Abstract
Background
During summer 2009, a US Navy ship experienced an influenza‐like illness outbreak with 126 laboratory‐confirmed cases of pandemic influenza A (H1N1) 2009 virus among the approximately 2000‐person crew.
Methods
During September 24–October 9, 2009, a retrospective seroepidemiologic investigation was conducted to characterize the outbreak. We administered questionnaires, reviewed medical records, and collected post‐outbreak sera from systematically sampled crewmembers. We used real‐time reverse transcription‐PCR or microneutralization assays to detect evidence of H1N1 virus infection.
Results
Retrospective serologic data demonstrated that the overall H1N1 virus infection attack rate was 32%. Weighted H1N1 virus attack rates were higher among marines (37%), junior‐ranking personnel (34%), and persons aged 19–24 years (36%). In multivariable analysis, a higher risk of illness was found for women versus men (odds ratio [OR] = 2·2; 95% confidence interval [CI]: 1·1–4·4), marines versus navy personnel (OR = 1·7; 95% CI, 1·0–2·9), and those aged 19–24 versus ≥35 years (OR = 3·9; 95% CI, 1·2–12·8). Fifty‐three percent of infected persons did not recall respiratory illness symptoms. Among infected persons, only 35% met criteria for acute respiratory illness and 11% for influenza‐like illness.
Conclusions
Approximately half of H1N1 infections were asymptomatic, and thus, the attack rate was higher than estimated by clinical illness alone. Enhanced infection control measures including pre‐embarkation illness screening, improved self‐reporting of illness, isolation of ill and quarantine of exposed contacts, and prompt antiviral chemoprophylaxis and treatment might be useful in controlling shipboard influenza outbreaks.
Keywords: Disease outbreaks, epidemiology, H1N1 subtype, influenza A virus, military personnel
Background
In military ship settings, close living and working environments can contribute to the rapid spread of illness among a healthy, young adult population.1 During April 2009, the initial cases of pandemic influenza A (referred to as H1N1) 2009 virus infection were identified in San Diego and Imperial Counties, California.2 This novel virus was also subsequently identified in cases in Mexico, Canada, and throughout the world.3 While training off the Southern California coast during June 29–July 10 and July 21–August 3, 2009, sailors and marines aboard the USS Bonhomme Richard, a US Navy ship, experienced an outbreak of respiratory illness with reports of fever, cough, and body aches. The ship's medical staff collected throat and nasal swabs from patients with respiratory illness who were isolated in the medical department. Respiratory illness cases peaked during the first week of July. On July 4, 2009, the Naval Health Research Center (NHRC) detected H1N1 virus in respiratory specimens by using real‐time reverse transcription‐PCR (rRT‐PCR).
During the outbreak, 160 persons with symptoms of respiratory illness sought medical care at the shipboard clinic. Ill personnel with signs and symptoms indicative of influenza were empirically treated with oseltamivir if they sought care at the ship's clinic ≤24 hours after symptom onset. Medical officers isolated ill persons in the medical department for 5 days after symptom onset. Illness was self‐limited and mild, and no persons required hospitalization. Oseltamivir chemoprophylaxis was offered to the medical staff. Monovalent H1N1 vaccine was not yet available. Medical staff wore N95 respirators, donned gowns and gloves, and practiced standard precautions while caring for patients. To mitigate the spread of disease, additional shipboard control measures were instituted on July 2, 2009, including respiratory and hand hygiene education, and infection control messages delivered through the ship's public address system, television, and flyers. The ship's medical crew also distributed hand sanitizer in dining areas during meals.
While in port between the two at‐sea periods, sailors and marines reported to the ship and returned home daily. Ill sailors and marines were ordered to stay home until symptoms resolved. During June 29–August 3, NHRC identified 126 (79%) H1N1 virus infections among 160 symptomatic persons tested by rRT‐PCR aboard the ship.
Because this was among the first outbreaks of H1N1 on a ship, we conducted an investigation to characterize the outbreak, to estimate the overall attack rate, and to assess shipboard risk factors for H1N1 virus infection.
Methods
Beginning September 24, 2009, a retrospective seroepidemiologic investigation was conducted among crew members who were aboard the ship during the 2 at‐sea training exercises. Each officer and every third enlisted sailor or marine was selected from the ship's roster for voluntary participation. Participants were provided a questionnaire that collected demographic data; information on dates, symptoms, and outcomes of illness; and exposure to ill persons in assigned work and sleep locations aboard the ship. Available shipboard medical records were reviewed for all participants.
Specimen collection
The ship's medical staff collected throat and nasal swabs from patients who presented with respiratory illness and were isolated in the medical department. The swabs were placed into viral transport media, refrigerated, and transported to the NHRC laboratory during the at‐sea training exercises (June 29–August 3, 2009).
Banked blood samples, collected at time of entry into military service and stored by the Department of Defense Serum Repository, were used as the pre‐outbreak baseline samples (median collection date, September 2008; range, February 2006–June 2009). During September 25–October 9, 2009, we collected blood samples in 5‐ml serum separator tubes from participants approximately 60 days (range, 53–68) after the training exercises. Samples were centrifuged for 10 minutes, and sera were aliquoted and frozen at −20°C. The sera drawn during this investigation were matched individually with pre‐outbreak sera to measure H1N1 virus neutralizing antibody titers.
rRT‐PCR amplification
RNA was extracted from combined throat and nasal swab specimens using the QIAGEN® OneStep RT‐PCR Kit (QIAGEN N.V., Hilden, Germany) following the manufacturer's instructions. Influenza virus genome was detected using an rRT‐PCR assay to detect influenza A and B viral RNA, and to identify influenza A virus subtypes (H1N1, seasonal H1, H3, and H5) using the Centers for Disease Control and Prevention (CDC) assay.4 Briefly, 1‐step rRT‐PCR was performed in a final volume of 25 μl containing 5 μl of extracted RNA, 12·5 μl of buffer mix and 0·5 μl of Superscript™ III/Platinum® Taq‐Enzyme mix (Invitrogen, Carlsbad, CA, USA), 0·8 μm for each primer and 0·2 μm of probe. An ABI 7500HT Fast Real‐Time thermocyler (Applied Biosystems, Foster City, CA, USA) was used for rRT‐PCRs.. The thermocycling parameters for targets consisted of 50°C for 30 minutes, 95°C for 2 minutes, and 45 cycles with 95°C for 15 seconds, and 55°C for 30 seconds (CDC, Atlanta, GA, USA).
Virus propagation and microneutralization assay
Serum antibody microneutralization assay was performed according to procedures described previously.5, 6, 7 Briefly, serum was inactivated at 56°C for 30 minutes and then 2‐fold serum dilutions made in the range 1:5–1:640. Positive control serum was serially diluted to 1:1280; negative control was initially undiluted and then serially diluted 2‐fold down. After sera (both samples and controls) were serially diluted, 50 μl of working dilution (200TCID50/50 μl) was added to the wells. Cell controls were included in each plate for data analysis. The seasonal influenza A (H1N1) (A/Brisbane/H1N1) virus was propagated in embryonated chicken eggs, and the H1N1 virus A/Mexico/2009 was propagated in MDCK tissue cells. After 1 hour incubation at 37°C in 5% CO2, 30 ml of virus growth medium (Dulbecco's Modification of Eagle's Medium containing 0·25% bovine serum albumin, 25 mm HEPES buffer, 100 U/ml penicillin, 100 μg/ml streptomycin, and 3 μg/ml tosyl phenylalanyl chloromethyl ketone‐trypsin) was added, and flasks were incubated for 16 hours at 37°C in 5% CO2. MDCK flasks at 75–95% confluence were harvested and cell suspensions combined and centrifuged at 1,500 rpm for 5 minutes. The culture supernatant was then replaced with 15 ml of fresh virus growth medium, and the cultures incubated 18–24 hours or until cells exhibited approximately 50% cytopathic effect and supernatants had a hemagglutination activity of at least 32 hemagglutination units using a 0·5% suspension of turkey erythrocytes.
After an overnight incubation, plates were decanted and washed once with 200‐μl sterile phosphate‐buffered saline. Plates were then fixed and anti‐influenza A nucleoprotein was added at a 1:1000 and incubated at room temperature for 1 hour. After washing, goat anti‐IgG conjugated horseradish peroxidase at a 1:2000 dilution was added and incubated at room temperature for 1 hour. The absorbance in washed and blocked plates was read at 490‐nm wavelength. A seropositive was defined as a 4‐fold increase in H1N1 virus neutralizing antibody titer between baseline and convalescent sera.
Case definitions
A H1N1 case was defined as infection in a sailor or marine assigned to the ship during the outbreak (June 29–August 3), with or without respiratory symptoms, and with laboratory confirmation by ≥1 test as follows: rRT‐PCR by testing upper respiratory tract specimens or a 4‐fold rise in H1N1 microneutralization titer between pre‐outbreak and post‐outbreak sera. We defined cases of acute respiratory illness (ARI) as self‐report of cough or sore throat.8 Influenza‐like illness (ILI) was defined as documented fever ≥100°F (37·8°C) and self‐report of cough or sore throat in the absence of a known cause other than influenza. An asymptomatic case of H1N1 was defined as an infection in a person without reported respiratory illness.
Statistical analysis
Data analysis was conducted with sas ® statistical software, version 9.2 (SAS Institute, Inc., Cary, NC, USA). Observations were stratified by service. Weighted population estimates and confidence intervals were calculated using variance estimates based on the sampling design to reflect the population of officers to enlisted personnel and Navy to Marine Corp personnel. Descriptive analysis was performed for categorical data. Bivariate analysis using Wald chi‐square was performed, and odds ratios (ORs) with 95% confidence intervals (95% CIs) were calculated. We conducted a multivariable analysis to determine whether any covariates were independently associated with H1N1 infection. Weighted unadjusted and adjusted multivariable logistic regression was used to estimate odds of illness. Adjustments were made for age, sex, race/ethnicity, service, rank, and shipboard location of exposure to ill persons. Statistical testing was two‐sided and considered significant at P < 0·05.
Human subjects
The investigation was considered to be a public health response, non‐research activity by the CDC and NHRC institutional review boards.
Results
Sample demographics and characteristics
Of 961 eligible persons, 193 (20%) did not respond to requests for participation; 60 (6%) were unavailable because of personnel turnover; 43 (4%) stated that they were not aboard during the outbreak; and 52 (5%) declined participation. A total of 613 (64%) persons participated in the investigation during September 24–October 13, 2009. Available medical charts from the shipboard medical department were reviewed and abstracted for 589 participants. In total, 541 serum samples were collected. Paired pre‐ and post‐outbreak sera were available for 500 participants. Infected persons who reported illness either prior to or after the training exercises were excluded from the analysis. The final analytic sample included 489 participants with complete survey and serology information, representing 53% of eligible participants. The population parameters were based on weighted estimates of the sample participants.
The majority of participants (90%) were men and 54% were marines (Table 1). The majority of participants were junior enlisted persons (86%), followed by commissioned officers (9%) and senior enlisted non‐commissioned officers (5%). The median participant age was 24 years (range, 19–58). Non‐Hispanic whites comprised 52% of the serosurvey participants. Previous seasonal influenza vaccination (2008–2009) was documented in medical records for 382 (78%) participants.
Table 1.
Characteristics of shipboard participants and population: pandemic influenza A (H1N1) serosurvey
| Characteristic | Sample (n = 489) | Weighted population percentage estimates, % | 95% confidence interval |
|---|---|---|---|
| Sex | |||
| Women | 57 | 10 | (8–13) |
| Men | 432 | 90 | (87–92) |
| Service | |||
| US Navy | 259 | 46 | (44–48) |
| US Marine Corps | 230 | 54 | (52–56) |
| Rank | |||
| Jr. enlisted (E1–E6) | 377 | 86 | (84–88) |
| Sr. enlisted (E7–E9) | 23 | 5 | (3–7) |
| Officer (Officer/Chief Warrant Officer) | 89 | 9 | (9–9) |
| Age groups (years)* | |||
| 19–24 | 276 | 64 | (59–68) |
| 25–34 | 144 | 28 | (24–33) |
| ≥35 | 59 | 8 | (6–10) |
| Median age (years) (range) | 24·7 (19–58) | ||
| Race/ethnicity† | |||
| White, non‐Hispanic | 228 | 52 | (47–57) |
| Hispanic | 104 | 27 | (22–31) |
| Black, non‐Hispanic | 32 | 7 | (5–10) |
| Non‐Hispanic, other race (includes Asian, Pacific Islander, American Indian, and other) | 58 | 14 | (10–17) |
Missing: *10; †67.
Epidemic curve
To differentiate possible community‐acquired H1N1 virus infection from infections acquired during the at‐sea period, patients reporting illness during the first 2 days at sea (June 29–June 30, 2009) were excluded from analysis. The presumed community‐acquired cases are noted in white (Figure 1). Three of the presumed community‐acquired cases, 1 with ILI and 2 with ARI were confirmed as H1N1 by serology. The three ill persons reported symptom onset during June 20–June 25, 2009, prior to embarkation. None had sought medical care and all reported for duty. Among participants reporting illness onset dates during the at‐sea exercises, the majority of H1N1 cases (43/68; 61%) occurred during the first at‐sea period (June 29–July 10). The number of new cases peaked on July 8 (n = 9). Cases declined while in port between the two at‐sea periods. Seventeen percent of cases occurred during the second at‐sea period.
Figure 1.
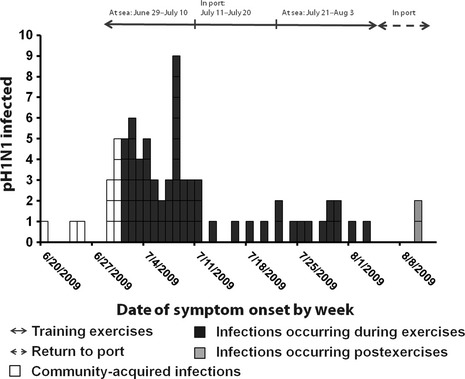
Epidemic curve of symptomatic, infected pandemic influenza A (H1N1) 2009 cases by symptom onset date.
2009 H1N1 attack rates and clinical manifestations
Among the sample of 489 sailors and marines, 142 persons had H1N1 virus infection; 123 were determined by serology alone, 16 by PCR alone, and 3 by both PCR and serology. The weighted shipboard H1N1 virus infection attack rate was 32% (Table 2). Marines had higher weighted attack rates compared with sailors (37% versus 25%; P = 0·01). H1N1 attack rates were highest among junior enlisted personnel compared with officers (34% versus 16%; P = 0·01), and among persons aged 19–24 years compared with ≥35 years (36% versus 15%; P = 0·01). The proportion of H1N1‐infected persons with documented 2008–2009 seasonal influenza vaccination was similar to those without documented vaccination (27% versus 36%; P = 0·09).
Table 2.
Pandemic influenza A (H1N1) 2009 attack rates, unadjusted, and adjusted odds ratios by participant characteristics (Wald chi‐square)
| Characteristics | Sample population | H1N1‐infected | Weighted population attack rate, % (95% confidence interval) | Unadjusted odds ratio, (95% confidence interval) n = 489 | P–value | Adjusted odds ratio, (95% confidence interval)¶ n = 386 | P–value |
|---|---|---|---|---|---|---|---|
| All cases | 489 | 142 | 32 (27–36) | – | – | – | – |
| Sex | |||||||
| Men | 432 | 120 | 31 (26–35) | Reference | Reference | ||
| Women | 57 | 22 | 41 (27–54) | 1·55 (0·86–2·81) | 0·15 | 2·19 (1·09–4·42) | 0·03 |
| Service | |||||||
| US Navy | 259 | 61 | 25 (20–31) | Reference | Reference | ||
| US Marine Corps | 230 | 81 | 37 (31–44) | 1·75 (1·16–2·62) | 0·01 | 1·75 (1·05–2·91) | 0·03 |
| Rank | |||||||
| Jr. enlisted (E1–E6) | 377 | 124 | 34 (29–39) | Reference | Reference | ||
| Sr. enlisted (E7–E9) | 23 | 5 | 21 (4–38) | 0·52 (0·18–1·44) | 0·21 | 1·37 (0·36–5·27) | 0·65 |
| Officer (Officer /Chief Warrant Officer) | 89 | 13 | 16 (7–26) | 0·38 (0·19–0·78) | 0·01 | 0·58 (0·24–1·38) | 0·22 |
| Age, (years)* | |||||||
| 19–24 | 276 | 97 | 36 (30–42) | Reference | Reference | ||
| 25–34 | 144 | 37 | 26 (19–34) | 0·62 (0·39–0·99) | 0·04 | 0·70 (0·39–1·26) | 0·24 |
| ≥35 | 59 | 6 | 15 (4–26) | 0·32 (0·13–0·77) | 0·01 | 0·26 (0·08–0·85) | 0·03 |
| Race/ethnicity† | |||||||
| White, non‐Hispanic | 228 | 62 | 30 (23–36) | Reference | Reference | ||
| Hispanic | 104 | 36 | 36 (26–45) | 1·30 (0·77–2·19) | 0·32 | 1·32 (0·74–2·34) | 0·34 |
| Black, non‐Hispanic | 32 | 12 | 41 (23–58) | 1·60 (0·72–3·57) | 0·25 | 1·88 (0·78–4·50) | 0·16 |
| Non‐Hispanic, Other (includes Asian, Pacific Islander, American Indian, and other) | 58 | 14 | 24 (12–35) | 0·74 (0·4–1·48) | 0·39 | 0·85 (0·40–1·83) | 0·68 |
| Exposure sites | |||||||
| Reported exposure in sleeping areas‡ | 138 | 61 | 39 (31–47) | 1·62 (1·06–2·47) | 0·03 | 1·21 (0·67–2·18) | 0·53 |
| Reported exposure in workplace§ | 133 | 57 | 35 (28–42) | 1·25 (0·82–1·91) | 0·31 | 0·96 (0·53–1·76) | 0·90 |
| Vaccination history (2008–2009 seasonal influenza) | |||||||
| Unknown | 107 | 38 | 36 (26–45) | Reference | – | – | |
| Documented | 382 | 104 | 27 (23–32) | 0·68 (0·43–1·07) | 0·09 | – | – |
Missing: *10; †67; ‡26; §33, ¶Adjusted for sex, service, rank, age, race/ethnicity, and shipboard location of exposure to ill persons.
Fifty‐three percent of H1N1‐infected persons were asymptomatic, and 47% reported respiratory symptoms (Table 3). Among all 142 H1N1‐infected persons, 17 (weighted proportion, 11%) reported ILI symptoms and 51 (weighted proportion, 35%) reported ARI symptoms. Seventeen (51%) of ILI cases were infected with H1N1. Twenty‐nine percent of all H1N1‐infected persons received medical care during the outbreak. Of 68 symptomatic‐infected persons, 43 (63%) sought medical care.
Table 3.
Reported signs and symptoms among infected pandemic influenza A (H1N1) 2009 persons
| Number H1N1‐infected n = 142 | Weighted percent, (95% confidence interval) | Number non‐infected n = 347 | Weighted percent, (95% confidence interval) | P–value | |
|---|---|---|---|---|---|
| Asymptomatic | 74 | 53 (45–62) | 233 | 69 (64–74) | – |
| Symptomatic | 68 | 47 (38–55) | 114 | 31 (26–36) | <0·01 |
| Acute respiratory illness | 51 | 35 (27–44)* | 99 | 26 (21–31) | 0·05 |
| Influenza‐like illness | 17 | 11 (6–17)* | 15 | 5 (2–7) | <0·01 |
| Sought medical care | 43 | 29 (21–36) | 48 | 13 (9–17) | <0·01 |
| Reported symptoms | |||||
| Fatigue | 61 | 41 (33–49) | 59 | 17 (12–20) | <0·01 |
| Body ache | 57 | 39 (30–47) | 43 | 12 (9–16) | <0·01 |
| Cough | 56 | 38 (30–46) | 52 | 15 (11–18) | <0·01 |
| Fever or felt febrile | 55 | 38 (30–46) | 48 | 14 (10–17) | <0·01 |
| Sore throat | 43 | 30 (22–38) | 48 | 14 (10–17) | <0·01 |
| Runny nose | 41 | 29 (20–35) | 38 | 11 (7–15) | <0·01 |
| Sneezing | 36 | 24 (17–31) | 31 | 9 (6–13) | <0·01 |
| Nausea | 26 | 18 (12–25) | 18 | 5 (3–8) | <0·01 |
| Difficulty breathing | 17 | 12 (6–17) | 12 | 3 (1–5) | <0·01 |
| Vomiting | 12 | 8 (4–13) | 8 | 2 (0–4) | <0·01 |
| Diarrhea | 10 | 7 (3–11) | 13 | 4 (2–6) | 0·18 |
*Numbers sum to 47% due to rounding.
Bivariate and multivariable logistic regression
Using bivariate analysis of all cases, no significant difference in odds of H1N1 virus infection by sex or race/ethnicity was indicated (Table 2). Marines were at higher odds of H1N1 virus infection, compared with sailors (unadjusted odds ratio [UOR], 1·75; 95% CI, 1·16–2·62). Officers had significantly lower odds of H1N1 virus infection, compared with junior enlisted personnel (UOR, 0·38; 95% CI, 0·19–0·78). Participants aged ≥35 years had lower odds of H1N1 virus infection than those aged 19‐24 years (UOR, 0·32; 95% CI, 0·13–0·77). Reported exposure to an ill person in sleeping areas was significant for increased odds of H1N1 virus infection in the bivariate analysis (P = 0·03).
In the adjusted model, women had higher odds of H1N1 virus infection (adjusted odds ratio [AOR], 2·19; 95% CI, 1·09–4·42, Table 2). Odds of infection remained higher for marines compared with sailors (AOR, 1·75; 95% CI, 1·05–2·91). Persons aged ≥35 years continued to have lower odds of infection, compared with those aged 19–24 years (AOR, 0·26; 95% CI, 0·08–0·85). No statistically significant differences were noted in the adjusted analysis by rank, race/ethnicity, or exposures to ill persons in sleep or work spaces.
Discussion
This investigation is the first on a naval vessel to use seroepidemologic analysis on matched pre‐ and post‐outbreak sera and a questionnaire to estimate infection attack rates, including asymptomatic or subclinical infections. Availability of baseline sera afforded the opportunity to serologically confirm the subclinical infections in characterizing this outbreak. Approximately half of the H1N1 infections were asymptomatic. Because one‐third of H1N1‐infected persons had symptoms of ARI without measured fever or reported feverishness, testing ARI cases might yield more accurate estimates of infection during similar outbreaks.
The ILI case definition required a measured fever ≥100°F (37·8°C). Eleven percent of survey participants met ILI case definition criteria for respiratory illness with 96% specificity (Table 3). Including tactile or self‐reported fever in the ILI case definition would substantially increase sensitivity (59‐infected persons, 41%), but lower specificity (84%). The sensitivity of ILI case definition that includes tactile or self‐reported fever was similar to sensitivity (36%) of the ARI case definition.
The three symptomatic crew members, confirmed retrospectively as H1N1‐infected by serology, reported for duty the week before embarkation and might have introduced H1N1 virus to the ship. The shipboard outbreak of H1N1 spread rapidly during the first at‐sea period among an otherwise healthy, young adult military population during the first pandemic wave. Other investigations of naval vessel influenza outbreaks have documented illness among shipboard personnel while ashore, and subsequent transmission while at sea.9, 10
Marines had higher odds of H1N1 virus infection. Multiple factors might have contributed to this finding, including crowding or exposure to greater numbers of people in common areas during the at‐sea training exercises, which created conditions that can increase disease transmission. Officers and senior enlisted personnel have separate, less crowded dining and sleeping areas. Although not statistically significant in the adjusted analysis, junior‐ranking enlisted persons, whose sleeping quarters are shared with many others, and those reporting exposure to ill persons in sleeping areas had higher odds of illness. Taken together, these findings provide evidence that physical proximity and crowding in sleeping spaces might have contributed to H1N1 virus transmission.10 Seroepidemiologic surveys for H1N1 virus antibodies in England and Singapore reported higher proportions of infection among school‐age children and young adults, consistent with increased susceptibility among younger persons and potential for transmission in semiclosed or military settings.11, 12, 13, 14
In multivariable analysis, women were at higher odds of H1N1 illness than men. The reasons for this are uncertain and might be attributed to occupational or social factors not reported in our questionnaire. During a H1N1 outbreak at a public university, women were at increased risk of ILI and H1N1 infection, as determined by chart abstraction and RT‐PCR, than were male students (relative risk 1·5; 95% CI 1·3–1·9).15
The 32% H1N1 attack rate among personnel was consistent with previously documented H1N1 and other influenza outbreaks among naval populations. During 1996, a US Navy vessel experienced an outbreak of ILI attributed to influenza A (H3N2) virus, with a symptomatic illness attack rate of 42% among the 548‐person crew.1 A Peruvian ship with a crew of 355 experienced a H1N1 outbreak with a symptomatic attack rate of 22% determined by RT‐PCR.10 In a retrospective serologic investigation, 39·3% of crew members aboard an Italian military ship had H1N1 virus infection. Of those who denied ARI, 36·9% had evidence of H1N1 virus infection as determined using complement fixation or hemagglutination‐inhibition antibody assays.13 Outbreaks of influenza have been documented in similarly congregate settings, including cruise ship travelers and university students.15, 16
A community‐level H1N1 serosurvey on Reunion Island in the Indian Ocean analyzed paired serum specimens by using hemagglutination‐inhibition assay and reported a cumulative incidence rate of 14% among adults aged 20–59 years, and an overall seroconversion rate of 65% among 1687 participants.17 In the United States, a 13% H1N1 clinical attack rate was calculated using a sample of reports of clinical symptoms among household members of laboratory‐confirmed and probable H1N1 cases (those testing positive for influenza A using RT‐PCR, with a body temperature >37·8°C and cough or sore throat), extracted from state health department reports and submitted to CDC during April 29–May 25, 2009.18 A nationally representative serologic survey performed in New Zealand reported an overall infection attack rate of 26·7%, with 45·2% of those infected denying illness symptoms.19 Cross‐sectional surveys performed in England and Greece reported attack rates ranging from 6 to 31% among adults aged ≥18 years and stratified by age group.11, 20
During this US military ship outbreak, a substantial proportion of infected persons had asymptomatic illness or had not sought medical care and thus may have had only mild illness. Adherences to infection control interventions, as well as an interim port stay with exclusion of ill persons, likely contributed to limiting respiratory disease transmission during the 36‐day training exercises. An outbreak during an extended deployment, during which isolation or social distancing might be less feasible, might result in a greater number of illnesses and potential negative impacts to ship operations.
Limitations
Because the investigation was retrospective, participant recall might have been incomplete or inaccurate. Recall might have been different in those who had symptoms of respiratory illness compared with those who did not. No active surveillance system was in place to identify all ill persons, although symptomatic crew members could have been ordered to the medical clinic by supervisors. Additionally, sera were not collected at the beginning or 2 weeks after the end of the training exercises for the estimation of H1N1 virus antibodies. However, pre‐outbreak sera, excluding certain seropositives in baseline sera, were matched with post‐outbreak specimens. Personnel turnover (6%) or non‐response of selected participants (20%) made selection bias possible.
We attempted to differentiate community‐acquired illness from illness associated with shipboard exposure. We defined community‐acquired cases as those occurring within 2 days of the ship's departure from San Diego; however, using a 2‐day time frame might have resulted in misclassification and underestimation of community‐acquired cases as ship acquired. Misclassification of asymptomatic persons infected outside of the training exercises that were attributed to shipboard exposure was possible. If symptomatic, infections were assumed to be H1N1, but might have been due to another respiratory pathogen.
In conclusion, this investigation found an H1N1 attack rate of 32% aboard the Navy vessel, and a large proportion of asymptomatic infection (53%). Attack rates were highest among marines, younger persons, and women. In the week preceding the exercises, three infected individuals reported to work and might have triggered the outbreak. In addition to annual influenza vaccination, to prevent or mitigate future influenza outbreaks in ship settings, we recommend considering the implementation of a screening process to restrict persons with acute respiratory illness from boarding. While at sea, onboard active surveillance should include mechanisms to improve self‐report of illness, incorporate active surveillance of close contacts of ill persons in workspaces or sleeping areas to enable prompt isolation of additional ill persons, potential quarantine of exposed contacts, prompt empiric antiviral treatment of symptomatic persons, and consideration of antiviral chemoprophylaxis of exposed persons.21, 22 Maintaining laboratory surveillance programs for early detection of respiratory illness outbreaks, including influenza testing, will enable prompt intervention for disease mitigation, control, and prevention.
Disclaimer
The opinions expressed by authors do not necessarily reflect the official policies or positions of the Centers for Disease Control and Prevention, the Department of the Navy, the Department of Defense, or the US Government.
Financial support
This work was funded in part by a grant from the US Department of Defense's Armed Forces Health Surveillance Center, Division of the Global Emerging Infections Surveillance and Response System.
Conflicts of interest
None.
Acknowledgements
We thank Daniel B. Fishbein, Michele Ginsberg, Kenneth A. Katz, Samantha Tweeten, Denise Borntrager, Monica Sovero, Erin Murray, Diana Bensyl, Xu Xiyan, Angie Eick, Tom Gao, CAPT John Funk, Col Gregg Olson, LCDR Noelle Griffith, HMCM Brad Kowitz, HMCS Carol Merricks, and US Sailors & Marines aboard the USS Bonhomme Richard for their support with this investigation.
Please cite this paper as: Khaokham et al (2013) Seroepidemiologic investigation of an outbreak of pandemic influenza A H1N1 2009 aboard a US Navy Vessel—San Diego, 2009. Influenza and Other Respiratory Viruses 7(5), 791–798
References
- 1. Earhart KC, Beadle C, Miller LK et al Outbreak of influenza in highly vaccinated crew of U.S. Navy ship. Emerg Infect Dis 2001; 7:463–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. CDC . Swine influenza A (H1N1) infection in two children–Southern California, March‐April 2009. MMWR Morb Mortal Wkly Rep 2009; 58:400–402. [PubMed] [Google Scholar]
- 3. Dawood FS, Jain S, Finelli L et al Emergence of a novel swine‐origin influenza A (H1N1) virus in humans. N Engl J Med 2009; 360:2605–2615. [DOI] [PubMed] [Google Scholar]
- 4. Shu B, Wu K‐H, Emery S et al Design and performance of the CDC real‐time reverse transcriptase PCR swine flu panel for detection of 2009 A (H1N1) pandemic influenza virus. J Clin Microbiol 2011; 49:2614–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tang JW, Shetty N, Lam TT. Features of the new pandemic influenza A/H1N1/2009 virus: virology, epidemiology, clinical and public health aspects. Curr Opin Pulm Med 2010; 16:235–241. [DOI] [PubMed] [Google Scholar]
- 6. Xu R, Ekiert DC, Krause JC, Hai R, Crowe JE Jr, Wilson IA. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science 2010; 328:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hancock K, Veguilla V, Lu X et al Cross‐reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med 2009; 361:1945–1952. [DOI] [PubMed] [Google Scholar]
- 8. World Health Organization . WHO Regional Office for Europe guidance for sentinel influenza surveillance in humans. 2011. Available at http://www.euro.who.int/__data/assets/pdf_file/0020/90443/E92738.pdf (Accessed 16 September 2011).
- 9. Ksiazek TG, Olson JG, Irving GS, Settle CS, White R, Petrusso R. An influenza outbreak due to A/USSR/77‐like (H1N1) virus aboard a US Navy ship. Am J Epidemiol 1980; 112:487–494. [DOI] [PubMed] [Google Scholar]
- 10. CDC . Outbreak of 2009 pandemic influenza A (H1N1) on a Peruvian Navy ship ‐ June‐July 2009. MMWR Morb Mortal Wkly Rep 2010; 59:162–165. [PubMed] [Google Scholar]
- 11. Miller E, Hoschler K, Hardelid P, Stanford E, Andrews N, Zambon M. Incidence of 2009 pandemic influenza A H1N1 infection in England: a cross‐sectional serological study. Lancet 2010; 375:1100–1108. [DOI] [PubMed] [Google Scholar]
- 12. Lim W‐Y. Risk factors for pandemic (H1N1) 2009 Seroconversion among Adults, Singapore, 2009. Emerg Infect Dis 2011; 17:1455–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tarabbo M, Lapa D, Castilletti C et al Retrospective investigation of an influenza A/H1N1pdm outbreak in an Italian military ship cruising in the Mediterranean Sea, May‐September 2009. PLoS ONE 2011; 6:e15933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen MIC, Lee VJM, Lim W‐Y et al 2009 influenza A(H1N1) seroconversion rates and risk factors among distinct adult cohorts in Singapore. JAMA 2010; 14:303. [DOI] [PubMed] [Google Scholar]
- 15. Iuliano AD, Reed C, Guh A et al Notes from the field: outbreak of 2009 pandemic influenza A (H1N1) virus at a large public university in Delaware, April–May 2009. Clin Infect Dis 2009; 49:1811–1820. [DOI] [PubMed] [Google Scholar]
- 16. Uyeki TM, Zane SB, Bodnar UR et al Large summertime influenza A outbreak among tourists in Alaska and the Yukon Territory. Clin Infect Dis 2003; 36:1095–1102. [DOI] [PubMed] [Google Scholar]
- 17. Dellagi K, Rollot O, Temmam S et al Pandemic influenza due to pH1N1/2009 virus: estimation of infection burden in Reunion Island through a prospective serosurvey, Austral winter 2009. PLoS ONE 2011; 6:e25738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cauchemez S, Donnelly CA, Reed C et al Household transmission of 2009 pandemic influenza A (H1N1) virus in the United States. N Engl J Med 2009; 361:2619–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bandaranayake D, Huang QS, Bissielo A et al Risk factors and immunity in a nationally representative population following the 2009 influenza A(H1N1) pandemic. PLoS ONE 2010; 5:e13211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maltezou HC, Katerelos P, Mavrouli M et al Seroepidemiological study of pandemic influenza H1N1 following the 2009‐2010 wave in Greece. Vaccine 2011; 29:6664–6669. [DOI] [PubMed] [Google Scholar]
- 21. Fiore AE, Fry A, Shay D, Gubareva L, Bresee JS, Uyeki TM. Antiviral agents for the treatment and chemoprophylaxis of influenza — recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recomm Rep 2011; 60:1–24. [PubMed] [Google Scholar]
- 22. Harper SA, Bradley JS, Englund JA et al Seasonal influenza in adults and children–diagnosis, treatment, chemoprophylaxis, and institutional outbreak management: clinical practice guidelines of the Infectious Diseases Society of America. Clin Infect Dis 2009; 48:1003–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]


