and N79 (Table 1). Eighty‐eight (40%) PB1F2 sequence analyzed were from Indonesia, 46 (20·9%) from Vietnam, 32 (14·5%) from China, 22 (10%) from Hong Kong, 12 (5·5%) from Thailand, and 13 (5·9%) from other Asian countries, while only two (0·9%) were from Egypt. Of the 22 strains from Hong Kong, 18 were isolated in 1997 epidemic. Analysis of the amino acid sequence revealed that of a total 90 amino acid (full‐length PB1F2 protein), 41 amino acids (45·5%) were conserved in 99·5% of the isolates (Figure 1A). Also, we identified an 18 amino acid (52–69) conserved domain (Figure 1A) in 97% of the human H5N1 isolates analyzed in this study. The amino acid positions that showed more than 10% variation were 11, 21, 37, 44, 50, 74, 83, and 84 having variability of 25·5%, 17·27%, 13·6%, 10·5%, 30%, 16·4%, 20%, and 12·4%, respectively (Figure 1A).
Global database analysis of the PB1F2 sequence of influenza A H5N1 isolates revealed that N66S mutation was present in only 35 (3·8%) strains that included 17 from avian species, 12 from environment, and six from human hosts (Table 3). Among the 17 avian H5N1 strains, only one strain, A/chicken/Shantou/904/2001, was highly pathogenic avian influenza (HPAI). Also, none of the 12 H5N1 isolates from environment had the HA cleavage site, signifying the fact that this mutation was prevalently present in low‐pathogenic H5N1 viruses isolated from environment. However, all the six human isolates having N66S mutation in PB1F2 protein isolated from Hong Kong (five in 1997; one in 2001) were found to be highly pathogenic (Table 3). N66S mutation in the PB1F2 protein has been earlier reported in H5N1 strains isolated in 1997 Hong Kong outbreak and H1N1 virus of the 1918 Spanish flu and found to contribute to the increased virulence in mice. 17
Table 3.
No. of strains showing N66S and N84S mutation in the PB1F2 protein from a total of 919 H5N1 viruses isolated from various hosts
| Mutation | Host | Total no. of strains | No. of strains harboring mutation (%) | Polybasic HA cleavage site* (%) |
|---|---|---|---|---|
| N66S | 919 | 35 (3·8) | 7 (20) | |
| Human | 220 | 6 (2·7) | 6 (100) | |
| Avian | 666 | 17 (2·5) | 1 (5·5) | |
| Environment | 16 | 12 (70·5) | 0 | |
| N84S | 919 | 86 (9·35) | 74 (86) | |
| Human | 220 | 27 (12·2) | 27 (100) | |
| Avian | 666 | 58 (8·7) | 46 (79·3) | |
| Environment | 16 | 1 (6·25) | 1 (100) |
*There was variation seen in the HA cleavage site, most of the strains harbored RERRKKR site, while few strains had RERRRKKR; in some strains from human it was RESRRKKR, and in few avian strains it was GERRRKKR.
A total of 32 PB1F2 sequences of H5N1 strains isolated from India were available in the flu database, of which eight were isolated from an outbreak in Western part of India in 2006, and the rest 24 were from east and northeastern part of India isolated between the years 2007 and 2010. Of 32 strains available in the data base, seven viruses were reported in our previous studies including a unique variant isolated from Manipur, a northeastern state of India, which had 13 unique amino acid substitutions in all the eight gene segments. 18 It had an N‐terminal‐truncated 52 amino acid (C52aa) PB1F2 protein. Based on the available PB1F2 protein sequences of H5N1 isolates in the influenza virus database, our analysis showed that only 15 (1·63%) H5N1 isolates had an N‐terminal‐truncated PB1F2 protein consisting of 52 amino acids, of which four (26·6%) were from India (Figure 2). These four sequences with truncation in PB1F2 protein grouped together when an evolutionary relationship was determined as depicted in Figure 2. Alignment of the amino acid sequences of the Indian strains revealed that five of the 32 strains showed a mutation at the 84 position from aspargine to serine (designated as N84S). Phylogenetic tree drawn using the Neighbor‐Joining method (mega 5.0) of 32 Indian strains with few reference strains showed that five strains with N84S mutation branched out as shown in the Figure 2. Global data analysis of the N84S mutation in PB1F2 protein of H5N1 isolates revealed that 86 (9·35%) strains harbored this mutation Table 3. The 86 strains included 58 from avian strain, 27 from human, and one from environment (Table 3). Among the 58 avian strains having N84S mutation, 46 (79%) were from HPAI, because they harbored the polybasic HA cleavage site. All the strains isolated from human host and the one environmental isolate harbored the polybasic HA cleavage site in their HA gene sequence, indicating that they were HPAI. Although this mutation is present only in 9·35% of the total H5N1 strains available in the database, of importance is the fact that it is also present in 1996 Guangdong strain (A/Goose/Guangdong/1/96 (H5N1) and in the 13 HPAI (human‐7; chicken‐6) strains of the 1997 Hong Kong outbreak (Figure 2). This observation from our study highlights the fact that this mutation might have significant role in the pathogenicity of the HPAI H5N1 viruses; however, more studies are warranted to understand the effect of the mutation both in vitro and in vivo studies.
Figure 2.
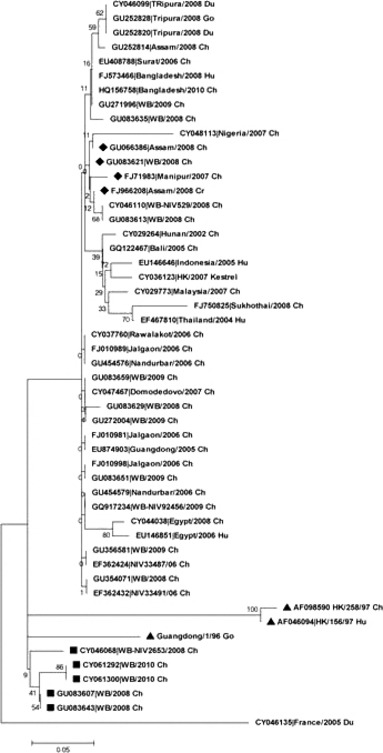
Phylogenetic tree was constructed using Neighbor‐Joining method, the bootstrap consensus tree was inferred from 500 replicates, and this was conducted in mega5. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method. ■– Five Indian H5N1 strains that branched out together and possessed N84S mutation. ▲– A/Goose/Guangdong/1/96, A/Chicken/Hong Kong/258/97, and A/Hong Kong/156/97 strains harboring N84S mutation and grouping with the aforementioned five Indian strains. ◆ Four Indian strains harboring a N‐terminal‐truncated PB1F2 fragment (C52). Ch, chicken; Go, goose; Du, duck; Hu, human; WB, West Bengal, India; HK, Hong Kong; NIV, National Institute of virology, Pune, India.
H2N2 subtype was responsible for the 1957 influenza pandemic, which was called the “Asian flu.” It was initially identified in Hong Kong and involved 250 000 people within a brief span of time. A total of 83 PB1F2 protein sequences of H2N2 were available in the database, of which 54 (65·9%) were isolated from human, 22 (26·8%) from avian species, one (1·2%) from environment, six (7·31%) from guinea pigs, and none from the swine (Table 1). All the 54 viruses from human host were isolated during the Asian flu from 1957 to 1968, and of them, 52 (96·2%) showed complete PB1F2 fragment, while 1 (A/Albany/4/1967(H2N2)) had 101 amino acid fragment and other (A/Cottbus/1/1964(H2N2)) had a C‐terminal‐truncated 57aa fragment designated as N57aa. However, all the 29 viruses from the avian species, environment, and guinea pig had a complete 90aa PB1F2 fragment. Multiple alignment of the H2N2 sequences revealed 52 of the 90 amino acid positions were conserved in 96·6% of the strains. Phylogenetic analysis of the H2N2 PB1F2 protein has been described in Figure 3, which indicates that the protein has evolved over the time period and grouped in time specific manner. However, it is important to understand that these observations are based on the only 83 PB1F2 sequences that were available in the database, and we believe the number of strains is fewer than the other subtypes. Mutational analysis for the N66S mutation in the 83 H2N2 strains revealed that 24 (28·91%) harbored the mutation (Table 4). This mutation was present only in 3·70% of the human strains, while in avian strains, it was present in 72·7% of the strains. After 11 years of the Asian flu pandemic, the circulating H2N2 was replaced by H3N2 subtype of influenza A virus in 1968 and a new pandemic strain emerged as Hong Kong flu. This subtype retained the NA antigen (N2) from the previous pandemic, hence was thought to have variable and sporadic impact in different parts of the world. There were 2566 PB1F2 sequences belonging to the H3N2 subtype in the flu database, of which 2419 (94·27%) were from humans, isolated from the years 1968 till 2011. A total 2048 (84·6%) strains had a complete PB1F2 fragment that included 1979 (81·8%) with 90aa, 44 (1·81%) with 87aa, and 25 (1·03%) with 101aa fragment (Table 1). Three hundred and twenty‐four (13·4%) strains had an N‐terminal‐truncated 52 amino acid (C52aa) fragment. Forty‐seven strains with varied size protein included fragments having C‐terminal‐truncated protein designated as N79 (79aa; 21), N63 (63aa; 17), N57 (57aa; 4), N76 (76aa; 1), and N81 (81aa; 2), while two strains had N‐terminal‐truncated C38 and C42 fragments. The database revealed that of 37 strains isolated during the pandemic in the year of 1968, 34 (91·8%) harbored full‐length PB1F2 protein. N66S mutational analysis for the H3N2 strains revealed that this mutation was present in only 3·96% of the strains (Table 4). Further analysis revealed that N66S mutation was present only 1·24% of the strains from human and 77·94% of the avian strains.
Figure 3.
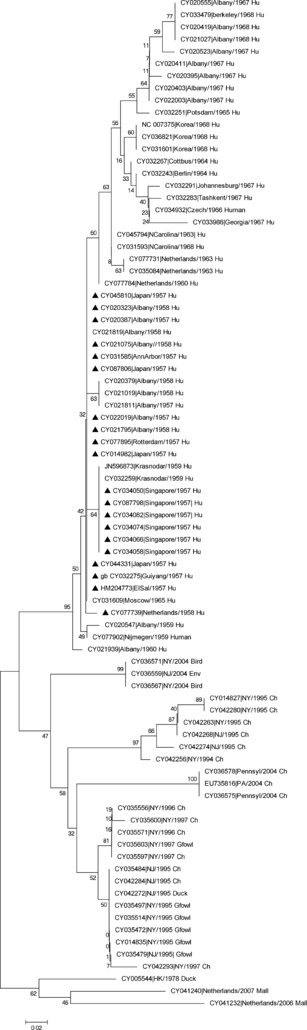
Phylogenetic tree was constructed using Neighbor‐Joining method, the bootstrap consensus tree was inferred from 500 replicates, and this was conducted in mega5. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method. ▲ H2N2 strains isolated during the pandemic 1957–1958. Ch, chicken; Mall, mallard; Gfowl, guinea fowl; Hu, human.
Table 4.
Strains of H2N2 and H3N2 subtypes showing N66S mutation in the PB1F2 protein from a various hosts
| Mutation | Host | Total no. of strains | N66S mutation (%) |
|---|---|---|---|
| H2N2 | 83 | 24 (28·91) | |
| Human | 54 | 2 (3·70) | |
| Avian | 22 | 16 (72·7) | |
| Environment | 1 | 1 (100) | |
| Guinea pig | 6 | 5 (83·3) | |
| H3N2 | 2566 | 89 (3·96) | |
| Human | 2419 | 30 (1·24) | |
| Avian | 68 | 53 (77·94) | |
| Swine | 76 | 3 (3·94) | |
| Environment | 16 | 2 (12·5 | |
| Dog | 1 | 1 (100) |
A total of 1530 PB1F2 sequences for H1N1 influenza virus strains were available in the database which were isolated in the past 80 years (1931 till November 2011). Analysis revealed that 1155 (75·5%) were from humans (Table 1), of which only 73 (6·56%) strains had a complete (90aa or 87aa) PB1F2 fragment. Among the 73 strains harboring full‐length PB1F2 fragment, 62 were isolated between the years 1933 and 1949 (data not shown). This observation is in conjunction with earlier report. 14 Beyond the year 1949, most of the PB1F2 proteins were truncated, because a stop codon was introduced at position 58, leading to C‐terminal‐truncated protein. One thousand and seventy‐three (92·7%) strains had a C‐terminal‐truncated PB1F2 protein fragment having 57 amino acids, while six had N‐terminal‐truncated PB1F2 protein having 52 amino acids. Our analysis revealed that from 1978 onwards, there were only 11 H1N1 strains with complete (90 or 87aa) PB1F2 protein (isolated between the years 1978 and 2009). These strains included CY021723 (A/California/10/1978), CY028730 (A/California/45/1978), CY019745 (A/Memphis/1/1979), CY026417 (A/Albany/8/1979), CY021915 (A/USSR/46/1979), DQ415295 (A/TW/3355/1997), JF758484 (A/South Dakota/03/2008), GQ457565 (A/Saskatchewan/5350/2009), GQ457567 (A/Saskatchewan/5351/2009), GQ457566 (A/Saskatchewan/5131/2009), and CY079535 A/Switzerland/9356/2009). Further study would be very interesting on these PB1F2 genes. Among the 261 (17%) strains from swine, 44 (16·85%) had a complete PB1F2 fragment, while unlike in human isolates, 202 (77·4%) had an N‐terminal‐truncated 52 amino acid fragment and only seven C‐terminal‐truncated 57aa strains. There were 112 (7·3%) from avian species among which 99 (88·4%) had complete 90aa PB1F2 fragment, while other 13 (11·6%) had an N‐terminal‐truncated 52aa fragment. Zell et al. in 2007 studied PB1 segment of 2226 viral strains and found that regardless of sequence origin, 87% of the strains encoded for PB1F2 protein and their size was >78aa. 15 They concluded with their data analysis of PB1F2 sequences that evolutionary selection seems to force the C‐terminal truncation and N‐terminal preservation of PB1F2. The recent 2009 H1N1 pandemic strain is a reassortant virus comprising of genes derived from avian, swine, and human viruses, and it expresses only a C‐terminal‐truncated 11 amino acid PB1F2 protein. 15 Its open reading frame has three stop codons that prevents the expression of full‐length protein. 15
Discussion
PB1F2 is a recently discovered protein in influenza viruses and is thought to have multitude of different function, which are thought to play important role in viral pathogenesis. However, the precise role of PB1F2 protein is still unclear in the life cycle of the virus. This study deals with comprehensive global data analysis of the PB1F2 protein of various influenza A subtypes, and our data are based on the analysis of the all the available PB1F2 sequences from the influenza virus data base that may not represent all the influenza A viruses found in nature.
In many of the subtypes of influenza viruses isolated from different hosts, PB1F2 is truncated either from the C‐terminal or N‐terminal end, thus questioning about their evolutionary utility in different hosts. Analysis of the PB1F2 protein sequence of influenza subtype H5N1 revealed that approximately 96% of the H5N1 strains possessed complete PB1F2 fragment, suggesting that as PB1F2 is positively selected in these strain, it would definitely be necessary for the virus. Of immense interest is the fact that 10 (11·1%) amino acids were found conserved in 666 H5N1 PB1F2 sequences isolated from the avian host and in contrast much more (41 amino acids, 45·5%) amino acids were found conserved in the 220 H5N1 PB1F2 sequences isolated from human host. Similar results were obtained in another study where among all the 11 segments of influenza A viruses, only PB1F2 protein showed diversity between H5N1 strains from human and avian hosts. 19
PB1F2 has an intrinsic property to localize to the mitochondria causing alteration in its morphology and ultimately dissipating the mitochondrial membrane potential and inducing apoptosis. 7 Fine mapping of the protein revealed that there is a mitochondrial targeting sequence (MTS) in the C‐terminal end that enables interaction with mitochondria. 20 , 21 The 18 amino acids conserved domain (52–69aa positions) described in the H5N1 strains from the human host in this study is part of the MTS, and probably these amino acids would play role to maintain proapoptotic function of this protein. Analysis was also performed for the amino acid positions (Leu64, Glut69, Lys73, Arg75, and Val76) that have been earlier reported to be essential and most important for the mitochondrial targeting. 20 , 21 , 22 The analysis revealed that the aforementioned five amino acids were conserved in 92·1% of the H5N1 strains from chicken, 88·4% in ducks, and 90·9% in humans. Among the five amino acid positions, maximum variability was in the position Arg 75. We believe this data would be very useful in enriching our understanding regarding the structure and function of PB1F2 protein. It might also help in developing vaccine targets and identifying both host‐specific and also high‐mortality genetic markers.
Analysis of the pathogenic mutation N66S revealed that it was present only in 35 (3·8%) of 919 H5N1 strains. It was present in six (2·7%) of the 220 H5N1 strains analyzed from human host, in 17 (2·5%) of the 666 strains from the avian host, and in 12 (low‐pathogenic H5N1) of the 17 (70%) strains isolated from environment. This mutation was not present in any of the viruses recently isolated from human in Indonesia, Vietnam, or Egypt. Therefore, considering this mutation as markers for high pathogenicity is questionable in the current scenario. In search of the pathogenic marker for H5N1 viruses, we identified a mutation (N84S) at the C‐terminal end of the PB1F2 protein. This mutation was present in 86 (9·35%) of all the 919 H5N1 strains. Although both the amino acids are polar, uncharged, and hydrophilic, we hypothesize this mutation might bring about the change in mRNA folding and rate of protein translation, which may bring about changes in the way the protein functions. However, this data need to be validated by both in vivo and in vitro experiments.
Analysis of data available from the flu database revealed that in the last decade, protein sequences of only 13 strains belonging to H2N2 subtype have been added. This highlights the fact that viruses of H2N2 subtype are not circulating in humans and other hosts and also there is waning immunity against this subtype in the population. 23 In this scenario, the likelihood of re‐emergence of this virus looms, which makes it increasingly important to study the virus and develop strategy to mitigate it in case of emergence. The analysis of PB1F2 sequences of H2N2 subtype revealed that most of the strains harbored full‐length PB1F2 protein, and more than half of its amino acids were conserved in the strains. Unlike the H2N2 subtype, viruses of the H3N2 have been circulating since the pandemic of 1968. Analysis of the database showed that 84·8% of the strains had a complete PB1F2 protein, and in the rest, the N‐terminal end of the protein was truncated. Analysis for the N66S mutation in the strains belonging to the H2N2 and H3N2 subtypes revealed that this mutation is enriched in the avian strains. Hence, it could be considered as pathogenic marker in the avian species belonging to these two subtypes. PB1F2 sequences analysis of the H1N1 subtype revealed that beyond the year 1949, these strains harbored a truncated form of the protein. In human host, N‐terminal end of the protein was preserved, while in another mammalian host, that is, swine, it was C‐terminal which was retained. Absence of PB1F2 from 2009 H1N1 pandemic shows that it is not an essential protein for the fitness of the 2009 pandemic H1N1 strains. At present, this pandemic strain is not highly pathogenic, but in future, there is possibility it might reassort with a circulating strain like seasonal H3N2 that expresses a full‐length PB1F2 protein and become more virulent; therefore, it is very important to study this protein.
There is immense interest and urgency in understanding the viral factors involved in infection with influenza A viruses. It is also very important to be prepared for emergence of viral strain that may be a reassortant or a mutated strain of the presently circulating influenza viruses or an entirely new reassortant novel strain in causing pandemic. This is mainly important in the current scenario wherein the pandemic of 2009 took the healthcare workers and scientist by surprise and exposed our weak armamentarium and preparedness toward tackling the influenza virus infection. In this study, we found varying sizes of the PB1F2 protein in different influenza A virus subtypes. The genetic divergence of the protein in various hosts highlighted the host‐specific evolution in the virus. Studies are required to correlate this sequence variability of PB1F2 with the virulence and pathogenicity of the virus, which might aid in predicting the pathogenicity of the emerging viral strains in the human population and aid in developing effective vaccines and therapeutics.
Acknowledgement
The study was supported by the Indian Council of Medical Research, Government of India.
References
- 1. Taubenberger JK, Reid AH, Lourens RM et al. Characterization of the 1918 influenza virus polymerase genes. Nature 2005; 437:889–893. [DOI] [PubMed] [Google Scholar]
- 2. Palese P, Shaw ML. Orthomyxoviridae: the viruses and their replication; in Knipe DM, Howley PM. (eds): Fields Virology. Philadelphia, PA: Lippincott Williams & Wilkins, 2007; 1647–1689. [Google Scholar]
- 3. Kawaoka Y, Krauss S, Webster RG. Avian‐to‐human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J Virol 1989; 63:4603–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Karasin AI, Schutten MM, Cooper LA et al. Genetic characterization of H3N2 influenza viruses isolated from pigs in North America, 1977–1999: evidence for wholly human and reassortant virus genotypes. Virus Res 2000; 68:71–85. [DOI] [PubMed] [Google Scholar]
- 5. Tumpey TM, Basler CF, Aguilar PV et al. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 2005; 310:77–80. [DOI] [PubMed] [Google Scholar]
- 6. Zamarin D, Ortigoza MB, Palese P. Influenza A virus PB1‐F2 protein contributes to viral pathogenesis in mice. J Virol 2006; 80:7976–7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen W, Calvo PA, Malide D et al. A novel influenza A virus mitochondrial protein that induces cell death. Nat Med 2001; 7:1306–1312. [DOI] [PubMed] [Google Scholar]
- 8. Košík I, Krejnusová I, Bystrická M et al. N‐terminal region of the PB1‐F2 protein is responsible for increased expression of influenza A viral protein PB1. Acta Virol 2011; 55:45–53. [DOI] [PubMed] [Google Scholar]
- 9. Conenello GM, Tisoncik JR, Rosenzweig E et al. A single N66S mutation in the PB1‐F2 protein of influenza A virus increases virulence by inhibiting the early interferon response in vivo. J Virol 2011; 85:652–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McAuley JL, Hornung F, Boyd KL et al. Expression of the 1918 influenza A virus PB1‐F2 enhances the pathogenesis of viral and secondary bacterial pneumonia. Cell Host Microbe 2007; 2:240–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mazur I, Anhlan D, Mitzner D, Wixler L, Schuber U, Ludwig S. The proapoptotic influenza A virus protein PB1‐F2 regulates viral polymerase activity by interaction with the PB1 protein. Cell Microbiol. 2008; 10:1140–1152. [DOI] [PubMed] [Google Scholar]
- 12. Le Goffic R, Leymarie O, Chevalier C et al. Transcriptomic analysis of host immune and cell death responses associated with the influenza A virus PB1‐F2 protein. PLoS Pathog 2011; 7:e1002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Krumbholz A, Philipps A, Oehring H. Current knowledge on PB1‐F2 of infuenza A viruses. Med Microbiol Immunol 2011; 200:69–75. [DOI] [PubMed] [Google Scholar]
- 14. Zell R, Krumbholz A, Eitner A et al. Prevalence of PB1‐F2 of influenza A viruses. J Gen Virol 2007; 88:536–546. [DOI] [PubMed] [Google Scholar]
- 15. Wang TT, Palese P. Unraveling the mystery of swine influenza virus. Cell 2009; 137:983–985. [DOI] [PubMed] [Google Scholar]
- 16. Squires B, Macken C, Garcia‐Sastre A et al. BioHealthBase: informatics support in the elucidation of influenza virus host pathogen interactions and virulence. Nucleic Acids Res 2008;36:D497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Conenello GM, Zamarin D, Perrone LA et al. A single mutation in the PB1‐F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog 2007; 3:1414–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mishra AC, Cherian SS, Chakrabarti AK et al. A unique influenza A (H5N1) virus causing a focal poultry outbreak in 2007 in Manipur, India. Virol J 2009; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heiny AT, Miotto O, Srinivasan KN et al. Evolutionarily conserved protein sequences of influenza A viruses, avian and human, as vaccine targets. PLoS ONE 2007; 2:e1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gibbs JS, Malide D, Hornung F, Bennink JR, Yewdell JW. The influenza A virus PB1‐F2 protein targets the inner mitochondrial membrane via a predicted basic amphipathic helix that disrupts mitochondrial function. J Virol 2003; 77:7214–7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yamada H, Chouna R, Higashi Y, Kurihara N, Kido H. Mitochondrial targeting sequence of the influenza A virus PB1‐F2 protein and its function in mitochondria. FEBS Lett 2004; 578:331–336. [DOI] [PubMed] [Google Scholar]
- 22. Danishuddin M, Khan SN, Khan AU. Molecular interactions between mitochondrial membrane proteins and the C‐terminal domain of PB1‐F2: an in silico approach. J Mol Model 2010; 16:535–541. [DOI] [PubMed] [Google Scholar]
- 23. Nabel GJ, Wei C‐J, Ledgerwood JE. Vaccinate for the next H2N2 pandemic now. Nature 2011; 471:157–158. [DOI] [PubMed] [Google Scholar]


