Abstract
Purpose
Limb-bud and heart (LBH) levels are correlated with adverse survival in several malignancies; however, their significance in hepatocellular carcinoma (HCC) remains unclear. The objective of this study was to determine the association between LBH status and clinical outcomes.
Methods
We selected 226 patients with HCC who were treated surgically between 2003 and 2010 at a single academic center. Immunohistochemistry (IHC) was used to detect the protein expression of LBH in HCC samples. Receiver operating characteristic (ROC) curve analysis, Spearman’s rank correlation, Kaplan–Meier plots, and the Cox proportional hazards regression model were used to analyze the data.
Results
A high expression of LBH was detected in 20 (8.8%) of 226 HCC samples. Correlation analysis demonstrated that LBH in HCC was significantly correlated with aspartate aminotransferase (AST)/alanine aminotransferase (ALT) levels and clinical stages (P<0.05). In the Kaplan–Meier analysis, the mean survival time of patients with low levels of LBH was longer than that for those with high levels of LBH (P<0.05). The 3-year overall survival rate was 20% for patients with HCC and high levels of LBH versus 67% for patients with HCC and low levels of LBH. In the multivariate analysis, AST/ALT level, clinical stage, tumor relapse, and the level of LBH were the independent prognostic factors for overall survival (P<0.05).
Conclusion
Overexpression of LBH might contribute to the development and progression of HCC. LBH could be a novel prognostic marker for HCC.
Keywords: LBH, hepatocellular carcinoma, immunohistochemistry, prognosis
Introduction
The 2017 cancer statistics report showed that over the past 25 years, the total cancer mortality in the US has fallen by 25%; however, the incidence and mortality of hepatocellular carcinoma (HCC) are increasing. HCC is a deadly cancer that occurs three times more frequently in men than in women, partly reflecting higher levels of hepatitis B and C virus infection rates, historical rates of smoking, and alcohol consumption in men.1,2 Surgery is the main radical treatment for HCC; however, 60%–70% of patients with HCC relapse within 5 years after radical resection.3 HCC treatment outcomes are affected by several variables, including liver function and clinical stage.4 Studies on the etiology of HCC defined viral infections, obesity, and diabetes as factors for HCC.5,6 However, the genes driving the development of HCC need to be fully elucidated.
The limb-bud and heart (LBH) protein acts as a key transcriptional regulator in the mitogen-activated protein kinase signaling pathway to mediate cellular functions.7,8 LBH was isolated from a human embryonic heart cDNA library. The LBH cDNA is 2,927 bp long, encoding a protein product of 105 amino acids. Al-Ali et al9 reported that, similar to other intrinsically disordered proteins (IDPs), LBH may undergo a “disorder-to-order” transition upon binding to its target molecules and depending on the target-induced conformational changes, might acquire different functional activities. Rieger et al first noted an association of LBH with cancer in 2010, observing that the LBH gene played an important transcriptional role in wingless/int-1 (Wnt) signaling pathways, which suppress mammary epithelial cell differentiation and could contribute to Wnt-induced tumorigenesis. Thus, LBH was highlighted as a potential new marker for therapeutically challenging basal-like breast cancer.7 However, there has been no relevant report on the clinicopathological significance and mechanism of LBH in HCC. In the present study, we used immunohistochemistry (IHC) to evaluate the protein level of LBH in HCC and statistical analysis methods to identify the association between LBH and the prognosis of HCC.
Methods
Patients and tissue specimens
In this study, 226 patients with HCC treated surgically between 2003 and 2010 who were patients of the Department of Pathology of Sun Yat-sen University Cancer Center (Guangzhou, China) were selected. Case selection was based on the pathological diagnosis of HCC, including complete follow-up data. These HCC cases included 203 (89.8%) men and 23 (10.2%) women. Average follow-up time was 60.23 months (median: 58.5 months; range: 1.0–185 months). In all, 47 patients (20.8%) were diagnosed at late stages (III and IV) and the other 179 patients (79.2%) were diagnosed at early stages (I and II). Patients whose cause of death remained unknown were excluded from our study. Table 1 details the patients’ clinicopathological features (age, gender, history of hepatitis, alpha-fetoprotein [AFP], cirrhosis, tumor size, clinical stage, and relapse). Tumor differentiation was based on the Edmondson and Steiner criteria. Tumor stage was defined according to the tumor–node–metastasis (TNM) classification system proposed by the American Joint Committee on Cancer/International Union Against Cancer.
Table 1.
Correlation of LBH expression with patients’ clinicopathological features in HCC
| Variable | LBH protein
|
|||
|---|---|---|---|---|
| All cases, n | Low expression, n (%) | High expression, n (%) | P-valuea | |
| Age (years) | ||||
| <51b | 107 | 96 (89.7) | 11 (10.3) | |
| ≥51 | 119 | 110 (92.5) | 9 (7.5) | |
| Gender | 0.115 | |||
| Male | 203 | 183 (90.1) | 20 (9.9) | |
| Female | 23 | 23 (100) | 0 (0) | |
| HbsAg | 0.156 | |||
| Positive | 207 | 187 (90.3) | 20 (9.7) | |
| Negative | 19 | 19 (100) | 0 (0) | |
| AFP (ng/mL) | 0.061 | |||
| <81c | 112 | 106 (94.6) | 6 (5.4) | |
| ≥81 | 112 | 98 (87.5) | 14 (12.5) | |
| Liver cirrhosis | 0.301 | |||
| No | 90 | 80 (88.9) | 10 (11.1) | |
| Yes | 127 | 118 (92.9) | 9 (7.1) | |
| Tumor size (cm) | 0.200 | |||
| <5.5d | 109 | 102 (93.6) | 7 (6.4) | |
| ≥5.5 | 115 | 102 (88.7) | 13 (11.3) | |
| AST/ALT | 0.042* | |||
| <1.4e | 115 | 109 (94.8) | 6 (5.2) | |
| ≥1.4 | 111 | 97 (87.4) | 14 (12.6) | |
| Relapse | 0.272 | |||
| No | 138 | 131 (94.9) | 7 (5.1) | |
| Yes | 66 | 60 (90.9) | 6 (9.1) | |
| Clinical stage | 0.027* | |||
| I–II | 179 | 167 (93.3) | 12 (6.7) | |
| III–IV | 47 | 39 (83.0) | 8 (17.0) | |
Notes:
Chi-square test.
Mean age.
Mean.
P<0.05.
Abbreviations: LBH, limb-bud and heart; HbsAg, hepatitis B surface antigen; HCC, hepatocellular carcinoma; AFP, alpha-fetoprotein; AST, aminotransferase; ALT, alanine aminotransferase.
IHC
Immunohistochemical staining of the LBH protein was detected using the standard EnVision procedure. The paraffin-embedded tissue blocks were cut into 3-μm thick sequential sections. The slides were dried and deparaffinized in xylene, rehydrated through graded alcohol, immersed in 3% hydrogen peroxide for 10 min to block endogenous peroxidase activity, and subjected to antigen retrieval by pressure cooking for 3 min in citrate buffer (pH =6.0). The slides were then incubated with 5% bovine serum albumin (BSA) for 15 min to reduce nonspecific reactions. Subsequently, the slides were incubated with a rabbit anti-LBH polyclonal antibody (ab122223, 1:400 dilution; Abcam, Cambridge, MA, USA) for 50 min at 37°C. The slides were sequentially incubated with a secondary antibody (K5007; Dako Denmark A/S, Glostrup, Denmark) for 30 min in the incubator at 37°C and stained with 3,3′-diaminobenzidine (DAB). Finally, the sections were counterstained with Mayer’s hematoxylin, dehydrated, and mounted. A negative control was obtained by replacing the primary antibody with normal rabbit IgG.
IHC evaluation
LBH protein levels were evaluated by microscopic examination of the stained tissue microarray (TMA) slides. Each TMA spot was ranked using an intensity evaluation from 1 to 3 (A) and with the proportion of tumor cells as 5% increments from a range of 0–100 (B). A final score (range 0–300) was achieved using each intensity and the proportion of the area stained (score = A × B).10 LBH expression was scored by three independent senior pathologists who were blinded to the patients’ clinicopathological data. Their diagnoses were completely consistent in ~75% of the cases, which proved that this method was highly repeatable. If two or all of them agreed with the diagnosis results, the value was selected. If their opinions were different, all three of them repeated the diagnosis and came to a consensus by discussion.
Statistical analysis
SPSS software (version 21.0; SPSS Inc., Chicago, IL, USA) was used for the statistical analysis. Receiver operating characteristic (ROC) analysis was performed to determine the cutoff value for LBH. The correlation between LBH protein levels and clinicopathological parameters in patients with HCC was analyzed using the Chi-square test. The survival of patients with HCC was evaluated using the Kaplan–Meier method with the log-rank test. Multivariate analyses were performed using the Cox proportional hazards model. All P-values were reported by two-sided analyses, and P<0.05 was considered as statistically significant.
Ethics approval and consent to participate
This study was approved by the Institute Research Medical Ethics Committee of Sun Yat-sen University Cancer Center. No informed consent (written or verbal) was obtained for use of retrospective data from the patients within this study, most of whom were deceased, since this was not deemed necessary by the ethics committee, who waived the need for consent. All samples were anonymized.
Results
Expression of LBH in HCC
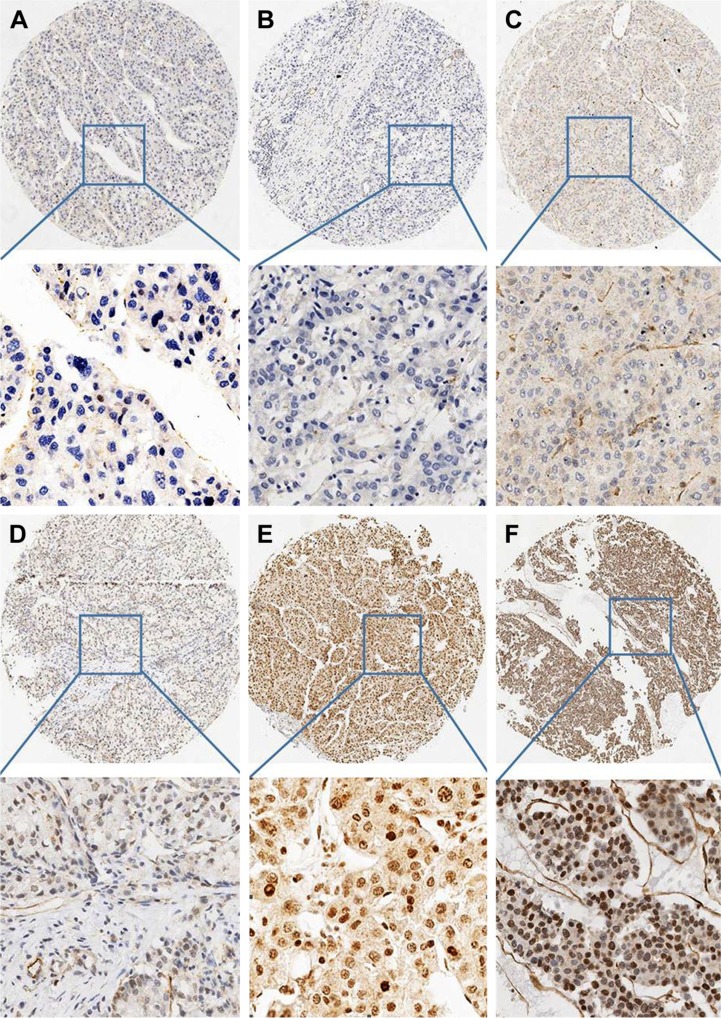
The IHC results showed that LBH protein was mainly located in the nuclei in HCC (Figure 1) and that 8.8% (20/226) of the HCC specimens were positive for LBH.
Figure 1.
Expression of LBH in HCC.
Notes: (A and B) HCC cases showing nearly negative expression of LBH protein (magnification ×4, ×20). (C and D) HCC cases demonstrating low expression of LBH (magnification ×4, ×20). (E and F) High expression of LBH in HCC (magnification ×4, ×20).
Abbreviations: LBH, limb-bud and heart; HCC, hepatocellular carcinoma.
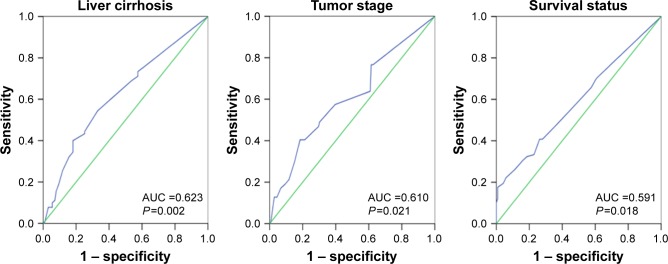
Selection of a cutoff value for LBH expression
ROC curves were used to determine the cutoff score for positive expression of the LBH protein. The sensitivity and specificity for liver cirrhosis, tumor stage, and survival status were plotted. The ROC curve analysis revealed that the cutoff value for the expression of LBH protein was 85 (area under the curve [AUC] =0.591, P=0.018; Figure 2).
Figure 2.
ROC curves were used to determine the cutoff score for positive expression of the LBH protein.
Note: The sensitivity and specificity for each outcome were plotted: liver cirrhosis, tumor stage, and survival status.
Abbreviations: ROC, receiver operating characteristic; LBH, limb-bud and heart; AUC, area under the curve.
Association of LBH expression with HCC patients’ clinicopathological parameters
Chi-square analysis revealed that a high level of LBH was more common in patients with higher aspartate aminotransferase (AST)/alanine aminotransferase (ALT) levels (P=0.042) and those at later clinical stages (P=0.027). There was no significant correlation between LBH levels and other clinicopathological parameters, such as patient age, gender, hepatitis history, liver cirrhosis, AFP level, tumor size, and tumor relapse (P>0.05). The details are shown in Table 1.
Relationship between clinicopathological features, LBH levels, and patient survival: univariate survival analysis
Univariate survival analysis demonstrated a significant effect of well-known clinicopathological prognostic parameters, such as tumor size (P=0.001), serum AST/ALT level (P=0.002), clinical stage (P=0.004), and relapse (P=0.000). LBH expression (P=0.000) could be used as an important factor to assess the prognosis of HCC. Kaplan–Meier survival analysis showed that the mean survival time of the low-LBH group was significantly longer than that of the high LBH group (111.4 months vs 30.2 months, P=0.000; Table 2 and Figure 3). Among the 206 patients with HCC and low LBH levels, the 3-year mean survival rate was 67%; however, in the high LBH group (20 patients), the mean 3-year survival rate was only 20% (Table 3).
Table 2.
Univariate analysis of LBH expression and various clinicopathological parameters in 226 patients with HCC (log-rank test)
| Variable | All cases, n | Mean survival (months) | P-value |
|---|---|---|---|
| Age (years) | 0.52 | ||
| <51 | 107 | 80 | |
| ≥51 | 119 | 105 | |
| Gender | 0.348 | ||
| Male | 203 | 99.6 | |
| Female | 23 | 86.7 | |
| HbsAg | 0.744 | ||
| Positive | 207 | 100.3 | |
| Negative | 19 | 82.8 | |
| AFP (ng/mL) | 0.161 | ||
| <81 | 112 | 86.4 | |
| ≥81 | 112 | 92.7 | |
| Liver cirrhosis | 0.193 | ||
| No | 90 | 96.6 | |
| Yes | 127 | 85.8 | |
| Tumor size (cm) | 0.001* | ||
| <5.5 | 109 | 93.7 | |
| ≥5.5 | 115 | 87 | |
| AST/ALT | 0.002* | ||
| <1.4 | 115 | 92.4 | |
| ≥1.4 | 111 | 84.2 | |
| Relapse | 0.000* | ||
| No | 138 | 123.6 | |
| Yes | 66 | 67.2 | |
| Clinical stage | 0.004* | ||
| I–II | 179 | 108.4 | |
| III–IV | 47 | 62.7 | |
| LBH expression | 0.000* | ||
| Low | 206 | 111.4 | |
| High | 20 | 30.2 |
Note:
P<0.05.
Abbreviations: LBH, limb-bud and heart; HbsAg, hepatitis B surface antigen; HCC, hepatocellular carcinoma; AFP, alpha-fetoprotein; AST, aminotransferase; ALT, alanine aminotransferase.
Figure 3.
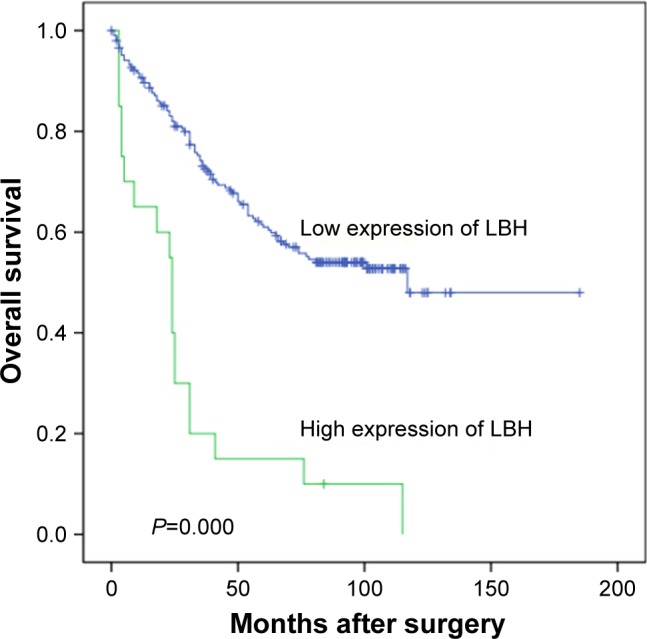
Kaplan–Meier survival analysis of LBH expression in total patients.
Notes: Probability of survival of all patients with HCC: low expression, n=206; high expression, n=20.
Abbreviations: LBH, limb-bud and heart; HCC, hepatocellular carcinoma.
Table 3.
Three-year survival rate in 226 patients with HCC
| Variable | All cases, n | Survival <3 years, n (%) | Survival >3 years, n (%) | P-value |
|---|---|---|---|---|
| LBH expression | 0.000* | |||
| Low expression | 206 | 68 (33) | 138 (67) | |
| High expression | 20 | 16 (80) | 4 (20) |
Note:
P<0.05.
Abbreviations: HCC, hepatocellular carcinoma; LBH, limb-bud and heart.
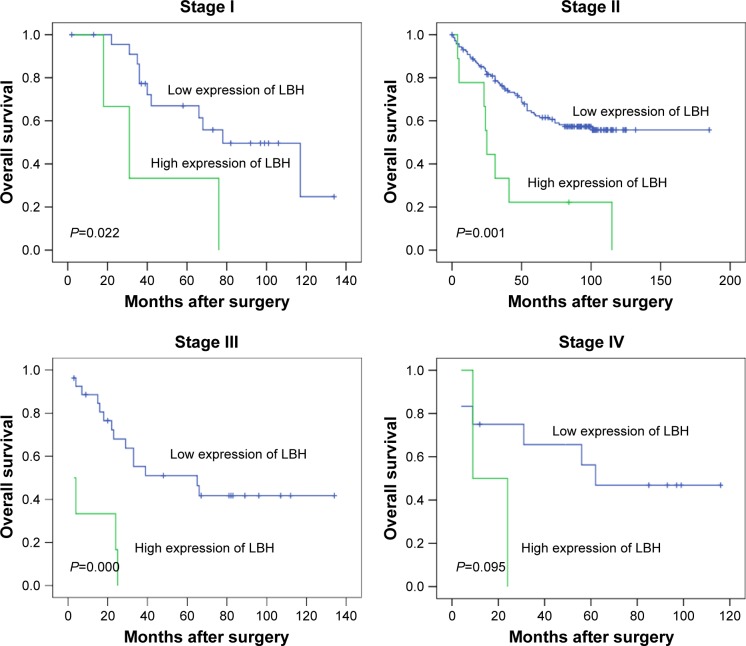
Further analysis was performed with regard to LBH levels in subsets of patients at different stages of HCC. The results demonstrated that a high level of LBH was a prognostic factor in patients with stage I (P=0.022), stage II (P=0.001), and stage III (P=0.000) HCC. However, they could not differentiate the outcome of patients with stage IV HCC (P=0.095). The details are shown in Figure 4.
Figure 4.
Kaplan–Meier survival analysis of LBH expression in subsets of patients at different clinical stages (log-rank test).
Notes: Stage I: low expression, n=24; high expression, n=3. Stage II: low expression, n=143; high expression, n=9. Stage III: low expression, n=27; high expression, n=6. Stage IV: low expression, n=12; high expression, n=2.
Abbreviation: LBH, limb-bud and heart.
Independent prognostic factors of HCC: multivariate Cox regression analysis
The factors that had a significant difference in the univariate analysis were subjected to Cox regression analysis. The results showed that a high level of LBH was an independent prognostic factor associated with poor overall patient survival (hazard ratio [HR]: 3.075; 95% confidence interval [CI]: 1.644–5.749, P=0.000; Table 4). At the same time, the study also found that the serum AST/ALT level (P=0.004), tumor relapse (P=0.001), and clinical stage (P=0.012) were independent prognostic factors for overall patient survival.
Table 4.
Cox multivariate analyses of prognostic factors for overall survival
| Variable | HR | 95% CI | P-value |
|---|---|---|---|
| AST/ALT (≥1.4 vs <1.4) | 1.884 | 1.222–2.905 | 0.004* |
| Tumor size (≥5.5 cm vs <5.5 cm) | 1.374 | 0.893–2.113 | 0.149 |
| Clinical stage (III–IV vs I–II) | 1.848 | 1.143–2.990 | 0.012* |
| Relapse (yes vs no) | 2.102 | 1.346–3.285 | 0.001* |
| LBH expression (high vs low) | 3.075 | 1.644–5.749 | 0.000* |
Note:
P<0.05.
Abbreviations: HR, hazard ratio; CI, confidence interval; AST, aminotransferase; ALT, alanine aminotransferase; LBH, limb-bud and heart.
Discussion
LBH is a member of a highly conserved family of small acidic proteins in vertebrates, which are localized at the nucleus.11 LBH is a key transcriptional regulator, with pivotal roles in embryonic development, early mouse heart development, and human diseases.9,12 However, the molecular mechanisms upstream of LBH and its role in tumor development remain unknown.
In the present study, we found that high levels of LBH could be detected in 20/226 (8.8%) of HCC samples. Correlation analysis demonstrated that LBH protein levels in HCC were significantly correlated with serum AST/ALT levels and clinical stage. In the Kaplan–Meier analysis, the mean survival time of patients with low levels of LBH was longer than that of those with high levels of LBH. The results suggested that the LBH level might be a new treatment classification for HCC patients.
In the multivariate analysis, serum AST/ALT level, tumor stage, tumor relapse, and the LBH level were the independent prognostic factors for overall survival. Indeed, Rieger et al7 also found that LBH was aberrantly overexpressed in mammary tumors of MMTV-Wnt1 transgenic mice, as well as in highly aggressive basal-like subtypes of human breast cancers. This result is consistent with ours. However, the study of Liu et al13 showed that LBH was considerably downregulated in nasopharyngeal cancer tissues. LBH functions as a tumor suppressor of nasopharyngeal carcinoma by inducing G1/S cell cycle arrest. Another study showed that overexpression of LBH significantly inhibited cell growth in fibroblast-like synoviocytes.14 The conclusion from these studies is that LBH can function as both an oncogene and a tumor suppressor gene.
Ai et al reported that overexpression of LBH activated the transcriptional activities of activator protein 1 (AP-1; also known as JUN) and serum response element (SRE), which are potential targets of extracellular signal-regulated kinase (ERK), JUN N-terminal kinase (JNK), and p38 in cellular signaling and functions. LBH might act as a transcriptional activator in mitogen-activated protein kinase signaling pathways to mediate cellular functions.8 LBH is the specific receptor of transforming growth factor beta (TGF-β) and is an essential factor to promote TGF-β function.13,15,16 LBH could reduce the expression of cyclin-dependent kinase (CDK) to affect cell proliferation.13 A primary breast cancer study showed that LBH expression was highest in the Claudin low subtype and a higher LBH expression correlated with worse survival.16,17 Deregulation of LBH in human basal breast cancer appears to be Wnt/b-catenin dependent, as Dickkopf WNT signaling pathway inhibitor 1 (DKK1) and Wnt7a inhibit LBH expression in breast tumor cells. Overexpression studies indicated that LBH suppresses mammary epithelial cell differentiation, an effect that could contribute to Wnt-induced tumorigenesis.7 Collectively, LBH induces the expression of key epithelial stem cell transcription factors. These studies identified LBH as an essential regulator of basal mammary stem cell expansion/maintenance, raising important implications for its potential role in breast cancer pathogenesis.18 This is consistent with our findings. Tumor cells that overexpressed LBH were chaotically multiplying, and the prognosis of this group of patients was poor. The 3-year overall survival rate was 20% for the high LBH group versus 67% for the low LBH group. Taken together, our clinical data and expression studies provided evidence that LBH could be used as an independent prognostic factor to assess the overall survival of patients with HCC.
Conclusion
Overexpression of LBH is associated with poor prognosis in human HCC. LBH could be used as an independent prognostic factor in patients with HCC and is expected to be a molecular target for anticancer therapy.
Acknowledgments
This work was supported by the Guangdong Natural Science who funded this work under the funds for distinguished young scholar (No 2015A030306001).
Footnotes
Author contributions
YX and JWC were responsible for the study design. JWC, CQH and KC performed the experiments and drafted the manuscript; SML, XZ, JC and MYC participated in the data analysis and interpretation. All authors read and approved the final manuscript. All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Njei B, Rotman Y, Ditah I, Lim JK. Emerging trends in hepatocellular carcinoma incidence and mortality. Hepatology. 2015;61(1):191–199. doi: 10.1002/hep.27388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pascual S, Herrera I, Irurzun J. New advances in hepatocellular carcinoma. World J Hepatol. 2016;8(9):421. doi: 10.4254/wjh.v8.i9.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mazzoccoli G, Tarquini R, Valoriani A, Oben J, Vinciguerra M, Marra F. Management strategies for hepatocellular carcinoma: old certainties and new realities. Clin Exp Med. 2016;16(3):243–256. doi: 10.1007/s10238-015-0368-z. [DOI] [PubMed] [Google Scholar]
- 5.Dhar D, Seki E, Karin M. NCOA5, IL-6, type 2 diabetes, and HCC: The deadly quartet. Cell Metab. 2014;19(1):6–7. doi: 10.1016/j.cmet.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu D, Cui L, Wang Y, et al. Hepatitis B e antigen and its precursors promote the progress of hepatocellular carcinoma by interacting with NUMB and decreasing p53 activity. Hepatology. 2016;64(2):390–404. doi: 10.1002/hep.28594. [DOI] [PubMed] [Google Scholar]
- 7.Rieger ME, Sims AH, Coats ER, Clarke RB, Briegel KJ. The embryonic transcription cofactor LBH is a direct target of the Wnt signaling pathway in epithelial development and in aggressive basal subtype breast cancers. Mol Cell Biol. 2010;30(17):4267–4279. doi: 10.1128/MCB.01418-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ai J, Wang Y, Tan K, et al. A human homolog of mouse Lbh gene, hLBH, expresses in heart and activates SRE and AP-1 mediated MAPK signaling pathway. Mol Biol Rep. 2008;35(2):179–187. doi: 10.1007/s11033-007-9068-4. [DOI] [PubMed] [Google Scholar]
- 9.Al-Ali H, Rieger ME, Seldeen KL, Harris TK, Farooq A, Briegel KJ. Biophysical characterization reveals structural disorder in the developmental transcriptional regulator LBH. Biochem Biophys Res Commun. 2010;391(1):1104–1109. doi: 10.1016/j.bbrc.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li M, Luo RZ, Chen JW, et al. High expression of transcriptional coactivator p300 correlates with aggressive features and poor prognosis of hepatocellular carcinoma. J Transl Med. 2011;9:5. doi: 10.1186/1479-5876-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Briegel KJ, Joyner AL. Identification and characterization of Lbh, a novel conserved nuclear protein expressed during early limb and heart development. Dev Biol. 2001;233(2):291–304. doi: 10.1006/dbio.2001.0225. [DOI] [PubMed] [Google Scholar]
- 12.Briegel KJ, Baldwin HS, Epstein JA, Joyner AL. Congenital heart disease reminiscent of partial trisomy 2p syndrome in mice transgenic for the transcription factor Lbh. Development. 2005;132(14):3305–3316. doi: 10.1242/dev.01887. [DOI] [PubMed] [Google Scholar]
- 13.Liu Q, Guan X, Lv J, Li X, Wang Y, Li L. Limb-bud and Heart (LBH) functions as a tumor suppressor of nasopharyngeal carcinoma by inducing G1/S cell cycle arrest. Sci Rep. 2015;5:7626. doi: 10.1038/srep07626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ekwall AK, Whitaker JW, Hammaker D, Bugbee WD, Wang W, Firestein GS. The rheumatoid arthritis risk gene LBH regulates growth in fibroblast-like synoviocytes. Arthritis Rheumatol. 2015;67(5):1193–1202. doi: 10.1002/art.39060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moroy T, Geisen C, Cyclin E. Int J Biochem Cell Biol. 2004;36(8):1424–1439. doi: 10.1016/j.biocel.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Ana Tufegdzic V, Oscar MR, Stephin JV, et al. Context-specific effects of TGF-β/SMAD3 in cancer are modulated by the epigenome. Cell Rep. 2015;13(11):2480. doi: 10.1016/j.celrep.2015.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindley LE, Curtis KM, Sanchez-Mejias A, Rieger ME, Robbins DJ, Briegel KJ. The WNT-controlled transcriptional regulator LBH is required for mammary stem cell expansion and maintenance of the basal lineage. Development. 2015;142(5):893–904. doi: 10.1242/dev.110403. [DOI] [PMC free article] [PubMed] [Google Scholar]