Abstract
Ovarian cancer is a gynecological malignant tumor with a high mortality rate among women, owing to metastatic progression and recurrence. Acquisition of invasiveness is accompanied by the loss of epithelial features and a gain of a mesenchymal phenotype, a process known as epithelial–mesenchymal transition (EMT). Transforming growth factor-β (TGF-β) has been implicated in the regulation of EMT. In the present study, we aimed to investigate the role of long noncoding RNA H19 and microRNA-370 (miR-370-3p) in TGF-β-induced EMT. Ovarian cancer cell lines SKOV-3 and OVCAR3 were incubated with different concentrations of TGF-β, and the results showed that TGF-β treatment upregulated H19 and downregulated miR-370-3p. In addition, an H19 knockdown or miR-370-3p overexpression suppressed TGF-β-induced EMT, while H19 overexpression or a miR-370-3p knockdown promoted TGF-β-induced EMT. Mechanistically, H19 could directly bind to miR-370-3p and effectively act as its competing endogenous RNA. Furthermore, we demonstrated that this activity of H19 was involved in its promotion of TGF-β-induced EMT. Thus, our results may provide novel insights into the process of TGF-β-induced EMT.
Keywords: transforming growth factor-β, long noncoding RNA H19, microRNA-370-3p, competing endogenous RNA, epithelial–mesenchymal transition
Introduction
Ovarian cancer is the seventh most common cancer and the eighth most common cause of cancer-related death among women, owing to metastatic progression and recurrence.1 Approximately 90% of ovarian cancers are surface epithelial-stromal tumors, also known as ovarian epithelial carcinomas, and 70% of the subtypes of these tumors are high-grade serous carcinomas.2 Although the capacity of cancer cells for invasion and metastasis is regulated by a variety of cellular and signaling proteins, it is known that acquisition of the invasiveness and metastatic ability by cancers is accompanied by the loss of epithelial features and a gain of a mesenchymal phenotype, a process known as epithelial–mesenchymal transition (EMT).3 It is therefore important to identify factors that inhibit EMT.
Long noncoding RNAs (lncRNAs) are defined as non-protein-coding transcripts that are longer than 200 nucleotides. Some lncRNAs such as MALAT1 and HOXA11 have been demonstrated to play key roles in the regulation of EMT in ovarian cancers.4,5 The imprinted lncRNA H19 has been implicated in the pathogenesis of diverse human cancers.6,7 It is highly expressed in ovarian cancer tissues and cell lines and has been shown to promote migration and invasiveness of tumor cells.8,9 In addition, H19 contributes to cisplatin resistance by regulating the glutathione metabolism in high-grade serous ovarian cancers.10 These results indicate that H19 functions as an oncogene in ovarian cancer, but the underlying mechanisms remain poorly understood.
MicroRNAs (miRNAs), small noncoding RNA molecules ~22 nucleotides long, modulate the biological behavior of cancer cells.9,11 In contrast, H19 functions as a competitive endogenous RNA (ceRNA) to regulate metastasis by decreasing the activity of miRNAs such as miR-200s, miR-630, miR-17-5p, and miR-29a;12–15 H19 has also been shown to be regulated by miRNAs such as miR-141.16 Thus, the relation between H19 and miRNAs is a complex one. Many miRNA-binding sites are present in the sequence of H19. MiR-370-3p is one of the miRNAs that have a binding site in the H19 sequence. MiR-370-3p is known to inhibit the proliferation of human glioma cells and to induce cell cycle arrest in these cells;17 however, its role in ovarian cancer is not clear. In addition, the relation between H19 and miR-370-3p is poorly understood.
Transforming growth factor (TGF)-β is an important inducer of EMT.18–20 It is known that TGF-β-induced EMT is involved in ovarian cancer progression and metastasis.21,22 Although some factors, such as miRNAs and proteins, can inhibit TGF-β-induced EMT in ovarian cancer,23,24 it is still urgently necessary to find key regulators of EMT for inhibiting tumor progression. In the present study, we focused on the role of H19 and miR-370-3p in the regulation of TGF-β-induced EMT; we also aimed to clarify the relation between H19 and miR-370-3p in this regulatory process. Our results revealed that H19 promoted TGF-β-induced EMT by acting as a ceRNA of miR-370-3p and might provide novel insights into the process of TGF-β-induced EMT and contribute to elucidation of the regulatory network in TGF-β-induced EMT in ovarian cancer.
Materials and methods
Cell culture and treatment
Human ovarian epithelial adenocarcinoma cell lines, SKOV-3 and OVCAR3, were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and maintained in the RPMI 1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) with 10% fetal bovine serum (FBS; Sigma-Aldrich Co., St Louis, MO, USA), 100 U/mL penicillin, and 100 µg/mL streptomycin (Beyotime Biotechnology, Shanghai, China) in a humidified atmosphere containing 5% CO2 at 37°C. The cells were cultured until ~80% confluence and starved in a serum-free RPMI 1640 medium overnight. Then, SKOV-3 and OVCAR3 cells were removed from the culture medium and treated with different factors.
Recombinant human TGF-β1 was reconstituted at 20 µg/mL in sterile 4 mM HCl containing 1 mg/mL bovine serum albumin (BSA). To detect the effect of TGF-β1 on the expression of H19 and miR-370-3p, the cells in the control group were treated with a vehicle (isometric 4 mM HCl containing 1 mg/mL BSA), and the TGF-β1-treated groups were incubated with different concentrations of TGF-β1 (2.5, 5, or 10 ng/mL; Peprotech, Princeton, NJ, USA). After 48 h of treatment, the cells were harvested for quantitative reverse-transcription PCR (qRT-PCR) to evaluate the expression levels of H19 and miR-370-3p. To detect the effect of H19 on EMT or miR-370-3p expression, SKOV-3 and OVCAR3 cells were pretransfected with a negative control (NC) small interfering RNA (siRNA, “si-NC”), H19 siRNA (“si-H19”), an empty vector (pcDNA3.0), or an H19 overexpression vector (pcDNA-H19). To detect the effect of miR-370-3p on EMT, cell lines SKOV-3 and OVCAR3 were pretransfected with an NC miRNA mimic (miR-NC), miR-370-3p mimic, NC inhibitor, or a miR-370-3p inhibitor. To test whether H19 regulates EMT through miR-370-3p, cell lines SKOV-3 and OVCAR3 were pretransfected with H19 siRNA plus the NC inhibitor, H19 siRNA plus the miR-370-3p inhibitor, pcDNA-H19 plus the NC miRNA mimic, or pcDNA-H19 plus the miR-370-3p mimic. After transfection for 24 h, the transfected cells were treated with 5 ng/mL of TGF-β1 for 48 h and then harvested for qRT-PCR, Western blotting, and immunofluorescent staining.
Synthesis and transfection of siRNA and miRNA mimics
H19 siRNA, targeting sequence 5′-CAAGACGCCAGGUCCGGUG-3′, and NC siRNA were designed and synthesized by GenePharma (Shanghai, China). The miR-370-3p mimic and miR-370-3p inhibitor were also synthesized by Gene-Pharma. SKOV-3 and OVCAR3 cells were transfected with a miRNA mimic or siRNA at a final concentration of 10 nM, using Lipofectamine RNAiMAX (Thermo Fisher Scientific).
qRT-PCR
After the different groups of cells were treated with the respective reagents, SKOV-3 and OVCAR3 cells were harvested, and total RNA was extracted using the TRIzol reagent (Thermo Fisher Scientific). To analyze the expression of miR-370-3p, RT-PCR was carried out with specific stem-loop RT primers by means of the Mir-X™ miRNA First Strand Synthesis Kit (Takara, Tokyo, Japan), and quantitative PCR was conducted using the Mir-X™ miRNA qRT-PCR SYBR® Kit (Takara) on an Applied Biosystems 7500 system. U6 served as the internal control. To quantify the expression of H19, Snail, E-cadherin, and vimentin, RT-PCR was carried out using the mProm-IITM Reverse Transcription System (Thermo Fisher Scientific), and quantitative PCR was conducted with SYBR Green qPCR SuperMix (Thermo Fisher Scientific) on the Applied Biosystems 7500 system. The internal control was 18S ribosomal RNA. The primer sequences used for the qRT-PCR are shown in Table 1. Gene expression was measured in triplicate and quantified by the 2−ΔΔCT method with normalization to a control.
Table 1.
Primers for qRT-PCR
| Primer name | Sequence (5′–3′) |
|---|---|
| miR-370-3p-F | ACACTCCAGCTGGGGCCTGCTGGGGTGGAACCT |
| miR-370-3p-R | CTCAACTG GTGTCGTGGA |
| H19-F | GCGGGTCTGTTTCTTTACTTC |
| H19-R | GTGGTTGTAAAGTGCAGCAT |
| U6-F | CTCGCTTCGGCAGCACA |
| U6-R | AACGCTTCACGAATTTGCGT |
| E-cadherin-F | CCCACCACGTACAAGGGTC |
| E-cadherin-R | CTGGGGTATTGGGGGCATC |
| Vimentin-F | CGCCAGATGCGTGAAATGG |
| Vimentin-R | ACCAGAGGGAGTGAATCCAGA |
| Snail-F | GCTGCAGGACTCTAATCCAGA |
| Snail-R | ATCTCCGGAGGTGGGATG |
| 18srRNA-F | CCTGGATACCGCAGCTAGGA |
| 18srRNA-R | GCGGCGCAATACGAATGCCCC |
Abbreviations: F, forward primer; qRT-PCR, quantitative reverse-transcription PCR; R, reverse primer.
Western blot
After the treatments, SKOV-3 and OVCAR3 cells were harvested and lysed for extracting the total cellular protein with RIPA buffer (Beyotime Biotechnology). The samples were centrifuged at 12,000 rpm for 30 min, and the supernatants were collected. The protein concentration in the lysates was quantified with a bicinchoninic acid protein assay kit (Beyotime Biotechnology). Thirty micrograms of each total protein sample was loaded onto a 10%–12% polyacrylamide gel for SDS-PAGE, and afterwards electrophoretically transferred onto a polyvinylidene fluoride membrane (EMD Millipore, Billerica, MA, USA). After blockage with 5% skim milk, the membranes were incubated overnight with the following primary antibodies at 4°C: anti-E-cadherin (1:500; Cell Signaling Technology, Danvers, MA, USA), anti-vimentin (1:800; Cell Signaling Technology), and anti-Snail (1:500; Cell Signaling Technology). The cells were then washed thrice with phosphate-buffered saline containing 0.1% of Tween 20 (PBST) and incubated with horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG antibodies at 37°C for 30 min (secondary antibody), followed by washing thrice with PBST again. The detected antigen–antibody complexes were visualized by means of a chemiluminescence (Immobilon ECL) reagent (EMD Millipore). Western blotting was performed in triplicate. All films were scanned, and the densitometric analysis was carried out using the Image Pro-Plus 6.0 software (Media Cybernetics, Silver Spring, MD, USA). For quantification of specific bands, a square of the same size was drawn around each band to measure the density, and then the value was adjusted for the density of the background near that band. The results of densitometric analysis were expressed as a relative ratio of the target protein to a reference protein.
Immunofluorescent staining
SKOV-3 and OVCAR3 cells were grown on cover slides, which were placed in 24-well plates (three wells for each group). After the cells adhered to the plate, the groups were treated with various reagents and conditions. At the end of the treatment, the cells were washed twice with cool PBS and fixed in 4% paraformaldehyde for 20 min at 4°C. They were then permeabilized with 0.1% Triton X-100 at 4°C for 15 min, and nonspecific binding was blocked with 1% FBS in a confining liquid for 1 h at 37°C. Next, the cells were incubated overnight with the following primary antibodies at 4°C: anti-E-cadherin (1:100; Cell Signaling Technology), anti-vimentin (1:100; Cell Signaling Technology), and anti-Snail (1:50; Cell Signaling Technology). After that, the cells were stained with a secondary antibody, an IgG-Cy5 conjugate (1:1,000, #4412; Cell Signaling Technology), in the dark for 1.5 h and then washed thrice with PBS for 3 min on a rocking platform. The cell nuclei were labeled with 4′,6-diamidino-2-phenylindole (DAPI) for 20 min. Images were captured by means of a confocal laser scanning microscope (TCS SP2; Leica Microsystems, Wetzlar, Germany).
Luciferase reporter assay
To detect the miR-370-3p-binding site in the H19 sequence, we used a number of bioinformatics tools (lncRNABase and RNAhybrid). The full H19 wild-type sequence was amplified with primers H19-Sgfl-F and H19-NotI-R (Table 2) and then inserted downstream of the luciferase gene in the psi-CHECK2 luciferase vector (Promega Corporation, Fitchburg, WI, USA). A point mutation in the putative miR-370-3p-binding sequence in H19 was introduced by PCR-directed mutagenesis with the primers shown in Table 2. SKOV-3 and OVCAR3 cells were cotransfected with a psi-CHECK2 luciferase vector containing the wild-type or mutant H19 sequence, along with miR-NC, miR-370-3p, the NC inhibitor, or miR-370-3p inhibitor using Lipofectamine 2000 (Thermo Fisher Scientific). After 48 h, the cells were harvested, and luciferase activity was estimated via the dual-luciferase reporter assay system (Promega Corporation). Firefly luciferase activity was normalized to the Renilla luciferase activity. Results were obtained from three independent experiments performed in triplicate.
Table 2.
Primers for luciferase reporter construction
| Primer name | Sequence (5′–3′) |
|---|---|
| H19-Sgfl-F | ACATGCGATCGCAGTTAGAAAAAGCCCGGGCTAGGACCGAG |
| H19-NotI-R | ATAAGAATGCGGCCGCGCTGTAACAGTGTTTATTGATGATG |
| mutH19-1F | TCGCTCTGTGCCCCTCCCCACCAGGATGGTCATCAAAGCCCTGGACTCATCATCAATAAAC |
| mutH19-1R | GTTTATTGATGATGAGTCCAGGGCTTTGATGACCATCCTGGTGGGGAGGGGCACAGAGCGA |
| mutH19-2F | AAGACAGGCAGTGCTCGGGGAGTTGTCATCAAACGTCACCAGGAGGGCGAAGCGGCC |
| mutH19-2R | GGCCGCTTCGCCCTCCTGGTGACGTTTGATGACAACTCCCCGAGCACTGCCTGTCTT |
Abbreviations: F, forward primer; mut, mutant; R, reverse primer.
Statistical analyses
These analyses were performed in the SPSS 19.0 software (IBM Corporation, Armonk, NY, USA). All data were expressed as the mean ± standard deviation (SD). The differences between groups were assessed by Student’s t-test. Statistical significance was assumed if P<0.05.
Results
The expression profiles of H19 and miR-370-3p during TGF-β1-induced EMT in SKOV-3 and OVCAR3 cells
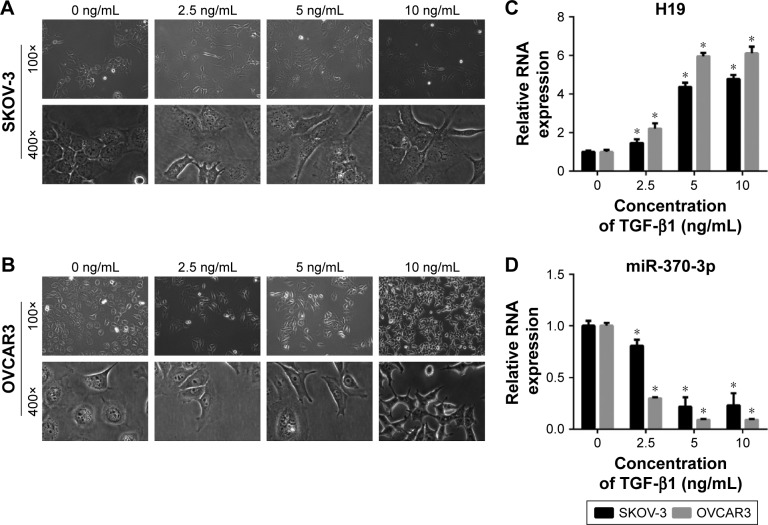
To determine whether H19 and miR-370-3p participate in TGF-β1-induced EMT in SKOV-3 and OVCAR3 cells, we first treated SKOV-3 and OVCAR3 cells with different concentrations of TGF-β1 (0, 2.5, 5, or 10 ng/mL). After incubation with TGF-β1 for 48 h, the SKOV-3 and OVCAR3 cells underwent EMT, as confirmed by the morphological change to spindle-shaped cells (Figure 1A and B). We then assessed the expression levels of H19 and miR-370-3p in these TGF-β1-stimulated SKOV-3 and OVCAR3 cells. It was found that H19 expression gradually increased, while miR-370-3p expression gradually decreased, as the concentration of TGF-β1 increased (Figure 1C and D). The upregulation of H19 and the downregulation of miR-370-3p indicated that they might affect EMT in SKOV-3 and OVCAR3 cells. In addition, we found that the effects of 5 and 10 ng/mL of TGF-β1 on H19 and miR-370-3p expression were similar, and accordingly, 5 ng/mL was chosen as the EMT-inducing concentration of TGF-β1 for all subsequent assays.
Figure 1.
The expression profiles of H19 and miR-370-3p during transforming growth factor-β1 (TGF-β1)-induced EMT in SKOV-3 and OVCAR3 cells. These cells were treated with 0, 2.5, 5, or 10 ng/mL of TGF-β1. After incubation with TGF-β1 for 48 h, the morphological change of SKOV-3 (A) and OVCAR3 cells (B) was detected under a light microscope (100× and 400×), and the expression profiles of H19 (C) and miR-370-3p (D) were determined by quantitative reverse-transcription PCR carried out in triplicate. Data are expressed as mean ± SD, *P<0.05.
The effect of H19 knockdown or overexpression on TGF-β1-induced EMT
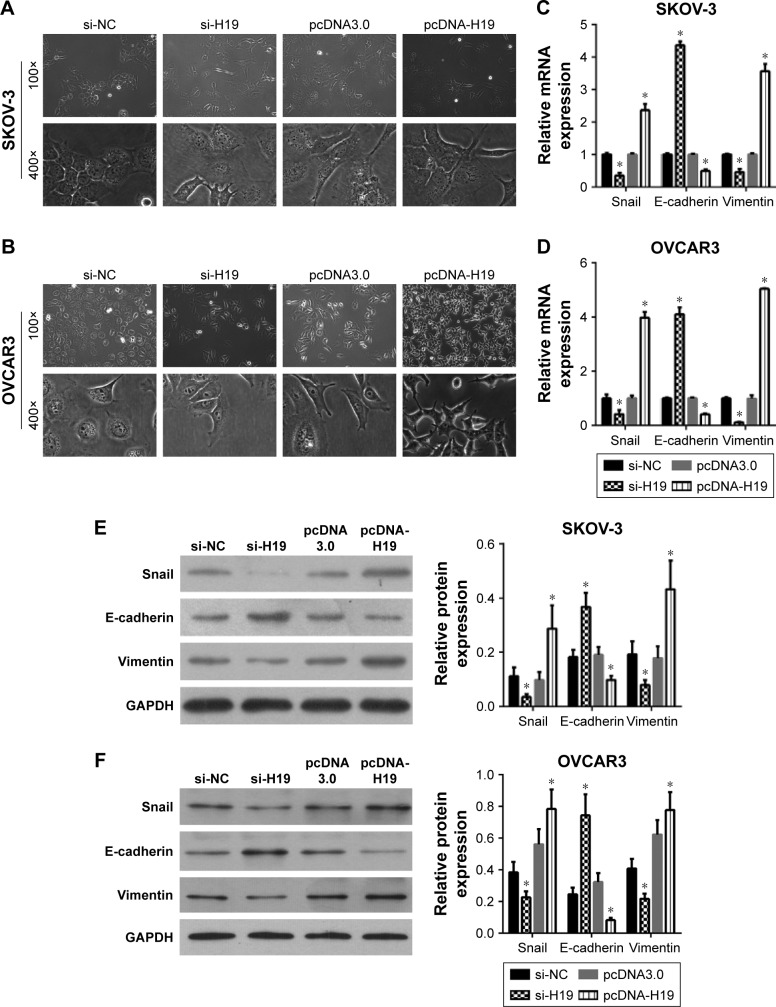
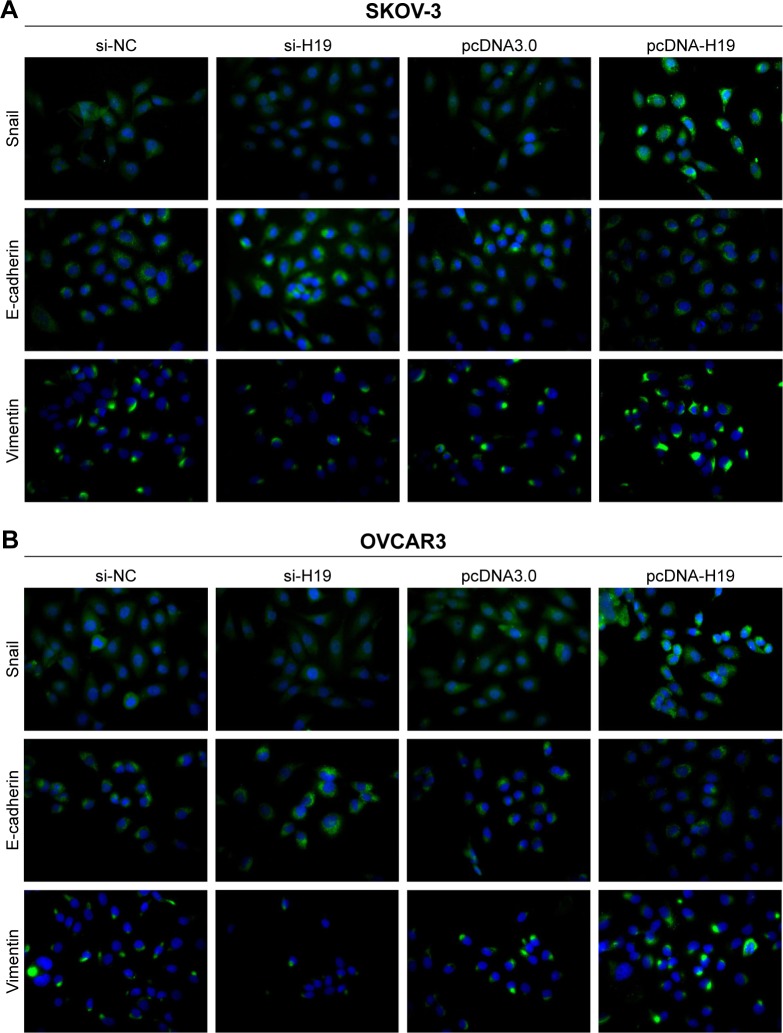
To investigate the role of H19 in TGF-β1-induced EMT, H19 was silenced or overexpressed by transfecting the SKOV-3 and OVCAR3 cells with H19 siRNA (si-H19) or pcDNA-H19 (overexpression plasmid). The results of qRT-PCR showed that H19 was successfully silenced in SKOV-3 and OVCAR3 cells transfected with si-H19, compared to those transfected with si-NC; H19 was successfully overexpressed in SKOV-3 and OVCAR3 cells transfected with pcDNA-H19, as compared to those transfected with pcDNA3.0 (empty vector; Figure 2). The effect of H19 knockdown or overexpression on TGF-β1-induced EMT was then estimated by measuring the changes in morphology and in the expression of Snail, E-cadherin, and vimentin. After pretransfection with NC siRNA, H19 siRNA, pcDNA3.0, or pcDNA-H19 for 24 h, the SKOV-3 and OVCAR3 cells were treated with 5 ng/mL of TGF-β1 for 48 h and harvested for qRT-PCR, Western blotting, and immunofluorescent staining. The morphological change to spindle-shaped cells was suppressed by H19 knockdown and promoted by H19 overexpression in SKOV-3 (Figure 3A) and OVCAR3 cells (Figure 3B). The results of qRT-PCR, Western blotting, and immunofluorescent staining revealed that the H19 knockdown significantly increased the protein level of the epithelial marker, E-cadherin, and correspondingly decreased the levels of mesenchymal markers, Snail and vimentin, whereas overexpression of H19 had the opposite effect on these EMT markers (Figures 3C–F, 4A and B).
Figure 2.
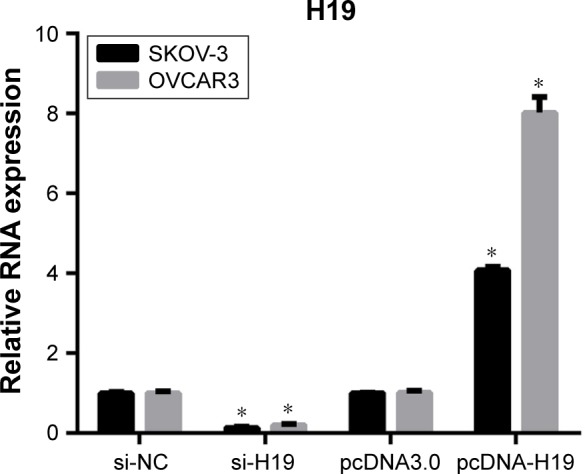
H19 expression in SKOV-3 and OVCAR3 cells after transfection with negative control (NC) siRNA, H19 siRNA, the empty vector (pcDNA3.0), or H19 overexpression vector (pcDNA-H19) for 24 h, followed by treatment with 5 ng/mL of TGF-β1 for 48 h according to quantitative reverse-transcription PCR conducted in triplicate. Data are expressed as mean ± SD, *P<0.05.
Abbreviation: TGF-β1, transforming growth factor-β1.
Figure 3.
The effects of an H19 knockdown or overexpression on TGF-β1-induced EMT. SKOV-3 and OVCAR3 cells were pretransfected with NC siRNA, H19 siRNA, pcDNA3.0, or pcDNA-H19 for 24 h, followed by treatment with 5 ng/mL of TGF-β1 for 48 h. The morphological change of SKOV-3 (A) and OVCAR3 cells (B) was detected under a light microscope (100× and 400×). The mRNA expression profiles of Snail, E-cadherin, and vimentin in SKOV-3 (C) and OVCAR3 cells (D) were determined by quantitative reverse-transcription PCR (qRT-PCR). The protein expression profiles of Snail, E-cadherin, and vimentin in SKOV-3 (E) and OVCAR3 cells (F) were evaluated by Western blotting (left: a representative graph, right: statistical results of Western blot quantification). qRT-PCR and Western blot assays were performed in triplicate and data are expressed as mean ± SD, *P<0.05.
Abbreviations: EMT, epithelial–mesenchymal transition; NC, negative control; TGF-β1, transforming growth factor-β1.
Figure 4.
The effect of the H19 knockdown or overexpression on protein expression profiles of Snail, E-cadherin, and vimentin in SKOV-3 (A) and OVCAR3 cells (B) according to immunofluorescent staining. SKOV-3 and OVCAR3 cells were pretransfected with NC siRNA, H19 siRNA, pcDNA3.0, or pcDNA-H19 for 24 h, followed by treatment with 5 ng/mL of TGF-β1 for 48 h.
Note: Magnification is 200×.
Abbreviations: NC, negative control; TGF-β1, transforming growth factor-β1.
Effects of miR-370-3p knockdown or overexpression on TGF-β1-induced EMT
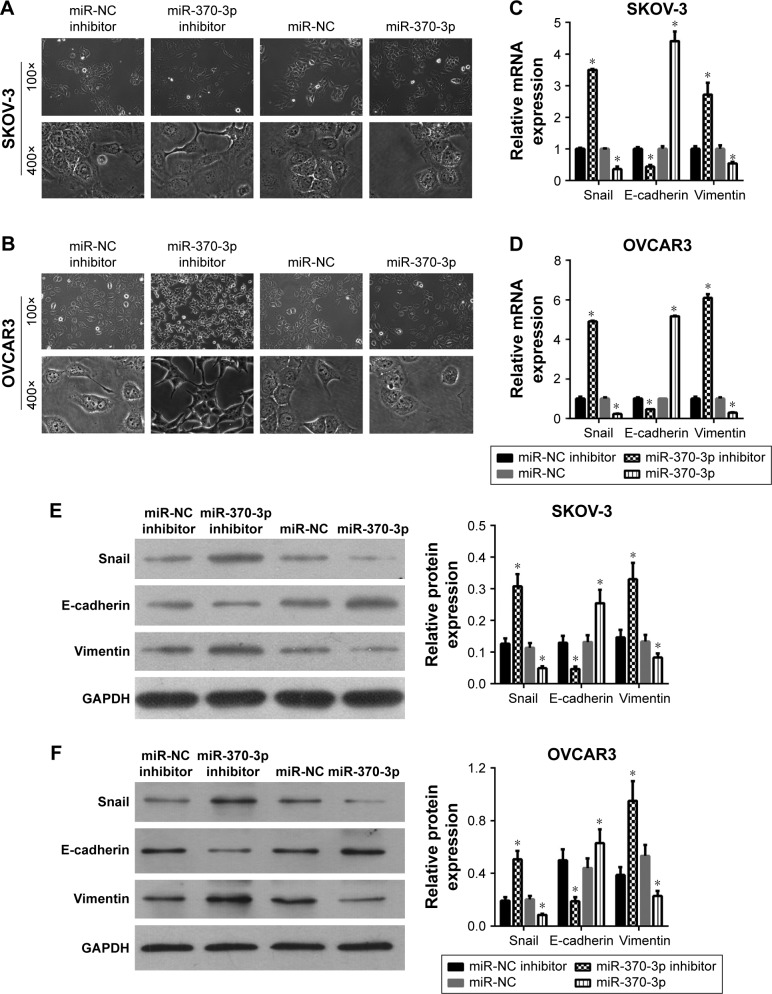
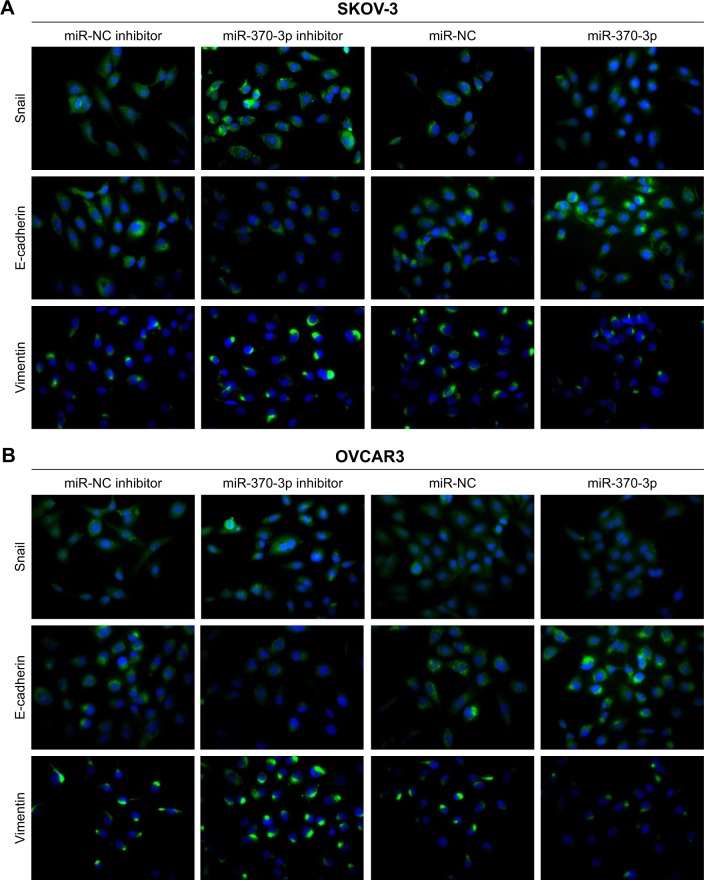
To investigate the role of miR-370-3p in TGF-β1-induced EMT, miR-370-3p was silenced or overexpressed by transfection with the miR-370-3p inhibitor or mimic. The results of qRT-PCR showed that miR-370-3p was successfully silenced in SKOV-3 and OVCAR3 cells transfected with the miR-370-3p inhibitor, as compared to those transfected with the miR-NC inhibitor, and miR-370-3p was successfully overexpressed in cells transfected with the miR-370-3p mimic, as compared to those transfected with miR-NC (Figure 5). The effect of the miR-370-3p knockdown or overexpression on TGF-β1-induced EMT was then estimated by measuring the changes in morphology and in the expression of Snail, E-cadherin, and vimentin. After pretransfection with miR-NC, miR-370-3p, NC inhibitor, or miR-370-3p inhibitor for 24 h, SKOV-3 and OVCAR3 cells were treated with 5 ng/mL of TGF-β1 for 48 h and harvested for qRT-PCR, Western blotting, and immunofluorescent staining. MiR-370-3p overexpression suppressed the morphological change to spindle-shaped cells, whereas the miR-370-3p knockdown promoted this morphological change (Figure 5). The results of the qRT-PCR, Western blotting, and immunofluorescent staining indicated that miR-370-3p overexpression significantly increased the protein level of epithelial marker (E-cadherin) and decreased the levels of the mesenchymal markers (Snail and vimentin), whereas silencing of miR-370-3p had the opposite effects on these EMT markers (Figures 6C–F, 7A and B).
Figure 5.
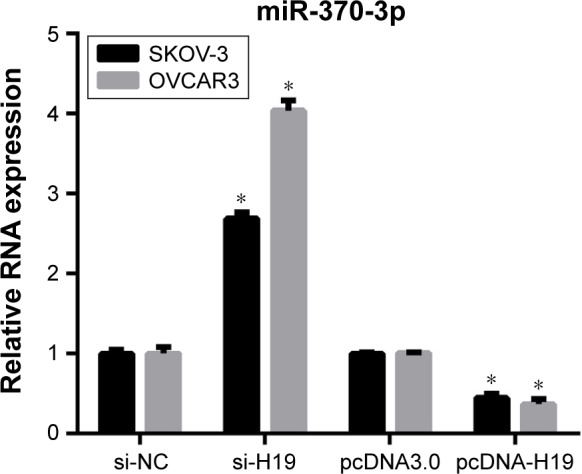
MiR-370-3p expression levels in SKOV-3 and OVCAR3 cells as detected by quantitative reverse-transcription PCR conducted in triplicate. SKOV-3 and OVCAR3 cells were pretransfected with miR-NC, miR-370-3p, NC inhibitor, or miR-370-3p inhibitor for 24 h, followed by treatment with 5 ng/mL of TGF-β1 for 48 h. Data are expressed as mean ± SD, *P<0.05.
Abbreviations: NC, negative control; TGF-β1, transforming growth factor-β1.
Figure 6.
Effects of the miR-370-3p knockdown or overexpression on TGF-β1-induced EMT. SKOV-3 and OVCAR3 cells were pretransfected with miR-NC, miR-370-3p, NC inhibitor, or miR-370-3p inhibitor for 24 h, followed by treatment with 5 ng/mL of TGF-β1 for 48 h. The morphological change of SKOV-3 (A) and OVCAR3 cells (B) was detected under a light microscope (100× and 400×). The mRNA expression profiles of Snail, E-cadherin, and vimentin in SKOV-3 (C) and OVCAR3 cells (D) were assessed by quantitative reverse-transcription PCR. The protein expression profiles of Snail, E-cadherin, and vimentin in SKOV-3 (E) and OVCAR3 cells (F) were determined by Western blotting (left: a representative graph, right: statistical results of Western blot quantification). The qRT-PCR and Western blot assays were performed in triplicate, and data are expressed as mean ± SD, *P<0.05.
Abbreviations: EMT, epithelial–mesenchymal transition; NC, negative control; TGF-β1, transforming growth factor-β1.
Figure 7.
The effects of the miR-370-3p knockdown or overexpression on protein expression profiles of Snail, E-cadherin, and vimentin in SKOV-3 (A) and OVCAR3 cells (B) as detected by immunofluorescent staining. SKOV-3 and OVCAR3 cells were pretransfected with miR-NC, miR-370-3p, the NC inhibitor, or miR-370-3p inhibitor for 24 h, followed by treatment with 5 ng/mL of TGF-β1 for 48 h.
Note: Magnification is 200×.
Abbreviations: NC, negative control; TGF-β1, transforming growth factor-β1.
H19 stimulates TGF-β1-induced EMT by regulating miR-370-3p expression
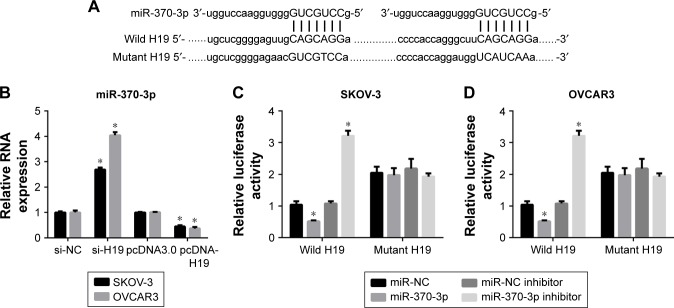
Two binding sites for miR-370-3p were found in the sequence of H19 (Figure 8A). Therefore, we predicted that H19 regulates miR-370-3p expression by functioning as its ceRNA. To verify our prediction, we first measured the miR-370-3p expression in SKOV-3 and OVCAR3 cells pretransfected with NC siRNA, H19 siRNA, pcDNA3.0, or pcDNA-H19 for 24 h, followed by treatment with 5 ng/mL of TGF-β1 for 48 h. The results showed that the H19 knockdown significantly increased the expression of miR-370-3p, whereas the overexpression of H19 had the opposite effect (Figure 8B). Next, to confirm the relation between H19 and miR-370-3p, luciferase reporter vectors containing wild-type or mutant versions of the predicted miR-370-3p-binding sites in the H19 sequence were cotransfected with miR-NC, miR-370-3p, NC inhibitor, or miR-370-3p inhibitor into SKOV-3 and OVCAR3 cells. The luciferase assays were performed 48 h after the transfection. A significant decrease in the luciferase activity of the reporter consisting of the wild-type H19 sequence-containing vector was observed in the presence of miR-370-3p in both SKOV-3 (Figure 8C) and OVCAR3 cells (Figure 8D), as compared to the reporter consisting of the NC-containing vector. Furthermore, a significant increase in the luciferase activity of the reporter consisting of the wild-type H19 sequence-containing vector was observed in the presence of the miR-370-3p inhibitor in both SKOV-3 (Figure 8C) and OVCAR3 cells (Figure 8D), as compared to the reporter consisting of the NC inhibitor-expressing vector. Such significant changes in reporter activity were not seen in assays for the reporter consisting of the vector containing the mutant H19 sequence in both SKOV-3 (Figure 8C) and OVCAR3 cells (Figure 8D), even in the presence of miR-370-3p or its inhibitor. These results suggested that miR-370-3p could bind to the sequence of H19, which functioned as a ceRNA of miR-370-3p.
Figure 8.
H19 acts as a competing endogenous RNA (ceRNA) that sequesters miR-370-3p. (A) Predicted duplex formation between the wild-type or mutant H19 and miR-370-3p. (B) MiR-370-3p expression levels in SKOV-3 and OVCAR3 cells after pretransfection with NC siRNA, H19 siRNA, pcDNA3.0, or pcDNA-H19 for 24 h, followed by treatment with 5 ng/mL of TGF-β1 for 48 h. (C and D) Luciferase activity of reporters with wild-type or mutant H19 sequences cotransfected with miR-NC, miR-370-3p, NC inhibitor, or miR-370-3p inhibitor into SKOV-3 (C) and OVCAR3 cells (D). Results were obtained in three independent experiments conducted in triplicate. Data are expressed as mean ± SD, *P<0.05.
Abbreviations: NC, negative control; TGF-β1, transforming growth factor-β1.
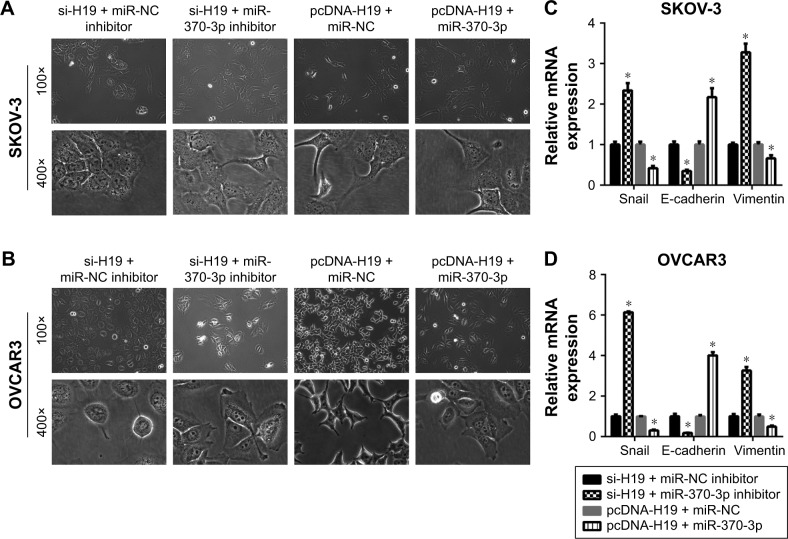
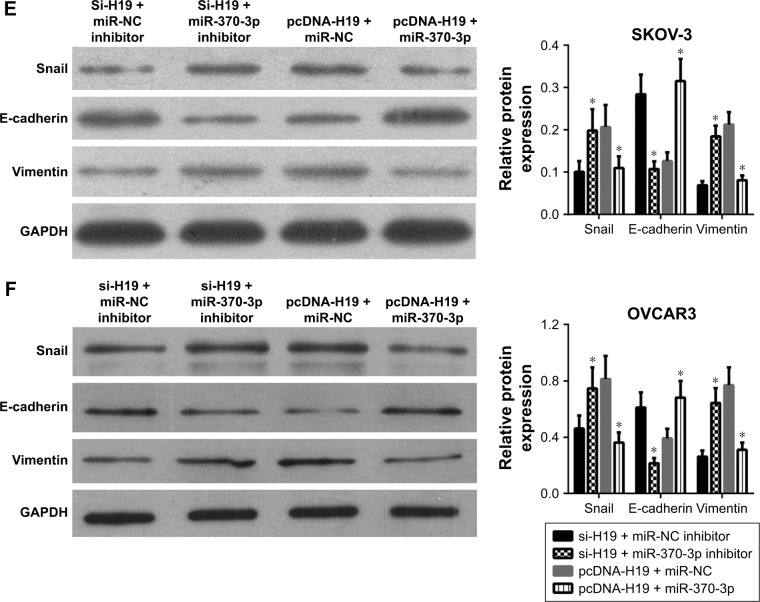
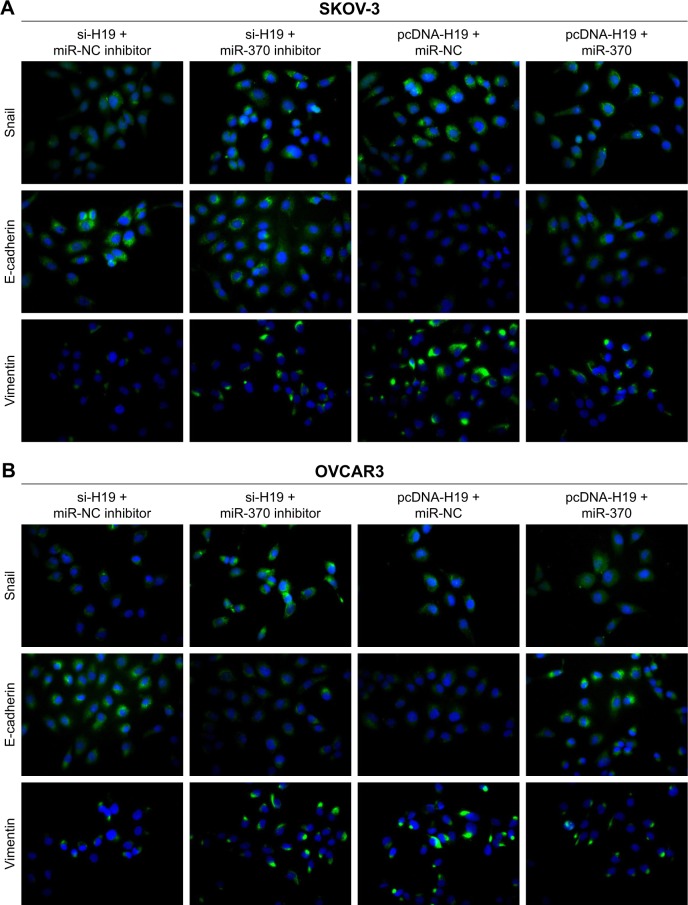
To verify the role of the relation between H19 and miR-370-3p in the regulation of TGF-β1-induced EMT, SKOV-3 and OVCAR3 cells were pretransfected with H19 siRNA plus the NC inhibitor, H19 siRNA plus the miR-370-3p inhibitor, pcDNA-H19 plus the NC miRNA mimic, or pcDNA-H19 plus the miR-370-3p mimic. After transfection for 24 h, the SKOV-3 and OVCAR3 cells were treated with 5 ng/mL of TGF-β1 for 48 h and harvested for analyzing the changes in the morphology and in the expression of Snail, E-cadherin, and vimentin. The results revealed that transfection with H19 siRNA plus the miR-370-3p inhibitor promoted TGF-β1-stimulated morphological changes to spindle-shaped cells among SKOV-3 and OVCAR3 cells, whereas transfection with H19 siRNA plus the NC inhibitor did not show this effect (Figure 9A and B). The results of qRT-PCR, Western blotting, and immunofluorescent staining showed that cells transfected with H19 siRNA plus the miR-370-3p inhibitor had a significantly lower protein level of epithelial marker E-cadherin and significantly higher levels of mesenchymal markers Snail and vimentin, as compared to the cells transfected with H19 siRNA plus the NC inhibitor (Figures 9C–F, 10A and B). Furthermore, transfection with pcDNA-H19 plus the miR-370-3p mimic also attenuated the effects of transfection with pcDNA-H19 plus the NC miRNA mimic on the cellular morphology and the levels of EMT markers in TGF-β1-stimulated SKOV-3 and OVCAR3 cells (Figures 9C–F, 10A and B). These results indicated that the expression level of miR-370-3p could affect the function of H19 in the regulation of cellular morphology and levels of EMT markers in TGF-β1-stimulated SKOV-3 and OVCAR3 cells.
Figure 9.
The effects of the miR-370-3p knockdown or overexpression on the role of H19 in TGF-β1-induced EMT. SKOV-3 and OVCAR3 cells were pretransfected with H19 siRNA plus the NC inhibitor, H19 siRNA plus the miR-370-3p inhibitor, H19 overexpression vector pcDNA-H19 plus the NC miRNA mimic, or pcDNA-H19 plus the miR-370-3p mimic, followed by treatment with 5 ng/mL of TGF-β1 for 48 h. The morphological change of SKOV-3 (A) and OVCAR3 cells (B) was detected under a light microscope (100× and 400×). The mRNA expression profiles of Snail, E-cadherin, and vimentin in SKOV-3 (C) and OVCAR3 cells (D) were determined by qRT-PCR. The expression profiles of Snail, E-cadherin, and vimentin in SKOV-3 (E) and OVCAR3 cells (F) were characterized by Western blotting (left: a representative graph, right: statistical results of Western blot quantification). The qRT-PCR and Western blot assays were performed in triplicate, and data are expressed as mean ± SD, *P<0.05.
Abbreviations: EMT, epithelial–mesenchymal transition; NC, negative control; qRT, quantitative reverse-transcription; TGF-β1, transforming growth factor-β1.
Figure 10.
The effects of the miR-370-3p knockdown or overexpression on the involvement of H19 in protein expression profiles of Snail, E-cadherin, and vimentin in SKOV-3 (A) and OVCAR3 cells (B) as detected by immunofluorescent staining. SKOV-3 and OVCAR3 cells were pretransfected with H19 siRNA plus the NC inhibitor, H19 siRNA plus the miR-370-3p inhibitor, H19 overexpression vector pcDNA-H19 plus the NC miRNA mimic, or pcDNA-H19 plus the miR-370-3p mimic, followed by treatment with 5 ng/mL of TGF-β1 for 48 h.
Note: Magnification is 200×.
Abbreviations: NC, negative control; TGF-β1, transforming growth factor-β1.
Discussion
Since 70% of the subtypes of surface epithelial-stromal tumors are high-grade serous carcinomas and EMT is associated with migration and invasion,2 it is crucial to throw light on relevant molecular mechanisms behind EMT in surface epithelial-stromal tumors for the development of targeted therapies that could help improve survival. TGF-β is known to promote tumor progression at the late stages through multiple mechanisms, including EMT, in cancer cells.25 In addition, TGF-β is frequently used as a key inducer of EMT in many experimental models. In the present study, we first determined the role of H19 and miR-370-3p in TGF-β-induced EMT in human ovarian epithelial adenocarcinoma cell lines, SKOV-3 and OVCAR3. We also demonstrated the relation between H19 and miR-370-3p. These results expanded our understanding of the function of H19 and miR-370-3p in TGF-β-induced EMT during ovarian cancer progression. All our results indicate that the H19–miR-370-3p axis is a possible molecular target for the prevention of EMT in human ovarian cancer. Besides, our present findings will provide helpful clues to the design of novel treatments for human ovarian cancer. First, we found that H19 was upregulated in TGF-β1-stimulated SKOV-3 and OVCAR3 cells, indicating that it might participate in TGF-β-induced EMT in ovarian cancer. We also assessed the effects of H19 knockdown or overexpression on the morphology and the expression of Snail, E-cadherin, and vimentin in cancer cells, to demonstrate the role of H19 in TGF-β1-induced EMT. During EMT, epithelial cells lose their compact organization in colonies, acquire a spindle-shaped morphology, and transform themselves into more motile and invasive cells with migratory behavior.3 We found that the H19 knockdown suppressed the morphological change to spindle-shaped cells, whereas H19 overexpression promoted it. In addition, we found that the H19 knockdown increased the level of epithelial marker E-cadherin and decreased the levels of mesenchymal markers Snail and vimentin in TGF-β1-stimulated SKOV-3 and OVCAR3 cells. H19 overexpression had the opposite effects on these EMT markers. These results suggest that H19 may promote TGF-β-induced EMT in ovarian cancer. Our results are consistent with previous observations that H19 promotes tumor cell migration and invasiveness in ovarian cancer9 and that H19 promotes EMT in other cancer types.26,27
MiR-370-3p inhibits proliferation of human glioma cells and induces cell cycle arrest; these observations point to a tumor suppressor role.17 MiR-370-3p has two binding sites in the H19 sequence. In addition, its effects on TGF-β1-stimulated SKOV-3 and OVCAR3 cells were antagonistic to those of H19. Therefore, we predicted that H19 promotes TGF-β-induced EMT by acting as a ceRNA of miR-370-3p in ovarian cancer cells. We then proceeded to verify our pre-diction. First, the H19 knockdown significantly increased the expression of miR-370-3p, whereas overexpression of H19 had the opposite effect. Second, miR-370-3p decreased the luciferase activity of the reporter vector containing the wild-type H19 sequence. Third, the miR-370-3p expression level affected the function of H19 in the regulation of the cellular morphology of (and EMT markers in) TGF-β1-stimulated SKOV-3 and OVCAR3 cells. Nevertheless, H19 also decreased the activity of miR-200s, miR-630, miR-17-5p, and miR-29a by functioning as a ceRNA.12–15 Therefore, more studies will be needed to clarify the relation between miR-370-3p and other miRNAs that are downregulated by H19.
Conclusion
Our results show that H19 expression is increased while miR-370-3p expression is decreased in TGF-β1-stimulated SKOV-3 and OVCAR3 cells. H19 was found to promote TGF-β-induced EMT in SKOV-3 and OVCAR3 cells, whereas miR-370-3p suppressed it. Furthermore, the expression of miR-370-3p was decreased by H19; in addition, miR-370-3p could bind to the H19 sequence. Moreover, the overexpression of miR-370-3p inhibited the function of H19 in TGF-β-induced EMT. These results led to our conclusion that lncRNA H19 promotes TGF-β-induced EMT by acting as a ceRNA of miR-370-3p in ovarian cancer cells. These results expanded our understanding of the involvement of the H19–miR-370-3p axis in ovarian cancer progression, suggesting that the H19–miR-370-3p axis has a good potential as a therapeutic target in ovarian cancer. Nevertheless, our study has some limitations. First, the key findings of our study will have to be validated in additional ovarian cancer cell lines. Second, the target genes of miR-370-3p in the regulation of TGF-β-induced EMT need to be identified because miRNAs function by either degrading target mRNAs or by inhibiting the translation of these mRNAs.28,29
Acknowledgments
This study was supported by Guangdong Province Natural Science Foundation (No 2014A030313343).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Kobel M, Kalloger SE, Huntsman DG, et al. Differences in tumor type in low-stage versus high-stage ovarian carcinomas. Int J Gynecol Pathol. 2010;29(3):203–211. doi: 10.1097/PGP.0b013e3181c042b6. [DOI] [PubMed] [Google Scholar]
- 3.Vergara D, Merlot B, Lucot JP, et al. Epithelial-mesenchymal transition in ovarian cancer. Cancer Lett. 2010;291(1):59–66. doi: 10.1016/j.canlet.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 4.Li Q, Zhang C, Chen R, et al. Disrupting MALAT1/miR-200c sponge decreases invasion and migration in endometrioid endometrial carcinoma. Cancer Lett. 2016;383(1):28–40. doi: 10.1016/j.canlet.2016.09.019. [DOI] [PubMed] [Google Scholar]
- 5.Yim GW, Kim HJ, Kim LK, et al. Long non-coding RNA HOXA11 antisense promotes cell proliferation and invasion and predicts patient prognosis in serous ovarian cancer. Cancer Res Treat. 2017;49(3):656–668. doi: 10.4143/crt.2016.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jing W, Zhu M, Zhang XW, et al. The significance of long noncoding RNA H19 in predicting progression and metastasis of cancers: a meta-analysis. Biomed Res Int. 2016;2016:5902678. doi: 10.1155/2016/5902678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu FT, Pan H, Xia GF, Qiu C, Zhu ZM. Prognostic and clinicopathological significance of long noncoding RNA H19 overexpression in human solid tumors: evidence from a meta-analysis. Oncotarget. 2016;7(50):83177–83186. doi: 10.18632/oncotarget.13076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu Z, Song L, He J, Sun Y, Liu X, Zou X. Ectopic expressed long non-coding RNA H19 contributes to malignant cell behavior of ovarian cancer. Int J Clin Exp Pathol. 2015;8(9):10082–10091. [PMC free article] [PubMed] [Google Scholar]
- 9.Yan L, Zhou J, Gao Y, et al. Regulation of tumor cell migration and invasion by the H19/let-7 axis is antagonized by metformin-induced DNA methylation. Oncogene. 2015;34(23):3076–3084. doi: 10.1038/onc.2014.236. [DOI] [PubMed] [Google Scholar]
- 10.Zheng ZG, Xu H, Suo SS, et al. The essential role of H19 contributing to cisplatin resistance by regulating glutathione metabolism in high-grade serous ovarian cancer. Sci Rep. 2016;6:26093. doi: 10.1038/srep26093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vennin C, Spruyt N, Dahmani F, et al. H19 non coding RNA-derived miR-675 enhances tumorigenesis and metastasis of breast cancer cells by downregulating c-Cbl and Cbl-b. Oncotarget. 2015;6(30):29209–29223. doi: 10.18632/oncotarget.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X, Lin Y, Yang X, Wu X, He X. Long noncoding RNA H19 regulates EZH2 expression by interacting with miR-630 and promotes cell invasion in nasopharyngeal carcinoma. Biochem Biophys Res Commun. 2016;473(4):913–919. doi: 10.1016/j.bbrc.2016.03.150. [DOI] [PubMed] [Google Scholar]
- 13.Li M, Chen H, Zhao Y, Gao S, Cheng C. H19 functions as a ceRNA in promoting metastasis through decreasing miR-200s activity in osteosarcoma. DNA Cell Biol. 2016;35(5):235–240. doi: 10.1089/dna.2015.3171. [DOI] [PubMed] [Google Scholar]
- 14.Liu L, Yang J, Zhu X, Li D, Lv Z, Zhang X. Long noncoding RNA H19 competitively binds miR-17-5p to regulate YES1 expression in thyroid cancer. FEBS J. 2016;283(12):2326–2339. doi: 10.1111/febs.13741. [DOI] [PubMed] [Google Scholar]
- 15.Jia P, Cai H, Liu X, et al. Long non-coding RNA H19 regulates glioma angiogenesis and the biological behavior of glioma-associated endothelial cells by inhibiting microRNA-29a. Cancer Lett. 2016;381(2):359–369. doi: 10.1016/j.canlet.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 16.He P, Zhang Z, Huang G, et al. miR-141 modulates osteoblastic cell proliferation by regulating the target gene of lncRNA H19 and lncRNA H19-derived miR-675. Am J Transl Res. 2016;8(4):1780–1788. [PMC free article] [PubMed] [Google Scholar]
- 17.Peng Z, Wu T, Li Y, et al. MicroRNA-370-3p inhibits human glioma cell proliferation and induces cell cycle arrest by directly targeting beta-catenin. Brain Res. 2016;1644:53–61. doi: 10.1016/j.brainres.2016.04.066. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y. Epithelial to mesenchymal transition in renal fibrogenesis: pathologic significance, molecular mechanism, and therapeutic intervention. J Am Soc Nephrol. 2004;15(1):1–12. doi: 10.1097/01.asn.0000106015.29070.e7. [DOI] [PubMed] [Google Scholar]
- 19.Hills CE, Squires PE. TGF-beta1-induced epithelial-to-mesenchymal transition and therapeutic intervention in diabetic nephropathy. Am J Nephrol. 2010;31(1):68–74. doi: 10.1159/000256659. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y. New insights into epithelial-mesenchymal transition in kidney fibrosis. J Am Soc Nephrol. 2010;21(2):212–222. doi: 10.1681/ASN.2008121226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bagnato A, Rosano L. Epithelial-mesenchymal transition in ovarian cancer progression: a crucial role for the endothelin axis. Cells Tissues Organs. 2007;185(1–3):85–94. doi: 10.1159/000101307. [DOI] [PubMed] [Google Scholar]
- 22.Guarino M, Rubino B, Ballabio G. The role of epithelial-mesenchymal transition in cancer pathology. Pathology. 2007;39(3):305–318. doi: 10.1080/00313020701329914. [DOI] [PubMed] [Google Scholar]
- 23.Ye Z, Zhao L, Li J, Chen W, Li X. miR-30d blocked transforming growth factor beta1-induced epithelial-mesenchymal transition by targeting Snail in ovarian cancer cells. Int J Gynecol Cancer. 2015;25(9):1574–1581. doi: 10.1097/IGC.0000000000000546. [DOI] [PubMed] [Google Scholar]
- 24.Norita R, Suzuki Y, Furutani Y, et al. Vasohibin-2 is required for epithelial-mesenchymal transition of ovarian cancer cells by modulating transforming growth factor-beta signaling. Cancer Sci. 2017;108(3):419–426. doi: 10.1111/cas.13157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zavadil J, Bottinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24(37):5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 26.Huang C, Cao L, Qiu L, et al. Upregulation of H19 promotes invasion and induces epithelial-to-mesenchymal transition in esophageal cancer. Oncol Lett. 2015;10(1):291–296. doi: 10.3892/ol.2015.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang SH, Wu XC, Zhang MD, Weng MZ, Zhou D, Quan ZW. Erratum: upregulation of H19 indicates a poor prognosis in gallbladder carcinoma and promotes epithelial-mesenchymal transition. Am J Cancer Res. 2016;6(4):876–877. [PMC free article] [PubMed] [Google Scholar]
- 28.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 29.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20(5):515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]