Abstract
Please cite this paper as: Peci et al. (2012) Community‐acquired respiratory viruses and co‐infection among patients of Ontario Sentinel practices, April 2009 to February 2010. Influenza and Other Respiratory Viruses 7(4), 559–566.
Background Respiratory viruses are known to cocirculate but this has not been described in detail during an influenza pandemic.
Objectives To describe respiratory viruses, including co‐infection and associated attributes such as age, sex or comorbidity, in patients presenting with influenza‐like illness to a community sentinel network, during the pandemic A(H1N1)pdm09 in Ontario, Canada.
Methods Respiratory samples and epidemiologic details were collected from 1018 patients with influenza‐like illness as part of respiratory virus surveillance and a multiprovincial case–control study of influenza vaccine effectiveness.
Results At least one virus was detected in 668 (65·6%) of 1018 samples; 512 (50·3%) had single infections and 156 (15·3%) co‐infections. Of single infections, the most common viruses were influenza A in 304 (59·4%) samples of which 275 (90·5%) were influenza A(H1N1)pdm09, and enterovirus/rhinovirus in 149 (29·1%) samples. The most common co‐infections were influenza A and respiratory syncytial virus B, and influenza A and enterovirus/rhinovirus. In multinomial logistic regression analyses adjusted for age, sex, comorbidity, and timeliness of sample collection, single infection was less often detected in the elderly and co‐infection more often in patients <30 years of age. Co‐infection, but not single infection, was more likely detected in patients who had a sample collected within 2 days of symptom onset as compared to 3–7 days.
Conclusions Respiratory viral co‐infections are commonly detected when using molecular techniques. Early sample collection increases likelihood of detection of co‐infection. Further studies are needed to better understand the clinical significance of viral co‐infection.
Keywords: Co‐infection, 2009 pandemic H1N1, respiratory viruses
Background
A novel influenza virus, A(H1N1)pdm09 emerged in April 2009 and spread rapidly, primarily through human‐to‐human transmission. Several million people were infected globally. 1 An important feature of this virus was that it mostly affected younger people with 60% of patients under 18 years of age, suggesting possible pre‐existing immunity in the elderly due to previous exposure to antigenically related influenza strains. 2 , 3
The assumption made in most pandemic plans before 2009 was that the pandemic virus would be the dominant circulating respiratory virus. 4 Few studies performed extensive respiratory testing beyond influenza during the pandemic, and fewer still focused on community cases. Casalegno et al. documented cocirculation and co‐infection of A(H1N1)pdm09 with rhinovirus during the pandemic. 5 Watanabe et al. found a wide range of etiologic agents were identified among respiratory samples that were influenza negative, highlighting the need to diagnose other viral organisms that can co‐circulate with influenza. 6 Louie et al. investigated samples from laboratory‐confirmed fatal A(H1N1)pdm09 cases during the pandemic and bacterial pathogens were identified in 22 of 77 samples. 7
Prior to the 2009 pandemic, respiratory viral co‐infection was reported in 7–27% of respiratory samples submitted for viral diagnosis. 8 , 9 , 10 , 11 , 12 , 13 Higher proportions of influenza A, respiratory syncytial virus (RSV), parainfluenza viruses, and rhinovirus, compared with other circulating viruses have been detected in patients with co‐infections. 14 , 15 , 16 , 17 , 18
Co‐infection has not been fully explored due to limitations of several studies. Some studies focused on younger age groups, hospitalized patients or deceased individuals, which does not represent the general population. 8 , 9 , 11 , 14 , 18 , 19 , 20 Others have utilized a small sample size or limited their focus to certain viral pathogens, underestimating the role of other viruses in co‐infection. 9 , 15 , 16 , 20 , 21
This study enrolled community patients presenting with (ILI) to a community sentinel network, during the influenza pandemic A(H1N1)pdm09 in Ontario, Canada and documented the profile of respiratory viruses causing ILI symptoms. This study aimed to describe respiratory viruses including co‐infections and host‐associated attributes such as age, sex, and comorbidity.
Methods
Study population
Data were collected as part of a multiprovincial case–control sentinel network study that has been described elsewhere. 21 The sentinel network included 117 sentinels across the province of Ontario (with a population of 13·4 million) who volunteered to participate in the study. It was anticipated that each sentinel would submit an average of 1–2 samples per week from their clinical practice during the study period, April 21, 2009 to February 25, 2010. This period was chosen to span the full pandemic in Ontario. Eligible patients were Ontario residents, who presented to a sentinel’s office with influenza‐like illness (ILI) within seven days of symptom onset; number and selection of eligible patients was at the sentinel’s discretion. ILI was defined as acute onset of fever and cough and one or more of the following: sore throat, myalgia, arthralgia, headache or prostration. Standard information was collected including date of birth, sex, chronic conditions, symptom onset, and sample collection date. The main outcome was the number of respiratory viruses detected per sample. Samples were categorized as negative, single infection or co‐infection when no virus, one virus, or at least two viruses were detected, respectively. Age was determined as age at symptom onset and categorized as 0–4, 5–14, 15–29, 30–54, 55–64, and 65+years. Time to sample collection was calculated as the difference between sample collection and symptom onset dates and categorized as less than or equal to two days or 3–7 days. Chronic condition was defined as heart/lung/renal/metabolic/blood/immune conditions or conditions that compromise the management of respiratory secretions and increase risk of aspiration and categorized as yes/no. This study was approved by the University of Toronto’s Ethics Board and all patients gave verbal consent to participate.
Viral diagnosis
A nasal or nasopharyngeal sample was collected from each patient using Starswab® Multitrans collection swab and transported at 4°C for testing at Public Health Ontario Laboratory (PHOL)‐Toronto; in this study, each sample represents one patient. Viral RNA was extracted directly from samples using NucliSENS® easyMAG® (BioMérieux, Inc., Marcy l’Etoile, France). Samples were tested for influenza A and influenza B by influenza real‐time reverse transcriptase (rRT)‐PCR and also for influenza A, influenza B, enterovirus/rhinovirus, RSV, parainfluenza, adenovirus, coronaviruses, and metapneumovirus by a commercial multiplex PCR method [Luminex Respiratory Viral Panel (Luminex Molecular Diagnostics, Toronto, ON, Canada) or Seeplex RV (Seegene USA, Rockville, MD, USA)]. In the event of discrepant results between the two methods, positive results for influenza A by either method were considered positive.
rRT‐PCR was used for subtyping of influenza A samples; all influenza A specimens were subtyped, but not all attempts were successful.
Statistical analysis
Statistical analyses were performed using STATA software version 10.0 (StataCorp, College Station, TX, USA).
Descriptive analyses were conducted to derive the proportion of single, co‐infection and no infection as well as describe patient characteristics using Chi‐Square. Crude and adjusted multinomial logistic regression were employed to evaluate any association of single, co‐infection and no‐infection with patient characteristics including age, sex, chronic condition, and time to sample collection. Odds ratios (OR) with 95% confidence intervals (CI) were calculated.
Results
Respiratory viruses detected
A total of 1018 respiratory samples from 1018 patients with influenza‐like illness were included in this study after excluding 102 (9·1%) samples that did not meet study inclusion criteria (Figure 1). At least one respiratory virus was detected in 668 (65·6%) of the samples. Of the 831 detected viruses, influenza A was the most frequent accounting for 452 (54·4%) followed by enterovirus/rhinovirus 194 (23·3%) and RSV 120 (14·4%) (Table 1). Of 452 influenza A viruses, 408 (90·3%) were A(H1N1)pdm09, two (0·4%) were H3, and 42 (9·3%) could not be subtyped presumably due to low viral load. Peaks in detection for influenza A occurred in June and October 2010, for enterovirus/rhinovirus in September 2010, and for RSV in October 2010 (Figure 2).
Figure 1.
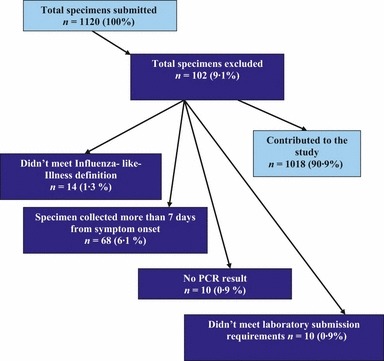
Study inclusion criteria, April 21, 2009 to February 25, 2010, sentinel network, Ontario, Canada.
Table 1.
Frequency of detected respiratory viruses among 1018 respiratory samples tested, April 21, 2009 to February 25, 2010, sentinel network, Ontario, Canada
| Virus detected | Counts n (%) | Total n (%) |
|---|---|---|
| Influenza A/(H1N1)pdm09 | 408 (49·1) | |
| Influenza A/H3 | 2 (0·2) | |
| Influenza A/not subtyped | 42 (5·1) | |
| Influenza A subtotal | 452 (54·4) | |
| Enterovirus/rhinovirus | 129 (15·5) | |
| Human Rhinovirus | 65 (7·8) | |
| Enterovirus/rhinoviruses subtotal | 194 (23·3) | |
| Respiratory Syncytial A | 12 (1·4) | |
| Respiratory Syncytial B | 108 (13·0) | |
| Respiratory Syncytial subtotal | 120 (14·4) | |
| Parainfluenza 1 | 19 (2·3) | |
| Parainfluenza 2 | 9 (1·1) | |
| Parainfluenza 3 | 15 (1·8) | |
| Parainfluenza 4 | 3 (0·4) | |
| Parainfluenza subtotal | 46 (5·5) | |
| Coronavirus 229E/NL63* | 4 (0·5) | |
| Coronavirus HKU1 | 1 (0·1) | |
| Coronavirus OC43 | 4 (0·5) | |
| Coronavirus subtotal | 9 (1·1) | |
| Adenovirus | 6 (0·7) | 6 (0·7) |
| Human metapneumovirus | 4 (0·5) | 4 (0·5) |
| Total viruses detected | 831 (100) | 831 (100) |
*Include Coronavirus 229 E and Coronvairus 229E/NL63.
Figure 2.
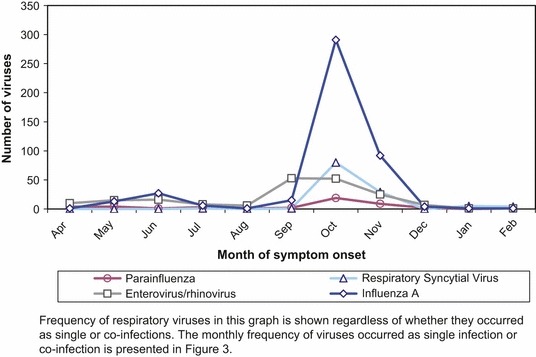
Number of detection of the most common respiratory viruses by month of symptom onset, April 21, 2009 to February 25, 2010 sentinel network, Ontario, Canada.
A single virus was detected in 512 (50·3%) samples. Of these, 304 (59·4%) were influenza A and 208 (40·6%) were other respiratory viruses, the most common being enterovirus/rhinovirus, detected in 149 (29·1%) samples (Table 2). Peaks for single infection occurred in June and September 2009, which were mainly due to the increase in influenza A and enterovirus/rhinovirus, respectively (Figure 3).
Table 2.
Number and percent of viruses detected as single infections, April 21, 2009 to February 25, 2010, sentinel network, Ontario, Canada
| Single infection viruses | Counts n (%) | Total n (%) |
|---|---|---|
| Influenza A | 27 (5·3) | |
| Influenza (H1N1)pdm09 | 275 (53·7) | |
| Influenza A/H3 | 2 (0·4) | |
| Influenza A subtotal | 304 (59·4) | |
| Enterovirus/rhinovirus | 116 (22·7) | |
| Human rhinovirus | 33 (6·4) | |
| Enterovirus/rhinovirus subtotal | 149 (29·1) | |
| Parainfluenza 1 | 12 (2·3) | |
| Parainfluenza 2 | 5 (1·0) | |
| Parainfluenza 3 | 11 (2·1) | |
| Parainfluenza 4 | 3 (0·6) | |
| Parainfluenza subtotal | 31 (6·1) | |
| Respiratory Syncytial A | 9 (1·8) | |
| Respiratory Syncytial B | 3 (0·6) | |
| Respiratory Syncytial subtotal | 12 (2·4) | |
| Coronavirus 229E/NL63* | 4 (0·8) | |
| Coronavirus HKU1 | 1 (0·2) | |
| Coronavirus OC43 | 2 (0·4) | |
| Coronavirus subtotal | 7 (1·4) | |
| Adenovirus | 5 (1·0) | 5 (1·0) |
| Human Metapneumovirus | 4 (0·8) | 4 (0·8) |
| Total viruses detected | 512 (100) | 512 (100) |
*Includes Coronavirus 229 E and Coronavirus 229E/NL63.
Figure 3.
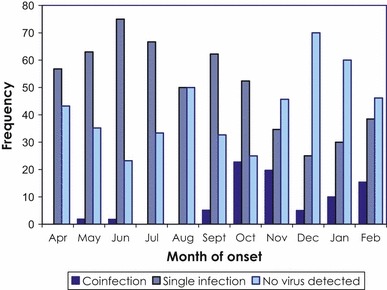
Frequency of single infection and co‐infections by month of symptom onset, April 21, 2009 to February 25, 2010, sentinel network, Ontario, Canada.
Viral co‐infection was detected in 156 (15·3%) of the samples of which 149 (95·5%) were dual infections and seven (4·5%) triple infections. One hundred and forty‐eight (94·9%) of the co‐infections were combination of A(H1N1)pdm09 and another respiratory virus and eight (5·1%) were non‐influenza combinations. The most common co‐infections were influenza A/RSV B and influenza A/enterovirus/rhinovirus, responsible for 64·1% and 21·8% of co‐infections, respectively (Table 3).The highest proportion of co‐infection was detected in October, corresponding with peak activity of influenza A and RSV (Figure 3).
Table 3.
Number and percent of viruses detected in samples where co‐infection was identified, April 21, 2009‐February 25, 2010 sentinel network, Ontario, Canada
| Co‐infection viruses | Counts n (%) |
|---|---|
| Influenza A*/Respiratory Syncytial B | 100 (64·1) |
| Influenza A/Enterovirus/rhinovirus† | 34 (21·8) |
| Parainfluenza 1/Enterovirus/rhinovirus | 5 (3·2) |
| Influenza A/Enterovirus/rhinovirus /Respiratory Syncytial | 4 (2·6) |
| Influenza A/Parainfluenza 3 | 3 (1·9) |
| Influenza A/Parainfluenza 1/Enterovirus/rhinovirus | 2 (1·3) |
| Influenza A/Parainfluenza 2 | 2 (1·3) |
| Respiratory Syncytial/Coronavirus OC 43 | 2 (1·3) |
| Influenza A/Adenovirus | 1 (0·6) |
| Influenza A/Respiratory Syncytial A | 1 (0·6) |
| Influenza A/Respiratory Syncytial B /Parainfluenza 2 | 1 (0·6) |
| Parainfluenza 2/Parainfluenza 3 | 1 (0·6) |
| Total | 156 (100) |
*All influenza A samples were influenza A/(H1N1) pdm09 except 15 samples which were unable to subtype due to low viral load.
†Enterovirus/rhinovirus includes human rhinovirus and enterovirus/rhinovirus.
Patient characteristics
The median age of patients in the study was 23 years with a range of 3 months–96 years of age (Table 4). The highest proportion of single and co‐infections was observed in children 5–14 and 0–4 years of age, respectively. The proportion of those with no infection detected steadily increased with age, peaking at the elderly, aged 65 years and over (Figure 4). Females were overrepresented, comprising 599 (58·8%) of the patients included in this study. Patients with no‐infection, single infection, and co‐infection did not differ with regards to sex. Two hundred and nineteen (21·6%) patients had a chronic condition. Of these, 37·4% had no virus detected, 44·8% had single infections, and 17·8% had co‐infections, whereas among the 795 participants without comorbidities, the distribution was 33·6%, 51·8%, and 14·6%, respectively; however, that was not statistically significant (Table 4). The median number of days from symptom onset to sample collection was two with a range of 0–7 days. Five hundred and eighty‐two (57·2%) and 436 (42·8%) samples were collected within 2 days and 3–7 days, respectively. Of the 582 samples collected within 2 days of onset, 32·9% had no virus detected, 48·5% had single infections, and 18·6% had co‐infections, whereas among those collected within 3–7 days, the distribution was 36·3%, 52·8%, and 11%, which was statistically significant.
Table 4.
Characteristics of patients and timing of specimen collection and testing, April 21, 2009 to February 25, 2010, sentinel network, Ontario, Canada
| Patient characteristics | No virus detected n (%) | Single infection n (%) | Co‐infection n (%) | Overall number | Chi‐square P‐value |
|---|---|---|---|---|---|
| Patients | n = 350 (34·4) | n = 512 (50·2) | n = 156 (15·3) | n = 1018 | |
| Age category (years) | |||||
| 0–4 | 28 (27·5) | 51 (50·0) | 23 (22·5) | 102 | <0·001* |
| 5–14 | 44 (22·3) | 115 (58·4) | 38 (19·3) | 197 | |
| 15–29 | 85 (30·1) | 148 (52·3) | 50 (17·7) | 283 | |
| 30–54 | 121 (40·7) | 145 (48·8) | 31 (10·4) | 297 | |
| 55–64 | 34 (44·2) | 38 (49·4) | 5 (6·5) | 77 | |
| 65+ | 38 (61·3) | 15 (24·2) | 9 (14·52) | 62 | |
| Median (Range) | 34 (0–92) | 20 (0–96) | 18 (0–88) | 23 (0–96) | |
| Sex | |||||
| Female | 207 (34·6) | 303 (50·6) | 89 (14·9) | 599 | 0·8 |
| Male | 143 (34·1) | 209 (49·9) | 67 (16·0) | 419 | |
| Chronic Condition | |||||
| No | 267 (33·6) | 412 (51·8) | 116 (14·6) | 795 | 0·4 |
| Yes | 82 (37·4) | 98 (44·8) | 39 (17·8) | 219 | |
| Unknown | 1 (25·0) | 2 (50·0) | 1 (25·0) | 4 | |
| Time to specimen collection** (days) | |||||
| 0–2 | 192 (32·9) | 282 (48·5) | 108 (18·6) | 582 | <0·01* |
| 3–7 | 158 (36·3) | 230 (52·8) | 48 (11·0) | 436 | |
| Median (Range) | 2 (0–7) | 2 (0–7) | 2 (0–7) | 2 (0–7) | |
*P‐value <0·05 is considered significant.
**Time to specimen collection = Collection date‐symptom onset date.
Figure 4.
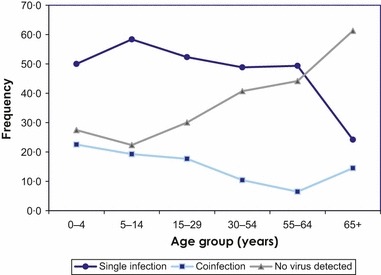
Age distribution of patients by virus detection status, April 21, 2009 to February 25, 2010, sentinel network, Ontario, Canada.
In crude and adjusted multinomial logistic regression, patients with single and co‐infections were compared to those with no infection. Compared to the elderly, patients under 65 years of age were more likely to have a single infection; the highest likelihood was observed in children 5–14 years of age (Table 5). Patients under 30 years of age were more likely to have co‐infections compared with patients 65 and over; this was most evident in the 0–4 age group. Presence of a chronic condition did not increase the likelihood of single infection but increased the likelihood of co‐infection; this did not achieve significance. Co‐infection was more likely detected in patients who had samples collected within 2 days as compared to 3–7 days; this did not apply for those with single infections. There was no sex difference.
Table 5.
Crude and adjusted odds ratio of patient characteristics by infection status using no infection as the reference (n = 1014)*
| Patient characteristics | Number of patients n = 1014 | Single infection n = 512 (50·2%) | Co‐infection n = 156 (15·3%) | ||||
|---|---|---|---|---|---|---|---|
| n (%) | Crude OR OR (95%CI) | Adjusted OR (95%CI) | n (%) | Crude OR OR (95%CI) | Adjusted OR (95%CI) | ||
| Age category (years) | |||||||
| 65+ | 62 | 15 (24·2) | 1·00 | 1·00 | 9 (14·5) | 1·00 | 1·00 |
| 0–4 | 102 | 51 (50·0) | 4·61 (2·16–9·81)* | 4·83 (2·22–10·48)* | 23 (22·6) | 3·46 (1·39–8·63)* | 4·38 (1·64–11·67)* |
| 5–14 | 197 | 115 (58·4) | 6·62 (3·31–13·21)* | 7·03 (3·44–14·35)* | 38 (19·3) | 3·64 (1·56–8·50)* | 4·79 (1·91–12·03)* |
| 15–29 | 283 | 148 (52·3) | 4·41 (2·29–8·48)* | 4·56 (2·32–8·96)* | 50 (17·7) | 2·48 (1·10–5·56)* | 3·32 (1·38–7·98)* |
| 30–54 | 297 | 145 (48·8) | 3·03 (1·59–5·78)* | 3·01 (1·56–5·80)* | 31 (10·4) | 1·08 (0·47–2·47) | 1·32 (0·55 –3·18) |
| 55–64 | 77 | 38 (49·4) | 2·83 (1·32–6·02)* | 2·83 (1·32–6·07)* | 5 (6·5) | 0·62 (0·18–2·03) | 0·73 (0·21–2·49) |
| Sex | |||||||
| Male | 419 | 209 (49·9) | 1·00 | 1·00 | 67 (16·0) | 1·00 | 1·00 |
| Female | 599 | 303 (50·6) | 0·99 (0·75–1·31) | 0·86 (0·64–1·15) | 89 (14·8) | 1·08 (0·74–1·59) | 0·82 (0·55–1·23) |
| Chronic condition | |||||||
| N | 795 | 412 (51·8) | 1·00 | 116 (14·6) | 1·00 | ||
| Y | 219 | 98 (44·8) | 0·77 (0·55–1·07) | 1·02 (0·71–1·45) | 39 (17·8) | 1·09 (0·70–1·69) | 1·56 (0·98–2·51)* |
| Time to specimen collection (days) | |||||||
| 3–7 days | 436 | 230 (52·7) | 1·00 | 1·00 | 48 (11·0) | 1·00 | 1·00 |
| 0–2 days | 582 | 282 (48·5) | 1·00 (0·76–1·32) | 0·93 (0·68–1·21) | 108 (18·6) | 1·85 (1·24–2·76)* | 1·66 (1·12–2·50)* |
OR, odds ratio; CI, confidence interval.
*Four patients were missing chronic condition information and were removed from the adjusted multinomial logistic regression model.
Discussion
In this study, 66% of samples tested during the 2009 pandemic in Ontario had at least one virus detected and 15% had co‐infections. These findings are consistent with reports from other studies with the range of co‐infection reported from 7–27%. 8 , 9 , 10 , 11 , 12 , 13 However, positivity and co‐infection rates vary widely between studies. There are various reasons for this finding: firstly, detection methods differ notably between studies, which impacts sensitivity, specificity and other technical parameters; secondly, viruses targeted differ from one study to another as does the study population. 21
This study was conducted during the influenza pandemic A(H1N1)pdm09 which was associated with an increased number of samples submitted and high detection of A(H1N1)pdm09. Not surprisingly, influenza was the primary virus detected in June and October 2010; however, other viruses dominated for other months during both influenza pandemic waves (Figure 2). This demonstrates the importance of monitoring circulating respiratory viruses when advising clinicians to prescribe antivirals empirically during a pandemic.
Despite the higher prevalence of enterovirus/rhinovirus (23·4%) than RSV (14·5%), co‐infection with A(H1N1)pdm09/RSV was more common than A(H1N1)pdm09/enterovirus/rhinovirus, accounting for 64·1% and 22·8% of the co‐infections, respectively. This may reflect the younger age of patients infected by A(H1N1)pdm09, who were therefore also at greater risk of RSV. In addition, RSV cocirculated with A(H1N1)pdm09 more than enterovirus/rhinovirus, which peaked before the second wave (Figure 2). There may also be preferential interactions among certain pathogens; viral interactions were not assessed in this study. 23 When other respiratory samples positive for enterovirus/rhinovirus were further evaluated at our laboratory, they all were confirmed as rhinovirus, not enterovirus. 24
Single infection was more commonly detected in those less than 65 years of age. It is known that respiratory infections are more common in children for several reasons, including an immature immune system, lack of pre‐existing immunity particularly to new emerging viruses, and greater viral exposure opportunities. 11 , 18 , 25 Younger patients shed higher levels of virus when infected and also may be brought for medical care earlier than older patients, facilitating detection in these groups. 26 In addition, lower detection of single and co‐infection in elderly may be explained by pre‐existing immunity to A(H1N1)pdm09 and other respiratory viruses. 2 Co‐infection was more common in persons less than 30 years of age as compared to older adults. These data are congruent with findings from a previously published study where co‐infection was more likely in younger than older adults. 18 The combined effect of predominance of A(H1N1)pdm09 and the greater likelihood of infection with other respiratory viruses among younger ages likely explains our age‐related findings of co‐infection during the pandemic, which may not be generalizable to a typical influenza season.
The presence of comorbidities did not increase the likelihood of having a single infection but increased the likelihood of co‐infections; this did not achieve statistical significance. Patients with chronic conditions are at higher risk of severe disease and consequently may be more likely to seek medical care. 26 Selection bias is unlikely to influence these results as the proportion of patients with comorbidities was similar (21·5%) to that in Ontario’s population (20·3%). 27
Sample collection within 2 days of symptom onset was found to independently increase the likelihood of detecting a viral co‐infection but not single infection. Long et al. reported an inverse relationship between duration of symptoms and viral detection rate due to greater viral shedding earlier in the disease process. 11
This study was designed to examine circulating viruses and co‐infection. The presence of more than two viruses in the same sample may not always indicate clinical infection. As viruses may be detected in asymptomatic patients, it is impossible to determine which viruses caused symptoms. Previous studies suggest co‐infection may manifest higher disease severity, which may shorten the time to medical care and viral detection; disease severity was not assessed in the current study. 19 , 28 As viral–bacterial co‐infections also occurred during the pandemic, it will be interesting for further studies to investigate their characteristics and impact on disease severity. 28 , 29
In summary, A(H1N1)pdm09 was frequently detected among community patients with ILI. However, other respiratory viruses cocirculated with A(H1N1)pdm09 during the pandemic, reinforcing the need to test for other viral agents even during a pandemic to appropriately guide clinical treatment decisions. Viral diagnosis, primarily A(H1N1)pdm09, was made more often in patients less than 65 years of age. Viral co‐infection was commonly detected in this study and was most likely detected in individuals less than 30 years of age. Earlier sample collection improves the detection of viral co‐infections. Understanding the contribution of other circulating respiratory pathogens during a pandemic may lead to improved individual diagnosis and recommendations for community‐based clinicians, and more effective prevention and treatment of respiratory infections, including use of influenza antivirals.
Conflict of interest
DMS was Principal Investigator on a clinical trial of pediatric influenza vaccine dose response for which influenza vaccine was provided free by Sanofi Pasteur. JBG has received research grants from GlaxoSmithKline Inc. and Hoffman‐La Roche Ltd to study antiviral resistance in influenza.
Acknowledgments
Authors wish to acknowledge the staff of Molecular Diagnostics and Virus Detection departments at Public Health Ontario Laboratory‐Toronto, the Ontario and national sentinel influenza vaccine effectiveness study and the College of Family Physicians of Canada funded by the Canadian Institute of Health Research.
References
- 1. Dawood FS, Jain S, Finelli L et al. Novel swine‐origin influenza A (H1N1) virus in humans. N Engl J Meg 2009; 360:2605–2615. [DOI] [PubMed] [Google Scholar]
- 2. WHO . http://www.who.int/csr/disease/swineflu/h1n1/sruveillance20090710en/index/html. In: Girard MP,Tam JS, Assossou OM, et al. A (H1N1) influenza virus pandemic: A review. Vaccine 2009; 28:4895–4902. [DOI] [PubMed]
- 3. Rothberg MB, Haessler SD. Complications of seasonal and pandemic influenza. Crit Care Med 2010; 38:e91–e97. [DOI] [PubMed] [Google Scholar]
- 4. WHO checklist for influenza pandemic preparedness planning 2005. Department of Communicable Disease Surveillance and Response Global Influenza Programme. Available online: http://http://www.who.int/influenza/resources/documents/FluCheck6web.pdf.
- 5. Casalegno JS, Bouscambert‐Duchamp M, Morfin F et al. Rhinovirus, A (H1N1)v, RVS: The race for hivernal pandemics, France 2009‐2010. Euro Surveill 2009;14, pii=19390. Available Online: http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19390. [PubMed] [Google Scholar]
- 6. Watanabe AS, Carraro E, Moreira L et al. Respiratory virus infections among hospitalized patients with suspected influenza A H1N1 2009 virus during the first pandemic wave in Brazil. CMAJ 2010; 182:257–264. [DOI] [PubMed] [Google Scholar]
- 7. Louie J, Jean C, Chen TH et al. Bacterial coinfections in lung tissue specimens from fatal cases of 2009 Pandemic Influenza A (H1N1). Morb Mortal Wkly Rep 2009; 58:1071–1074. [PubMed] [Google Scholar]
- 8. Choi EH, Lee HJ, Kim SJ et al. The association of newly identified respiratory viruses with lowers respiratory tract infections in Korean children, 2000‐2005. Clin Infect Dis 2006; 43:585–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Papadopoulos NG, Moustaki M, Tsolia M et al. Association of rhinovirus infection with increased disease severity in acute bronchiolitis. Am J Respir Crit Care Med 2002; 165:1285–1289. [DOI] [PubMed] [Google Scholar]
- 10. Aberle JH, Aberle SW, Pracher E et al. Single versus dual respiratory virus infections in hospitalized infants: Impact on clinical course of disease and interferon‐y response. Pediatr Infect Dis J 2005; 24:605–610. [DOI] [PubMed] [Google Scholar]
- 11. Long RB, Westin J, Olofsson S et al. Prospective evaluation of a novel multiplex real‐time PCR assay for detection of fifteen respiratory pathogens. Duration of symptoms significantly affects detection rate. J Clin Virol 2010; 47:589–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bonzel L, Tenenbaum T, Schroten H et al. Frequent detection of viral coinfection in children hospitalized with acute respiratory tract infection using a real‐time polymerase chain reaction. Pediatr Infect Dis J 2008; 27:589–594. [DOI] [PubMed] [Google Scholar]
- 13. Peng D, Zhao DC, Liu JT et al. Multipathogen infections in hospitalized children with acute respiratory infections. Virol J 2009; 6:155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mahony J, Chong S, Merante F et al. Development of a respiratory virus panel test for detection of twenty human respiratory viruses by use of multiplex PCR and a fluid microbead‐based assay. J Clin Microbiol 2007; 45:2965–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Machado CM, Vilas BoasLS, Mendes A et al. Low mortality rates related to respiratory virus infections after bone marrow transplant. Bone Marrow Transplant 2003; 31:695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tang JW, Lai FYL, Wong F et al. Incidence of common respiratory viral infections related to climate factors in hospitalized children in Hong Kong. Epidemiol Infect 2010; 138:226–235. [DOI] [PubMed] [Google Scholar]
- 17. Tregoning JS, Schwarze J. Respiratory viral infections in infants: causes, clinical symptoms, virology, and immunology. Clin Microbiol Rev 2010; 23:74–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ren L, Gonzalez R, Wang Z et al. Prevalence of human respiratory viruses in adults with acute respiratory tract infections in Beijing, 2005‐2007. Clin Microbiol Infect 2009; 15:1146–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Franz A, Adams O, Willems R et al. Correlation of viral load of respiratory pathogens and co‐infections with disease severity in children hospitalized for lower respiratory tract infection. J Clin Virol 2010; 48:239–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Glezen P, Greenberg SB, Atmar RL et al. Impact of respiratory virus infections on persons with chronic underlying conditions. JAMA 2010; 283:499–505. [DOI] [PubMed] [Google Scholar]
- 21. Von Linstow ML, Larsen HH, Olsen JE et al. Human metapneumovirus and respiratory syncytial virus in hospitalized Danish children with acute respiratory tract infection. Scand J Infect Dis 2004; 36:578–584. [DOI] [PubMed] [Google Scholar]
- 22. Skowronski DM, De Serres G, Crowcroft NS et al. Association between the 2008‐09 seasonal influenza vaccine and pandemic H1N1 illness during spring‐summer 2009: four observational studies from Canada. PLoS Med. 2010; 7:e1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brunstein JD, Cline CL, McKinney S et al. Evidence from multiplex molecular assays for complex multipathogen interaction in acute respiratory infections. J Clin Microbiol 2008; 46:97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Longtin J, Marchand‐Austing A, Winter AL et al. Rhinovirus outbreaks in long‐term care facilities, Ontario, Canada. Emerg Infect Dis 2010; 16:1463–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cilla G, Onate E, Perez‐Yarza EG et al. Viruses in community‐ acquired pneumonia in children aged less than 3 years old; High rate of viral confection. J Med Virol 2008; 80:1843–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li CC, Wang L, Eng HL et al. Correlation of pandemic (H1N1) 2009 viral load with disease severity and prolonged viral shedding in children. Emerg Infect Dis 2010; 16:1266–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Foisy G, Rosella LC, Sanderson R et al. Self‐reported pH1N1 influenza vaccination coverage for Ontario. Statistics Canada 2011; 22:1–5. [PubMed] [Google Scholar]
- 28. Palacios G, Hornig M, Cisterna D et al. Streptococcus pneumonia coinfection is correlated with the severity of H1N1 pandemic influenza. PLoS ONE 2009; 4:e8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jean C, Louie JK, Glaser CA et al. Invasive group A streptococcal infection concurrent with 2009 H1N1 influenza. Clin Infect Dis 2010; 50:e59–e62. [DOI] [PubMed] [Google Scholar]


