Abstract
Background
We investigated the roles of Toll‐like receptors (TLRs) in naturally occurring influenza.
Methods
A prospective, case – control study was conducted. Adults hospitalized with virologically confirmed influenza A infections (onset <48 hours, before treatment) were compared with age‐/gender‐matched controls. TLRs (2, 3, 4, 7, 8, 9) expression in monocytes and dendritic cells (DCs – total, myeloid, plasmacytoid) was quantitated using flow cytometry. Gene expression of RLRs (RIG‐1, MDA‐5) was evaluated using real‐time PCR. Concomitant signaling molecules expression, plasma cytokine/chemokine concentrations, and respiratory tract viral loads were measured. PBMCs were cultured and stimulated ex vivo with TLR‐specific ligands for cytokine responses.
Results
Forty two patients with influenza (24 A/H3N2, 18 A/H1N1pdm09) and 20 controls were studied. Patients' mean age was 68 ± 16 years; 81% had respiratory/cardiovascular complications. There were increased cellular expressions of TLR9, TLR8, TLR3, and TLR7 during influenza; TLR2 and TLR4 were suppressed. Results were similar for both virus strains. Higher TLR expression levels at presentation significantly correlated with lower viral loads (Spearman's rho: −0·46 to −0·69 for TLR9, TLR8, and TLR3; P‐values <0·05). Multivariate regression models (adjusted for age, comorbidity, disease severity, time from onset) confirmed their independent associations. Increased signaling molecules (phospho‐MAPKs, IκB) and inflammatory cytokines (IL‐6, sTNFR‐1, CCL2/MCP‐1; CXCL10/IP‐10, IFN‐γ) correlated with increased TLR expression. RLRs were upregulated simultaneously. PBMCs of patients with influenza showed significant, dynamic changes in their cytokine responses upon TLR stimulation, compared with controls.
Conclusions
Our results suggest that TLRs play an important role in early, innate viral inhibition in naturally occurring influenza. Inflammatory cytokine responses are concomitantly induced. These findings support investigation of TLR targeting as a novel intervention approach for prophylaxis against influenza.
Keywords: Influenza, Toll‐like receptors
Introduction
Both seasonal and pandemic influenza viruses can cause severe diseases, leading to hospitalizations and deaths.1 The current preventive strategies of annual vaccination and antiviral prophylaxis, however, are limited by virus strain specificity, suboptimal immunogenicity among certain patient groups, and antiviral resistance.2 Toll‐like receptors (TLRs) are pattern recognition receptors expressed by the antigen‐presenting dendritic cells, monocytes/macrophages, and epithelial cells, which trigger the innate immune responses against invading pathogens.3, 4, 5, 6 Broadly, viral nucleic acids (dsRNA, ssRNA, CpG oligodeoxynucleotides) are detected by the endosomal TLRs (3, 7, 8, 9), whereas components of bacteria (peptidoglycans/lipoproteins, lipopolysaccharides) are detected by the cell surface TLRs (2, 4).3, 4 Recent in vitro and in vivo studies on influenza pathogenesis have shown that TLR activation induces expression of type I interferons and pro‐inflammatory cytokines (e.g., IL‐6, TNF‐α), limiting viral replication and dissemination, mediating tissue inflammation, and link to adaptive immunity development.3, 7, 8, 9, 10, 11, 12 In vitro studies have also revealed an additional cytosolic system of retinoic acid‐inducible gene‐1 (RIG‐1)‐like receptor proteins (RLRs) that detect influenza viral RNAs, contributing to the innate responses.3, 7, 10, 13 Recently, data from animal studies have further shown that targeting the TLRs (with agonists) may rapidly upregulate the innate immunity to provide broad‐range, virus strain non‐specific protection against lethal influenza challenge.4, 11, 14, 15, 16, 17 However, our understanding on TLR's importance in natural influenza is very limited because of lack of clinical data. In this study, we hypothesized that in patients with natural influenza infection, the “virus‐sensing” TLRs are upregulated and are associated with virus inhibition and inflammatory cytokine induction. TLR expressions in monocytes and dendritic cells, signaling molecules and cytokine/chemokine levels, and the immune cells' responses toward TLR‐specific ligands were examined and compared between patients and controls. Relationships between TLR expressions and the concomitant respiratory tract viral loads were also examined. Such information not only can further our understanding on immunopathogenesis of naturally occurring influenza, it may also facilitate the planning of clinical studies using TLR targeting as a novel intervention approach against influenza in future.4
Methods
Patients and sampling
A prospective, case – control study was performed during the influenza seasonal peaks in 2010 and 2011 in Hong Kong; the predominant circulating virus strains were influenza A/H3N2 and A/H1N1pdm09, respectively.18 Adults aged ≥18 years hospitalized for laboratory‐confirmed influenza A infection, who presented <48 hours from illness onset, were recruited for study. Exclusion criteria included delayed presentation, underlying immunocompromised conditions (e.g., autoimmune diseases, HIV/AIDS) or receiving immunosuppressant (including corticosteroids), antiviral treatment before enrollment, and lack of consent. Age‐ and gender‐matched controls were recruited from the general outpatient clinics and the community for comparison, outside the seasonal peak periods. Exclusion criteria for controls included any immunocompromised condition and history of any febrile illness in the past 4 weeks.
Admission and management procedures for our patients with seasonal and pandemic H1N1 influenza have been described.19, 20 In brief, patients presenting with acute febrile respiratory illnesses would be considered for hospitalization if they had developed potentially serious medical conditions/complications, exacerbation of underlying illnesses, or severe constitutional and respiratory symptoms unmanageable at home. Nasopharyngeal samples were collected to test for influenza viruses, regardless of perceived etiology and disease severity.21 Patients with Influenza were identified and enrolled by the research team on a daily basis;20, 22 after obtaining informed written consent, peripheral venous blood samples were taken immediately for TLR assays, prior to antiviral treatment. Ethical approval was obtained from the Institutional Review Boards of the Chinese University of Hong Kong and the Hospital Authority of Hong Kong.
TLR expression analysis by flow cytometry
EDTA – blood samples collected were transported to a biosafety level II laboratory for immediate processing. After centrifugation at 4°C and plasma separation, PBMCs (peripheral blood mononuclear cells) were obtained using Ficoll‐Paque gradient centrifugation. Expression profile of TLRs 2, 3, 4, 7, 8, 9 on blood monocytes (CD14+), myeloid dendritic cells/mDC (CD16−CD14−CD85k+CD123−), and plasmacytoid dendritic cells/pDC (CD16−CD14−CD85k+CD123+) were analyzed by flow cytometry using established methods.23, 24 In brief, PE‐conjugated mouse anti‐human TLRs 2, 3, 4, 7, 8, 9‐specific monoclonal antibodies (Imgenex Corp., CA, USA) were used for TLR staining. PE‐conjugated purified mouse IgG1 (Imgenex, San Diego, CA, USA) was used as the corresponding isotypic control. To stain the DCs in PBMCs, cells were co‐stained with FITC‐conjugated CD14 and CD16, PC7‐conjugated CD85k, and APC‐conjugated CD123 (BD Biosciences, San Jose, CA, USA). “Total” DCs were identified as the CD14−, CD16−, and CD85k+ population, which were further differentiated into mDCs and pDCs by low and high CD123 expression, respectively. The FITC‐CD14+ gated population was set to identify the monocyte population. For TLRs 3, 7, 8, 9 intracellular staining, PBMCs were fixed in 4% formaldehyde for 15 minutes and permeabilized with 0·1% saponin for 30 minutes on ice. For TLRs 2 and 4 cell surface staining, PBMCs were first washed once with PBS and then incubated with 2% human serum to block any non‐specific epitopes. For both protocols, the cells were then incubated with the corresponding TLR antibodies and co‐stained with cell‐type surface markers. Finally, cells were washed with PBS and fixed with 1% paraformaldehyde for flow cytometric analysis. For each cell type, 105 viable cells were gated and analyzed with 4‐color FACSCalibur flow cytometer (BD Biosciences). All results were expressed in mean fluorescence intensity (MFI). In a subset of patients with influenza and controls, flow cytometric analysis of the intracellular signaling molecules including activated MAPKs (phospho‐p38 and phospho‐ERK) and NF‐κB (phospho‐IκB) were also performed, using established methods (Data S1).22, 25
RIG‐1 and MDA‐5 gene expression assay by quantitative real‐time PCR
Gene expressions of the RLRs (retinoic acid‐inducible gene‐1/RIG‐1 and the melanoma differentiation‐associated gene‐5/MDA‐5) in PBMCs were also studied.26 After RNA extraction, mRNA expressions were measured with the Applied Biosystems 48‐well StepOne™ Real‐Time PCR System; specific primer pairs (RIG‐1, forward: 5′‐TGCGAAGGAGATGGTTGGTCAGAA‐3′, reverse: 5′‐TTCCACCTGTTTACAGCGGGACTT‐3′; MDA‐5, forward: 5′‐AGGCACCATGGGAAGTGATTCAGA‐3′, reverse 5′‐ATTTGGTAAGGCCTGAGCTGGAGT‐3′) were added to the universal SYBR Green PCR master mix (Applied Biosystems, Foster City, CA, USA). Co‐amplification of the housekeeping GAPDH gene was used to normalize the amount of total RNA added. The mRNA expressions were calculated using threshold cycle relative quantitation, represented as “Relative Quantitation” (mRNA expression of RIG‐1 or MDA‐5/GAPDH) (Data S1).
Ex vivo stimulation of PBMCs using TLR‐specific ligands
PBMCs obtained were cultured and stimulated with TLR‐specific ligands to assess their response for cytokine/chemokine production.23, 24 Aliquots of 105 cells resuspended in culture medium RPMI 1640 (Gibco Laboratories, Grand Island, NY, USA) were dispensed in each well of a 96‐well plate (Nalge Nunc International, Penfield, NY, USA). The cells were then incubated with or without ligands individually: peptidoglycan (PGN, TLR2 ligand, Fluka Chemie GmbH, Buchs, Switzerland), polyinosinic–polycytidylic acid (Poly IC) (TLR3 ligand, Sigma‐Aldrich Co., St. Louis, MO, USA), ultra‐purified lipopolysaccharide (LPS, TLR4 ligand, Invivogen Corp., San Diego, CA, USA), R837/imiquimod (TLR7 ligand, Invivogen), cytosine–guanine repeat (CpG) DNA (TLR9 ligand, Invivogen) at 1 μg/ml, and single‐stranded RNA (ssRNA, TLR8 ligand, Invivogen) at 0·5 μg/ml for 24 hours at 37°C in a 5% CO2 atmosphere. The cell‐free supernatant was harvested for assay of selected cytokines/chemokines.
Assays of cytokine/chemokine concentration
Concentrations of 14 “pro‐inflammatory” or “adaptive” immunity‐related cytokines/chemokines, including IL‐6, TNF‐α, CXCL8/IL‐8, CCL2/MCP‐1, and IL‐1β; interferon (IFN)‐γ, CXCL10/IP‐10, CXCL9/MIG, CCL5/RANTES, and IL‐12p70 respectively; and IFN‐α2, CCL3/MIP‐1α, IL‐10, and sTNFR‐1 were measured using cytometric bead array (CBA)(BD Pharmingen, San Diego, CA, USA) with flow cytometry or ELISA. Details of their detection methods and clinical relevance for study have been reported and provided in Data S1.20, 22
Virological studies
All nasopharyngeal aspirates (or flocked swabs in some cases) collected at presentation were subjected to influenza and other respiratory virus detection using immunofluorescence or PCR assays as described; virus isolation was performed in parallel.19, 21 Subsequent virus subtyping (A/H3N2, A/H1N1pdm09) was performed by the National Influenza Centre, Centre for Health Protection, Hong Kong.18, 19, 21 In all enrolled cases, real‐time reverse‐transcription polymerase chain reaction assay (targeting M gene) was performed on the original respiratory specimens to determine viral RNA concentration (copies/μl RNA) as described.19, 21, 22
Data reporting and analysis
The level of expression (MFI) of individual TLR in each cell type was reported (median and interquartile range, IQR) and compared between patients with influenza and control subjects and between virus subtypes using Mann–Whitney U‐test. “Relative Quantitation” of mRNA expression of RLRs was similarly compared. The relationships between TLR expression levels and nasopharyngeal viral loads were analyzed using Spearman's rank correlation coefficient rho (r).20, 22 Independent factors affecting viral load were examined using multivariate linear regression (backward) model analysis.21 Relationships between TLRs, signaling molecules, and plasma cytokine levels were analyzed using Spearman's correlation. Cytokine responses of PBMCs after individual TLR‐specific ligand activation (reported as “fold increase”: cytokine concentration with ligand stimulation/without ligand) were compared between patients with influenza and controls using non‐parametric tests.20 In all analyses, P‐value <0·05 was considered to indicate statistical significance. All probabilities were 2‐tailed. Statistical analysis was performed using the PASW Statistics software, version 17.0 (SPSS Inc., Chicago, IL, USA).
Results
Altogether, 42 hospitalized patients with influenza (A/H3N2, n = 24; A/H1N1pdm09, n = 18) and 20 controls were studied. The mean ± SD of age of patients and controls was 67·7 ± 15·9 and 62·0 ± 13·5 years, respectively (P > 0·05); gender distribution was similar (male: 57% and 55%, respectively, P > 0·05). Among patients, comorbidities were present in 59·5%; 81·0% developed acute respiratory and/or cardiovascular complications; and 50·0% required supplemental oxygen therapy for hypoxemia. Four (9·5%) patients developed critical illness requiring ventilatory support and one died.
Levels of expression of the “viral‐sensing” TLR3, TLR7, TLR8, and TLR9 and “bacterial‐sensing” TLR2 and TLR4 in monocytes and DCs were compared between patients and controls. Patients' blood samples were collected at a median of 2 (IQR 1–2) days from symptom onset, prior to antiviral treatment. As shown in Figure 1, we found significant increase in expression of TLR9 and TLR8, but suppressed TLR2. There were also trends of increased expressions of TLR3 and TLR7 and lowered TLR4. Subgroup analyses on plasmacytoid and myeloid DCs showed similar results (Data S2). There was no significant difference in TLR expression profile and magnitude between A/H3N2 and A/H1N1pdm09 infections (in both monocytes and DCs, all P ≥ 0·1). We did not find significant correlation between TLR levels and sampling time within 2 days of illness. Expressions of RIG‐1 and MDA‐5 in patients with influenza were found to be significantly increased when compared with controls (Figure 2).
Figure 1.
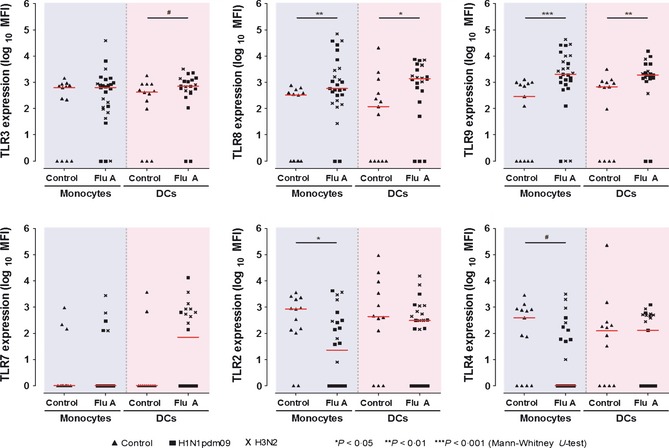
Expressions of Toll‐like receptors (TLRs 2, 3, 4, 7, 8, 9) in monocytes and dendritic cells (DCs) in influenza A patients and age‐gender matched controls, measured by quantitative flow cytometry. The mean fluorescence intensity (MFI) values are represented in logarithmic scale (median values indicated by horizontal bars in red). Each point represents the MFI in an individual study subject. ‘Monocytes’ [patients versus controls, MFI median (IQR)]: TLR8, 587·0 (202·0, 3374·0) versus 328·0 (1·0, 469·5), P = 0·006; TLR9, 2056·0 (819·0, 9996·0) versus 293·0 (1·0, 954·0), P < 0·001; TLR2, 23·0 (1·0, 296·5) versus 840·0 (119·0, 2159·5), P = 0·031; TLR4, 1·0 (1·0, 155·0) versus 388·0 (1·0, 842·5), P = 0·061. ‘Total Dendritic Cells, DCs’: TLR3, 711·0 (415·5, 1304·5) versus 420·0 (48·5, 859·0), P = 0·079; TLR8, 1389·0 (401·8, 4412·3) versus 118·0 (1·0, 852·0), P = 0·024; TLR9, 1906·0 (1411·5, 3817·0) versus 676·0 (1·0, 1046·0), P = 0·001; TLR7, 70·0 (1·0, 694·8) versus 1·0 (1·0, 1·0), P = 0·147. Detection of TLR7 (positive), 50·0% versus 18·2%, P = 0·078. Representative flow‐cytometry histograms (including isotypic control) and subgroup analyses on mDC and pDC are provided in Data S2. Available convalescentphase samples from 6 influenza patients showed normalizing TLR8, 9 and TLR2, 4 levels (also see Table 3).
Figure 2.
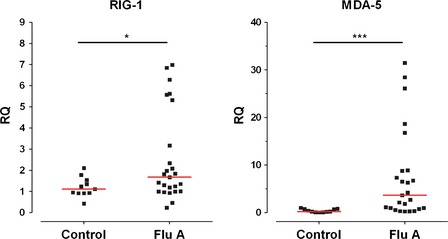
Level of gene expression of RIG‐1 and MDA‐5 in peripheral blood mononuclear cells in influenza A patients and age‐gender matched controls, measured by quantitative real‐time PCR. Each point represents the Relative Quantitation (RQ) in an individual study subject. *P = 0·038, ***P < 0·001, Mann‐Whitney U‐test ‘RQ’ (‘Relative Quantitation’) = mRNA (RIG‐1 or MDA‐5)/GAPDH.
The relationships between nasopharyngeal viral RNA concentrations (“viral load”) at the time of presentation and TLR expression levels in patients with influenza were examined. We found significant negative correlations between viral loads and TLR9, TLR8, and TLR3 expression levels, particularly in the DCs (Figure 3). Similar trends were shown for TLR7. Multivariate linear regression model analyses showed that low TLR level and severe disease (indicated by pneumonia and hypoxemia) were two independent factors associated with higher viral loads, adjusted for age, comorbidity, time elapsed from onset, and virus strain (Data S3). We did not find significant associations between RIG‐1 and MDA‐5 expressions with viral load. The effects of secondary bacterial infection (culture‐confirmed, n = 6) on TLR2 and TLR4 expressions are reported in Data S2.
Figure 3.
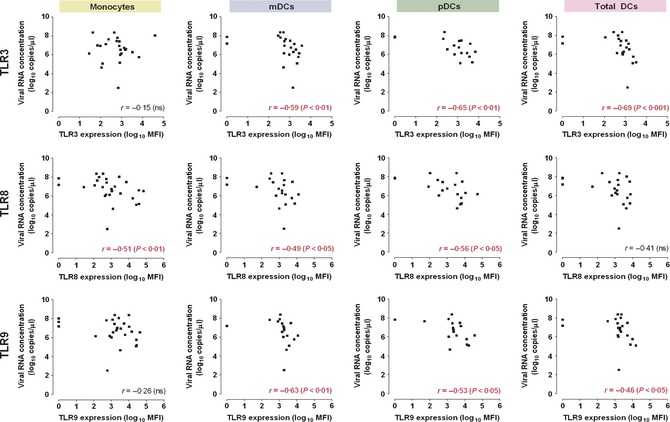
Negative correlations between expressions of TLRs and influenza ‘viral load’ in the respiratory tract. Both mean fluorescence intensity (MFI) values and viral RNA concentrations (measured by real‐time, quantitative reverse‐transcription PCR) are shown in logarithmic scale. mDC , myeloid dendritic cells; pDC, plasmacytoid dendritic cells. Trends of negative correlations between viral RNA concentration and TLR7 expression were also observed: ‘Total DC’, r = −0·34, P = 0·12; ‘pDC’, r = −0·31, P = 0·21; ‘mDC’, r = −0·34, P = 0·18. There was no significant correlation found between viral RNA concentration and TLR2 or TLR4 expression. r = Spearman's rank coefficient (rho).
Correlations between TLR or RLR expression levels, signaling molecules, and cytokines were examined. Only those cytokines/chemokines and signaling molecules shown to have significant increase in patients with influenza over controls were chosen for analysis (Table 1). We showed that increased cellular expression of TLRs such as TLR9 significantly correlated with increased plasma levels of inflammatory cytokines, including IL‐6, soluble TNF receptor‐1 (indicating TNF‐α release), CXCL8/IL‐8, CCL2/MCP‐1, as well as interferon‐γ, CXCL10/IP‐10, and CXCL9/MIG (Table 2). In addition, there were significant correlations with increased signaling molecules including phospho (p)‐IκB, pp38‐MAPK, and pERK. Associations between RLR expression and plasma levels of several cytokines were also observed.
Table 1.
Plasma cytokine/chemokine concentration and signaling molecule expression in patients with influenza, compared with age‐ and gender‐matched controls
| Cytokine/chemokine | Plasma concentration, median (IQR), pg/ml case versus control | P‐value |
|---|---|---|
| IL6 | 11·1 (7·3, 27·4) | <0·001* |
| 3·7 (2·7, 5·1) | ||
| CXCL8/IL‐8 | 13·1 (9·0, 21·4) | <0·001* |
| 5·8 (4·4, 7·5) | ||
| CCL2/MCP‐1 | 60·5 (38·2, 96·2) | <0·001* |
| 31·9 (22·7, 48·5) | ||
| sTNFR‐1 | 1589·7 (1246·1, 3139·4) | 0·01* |
| 748·9 (466·9, 1777·6) | ||
| CXCL10/IP‐10 | 1040·2 (700·5, 1602·3) | <0·001* |
| 133·5 (101·8, 269·3) | ||
| CXCL9/MIG | 317·4 (113·9, 608·6) | 0·01* |
| 124·1 (42·7, 256·4) | ||
| IFN‐γ | 10·840 (3·0, 18·2) | 0·004* |
| 3·0 (3·0, 3·0) | ||
| IL‐10 | 4·4 (3·7, 6·8) | <0·001* |
| 2·6 (1·9, 3·2) | ||
| RANTES | 647·6 (262·1, 1953·6) | n.s. |
| 974·3 (539·1, 2397·7) | ||
| IL‐12p70 | 1·9 (1·9, 1·9) | n.s. |
| 1·9 (1·9, 1·9) | ||
| TNF‐α | 3·7 (3·7, 3·7) | n.s. |
| 3·7 (3·7, 3·7) | ||
| IL‐1b | 4·9 (3·7, 5·4) | n.s. |
| 5·7 (4·0, 7·2) | ||
| CCL3/MIP | 5·4 (5·4, 5·4) | n.s. |
| 5·4 (5·4, 5·4) | ||
| IFN‐α2 | 1·2 (1·2, 1·2) | n.s. |
| 1·2 (1·2, 1·2) |
| Signaling molecules (monocytes) † | MFI, median (IQR) case versus control | P‐value |
|---|---|---|
| pIκB | 15·0 (0·0, 2724·0) | n.s. |
| 8·2 (1·6, 23·8) | ||
| pp38‐MAPK | 11·0 (0·0, 3468·0) | 0·054 |
| 0·0 (0·0, 3·1) | ||
| pERK | 62·0 (0·0, 1423·0) | 0·017 |
| 0·0 (0·0, 37·8) |
Mann–Whitney U‐test was used for comparison (*P < 0·05). Plasma cytokine/chemokine detection in influenza, the assays' reference ranges, and detection limits have been reported (see Data S1; the levels of IL‐12p70, TNF‐α, CCL3/MIP, and IFN‐α2 were at or below the assays' detection limits).20, 22 Subgroup analyses comparing A/H3N2 and A/H1N1pdm09 infections showed significantly lower CXCL10/IP‐10 and CXCL9/MIG levels in the latter, consistent with our previous report.20 Signaling molecules pIκB, pp38‐MAPK, and pERK in dendritic cells showed similar trends of activation (data not shown).
†Results on signaling molecules were available in a subset of 15 patients with influenza and 10 controls; MFI, mean fluorescence intensity.
Table 2.
Relationships between increased TLR or RLR expression, signaling molecule expression, and plasma concentration of cytokines/chemokines
| IL‐6 | CXCL8/IL‐8 | CCL2/MCP‐1 | sTNFR‐1 | CXCL10/IP‐10 | CXCL9/MIG | IFN‐γ | IL‐10 | pp38‐MAPK | pERK | pIκB | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| TLR3 (MN) | 0·106 | 0·165 | −0·011 | 0·182 | −0·64 | 0·097 | 0·232 | 0·010 | 0·491* | 0·536* | 0·523* |
| TLR3 (DC) | 0·210 | 0·288 | 0·208 | 0·254 | 0·057 | 0·121 | 0·174 | 0·137 | – | – | – |
| TLR7 (MN) | 0·000 | 0·106 | −0·086 | −0·111 | −0·050 | −0·053 | 0·004 | −0·157 | −0·341 | 0·142 | −0·102 |
| TLR7 (DC) | 0·305 | 0·331 | 0·342* | 0·206 | 0·108 | 0·161 | −0·006 | 0·100 | – | – | – |
| TLR8 (MN) | 0·213 | 0·319* | 0·110 | 0·190 | 0·183 | 0·106 | 0·410** | 0·251 | 0·558* | 0·629** | 0·510* |
| TLR8 (DC) | 0·236 | 0·407* | 0·289 | 0·329* | 0·096 | 0·102 | 0·114 | 0·193 | – | – | – |
| TLR9 (MN) | 0·405** | 0·489*** | 0·356* | 0·361* | 0·393** | 0·403** | 0·304* | 0·315* | 0·373 | 0·521* | 0·505* |
| TLR9 (DC) | 0·334* | 0·501** | 0·349* | 0·351* | 0·337* | 0·233 | 0·181 | 0·357* | – | – | – |
| RIG‐1 | −0·321 | −0·188 | 0·354* | −0·100 | 0·200 | 0·037 | 0·084 | 0·205 | – | – | – |
| MDA‐5 | 0·222 | 0·386* | 0·569** | 0·066 | 0·586*** | 0·204 | 0·399* | 0·483** | – | – | – |
MN, monocytes; DC, total dendritic cells; RLR assays were performed with PBMC only, and signaling molecule assays were not performed.
PS. There was no positive correlation found between TLR2 and TLR4 expressions and plasma cytokine/chemokine concentrations. Signaling molecules assays on DCs were performed in a small subset of patients, which showed similar results and trends (data not shown).
Values represent Spearman's rank coefficient (rho); *P ≤ 0·05, **P ≤ 0·01, ***P ≤ 0·001.
These in vivo findings were supported by the ex vivo experiment results which showed significant differences between patients with influenza and control subjects in their PBMCs' cytokine responses toward TLR‐specific ligand activation (Table 3). For instance, stimulation of the TLR9 signaling pathway had resulted in smaller increases in IL‐6, TNF‐α, CXCL10/IP‐10, and IFN‐α from baseline in patients compared with controls. On the other hand, the responsiveness for cytokine production was higher with TLR7 ligand binding in patients with influenza. In both cases, the responses normalized when patients' illness subsided (Table 3). No difference was found between patients and controls toward TLR2 or TLR4 ligand activation. Results were similar between A/H3N2 and A/H1N1pdm09 infections (data not shown).
Table 3.
Ex vivo cytokine/chemokine response of PBMC to TLR‐specific ligands in patients with influenza, compared with controls
| TLR‐specific ligand activation | IL‐6 | TNF‐α | CCL2/MCP‐1 | CXCL10/IP‐10 | IFN‐γ | IL‐10 | IFN‐α | |
|---|---|---|---|---|---|---|---|---|
| TLR3 (PolyIC) | FluA | 99·5 (26·4,390·6) | 101·0 (24·3,374·0) | 4·0 (1·0,17·6) | 1·0 (1·0,1·0) | 1·5 (1·0,2·1) | 39·5 (8·4,61·1) | 1·0 (0·9,1·5) |
| CTL | 84·5 (41·7,243·3) | 84·6 (29·2,241·3) | 1·2 (1·0,16·9) | 1·0 (1·0,1·0) | 1·8 (1·2,3·6) | 53·2 (21·7,85·5) | 1·3 (1·0,1·7) | |
| TLR7 (Imiquimod) | FluA | 7·1 (2·6,41·6)** | 1·3 (1·0,3·8)** | 3·7 (1·0,16·9) | 2·6 (1·0,12·4)*** | 1·0 (0·8,1·5) | 1·3 (1·0,2·1)* | 1·2 (0·9,2·3) |
| CTL | 4·2 (0·7,9·6) | 1·0 (0·8,1·9) | 1·5 (1·0,20·9) | 1·0 (1·0,1·2) | 1·4 (1·0,2·0) | 1·0 (1·0,1·2) | 1·2 (0·9,4·1) | |
| TLR8 (ssRNA) | FluA | 2·8 (1·1,15·3) | 1·5 (1·0,6·7) | 1·5 (1·0,5·6) | 1·0 (1·0,1·0) | 1·1 (0·9,1·8) | 1·0 (1·0,1·7) | 1·0 (0·9,1·1) |
| CTL | 2·9 (1·5,5·8) | 2·2 (1·0,6·9) | 1·0 (1·0,2·3) | 1·0 (1·0,1·0) | 1·0 (0·8,1·5) | 1·0 (1·0,1·1) | 1·0 (0·8,1·3) | |
| TLR9 (CpG DNA) | FluA | 1·5 (0·9,3·3)* | 1·0 (1·0,1·0)** | 2·2 (1·0,8·2) | 1·0 (1·0,3·9)*** | 1·2 (1·0,1·6) | 1·0 (1·0,1·0)*** | 1·2 (1·0,1·3)*** |
| CTL | 2·6 (1·6,7·0) | 1·6 (1·0,2·9) | 1·0 (1·0,16·9) | 9·2 (1·0,27·8) | 1·2 (1·0,2·2) | 1·5 (1·3,2·0) | 2·1 (1·3,10·5) | |
| TLR2 (PGN) | FluA | 122·8 (25·9,390·6) | 314·6 (84·5,1129·5) | 3·8 (1·0,16·9) | 1·0 (1·0,1·7) | 1·3 (1·0,2·0) | 47·7 (12·4,84·0) | 1·1 (0·9,1·4) |
| CTL | 89·8 (62·7,310·2) | 278·3 (65·4,434·7) | 1·3 (1·0,9·6) | 1·0 (1·0,1·0) | 1·4 (1·0,2·2) | 32·5 (15·1,89·1) | 1·5 (0·9,2·6) | |
| TLR4 (LPS) | FluA | 118·7 (26·3,394·0) | 125·4 (29·3,719·4) | 3·7 (1·0,14·8) | 1·0 (1·0,1·0) | 1·3 (1·0,2·5) | 37·3 (8·6,65·8) | 1·3 (1·0,1·5) |
| CTL | 84·5 (41·7,243·3) | 96·3 (21·6,453·7) | 1·2 (1·0,16·9) | 1·0 (1·0,1·8) | 1·5 (1·1,2·9) | 49·4 (31·7,97·9) | 1·1 (0·9,2·2) | |
LPS, lipopolysaccharide; PGN, peptidoglycan.
PS. Available convalescent‐phase samples from six patients with influenza: TLR9 ligand [median fold change in IL‐6, 4·8 (1·2,7·1); TNF‐α, 2·4 (1·0,4·2); CXCL10/IP10, 18·6 (0·8,34·5); IL‐10, 1·8 (1·1,2·7); IFN‐α, 2·1 (1·2,39·4)]; TLR7 ligand [median fold change in IL‐6, 2·1 (0·7,42·7); TNF‐α, 1·4 (1·0,3·9); CXCL10/IP‐10, 1·0 (1·0,5·3); IL‐10, 1·0 (0·9,1·7); time interval from acute‐phase samples, median 9 (7–14) days.
Values represent fold increase in cytokine release after TLR‐specific ligand activation (i.e., cytokine concentration post‐ligand stimulation/no ligand control medium); reported as median and interquartile range (IQR), round off to 1 decimal place. Comparisons between patients with influenza (FluA) and healthy controls (CTL); Mann–Whitney U‐tests: *P ≤ 0·1, **P ≤ 0·05, ***P ≤ 0·01.
Discussions
We found differential increase in expression of TLRs in patients with naturally occurring influenza. High TLR expression level at presentation was shown to correlate significantly with a lower viral load. Inflammatory cytokine responses were concomitantly induced. Our findings suggest that the TLR signaling pathways play active roles in controlling influenza and in mediating inflammation. Targeting the TLRs as a novel intervention approach for prophylaxis against influenza should warrant investigation.
Our results on natural human infections are consistent with earlier in vitro and animal studies (e.g., knockout mice) which showed that the “viral‐sensing” TLRs (3, 7, 8, 9) are upregulated in the antigen‐presenting cells, signaling the innate, virus inhibitory, and inflammatory responses in influenza.3, 7, 8, 9, 10, 11, 14, 15 TLR9 activation in DCs strongly induces the release of type‐1 IFNs and pro‐inflammatory cytokines and upregulates the co‐stimulatory molecules (e.g., CD80/86);3, 14 TLR7/8 activation induces IFNs, the pro‐inflammatory IL‐6, TNF‐α, CCL2/MCP‐1, and CXCL8/IL‐8 and promotes DC maturation;3, 11, 17 TLR3 in epithelial cells causes tissue inflammation in influenza pneumonia through IL‐6, TNF‐α, and CXCL8/IL‐8 induction and effector cell recruitment.3, 7, 8 Limited available reports had described upregulations of the “viral‐sensing” TLRs in association with the inflammatory cytokines in patients with severe A/H1N1pdm09 influenza.27, 28, 29 Interestingly, the “bacterial‐sensing” TLRs (2 and 4) were also reported to be suppressed.27, 28 Downregulation of these TLRs may impair phagocyte recruitment and bacterial elimination, contributing to the risk of secondary infections.9, 30, 31, 32 We found no significant difference in TLR expression pattern or magnitude between A/H3N2 and A/H1N1pdm09 infections, unlike their adaptive immune responses.20 This reflects the less specific nature of innate immunity, which can be advantageous when considering TLR targeting as a means of prophylaxis in influenza (discussed below). There is evidence to show that the RLRs are concomitantly upregulated and possibly play a contributory role in mediating the pro‐inflammatory cytokine responses.3, 7, 10, 15, 33 Although both RIG‐1 and MDA‐5 often showed upregulation in influenza and other respiratory viral infections, innate responses against influenza viruses are likely dependent on RIG‐1 and suppressible by the viral NS‐1 protein.7, 13, 26, 33
Importantly, our study is the first to show that the TLRs play a role in virus control in the early phase of natural influenza. Our data showed that higher expression levels of TLR3, TLR8, and TLR9 (and trends for TLR7) in the innate immune cells, particularly the DCs, significantly correlated with lower levels of virus replication (“viral load”) in the respiratory tract for both virus strains. Multivariate analyses confirmed their independent associations, accounted for disease severity, time interval from onset, virus strain, age, and comorbidity. Reduced viral load was associated with milder illness.5, 21 These observations were unlikely the sole result of adaptive immunity, as patients were studied within 48 hours of their illnesses.3, 5, 20 The alternative explanation of a higher level of viral replication inducing a smaller increase in these viral‐sensing TLRs also appeared unlikely.3, 4, 5, 7, 8 Our results are in line with recent mice experiments which showed that TLR activation rapidly produces virus inhibitory responses (predominantly through type I IFNs and IFN‐stimulated mechanisms), conferring protection against influenza challenge.3, 4, 10, 14 Tuvim et al. demonstrated that aerosolized TLR9 and TLR2/6 agonists given 3 days before or shortly after lethal challenge with A/H3N2 or A/H1N1pdm09 virus reduced mice lung viral titers and mortality.14 Lau et al.15 reported starting intranasal TLR3 agonist pretreatment 6 hours before lethal influenza challenges reduced lung viral titers and mortality; the protection was “broad range” (A/H1N1, A/H3N2, A/H1N1pdm09, A/H5N1, A/H9N2). Wong et al.16 reported pretreatment with TLR9 and TLR3 agonists upregulated the TLRs within hours and protected mice against lethal influenza (A/H1N1, A/H3N2, A/H5N1) infections for 7–14 days. Prophylactic TLR7/8 or TLR7 agonist administration also resulted in virus inhibition and improved mice survival.11, 17 Interestingly, the virus inhibitory effects of TLR agonists were found to be superior to exogenously administered interferons16, 17 and synergistic with oseltamivir.15 Our findings in naturally occurring influenza thus provide strong support to further investigate this novel approach of TLR targeting and activation as a means of preventive intervention against influenza in humans (e.g., as pre‐ or post‐exposure prophylaxis).4, 15
Our data also indicated that TLR signaling might have contributed to the induction of inflammatory responses in patients with influenza. Increased TLR (7, 8, and 9) expression was found to correlate significantly with the key intracellular signaling molecules (MAPKs, NF‐κB/IκB) and higher levels of pro‐inflammatory cytokines including IL‐6 and sTNFR‐1.3, 5, 7, 14, 20, 22, 27, 28, 32, 33 Consistent with earlier in vitro studies, associations with the “adaptive” cytokines (e.g., Th1‐related IFN‐γ, CXCL10/IP‐10, CXCL9/MIG) were also observed, particularly for TLR9 and TLR8, which signal through the MyD88 pathway.3, 4, 10, 34 TLR's active role in cytokine induction was supported by our ex vivo experiments, which showed significant differences in cellular cytokine responses between patients with influenza and controls upon TLR‐specific ligand stimulation (e.g., CpG DNA‐TLR9 and imiquimod‐TLR7; the resultant response pattern governed by the ligand tested, cell type studied, and disease stage at the time of sampling/“immune exhaustion”) and a dynamic change in their responsiveness during clinical recovery.20, 32 Perpetuating, uncontrolled pro‐inflammatory cytokine responses can lead to immunopathological damage in severe influenza (Data S1); and further stimulation of TLRs in a more advanced disease stage may exacerbate tissue inflammation.5, 6, 7, 8, 11, 17, 20, 22, 29 Whether TLR blockade alone can reduce inflammation is uncertain as compensatory mechanisms might exist.4, 5, 15 Nevertheless, recent studies have shown that TLR's role in regulating the adaptive responses can be harnessed to boost immunogenicity of influenza vaccines (e.g., TLR9 or TLR7 ligands as adjuvants). This may be particularly useful in the elderly and the immunologically naïve vaccinees.4, 33, 34, 35, 36, 37, 38, 39, 40 Our data in natural influenza provide additional support to this new vaccination approach.
The strengths of our study included the following: a comprehensive approach to examine TLR signaling (monocytes/DCs, cytokines/chemokines, signaling molecules; viral loads); evaluation of two circulating virus strains; and unique study design to allow immediate specimen collection (≤48 hours, pre‐treatment) and processing for TLR quantitative assay in various immune cells by multi‐parametric flow cytometry.20, 23 Ideally, the respiratory epithelial cells should also be studied (especially for TLR3 and RLRs);3, 5, 8, 16 however, obtaining such specimens (e.g., with bronchoalveolar lavage ) is generally infeasible in patients with influenza. Studying circulating cells exposed to the pulmonary bed has been shown to provide close estimates.6, 7, 20, 22, 32, 41 Although the inclusion of a “mild influenza” group might be helpful, earlier studies have indicated that inflammatory cytokine responses are barely detectable in such patients.20, 22, 27 We plan to compare TLR and RLR expressions between different naturally occurring respiratory viral infections (e.g., influenza versus RSV) in future, as available data suggest that these might be different.3, 26 Factors determining TLR expression in an individual should be further studied.3, 42 We report that in some old patients (>65 years of age, P = 0·025), despite successful cytokine and ligand stimulation studies, the number of DCs in circulation was too few and signals too weak (due to aging and/or site “migration”) to allow full‐range TLR analysis.6, 39 The conclusions were unlikely affected, however, as these were balanced between the age‐matched cases/controls (P = 0·768). Lastly, to avoid excessive multiple comparisons, only those significantly increased cytokines with known clinical relevance were chosen for further analyses.20, 22
In conclusion, the TLRs are shown to have important roles in signaling the virus inhibitory and pro‐inflammatory responses in the early phase of natural influenza. Our results, together with emerging reports on other pattern recognition receptors33, 36, 42, strongly support further clinical studies on the roles of innate immunity in controlling influenza and to explore potential applications of receptor targeting, especially as preventive interventions.2, 4, 37
Author contributions
Concept and design: Nelson Lee, Chun‐Kwok Wong. Drafting of the article: Nelson Lee. Analysis and interpretation of data: Nelson Lee, Chun‐Kwok Wong, David SC Hui, Martin CW Chan, Sharon KW Lee, Yu‐Jun Chu, Amy WY Ho. Laboratory work: Chun‐Kwok Wong, Sharon KW Lee, Yu‐Jun Chu, Amy WY Ho, Spea Ping Yip (immunology); Paul KS Chan, Karry LK Ngai (virology). Patient enrollment, collection of samples and data assembly: Nelson Lee, Rity YK Wong, Grace CY Lui, Bonnie CK Wong, Sunny H Wong (clinical). Critical revision for important intellectual content and final approval of article: Nelson Lee, Chun‐Kwok Wong, David SC Hui, Paul KS Chan.
Funding
Research Fund for the Control of Infectious Diseases (RFCID) from the Food and Health Bureau of the Hong Kong SAR Government, People's Republic of China (2009) (Ref. number 09080102).
Conflict of interests
N. Lee has received grant support from F. Hoffmann‐La Roche on principal investigator–initiated clinical influenza research (paid to the Chinese University of Hong Kong), honorarium for consultancy work from GlaxoSmithKline, and conference supports from Sanofi‐Aventis Hong Kong Ltd, MSD (Asia) Ltd., and Pfizer Hong Kong, but have no relation to this study. PKS Chan has received constancy fees and research funding from F. Hoffmann‐La Roche (paid to the Chinese University of Hong Kong) and has received support for attending academic conferences from GlaxoSmithKline, but have no relation to this study.
Supporting information
Data S1. Supplementary methods.
Data S2. Supplementary results on TLR expressions.
Data S3. Supplementary results on viral load and TLR expression correlations.
Lee et al (2013) Role of human toll‐like receptors in naturally occurring influenza a infections. Influenza and Other Respiratory Viruses 7(5), 666–675
Nelson Lee and CK Wong contributed equally to this article.
References
- 1. World Health Organization . Influenza, Available at http://www.who.int/topics/influenza/en (Accessed 1 May 2012).
- 2. Lambert LC, Fauci AS. Influenza vaccines for the future. N Engl J Med 2010; 363:2036–2044. [DOI] [PubMed] [Google Scholar]
- 3. Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev 2009; 227:75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hedayat M, Netea MG, Rezaei N. Targeting of Toll‐like receptors: a decade of progress in combating infectious diseases. Lancet Infect Dis 2011; 11:702–712. [DOI] [PubMed] [Google Scholar]
- 5. Peiris JS, Cheung CY, Leung CY, Nicholls JM. Innate immune responses to influenza A H5N1: friend or foe? Trends Immunol 2009; 30:574–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gill MA, Long K, Kwon T et al Differential recruitment of dendritic cells and monocytes to respiratory mucosal sites in children with influenza virus or respiratory syncytial virus infection. J Infect Dis 2008; 198:1667–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Le Goffic R, Pothlichet J, Vitour D et al Cutting Edge: Influenza A virus activates TLR3‐dependent inflammatory and RIG‐I‐dependent antiviral responses in human lung epithelial cells. J Immunol 2007; 178:3368–3372. [DOI] [PubMed] [Google Scholar]
- 8. Le Goffic R, Balloy V, Lagranderie M et al Detrimental contribution of the Toll‐like receptor (TLR) 3 to influenza A virus‐induced acute pneumonia. PLoS Pathog 2006; 2:e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hashimoto Y, Moki T, Takizawa T, Shiratsuchi A, Nakanishi Y. Evidence for phagocytosis of influenza virus‐infected, apoptotic cells by neutrophils and macrophages in mice. J Immunol 2007; 178:2448–2457. [DOI] [PubMed] [Google Scholar]
- 10. Koyama S, Ishii KJ, Kumar H et al Differential role of TLR‐ and RLR‐signaling in the immune responses to influenza A virus infection and vaccination. J Immunol 2007; 179:4711–4720. [DOI] [PubMed] [Google Scholar]
- 11. Wu CC, Hayashi T, Takabayashi K et al Immunotherapeutic activity of a conjugate of a Toll‐like receptor 7 ligand. Proc Natl Acad Sci U S A 2007; 104:3990–3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hidaka F, Matsuo S, Muta T, Takeshige K, Mizukami T, Nunoi H. A missense mutation of the Toll‐like receptor 3 gene in a patient with influenza associated encephalopathy. Clin Immunol 2006; 119:188–194. [DOI] [PubMed] [Google Scholar]
- 13. Pichlmair A, Schulz O, Tan CP et al RIG‐I‐mediated antiviral responses to single‐stranded RNA bearing 5′‐phosphates. Science 2006; 314:997–1001. [DOI] [PubMed] [Google Scholar]
- 14. Tuvim MJ, Gilbert BE, Dickey BF, Evans SE. Synergistic TLR2/6 and TLR9 activation protects mice against lethal influenza pneumonia. PLoS ONE 2012; 7:e30596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lau YF, Tang LH, Ooi EE, Subbarao K. Activation of the innate immune system provides broad‐spectrum protection against influenza A viruses with pandemic potential in mice. Virology 2010; 406:80–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wong JP, Christopher ME, Viswanathan S et al Activation of toll‐like receptor signaling pathway for protection against influenza virus infection. Vaccine 2009; 27:3481–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hammerbeck DM, Burleson GR, Schuller CJ et al Administration of a dual toll‐like receptor 7 and toll‐like receptor 8 agonist protects against influenza in rats. Antiviral Res 2007; 73:1–11. [DOI] [PubMed] [Google Scholar]
- 18. Centre for Health Protection . Department of Health, The Government of the Hong Kong SAR. Available at http://www.chp.gov.hk/en/view_content/14843.html. (Accessed 1 May 2012).
- 19. Lee N, Chan PK, Lui GC et al Complications and outcomes of pandemic 2009 Influenza A (H1N1) virus infection in hospitalized adults: how do they differ from those in seasonal influenza? J Infect Dis 2011; 203:1739–1747. [DOI] [PubMed] [Google Scholar]
- 20. Lee N, Wong CK, Chan PK et al Cytokine response patterns in severe pandemic 2009 H1N1 and seasonal influenza among hospitalized adults. PLoS ONE 2011; 6:e26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee N, Chan PK, Hui DS et al Viral loads and duration of viral shedding in adult patients hospitalized with influenza. J Infect Dis 2009; 200:492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee N, Wong CK, Chan PKS et al Hypercytokinemia and Hyperactivation of p38 Mitogen‐Activated Protein Kinase in Severe Human Influenza A Infections. Clin Infect Dis 2007; 45:723–731. [DOI] [PubMed] [Google Scholar]
- 23. Lun SWM, Wong CK, Ko FWS, Hui DSC, Lam CWK. Expression and functional analysis of toll‐like receptors of peripheral blood cells in asthmatic patients: implication for immunopathological mechanism in asthma. J Clin Immunol 2009; 29:330–342. [DOI] [PubMed] [Google Scholar]
- 24. Wong CK, Wong PTY, Tam LS, Li EK, Chen DP, Lam CWK. Activation profile of Toll‐like receptors of peripheral blood lymphocytes in patients with systemic lupus erythematosus. Clin Exp Immunol 2009; 159:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wong CK, Wong PTY, Tam LS, Li EK, Chen DP, Lam CWK. Activation profile of intracellular mitogen‐activated protein kinases in peripheral lymphocytes of patients with systemic lupus erythematosus. J Clin Immunol 2009; 29:738–746. [DOI] [PubMed] [Google Scholar]
- 26. Scagnolari C, Midulla F, Pierangeli A et al Gene expression of nucleic acid‐sensing pattern recognition receptors in children hospitalized for respiratory syncytial virus‐associated acute bronchiolitis. Clin Vaccine Immunol 2009; 16:816–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arankalle VA, Lole KS, Arya RP et al Role of host immune response and viral load in the differential outcome of pandemic H1N1 2009 influenza virus infection in Indian patients. PLoS ONE 2010; 5: pii: e13099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu Y, Chen H, Sun Y, Chen F. Antiviral role of Toll‐like receptors and cytokines against the new 2009 H1N1 virus infection. Mol Biol Rep 2012; 39:1163–1172. [DOI] [PubMed] [Google Scholar]
- 29. Mauad T, Hajjar LA, Callegari GD et al Lung pathology in fatal novel human influenza A (H1N1) infection. Am J Respir Crit Care Med 2010; 181:72–79. [DOI] [PubMed] [Google Scholar]
- 30. Didierlaurent A, Goulding J, Patel S et al Sustained desensitization to bacterial Toll‐like receptor ligands after resolution of respiratory influenza infection. J Exp Med 2008; 205:323–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Karlström A, Heston SM, Boyd KL, Tuomanen EI, McCullers JA. Toll‐like receptor 2 mediates fatal immunopathology in mice during treatment of secondary pneumococcal pneumonia following influenza. J Infect Dis 2011; 204:1358–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Heltzer ML, Coffin SE, Maurer K et al Immune dysregulation in severe influenza. J Leukoc Biol 2009; 85:1036–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hui KP, Lee SM, Cheung CY et al H5N1 influenza virus‐induced mediators upregulate RIG‐I in uninfected cells by paracrine effects contributing to amplified cytokine cascades. J Infect Dis 2011; 204:1866–1878. [DOI] [PubMed] [Google Scholar]
- 34. Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science 2010; 327:291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Geeraedts F, Goutagny N, Hornung V et al Superior immunogenicity of inactivated whole virus H5N1 influenza vaccine is primarily controlled by Toll‐like receptor signalling. PLoS Pathog 2008; 4:e1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schneider‐Ohrum K, Giles BM, Weirback HK, Williams BL, DeAlmeida DR, Ross TM. Adjuvants that stimulate TLR3 or NLPR3 pathways enhance the efficiency of influenza virus‐like particle vaccines in aged mice. Vaccine 2011; 29:9081–9092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kasturi SP, Skountzou I, Albrecht RA et al Programming the magnitude and persistence of antibody responses with innate immunity. Nature 2011; 470:543–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Behzad H, Huckriede AL, Haynes L et al GLA‐SE, a synthetic toll‐like receptor 4 agonist, enhances T‐cell responses to influenza vaccine in older adults. J Infect Dis 2012; 205:466–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Panda A, Qian F, Mohanty S et al Age‐associated decrease in TLR function in primary human dendritic cells predicts influenza vaccine response. J Immunol 2010; 184:2518–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schmitz N, Beerli RR, Bauer M et al Universal vaccine against influenza virus: linking TLR signaling to anti‐viral protection. Eur J Immunol 2012; 42:863–869. [DOI] [PubMed] [Google Scholar]
- 41. de Jong MD, Simmons CP, Thanh TT et al Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med 2006; 12:1203–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Everitt AR, Clare S, Pertel T et al IFITM3 restricts the morbidity and mortality associated with influenza. Nature 2012; 484:519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supplementary methods.
Data S2. Supplementary results on TLR expressions.
Data S3. Supplementary results on viral load and TLR expression correlations.


