Abstract
Isolation of newly transcribed RNA is an invaluable approach that can be used to study the dynamic life of RNA in cellulo. Traditional methods of whole-cell RNA extraction limit subsequent gene expression analyses to the steady-state levels of RNA abundance, which often masks changes in RNA synthesis and processing. This chapter describes a methodology with low cytotoxicity that permits the labeling and isolation of nascent pre-mRNA in cell culture. The resulting isolate is suitable for use in a series of downstream applications aimed at studying changes in RNA synthesis, processing, or stability.
Keywords: 4sU, 4sU-seq, mammalian cells, nascent RNA, decay, transcription, nascent pre-mRNA, mRNA processing, metabolic labeling, 4-thiouridine
1. Introduction
The majority of gene expression research focuses on RNA transcript abundance at a steady-state level, providing only a snapshot of the cellular state. This glimpse of transcript abundance in the cell limits the understanding of regulation to whether a gene is generally up or down regulated. This obscures whether a change in gene expression is due to differences in the rate of transcription, the rate of degradation, or both. Previous approaches aimed at elucidating the dynamics of co-transcriptional pre-mRNA processing focused on a variety of immunoprecipitation and cell fractionation techniques following a chosen pathway induction (LPS stimulation) [1–3]. Likewise, pulse-chase experiments using well-known transcription inhibitors such as Actinomycin D have been frequently used to measure mRNA stability and degradation [2,4]. While providing critical advances to the fundamental understanding of RNA dynamics, these methods are limited by cytotoxicity and a lack of kinetic resolution [4]. The emergence of next generation sequencing technologies, in conjunction with the uridine analog 4-thiouridine (4sU) as a metabolic label, has opened up an exciting avenue to studying genome-wide RNA kinetics at high resolution [5–7].
4sU can be used to metabolically label and track an RNA from synthesis to degradation by simply adding 4sU to mammalian cell culture media. 4sU is immediately taken up by cells, phosphorylated, and incorporated into any newly transcribed RNA. 4sU-labeled RNA can be tracked and isolated to study nascent RNA behavior using RNA sequencing. Alternatively, media replacement after a longer incubation period in 4sU media can be used to study the half-life and degradation of an RNA. Depending on the experimental approach taken, appropriate concentrations of 4sU should be selected for various cell types and incubation times to minimize off-target effects (see Table 1) [8].
Table 1.
Recommended 4sU concentrations [5]
| Duration of labeling [min] | Recommended 4sU concentration [μM] |
|---|---|
| 120 | 100–200 |
| 60 | 200–500 |
| 15–30 | 500–1,000 |
| <10 | 500–20,000 |
Once whole-cell RNA is extracted, the 4sU-labeled RNA can be biotinylated via its sulfylhydryl group and selectively isolated using streptavidin-coated magnetic beads. Given the strong biotin/streptavidin interaction, 4sU labeled RNA can be stringently washed. Eluted 4sU labeled RNA can then be used in subsequent qRT-PCR and RNA-seq experiments, with or without ribosomal RNA depletion. Given the fact that 4sU can be used to mark nascent transcripts, the use of the 4sU labeling protocol can lead to a wealth of new findings that directly relate to immediate changes in gene expression.
2. Materials and Reagents
All materials must be sterile, RNase-free, molecular biology grade. Large quantities of TRIzol reagent may be used for experiments. In case of contact with skin/eyes, have a polyethylene glycol 300 or 400 in industrial methylated spirits (70:30) solution prepared before proceeding.
2.1 4sU Labeling of Cells
4-thiouridine (Sigma, T4509 4-thiouridine) dissolved in sterile RNase-free water to 50mM. Store in small aliquots at −20°C, thawing only once.
2.2 Total RNA Extraction
TRIzol (Thermo Scientific)
75 % EtOH (ethanol)
RNase-free water
RNA Precipitation Solution: 0.8 M NaCl, 1.2 M NaCitrate
(optional) TE
2.3 Biotinylation of 4sU-Labeled RNA
EZ-Link Biotin-HPDP (Pierce). Make stock aliquots 1mg/ml dissolved in Dimethylformamide (see Note 1) and store at 4°C.
10x Biotinylation Buffer: 100 mM Tris pH 7.4, 10 mM EDTA. Store in aliquots of ~1 mL at 4°C.
5 M NaCl
75 % EtOH
(optional) Phase Lock Gel Heavy Tubes (2.0 mL) (Eppendorf)
2.4 Separation of Labeled and Unlabeled RNA Using Streptavidin-Coated Magnetic Beads
μMacs Streptavidin Kit (Miltenyi, Order No. 130-074-101) (see Note 2)
1x Washing Buffer: 100 mM Tris pH 7.5, 10 mM EDTA, 1 M NaCl, 0.1 % Tween20
100 mM Dithriothreitol (DTT) in RNase-free water
Magnetic Separator and Stand (2 each) (Miltenyi, Order No. 130-042-303, 130-042-602) Alternatively, one of each is included in the starter kit (Miltenyi, Order No. 130-091-287)
(optional) Qiagen RNeasy MinElute Cleanup Kit
2.5 Recovery of Unlabeled, Unbound RNA
Phenol/chloroform pH 6.7
Isopropanol
EtOH
3. Methods
3.1 4sU Labeling of Nascent RNA
Plate a number of cells of the desired cell type in either a 10 cm or 15 cm tissue culture plate that will reach 70–80 % confluency after 24 hours. For a 10 cm plate, use at least 10mL of culture medium. For a 15 cm plate, use at least 20 mL of culture medium.
For a 10 cm dish, a minimum of 5 mL of culture medium containing 4sU is needed. For a 15 cm dish, a minimum of 10 mL of culture medium containing 4sU is needed.
Once cells reach 70–80 % confluency, transfer 5 mL or 10 mL of culture medium from the plate to a clean 15 mL conical tube.
Add 4sU to the culture medium in the conical tube and pipette up and down with a serological pipette to mix thoroughly. Refer to Table 1 for general guidelines for 4sU concentrations (see Note 3).
Aspirate the remaining unlabeled culture medium from the plate. Add the culture medium containing 4sU to the cells (see Note 4).
Incubate cells with 4sU culture medium for the desired amount of time. Longer incubation period is recommended for RNA decay studies (see Note 5).
Quench the reaction by quickly aspirating the 4sU culture medium and adding 3 mL of TRIzol for 10 cm plate, or 5 mL of TRIzol for 15 cm dishes.
Ensure the entire plate is covered by TRIzol and allow to sit for 2–5 minutes for complete cell lysis.
Pipette the cell/TRIzol lysate to homogenize the cells and get all cells off the plate. Transfer the lysate to a 15 mL conical tube.
Immediately extract total RNA from TRIzol samples, or store at −80°C between 6 months to 1 year (see Note 6).
3.2 Total RNA Extraction
Transfer 1 mL of TRIzol sample to each of (3) 1.5 mL Eppendorf tubes.
Add 0.2 mL chloroform per mL TRIzol and shake vigorously for 15 seconds.
Incubate at room temperature for 2–3 minutes.
Centrifuge at 20,000 × g for 15 min at 4°C (see Note 7).
Transfer aqueous upper phase (containing the RNA) to a new tube.
Add ½ the reaction volume of both RNA precipitation buffer and isopropanol (e.g. to 3 mL of supernatant add 1.5 mL RNA Precipitation Solution and 1.5 mL isopropanol).
Invert to mix well.
Incubate at room temperature for 10 min.
Centrifuge at 20,000 × g for 10 min at 4°C.
Immediately remove supernatant.
Wash with an equal volume of 75 % EtOH.
Centrifuge at 20,000 × g for 10 min at 4°C.
Immediately remove supernatant.
Centrifuge again briefly to spin down remaining EtOH.
Remove remaining ethanol by pipetting using 200 μl pipette. Repeat step using 20 μl pipette (see Note 8).
Add 100 μl of 1x TE or RNase-free water (10 mM Tris, 1 mM EDTA) per 100 μg expected RNA yield.
If needed, dissolve RNA by heating to 65°C for 10 min.
Use a NanoDrop spectrophotometer to measure RNA yield. This RNA can be stored at −80°C for at least 3 months with minimal freeze-thaws.
3.3 Biotinylation of 4sU Labeled RNA
-
Labeling Reaction (use 60 – 100 μg total RNA):
2 μl Biotin-HPDP (1mg/ml DMF) per 1 μg RNA
1 μl 10X Biotinylation Buffer per 1 μg RNA
bring up to 7 μl with RNase-free water per 1 μg RNA
Rotate at room temperature in the dark for at least 1.5 hr (see Note 9).
Add an equal volume of Phenol/Chloroform pH 6.7.
Mix vigorously by vortex or by manually shaking.
Incubate for 2 – 3 minutes at room temperature until phases begin to separate and bubbles start to disappear.
Centrifuge at full speed (20,000 × g) for 5 min.
Carefully transfer upper phase into new tubes (see Note 10).
RNA precipitation: Add 1/10 the reaction volume of 5 M NaCl.
Add an equal volume of isopropanol, invert to mix well.
Centrifuge at 20,000 × g for 20 min.
Remove supernatant. Add an equal volume of 75 % EtOH.
Centrifuge at 20,000 × g for 10 min.
Remove EtOH completely and re-suspend the RNA pellet at approximately 1 μg/μl with RNase-free water or TE.
3.4 Separation of Labeled and Unlabeled RNA Using Streptavidin-Coated Magnetic Beads
Heat biotinylated RNA samples to 65°C for 10 min and immediately place on ice for 5 min.
Add up to 100 μg (max. 100 μl) of biotinylated RNA to 100 μl of streptavidin beads (see Note 11).
Incubate at room temperature with rotation for 15 min.
Place μMacs columns into magnetic stand. Process no more than 8 samples at a time (see Note 12).
Add 0.9 mL of washing buffer to columns to pre-run and equilibrate (see Note 13).
Apply bead-bound RNA to the columns.
For recovery of unlabeled/unbound RNA, collect this flow-through and see Section 3.5. Otherwise, discard the flow-through.
Place tubes or alternative collection apparatus underneath columns to catch the wash flow-through.
Wash 3x with 0.9 mL 65°C washing buffer. Optional: For recovery of unlabeled RNA, collect the first wash and see Section 3.5.
Wash 3x with 0.9 mL room temperature washing buffer.
Elute the labeled RNA by placing the 1.5 mL Eppendorf tubes underneath the columns and adding 100 μl 100 mM DTT to the columns (see Note 14).
Perform a second DTT elution into the same tubes 3–5 min later.
Immediately perform EtOH precipitation with 2.5V 100 % EtOH and 10 μg glycogen.
Precipitate overnight at −20°C.
Spin at 20,000 × g for 15 minutes.
Wash with 75 % EtOH.
Spin at 20,000 × g for 5 minutes.
Remove all EtOH using technique used in 3.2.15 and re-suspend in ~30 μl RNase-free water.
Spec labeled RNA with NanoDrop (see Note 15).
3.5 Recovery of Unlabeled, Unbound RNA (Optional)
For recovery of >90 % of unbound RNA, collect the flow-through and the first wash for subsequent precipitation.
Combine the two fractions and recover the unbound RNA by isopropanol/EtOH precipitation as performed after the biotinylation reaction (see Section 3.3). Omit the addition of NaCl; the washing buffer has sufficient NaCl.
3.6 Validation
Validate with RT-PCR/qPCR by comparing labeled RNA to total RNA or unlabeled RNA for genes/transcripts of interest.
Acknowledgments
Research in the Hertel laboratory is supported by NIH (R01GM062287 and R01GM110244 to K.J.H. and F31CA171791 to A.G.). Special thanks to Nate Hoverter for contribution of key graphics in Figure 1.
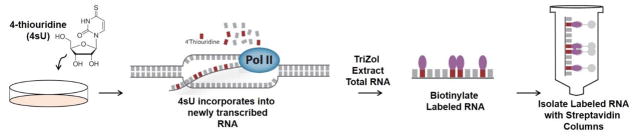
Figure 1.
Conceptual Workflow of 4sU labeling and isolation of newly transcribed RNA.
Footnotes
Gentle warming will ensure complete solubilization. Store aliquots at 4°C. Alternatively, store 20 mg/ml at −20°C. Do not use any polystyrene serological pipette in this process as DMF will degrade the plastic, leading to plastic residues that may inhibit biotinylation.
Per conversations with Miltenyi tech support, the beads are subject the expiration date on the box. Columns, however, are good for 3 years. At time of publication, beads are not sold separately.
Thaw 4sU only once, and just before use. Concentrations should be optimized based on cell line and desired labeling time to balance incorporation efficiency and possible inhibition of rRNA synthesis [8].
Handle labeled cells at room temperature as quickly as possible. Note that 4sU has crosslinking ability at 365 nm wavelength. Avoid light sources that may mimic this wavelength.
To study RNA decay you can perform a pulse chase experiment in which the duration of the 4sU labeling is increased and chased with cell media absent of 4sU. Timepoints can then be taken during the chase period to determine decay rates.
TRIzol samples may be freeze-thawed at least twice, thus allowing for 2 pull-down reactions on different dates from a single 15 cm plate depending on cell type. Otherwise, freeze in 2 aliquots to reduce freeze-thaws.
While not “best practice”, centrifugation at room temperature will not cause failure.
After these two steps, no further drying of the pellet is required. Over drying of pellet may risk making it difficult to dissolve, even with heating.
Rotation has been done under general lab lighting with success.
Alternatively, this step can be done using phase lock gel heavy tubes to avoid both the loss of material and phenol carry-over.
80 μl of beads for 80 μg RNA reaction is also sufficient.
When processing replicate samples, we find increased variability when the pulldown is done in different rounds. Therefore, it is recommended to perform the pulldown on replicates in the same round.
To initiate the flow through the column you can gently press on the top of the column with your gloved finger.
Here you have the option to finish the remainder of this section by eluting directly into 700 μl RLT buffer and complete RNA isolation/clean up using Qiagen RNeasy MinElute cleanup kit. However, residual kit buffer in the RNA may skew nanodrop OD readings.
For very short time points, this may be very low or unreliable detection.
References
- 1.Pandya-Jones A, Black DL. Co-transcriptional splicing of constitutive and alternative exons. RNA. 2009 Oct;15(10):1896–908. doi: 10.1261/rna.1714509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Core LJ, Waterfall JJ, Lis JT. Nascent RNA Sequencing Reveals Widespread Pausing and Divergent Initiation at Human Promoters. Science (80-) 2008;322(5909) doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brody Y, Neufeld N, Bieberstein N, Causse SZ, Böhnlein E-M, Neugebauer KM, et al. The In Vivo Kinetics of RNA Polymerase II Elongation during Co-Transcriptional Splicing. In: Misteli T, editor. PLoS Biol. 1. Vol. 9. Public Library of Science; 2011. Jan 11, p. e1000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tani H, Akimitsu N. RNA Biol. Taylor & Francis; 2012. Oct 1, Genome-wide technology for determining RNA stability in mammalian cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rädle B, Rutkowski AJ, Ruzsics Z, Friedel CC, Koszinowski UH, Dölken L. Metabolic labeling of newly transcribed RNA for high resolution gene expression profiling of RNA synthesis, processing and decay in cell culture. J Vis Exp. 2013 Jan 8;(78):e50195. doi: 10.3791/50195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rabani M, Levin JZ, Fan L, Adiconis X, Raychowdhury R, Garber M, et al. Nat Biotechnol. 5. Vol. 29. Nature Publishing Group, a division of Macmillan Publishers Limited. All Rights Reserved; 2011. May, Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells; pp. 436–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrass JD, Reid JEA, Huang Y, Hector RD, Sanguinetti G, Beggs JD, et al. Transcriptome-wide RNA processing kinetics revealed using extremely short 4tU labeling. Genome Biol BioMed Central. 2015 Dec 17;16:282. doi: 10.1186/s13059-015-0848-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burger K, Mühl B, Kellner M, Rohrmoser M, Gruber-Eber A, Windhager L, et al. 4-thiouridine inhibits rRNA synthesis and causes a nucleolar stress response. RNA Biol. 2013 Oct;10(10):1623–30. doi: 10.4161/rna.26214. [DOI] [PMC free article] [PubMed] [Google Scholar]