Abstract
Please cite this paper as: Dapat et al. (2012) Delayed emergence of oseltamivir‐resistant seasonal influenza A (H1N1) and pandemic influenza A(H1N1)pdm09 viruses in Myanmar. Influenza and Other Respiratory Viruses DOI: 10.1111/irv.12030.
The prevalence and timing of emergence of oseltamivir‐resistant seasonal and pandemic influenza A (H1N1) viruses in Myanmar in 2008 and 2009 are described in this report. In 2008, the oseltamivir‐resistant seasonal H1N1 virus was detected at a lower rate (6%) and emerged at least 2 months later when compared with neighboring countries. Similarly, the prevalence of pandemic H1N1 virus was low (3%) and the timing of emergence was late (August 2009) in Myanmar. Interestingly, we detected three isolates that were resistant to both amantadine and oseltamivir. Limited movement of people into the country is attributed to the delayed emergence of drug‐resistant seasonal and pandemic A(H1N1) viruses.
Keywords: Amantadine, antiviral drug resistance, H1N1 subtype, oseltamivir, pandemic influenza, seasonal influenza
Epidemiological data on the prevalence and timing of emergence of antiviral drug‐resistant influenza viruses in Myanmar are limited. In our previous report, we showed that influenza virus activity in Myanmar exhibited seasonality, which peaked during the rainy season between June and September. 1 We also reported on the increasing prevalence and circulation pattern of amantadine‐resistant A(H3N2) viruses in Myanmar, 1 which supports the finding that new H3N2 strains are seeded from Asia. 2 In this study, we characterized drug‐resistant seasonal and pandemic influenza A(H1N1) viruses in Myanmar in 2008 and 2009.
The study
Nasopharyngeal swabs were collected from outpatients at Sanpya Hospital, Yangon, and Department of Medical Research, Nay Pyi Taw, who presented with at least one of the following influenza‐like illness (ILI) symptoms, such as fever of >38°C, coughing, rhinorrhea, myalgia, arthralgia, or diarrhea. Informed consent and clinical information using standardized questionnaire were obtained from each patient. Samples were tested with rapid test kits (Quick Ex‐Flu; Denka Seiken Co. Ltd., Tokyo, Japan). Aliquots were stored at −80°C and were sent to Niigata University, Japan, for further analysis. Virus isolation, RNA extraction, and cDNA synthesis were performed as previously described. 1 Screening for drug resistance markers, S31N for amantadine, and H275Y for oseltamivir, by cycling probe real‐time PCR was performed as previously described. 3 Amantadine susceptibility was assessed using MDCK cell–based virus yield reduction assay, which measures 50% tissue culture infectious dose (TCID50). Fluorescent‐based NAI susceptibility assay was performed using oseltamivir carboxylate (F. Hoffman‐La Roche Ltd., Basel, Switzerland) and zanamivir (GlaxoSmithKline Research and Development Ltd., Hertfordshire, UK), which measures 50% inhibitory concentration (IC50). 4 Sequencing and phylogenetic analyses of HA, NA, and M genes were performed as previously described. 1 Nucleotide sequences were submitted to GenBank (HQ256584–HQ256677).
Epidemiology of influenza in 2008 and 2009
A total of 327 influenza A and 133 influenza B test kit–positive samples were collected in 2008. Of these, 302 virus isolates were obtained with 66 seasonal H1N1, 134 H3N2, and 102 type B viruses. In 2009, all 529 test kit–positive samples were influenza A. Of these, 486 isolates were obtained with 121 seasonal H1N1, 349 H3N2, and 16 A(H1N1)pdm09 viruses. In 2008, influenza virus activity peaked in September, when H1N1, H3N2, and influenza B viruses were detected in that same month (Figure 1). In 2009, influenza virus activity peaked in July, when seasonal influenza A (H1N1 and H3N2) viruses co‐circulated (Figure 1). This peak in influenza activity, which coincided during the rainy season, is consistent with our previous study. 1 Sporadic cases of A(H1N1)pdm09 infections occurred only from August to December 2009 (Figure 1), and it did not result in a community‐wide circulation. All H3N2 isolates that were analyzed in 2008 and 2009 were resistant to amantadine (data not shown). NAI assay was performed only on seasonal A(H1N1) and A(H1N1)pdm09, but not on A(H3N2) and influenza B isolates.
Figure 1.
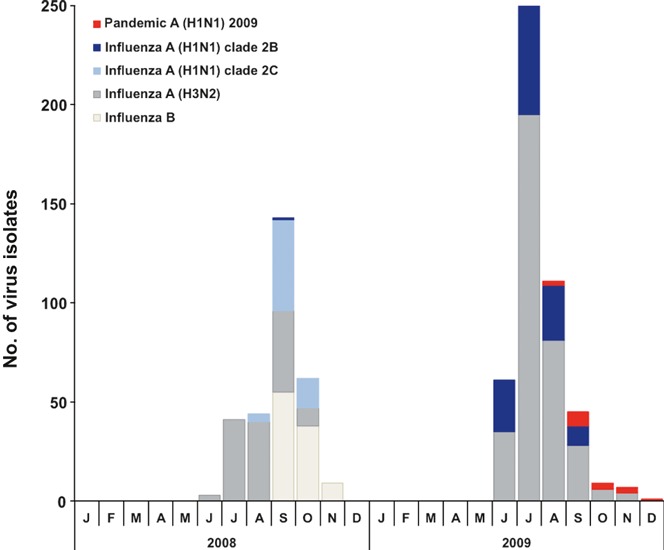
Monthly distribution of virus isolates of influenza A (subtypes H1 and H3) and influenza B viruses in Myanmar in 2008 and 2009.
Amantadine‐resistant H1N1 viruses (Clade 2C)
Two clades (2B and 2C) of seasonal H1N1 viruses with different drug susceptibility profiles were detected in 2008. Of 66 H1N1 viruses, 65 isolates were amantadine resistant with S31N mutation in M2 (Table 1). HA and NA phylogeny showed that these amantadine‐resistant viruses belong to clade 2C (Figure 2). Amantadine‐resistant clade 2C viruses were first detected in the third week of August, and its activity peaked in September 2008 (Figure 1). These viruses were closely related to the isolates from Hong Kong and China (Figure 2). The prevalence of amantadine resistance (98%) in Myanmar increased from 0%, when compared with the isolates in 2005. 1 This trend of increasing incidence of amantadine resistance had been reported elsewhere. 5 Clade 2C viruses remain susceptible to oseltamivir and zanamivir (Table 2).
Table 1.
Prevalence of amantadine‐ and oseltamivir‐resistant influenza A virus (H1N1) in Myanmar in 2008 and 2009 by cycling probe real‐time PCR
| No. of samples | No. (%) of amantadine resistant* | No. (%) of oseltamivir resistant** | No. (%) of amantadine‐ and oseltamivir‐resistant viruses | |
|---|---|---|---|---|
| 2008 Seasonal H1N1 | 66 | 65 (98) | 4 (6) | 3 (4) |
| 2009 Seasonal H1N1 | 121 | 0 | 120 (99) | 0 |
| 2009 Pandemic H1N1 | 16 | 16 (100) | 0 | 0 |
*S31N mutation in M2.
**H275Y mutation in NA.
Figure 2.
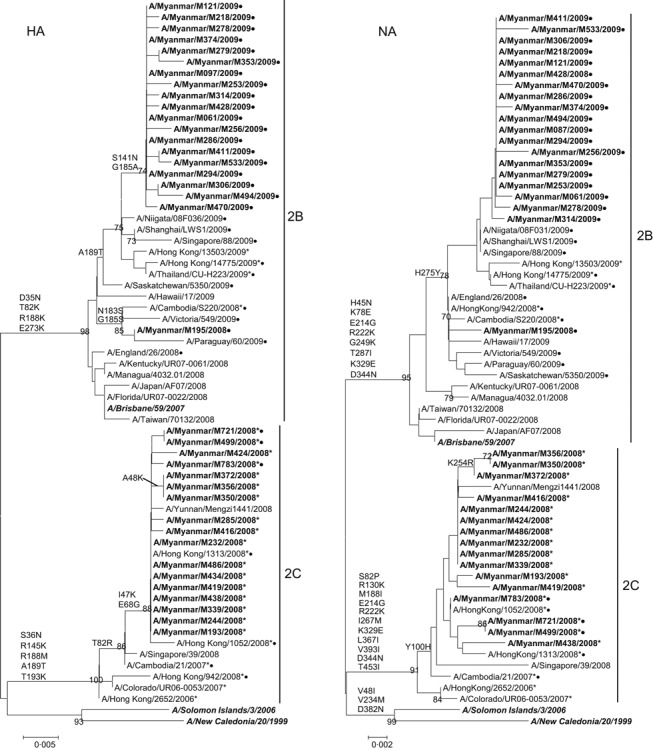
Phylogenetic analysis of HA and NA genes of seasonal influenza A (H1N1) virus isolates in Myanmar in 2008 and 2009. Trees were generated using neighbor‐joining method. Bootstrap values >70% of 1000 replicates, and amino acid changes that characterize a branch are indicated in the left side of the node. Amantadine‐resistant isolates with S31N mutation in M2 are marked with asterisks (*), and oseltamivir‐resistant isolates with H275Y (H275Y in N1 numbering) mutation in NA are marked with circles (•). Isolates from this study are boldfaced. Vaccine strains are boldfaced and italicized.
Table 2.
Oseltamivir and zanamivir IC50 values of influenza A (H1N1) isolates in Myanmar in 2008 and 2009
| Influenza isolates | No. of samples tested | M2 mutation | NA mutation | IC50 values (nm)* | |
|---|---|---|---|---|---|
| Oseltamivir (Median ± SD) | Zanamivir (Median ± SD) | ||||
| 2008 Seasonal H1N1 | 13 | S31N | None | 2·27 ± 0·84 | 1·24 ± 0·42 |
| 2008 Seasonal H1N1 | 1 | None | H275Y | 610·88 | 1·48 |
| 2008 Seasonal H1N1 | 3 | S31N | H275Y | 850·81 ± 144·12 | 2·11 ± 0·56 |
| 2009 Seasonal H1N1 | 17 | None | H275Y | 925·71 ± 294·83 | 1·67 ± 0·73 |
| 2009 Pandemic H1N1 | 16 | S31N | None | 2·27 ± 0·56 | 1·13 ± 0·34 |
*IC50 values were calculated from at least two independent experiments performed in duplicate.
Oseltamivir‐resistant H1N1 viruses (Clade 2B)
In 2008, one clade 2B virus and three clade 2C viruses of 66 viruses analyzed were resistant to oseltamivir. Thus, proportion of oseltamivir‐resistant H1N1 viruses (clade 2B) in Myanmar in 2008 is lower (6%) compared with other South‐East Asian countries such as Malaysia (44%), Philippines (91%), Singapore (30%), and Thailand (32%). 5 This clade 2B virus (A/Myanmar/M195/2008) was collected from a 2‐year‐old male outpatient on September 23, 2008. This timing of the first detection of clade 2B virus in Myanmar is at least 2 months late than Singapore (May), Malaysia (June), Philippines (June), and Thailand (June). 5
However, in 2009, the antiviral susceptibility profile changed when clade 2C viruses were replaced by clade 2B (Figure 2). These clade 2C viruses have elevated oseltamivir IC50 values (median IC50 of 925·71 ± 294·83 nm, n = 17), which is 400‐fold higher than sensitive strains (median IC50 of 2·27 ± 0·84 nm, n = 13) (Table 2). All seasonal H1N1 viruses collected in Myanmar remained sensitive to zanamivir.
Dual‐resistant (oseltamivir and amantadine‐resistant) H1N1 viruses
In October 2008, we detected three viruses with S31N and H275Y mutations that confer resistance to both amantadine and oseltamivir. NAI assay results showed that these viruses have elevated oseltamivir IC50 values (Table 2). The oseltamivir IC50 values of dual‐resistant viruses are equivalent to H275Y viruses without the S31N mutation. Phylogenetic analysis showed that the dual‐resistant viruses were closely related to amantadine‐resistant clade 2C strains (Figure 2). These dual‐resistant viruses may have emerged due to drug selection, genetic reassortment, or spontaneous mutation. The patients in Myanmar did not receive any anti‐influenza drugs; thus, drug selection pressure may not contribute to the emergence of drug‐resistant strains. Phylogeny of HA, NA, and M (data not shown) suggested that genetic reassortment may be ruled out because all three segments analyzed belong to clade 2C. In addition, these clade 2C viruses were typically oseltamivir‐sensitive and amantadine‐resistant and were genetically related to A/Hong Kong/2652/2006‐like strains, which were circulating in Asia in the previous seasons. Thus, the H275Y mutation may have occurred by spontaneous mutation. 6 These dual‐resistant viruses were detected earlier in Cambodia in October 2007 and Hong Kong in March 2008. 6
Pandemic influenza A(H1N1) 2009 isolates
The influenza A(H1N1)pdm09 virus emerged in Mexico and the United States in April 2009. 7 A few weeks later, the virus spread worldwide, and it was declared a pandemic virus in June 2009. In Myanmar, we detected only 16 sporadic cases of A(H1N1)pdm09 from August to December 2009. The influenza A(H1N1)pdm09 strains co‐circulated with seasonal H1N1 and H3N2 viruses. However, A(H1N1)pdm09 virus was not the predominant isolate, and the prevalence rate (3·3%) is lower than Singapore (52·9%), Thailand (21·0%), and Taiwan (72·8%). 8 , 9 , 10 The first A(H1N1)pdm09 virus (A/Myanmar/M280/2009) was collected on August 8, 2009, which is delayed for at least 2 months when compared with the timing of appearance in Singapore, Thailand, and Taiwan. 8 , 9 , 10
Genotyping of the A(H1N1)pdm09 viruses showed S31N mutation (Table 1). M2 gene sequencing confirmed the presence of S31N mutation, and no substitutions were identified at other amino acid positions that are associated with amantadine resistance. The results of the MDCK cell–based virus yield reduction assay also showed resistance to amantadine. The mean IC50 values of A(H1N1)pdm09 strains to neuraminidase inhibitors are low (Table 2), indicating susceptibility to oseltamivir and zanamivir. Phylogenetic analysis of the HA and NA genes of A(H1N1)pdm09 isolates revealed two clusters (Figure 3). Cluster 1 viruses are related to the vaccine strain, A/California/07/2009. All 16 A(H1N1)pdm09 isolates from Myanmar belonged to cluster 2 viruses (Figure 3), which are related to the reference strain, A/New York/10/2009. 11 These cluster 2 viruses harbored threonine (T) at position 220 in HA and aspartic acid (D) at position 248 in NA. 11
Figure 3.
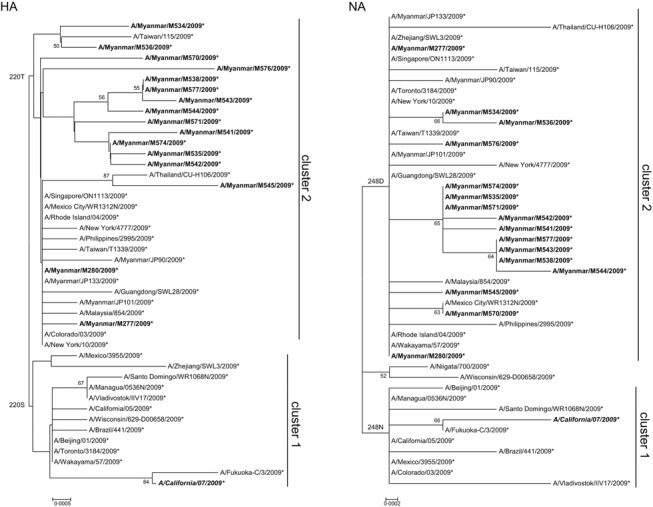
Phylogenetic analysis of HA and NA genes of 2009 pandemic influenza A (H1N1) isolates in Myanmar in 2008 and 2009. Trees were inferred using the neighbor‐joining method. Figures on the left side of the branches indicate the percentages of replicated trees using bootstrap analysis (1000 replicates). Amino acid changes that characterized a particular branch are indicated on the left side of the node. Myanmar isolates are boldfaced. Vaccine strains are boldfaced and italicized.
Conclusion
Our report demonstrated the dynamics of antiviral drug resistance of influenza A (H1N1) viruses and the detection of dual‐drug‐resistant isolates. This study highlights the importance of continued monitoring of antiviral drug resistance and its application in the formulation of treatment guidelines of influenza virus infection.
Of particular interest is the delayed timing of appearance of influenza A (H1N1) viruses in Myanmar. The oseltamivir‐resistant seasonal H1N1 viruses were detected several months later in Myanmar when compared with neighboring South‐East Asian countries. Similarly, the pandemic H1N1 viruses emerged at least 2 months later. Both seasonal and pandemic H1N1 viruses were detected sporadically, which is in contrast with the prevalence rate in other countries. The late emergence of H1N1 viruses may be attributed to lesser volume of traffic and mobility of people in Myanmar. According to the website of the Ministry of Hotels and Tourism, the number of visitors in Myanmar was 731 230 and 762 547 in 2008 and 2009, respectively. 12 , 13 Of these, only 193 319 (26·4%) and 243 278 (31·9%) visitors entered the country via international air travel in 2008 and 2009, respectively; the rest entered through the border. 14 These figures are relatively low when compared with the reported number of visitors in 2009 in Malaysia (23 646 191), Thailand (14 149 841), Singapore (9 681 259), Viet Nam (3 772 259), Cambodia (2 161 577), and Lao PDR (2 008 363). 15 This study showed the link between the delayed emergence of influenza A (H1N1) viruses and the lack of movement of people into the country.
Financial disclosure
This work was supported by the Special Coordination Funds for Promoting Science and Technology of the Ministry of Education, Culture, Sports, Science, and Technology (MEXT, Japan).
Competing interests
The authors have no conflict of interests to declare.
Acknowledgements
We are grateful to the clinicians and staff of the Chest Medical Unit of Sanpya Hospital, Respiratory Medicine Department of Yangon General Hospital, and Department of Medical Research, for their help in sample collection. We thank Akinori Miyashita and Ryozo Kuwano of the Department of Molecular Genetics, Brain Research Institute, Niigata University for support on DNA sequencing. We thank Akemi Watanabe and Kae Horie for technical assistance, and we thank Yasuko Yamamoto for administrative support.
References
- 1. Dapat C, Saito R, Kyaw Y et al. Epidemiology of human influenza A and B viruses in Myanmar from 2005 to 2007. Intervirology 2009; 52:310–320. [DOI] [PubMed] [Google Scholar]
- 2. Russell C, Jones T, Barr I et al. The global circulation of seasonal influenza A (H3N2) viruses. Science 2008; 320:340–346. [DOI] [PubMed] [Google Scholar]
- 3. Suzuki Y, Saito R, Sato I et al. Identification of oseltamivir resistance among pandemic and seasonal influenza A (H1N1) viruses by an His275Tyr genotyping assay using the cycling probe method. J Clin Microbiol 2011; 49:125–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hurt A, Barr I, Hartel G, Hampson A. Susceptibility of human influenza viruses from Australasia and South East Asia to the neuraminidase inhibitors zanamivir and oseltamivir. Antiviral Res 2004; 62:37–45. [DOI] [PubMed] [Google Scholar]
- 5. Hurt A, Ernest J, Deng Y et al. Emergence and spread of oseltamivir‐resistant A(H1N1) influenza viruses in Oceania, South East Asia and South Africa. Antiviral Res 2009; 83:90–93. [DOI] [PubMed] [Google Scholar]
- 6. Cheng PKL, Tommy WC, Ho EC et al. Oseltamivir‐ and amantadine‐resistant influenza viruses A (H1N1). Emerg Infect Dis 2009; 15:966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Garten R, Davis C, Russell C et al. Antigenic and genetic characteristics of swine‐origin 2009 A(H1N1) influenza viruses circulating in humans. Science 2009; 325:197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yap J, Tan CH, Cook AR et al. Differing clinical characteristics between influenza strains among young healthy adults in the tropics. BMC Infect Dis 2012; 12:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chittaganpitch M, Supawat K, Olsen SJ et al. Influenza viruses in Thailand: 7 years of sentinel surveillance data, 2004–2010. Influenza Other Respi Viruses 2011; 6:276–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang J, Lin C, Chen C et al. A new antigenic variant of human influenza A (H3N2) virus isolated from airport and community surveillance in Taiwan in early 2009. Virus Res 2010; 151:33–38. [DOI] [PubMed] [Google Scholar]
- 11. Fereidouni S, Beer M, Vahlenkamp T, Starick E. Differentiation of two distinct clusters among currently circulating influenza A(H1N1)v viruses, March–September 2009. Euro Surveill 2009; 14:1–3. [PubMed] [Google Scholar]
- 12. Ministry_of_Hotels_and_Tourism. Myanmar Tourism Statistics 2008 2008. Available at http://www.myanmartourism.org/tourismstatistics.htm (Accessed 23 May 2012).
- 13. Ministry_of_Hotels_and_Tourism. Myanmar Tourism Statistics 2009 . 2009. Available at http://www.myanmartourism.org/tourismstatistics.htm (Accessed 23 May 2012).
- 14. Ministry_of_Hotels_and_Tourism. Myanmar Tourism Statistics 2011 . 2011. Available at http://www.myanmartourism.org/tourismstatistics.htm (Accessed 23 May 2012).
- 15. Association_of_Southeast_Asian_Nations . ASEAN tourism marketing strategy 2012–2015. 2012. Available at http://www.aseansec.org/22073.htm (Accessed 23 May 2012).


