Abstract
Splicing is an important eukaryotic mechanism for expanding the transcriptome and proteome, influencing a number of biological processes. Understanding its regulation and identifying small molecules that modulate this process remains a challenge. We developed an assay based on time-resolved FRET (TR-FRET) to detect the interaction between the protein NHP2L1 and U4 RNA, which are two key components of the spliceosome. We used this assay to identify small molecules that interfere with this interaction in a high-throughput screening (HTS) campaign. Topotecan and other camptothecin derivatives were among the top hits. We confirmed that topotecan disrupts the interaction between NHP2L1 and U4 by binding to U4 and inhibits RNA splicing. Our data reveal new functions of known drugs which could facilitate the development of therapeutic strategies to modify splicing and alter gene function.
Keywords: Screening, NHP2L1, U4, splicing
Introduction
Most genes in the human genome comprise multiple exons interspersed with introns, necessitating splicing to form mature mRNA and protein products1. The process of alternative splicing is a central mechanism for the regulation of gene expression, allowing increased proteomic complexity in higher eukaryotes2.
Splicing is catalyzed by a large ribonucleoprotein complex called the spliceosome, which is formed by the interaction of small nuclear ribonucleoprotein particles (snRNPs) U1, U2, U4, U5 and U6, appropriate regions of the pre-mRNA and numerous non-snRNP proteins3.
NHP2L1 (15.5K) is one of the non-snRNP proteins4-5 that binds to and stabilizes a kink turn (K turn) in the U4 5′ stem loop (U4 5′-SL) and is required for the subsequent recruitment of the human PrP31 protein to U4/U66-7. This interaction between NHP2L1 and U4 has been shown to play a critical role in the late stage of spliceosome assembly3.
We developed an assay based on the time-resolved FRET (TR-FRET) method to detect the interaction between NHP2L1 and U4 5′SL. We used this assay to identify small molecules that interfere with this interaction in a high-throughput screening (HTS) campaign, and validated the results by orthogonal methods. Topotecan and other camptothecin derivatives were among the top hits, and we confirmed that topotecan disrupts the interaction between NHP2L1 and U4 5′SL by binding to U4 and documented that this disruption leads to inhibition of RNA splicing.
Materials and Methods
Reagents
Cy5-U4 5′SL (5′Cy5-AUCGUAGCCAAUGAGGUUUAUCCGAGGCGCGAU) and Biotin-U4 5′ SL (5′Biosg-AUCGUAGCCAAUGAGGUUUAUCCGAGGCGCGAU) were synthetized by Integrated DNA Technologies (IDT). Histidine-Tagged NHP2L1 (His-NHP2L1) was obtained as described below. Terbium-labeled anti-His-tag antibody (Tb-anti-His antibody), Bovine Serum Albumin (BSA) and dithiothreitol (DTT) were purchased from Invitrogen. Tris-HCl (pH 7.6, 1M) was purchased from TEKNOVA. NaCl (5M) and Dimethyl Sulfoxide (DMSO) were purchased from Fisher Scientific (Pittsburgh, PA). Triton X-100, (S)-camptothecin and 9-NO2-10-OH-(S)-camptothecin were purchased from sigma (St. Louis, MO). Topotecan, SN-38, 9-NH2-(S)-camptothecin, 10-OH-(S)-camptothecin and irinotecan were purchased from Cayman Chemical. N-Desmethyl topotecan and 7,11-diethyl-10-OH-(S)-camptothecin were purchased from Toronto Research Chemicals. tRNA was purchased from Roche Diagnostics. 384-well low volume black assay plates were obtained from Corning Incorporated.
Chemical library
The St. Jude Children’s Research Hospital FDA drug and bioactive library was assembled from four commercial suppliers of MicroSource (Gaylordsville, CT), Prestwick Chemical (San Diego, CA), Sigma (St. Louis, MO) and Selleckchem (Houston, TX) with a total of 10,173 compounds (4,617 unique small molecules) as previously described8-11.
Protein expression and purification
NHP2L1 was cloned into pET-15b vector by using XhoI and BamhI restrictions sites. This plasmid was used to transform BL21 (DE3) cells (Novagen) and grown in Luria-Bertani (LB) medium at 37°C in the presence of ampicillin (100μgml-1) to an optical density at 600 nM (OD600) of 0.6. Protein was induced by addition of 0.2 mM isopropyl-β-D-1-thiogalactopyranosine (IPTG) and cells were allowed to grow for a further 4h at 30°C. The cells were harvested and suspended in lysis buffer (20 mM Tris-HCl, pH 7.8, 500 mM NaCl and 10% glycerol). Cells were disrupted by using a microfluidizer. After centrifugation, the supernatant was applied onto 5 ml HiTrap Chelating HP column (GE Healthcare). The bound protein was eluted by a linear gradient of imidazole. The protein-containing fractions were collected and further purified by gel filtration using a HiLoad Superdex 200 column (GE Healthcare). Where required, His-tag was removed by thrombin treatment. Protein concentration was determined using the Bio-Rad protein assay and the Pierce BCA Protein assay.
Time-resolved fluorescence resonance energy transfer (TR-FRET) assay
Various concentrations of Cy5-U4 5′SL were incubated with 2nM Terbium-anti-his in 20μl binding buffer (20mM Tris-HCl pH 7.6, 150mM NaCl, 0.1% Triton X-100, 0.5mg/ml tRNA, 0.01% BSA, 2mM DTT) with or without 2nM His-NHP2L1 in low volume black 384-well plates with the final DMSO concentration of 0.5% for each well. After centrifugation, shake and room temperature incubation, the TR-FRET signals (fluorescence emission ratio of 10,000 × 665nm/620nm) for each well were collected with a PHERAstar FS (BMG Labtech, Durham, NC) using a 340 nm excitation filter, 100 μs delay time and 200μs integration time. The reactions were incubated between 15 to 300 minutes. Data were plotted with GraphPad Prism 6.07 (GraphPad Software, Inc., La Jolla, CA) by fitting into the one site total binding equation to derive the Kd value of the interaction between Cy5-U4 5′ SL and His-NHP2L1.
High-throughput screening (HTS) using TR-FRET assay
For the primary screen, stock compound (the St. Jude children’s research hospital bioactive and FDA drug library with a total of 10,173 compounds and 4,617 unique ones) solutions (10 mM in DMSO) or DMSO alone (vehicle control) were transferred to the individual wells in low volume black 384-well assay plates containing 20μl complete U4-NHP2L1 interaction mixture (2 nM Terbium-anti-His, 2 nM His-NHP2L1, and 2 nM Cy5-U4 5′SL in binding assay buffer) or partial U4-NHP2L1 interaction mixture (complete U4-NHP2L1 interaction mixture without His-NHP2L1) by using a V&P 384-well pintool at 30nl/well to give a final compound concentration of 15μM. The final DMSO concentration was 0.15% for each well. The DMSO control wells with complete U4-NHP2L1 interaction mixture and the DMSO wells with partial U4-NHP2L1 interaction mixture were used as negative (0% inhibition) and positive (100% inhibition) controls, respectively. After a 45 minutes room temperature incubation, the TR-FRET signals (fluorescence emission ratio of 10,000 × 665nM/620nm) for each well in individual assay plates were collected. Compounds with % Inhibition ≥ 40% were selected for the dose responsive analysis (10 concentrations, following a 1:3 serial dilution scheme with final concentrations ranged from 3.5 nM to 70 μM, in triplicate) under similar assay conditions as the primary screen with final DMSO concentration of 0.7% for all assay wells. The activity data for each small molecule were normalized to those of positive and negative controls and fit into sigmoidal dose-response curves, if applicable, to derive IC50 values with GraphPad Prism 6.07 (GraphPad Software, Inc., La Jolla, CA). To confirm the activity of topotecan and other camptothecin derivatives, powder compounds were solubilized in DMSO as 10 mM stocks and dilutions of individual chemicals (16 concentrations, following a 1:2 serial dilution scheme with final concentrations ranging from 3.1 nM to 100 μM, in triplicate) were used in the TR-FRET assay. The DMSO control (1.0%) wells with and without 2 nM His-NHP2L1 were used as negative control (0% inhibition) and positive control (100% inhibition), respectively.
Chemical similarity clustering
The hierarchical molecular network graph relates biological activity in the NHP2L1 primary assay to chemical structure. Leaf nodes represent the actual molecules tested and are color-coded according to % inhibition (blue = higher inhibition). Intermediate nodes represent scaffolds at various levels of chemical abstraction, and edges between nodes signify chemical similarity. To define the network, input molecules were first abstracted to Murcko scaffolds12 using the ‘Generate Fragments’ module in Pipeline Pilot (v. 9.2.0, Biovia, Inc). Murcko scaffolds were clustered according to Tanimoto similarity using the FCFP_4 fingerprint (cluster center selection method = ‘Maximum Dissimilarity’; maximum distance = 0.6; distance metric = Euclidean; recenter count = 10). In our experience, the above two-step procedure does the best job of clustering together compounds with high global similarity. However, Murcko fragmentation reduces molecules to contiguous ring systems plus chains that link two or more rings, and will sometimes fail to identify relationships between molecules that are not similar according to a global chemical fingerprint, but that nonetheless share a common substructure. To address this limitation, we further abstracted the Murcko scaffolds using the Schuffenhauer algorithm13, which hierarchically decomposes molecules to a single ring system according to a defined set of rules. The network graph relating molecules and intermediate scaffolds was visualized using Cytoscape (v.3.4.0)14.
Electrophoretic Gel Mobility Shift Assay (EMSA)
NHP2L1 and biotinylated U4 5′ SL were mixed in 10 μl of buffer reaction (containing 20 mM Tris-Hcl, pH 7.6, 150mM NaCl, 0.1% Triton X-100, 2mM DTT, 0.5g/L tRNA) and incubated at room temperature for 45 min. Reactions were loaded onto 6% DNA retardation gel (Invitrogen) and separated in 0.5 × TBE buffer (Invitrogen) at 100V for 60 minutes at room temperature and transferred to Nylon membrane. After cross-linking the complexes were detected using the chemiluminescent Nucleic Acid detection module (Thermo scientific) according to manufacturer’s instructions.
Biotinylation of NHP2L1
NHP2L1 was minimally biotinylated by reaction with EZ-Link Sulfo-NHS-LC-LC-Biotin (Thermo Scientific)15. The biotin reagent was added to the protein at a 0.5:1 molar ratio, and the reaction was incubated on ice for 3 hours. Unconjugated biotin was removed by processing the reaction sample through two Zeba Spin Desalting Columns (Thermo Scientific) that had been equilibrated with storage buffer (20 mM Bis-Tris pH 6.5, 100mM NaCl, 2mM TCEP, and 20% glycerol). The biotinylated protein was aliquoted, flash-frozen in liquid N2 and stored at −80° C for later use.
Surface Plasmon resonance (SPR) experiments
SPR experiments were performed at 20° C on a SensiQ Pioneer optical biosensor (SensiQ Technologies). Neutravidin (Thermo Scientific) was immobilized on carboxylated polysaccharide-coated gold chips (COOH5 chips; SensiQ Technologies) by routine amine coupling chemistry in immobilization buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 0.005% Tween20). Carboxyl groups on the hudrogel were activated with N-ethyl-N′-(3-dimethylaminopropyl) carbodiimide (EDC) and N-hydroxysuccinimide (NHS), and neutravidin was injected in 10 mM sodium acetate pH 5.0 until immobilization levels of ~10,000-11,500 RU/channel were achieved. Any remaining active sites were blocked with ethanolamine. Biotin-NHP2L1 and U4 5′SL-biotin (Integrated DNA Technologies) were injected on separate channels until 1330 RU and 1014 RU were captured respectively. One channel on the chip was immobilized with neutravidin without adding protein or RNA and was used as a reference cell. Topotecan was prepared in binding buffer (20 mM Bis-Tris pH 6.5, 200 mM NaCl, 2 mM TCEP, 0.01% Triton X-100, 10% glycerol, 5% DMSO) at a concentration of 50 μM and was injected in duplicate at a flow rate of 200 μL using the OneStep injection feature, which exploits Taylor dispersion to generate a concentration gradient that provide a full titration of analyst in a single injection16-17. Buffer-only (blank) injections were included in the experiment to account for instrumental noise. The data were processed, double-referenced, solvent corrected and analyzed using the software package Qdat (version 2.6.1.8, SensiQ Technologies). Equilibrium dissociation constants (KD) were determined by globally fitting the data to a 1:1 equilibrium affinity model.
Nuclear Magnetic Resonance (NMR) spectroscopy and resonance assignments
Uniformly labelled NHP2L1 with 15N or with 15N and 13C was produced in MOPS minimal medium supplemented with 15NH4Cl (Cambridge Isotope Laboratories, Inc.) or 15NH4Cl and 13C6H12O6 (Sigma-Aldrich). Cells were harvested and lysed and proteins were purified following the same procedures as mentioned above in the section “Protein expression and purification”. Samples were prepared for NMR spectroscopy in a buffer consisting of 100mM sodium chloride, 100mM sodium phosphate, pH 5.0, 10% D20, 2% DMSO-d6. 15N isotope labelled NHP2L1 alone or in presence of unlabelled U4 5′ SL (stoichiometric ratios NHP2L1:U4 5′ SL of 1:1) were titrated with increasing amounts of Topotecan to stoichiometric ratios (NHP2L1 or NHP2L1 +U4: topotecan) of 1:0, 1:2, 1:10, 1:20. Spectral changes were monitored by 1D 1H and 2D [1H, 15N] HSQC or TROSY spectra. Unlabelled U4 5′ SL was titrated with increasing amounts of Topotecan to stoichiometric ratios (U4: topotecan) of 1:0, 1:1, 1:2, 1:5, 1:10. Spectral changes were monitored by 1D 1H NMR spectra. NMR experiments were recorded at 25°C on either a Bruker Avance 600 MHz or 800 MHz spectrometer equipped with 5 mm triple resonance cryoprobe (TCI) using single axis pulse field gradient. Though the assignments of NHP2L1 were closed to the data in BMRB ID 724918, several resonances were shifted because of the pH (5.0) and the buffer conditions. Therefore we assigned the backbone resonances of NHP2L1 from the analysis of 2D [15N, 1H] HSQC along with HNCA and HNCACB spectra on 13C, 15N-labeled NHP2L1. 2D [15N, 1H] HSQC or TROSY spectra were used for monitoring chemical shift perturbations of NHP2L1 resonances, U4 5′ SL at 100μM concentration and various concentrations of topotecan using 64 scans. Only the 2D [15N, 1H] TROSY of the NHP2L1 complex with U4 and topotecan at molar ratio of 1:1:20 was measured using 400 scans. Binding of U4 with topotecan was monitored using both 1D and 2D [1H, 1H] TOCSY (60 ms mixing time) and NOESY (100 ms mixing time) spectra at 100 μM of U4 and varying concentrations of Topotecan (data not shown). 1D waterlogsy spectra were measured using 100 μM topotecan with and without NHP2L1 (5 μM) with 128 scans. All the NMR data were processed using Topspin 3.2 software and analyzed using Topspin or CARA19.
Cell culture
Human embryonic Kidney 293 cells (HEK293 cells) was obtained from the German Collection of Microorganisms and Cell Cultures. Cells were cultured in RPMI-1640 medium containing 2 mM glutamine and 10% (vol/vol) FBS at 37 °C with 5% CO2.
NHP2L1 overexpression
HEK293 cells were infected with lentiviral transduction particles over-expressing GFP or NHP2L1-GFP. GFP positive cell clones were isolated by cell sorting.
Splicing assay
The HEK293 cells were transfected with pTN23 splicing reporter using Gene Jammer (Stratagene). The plasmid pTN23 contains reporter genes for β-galactosidase and luciferase separated by an exon-intron-exon cassette20. Cells were harvested 24h after transfection with or without topotecan and lysed in reporter lysis buffer. Luciferase and β-galactosidase activities were measured using the dual light system (applied Biosystems). The ratio of luciferase to β–galactosidase activities was determined.
Western blot analysis
Cell lysates were separated by electrophoresis on a SDS-polyacrylamide gel, and the proteins were then electroblotted onto a Hybond P PVDF membrane. Protein expression was analyzed using different antibodies.
Antibodies
Primary antibodies were purchased for GAPDH and GFP (B2) from Santa Cruz Biotechnology. Horseradish Peroxidase-conjugated secondary antibodies were purchased from Dako.
Results
Detection of NHP2L1 and U4 interaction by using the time-resolved FRET assay
The interaction between NHP2L1 and U4 5′SL was previously characterized by using techniques such as nuclear magnetic resonance (NMR)6, crystallography3, 6, co-immunoprecipitation21 and electrophoretic mobility shift assay (EMSA)6, but those techniques are not suitable for a high throughput screen. We therefore developed a time resolved fluorescence resonance energy transfer (TR-FRET) assay suitable for high-throughput screening to identify small molecules capable of disrupting the interaction between NHP2L1 and U4 5′ SL. TR-FRET obeys the same principles as standard FRET assays: when a suitable pair of fluorophores are brought within close proximity of one another, excitation of the first fluorophore (the donor) results in energy transfer to the second fluorophore (the acceptor). However compared to conventional FRET techniques, TR-FRET has the advantage of much lower background22. We used TR-FRET to monitor the interaction between NHP2L1 and U4 5′SL using Terbium anti-his-tag antibody which recognizes histidine tagged NHP2L1 (his-NHP2L1) and cy5-labeled U4 5′ SL (Cy5-U4 5′ SL). The TR-FRET signals were stable from 30 min to 300 min (Supp. Fig. S1) and were not affected by up to 5% DMSO. We selected 45 minutes of reaction for the further tests. Using this method, we were able to detect specific TR-FRET signals by subtracting background from a reaction mixture without His-tag NHP2L1, (used as control) (Fig. 1A), which confirmed the specific interaction of NHP2L1 and U4.
Figure 1.
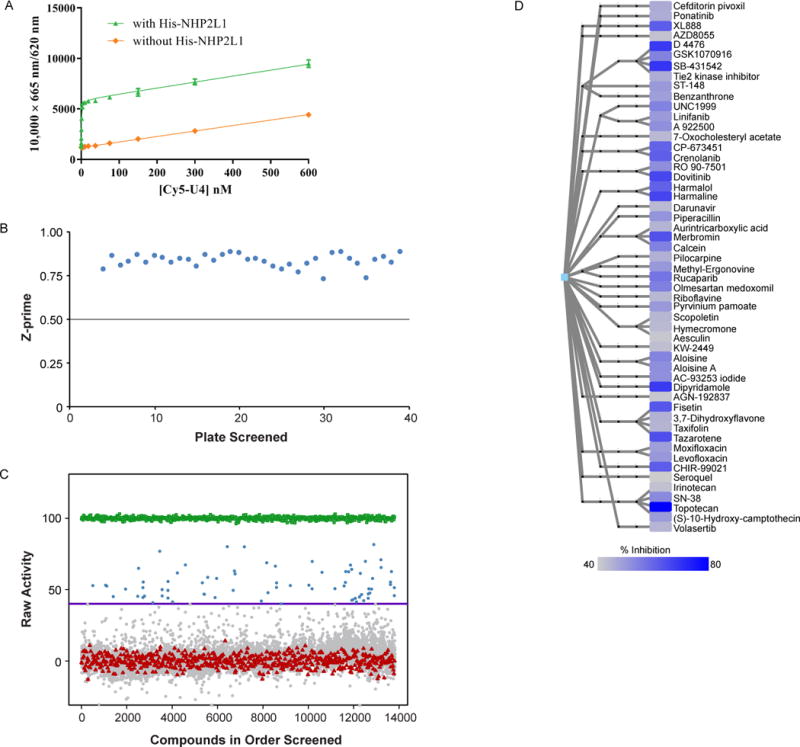
Specific interaction of NHP2L1 protein and U4 and Results of the primary screen of small molecule inhibitors of NHP2L1-U4 interaction. (A) Increasing concentrations of Cy5-U4 5′ SL were incubated with 2nM Terbium-anti-His and 2nM His-NHP2L1. The TR-FRET signals are depicted at each concentration of Cy5-U4 5′ SL, after an incubation of 45 min which led to Kd of 0.68 nM. (B) Scatterplot of plate Z-prime in the order of plates screened. Each dot represents the Z-prime value of the corresponding plate. The averaged Z prime values [0.83 ± 0.04 (0.72 to 0.88)], consistently exceeded plate acceptance criteria indicated by horizontal line (Z prime> 0.5). (C) Scatterplot of percentage inhibition of 10,173 compounds tested at single concentration (15μM) in the primary TR-FRET assay (Blue=hits [≥40% inhibition], grey=inactive [<40% inhibition], red=negative control, green=positive control). (D) Hierarchical chemical clustering of small molecules found to be inhibitors of the NHP2L1-U4 interaction. Leaf nodes represent the actual molecules tested and are color-coded according to percentage of inhibition (darker blue corresponds to greater inhibition, according to the color scale shown). Intermediate nodes represent scaffolds at various levels of chemical abstraction, and edges between nodes signify chemical similarity as described in the methods.
High throughput screen of small molecules interfering with NHP2L1 and U4 interaction
To identify small molecules interfering with NHP2L1 and U4 interaction a total of 10,173 bioactive compounds (4,617 unique molecules) described in the methods section, were first tested at a single concentration of 15μM in the primary screening. Consistency and reproducibility of each assay plate was measured by the Z′-factor which was 0.83 in average (Fig. 1B) beyond the minimum value (0.5) required for a valid assay23. Forty-six compounds (31 unique) achieved 50% inhibition and 75 compounds (53 unique) achieved 40% inhibition (Fig. 1C) with topotecan hydrochloride giving the highest percentage of inhibition 81.6% (Suppl. Table S1). Hierarchical cluster analysis was performed in order to visualize the distribution of these 53 compounds (Fig. 1D), revealing chemical diversity among these small molecules with an average number of neighbors per node=1.987. The 53 unique molecules (providing ≥ 40% inhibition at 15μM) were validated by dose responsive analysis covering ten, 1:3 serially diluted concentrations. The concentrations of each molecule required for 50% disruption of the complex NHP2L1-U4 (IC50) was determined (Suppl. Table S1). The dose response screening confirmed the primary screening hits with topotecan and other campthothecin derivatives (SN-38, 10-OH-(S)-camptothecin) among the most potent inhibitors (Fig. 2A and Suppl. Fig. S2A–D). We further tested additional campthothecin derivatives ((S)-camptothecin, 9-NH2-(S)-camptothecin, 9-NO2-10-OH-(S)-camptothecin, N-Desmethyl topotecan, 7, 11-diethyl-10-OH-(S)-camptothecin) in the dose responsive analysis, revealing that in addition to topotecan, several other campthotecin derivatives also interfere with NHP2L1 and U4 interaction. (Suppl. Fig. S3A–F and Suppl. Table S2).
Figure 2.
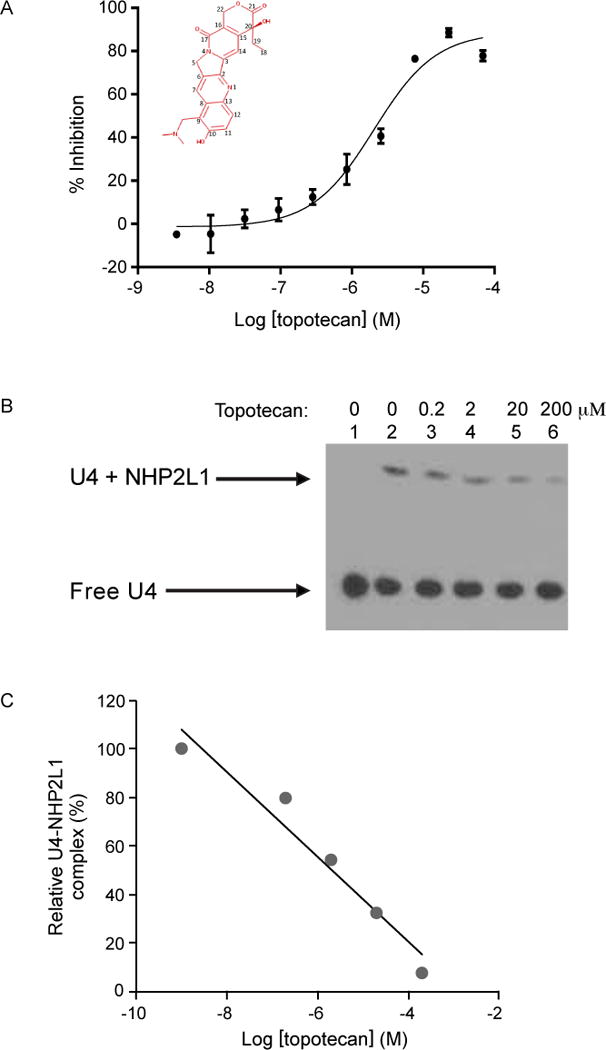
Analysis of the effect of topotecan on NHP2L1-U4 interaction. (A) Increasing concentrations of topotecan were incubated for 45 min with 2nM Tb-anti-His, 2nM His-NHP2L1, 2nM Cy5-U4 5′ SL. The TR-FRET signals were determined and converted into percentage of inhibition by reference to controls. A sigmoid curve was fitted to the resulting data using GraphPad Prism 6.07. IC50 value was determined from log [concentration]-percent inhibition curve constructed for topotecan. (B) EMSA was performed using biotin labeled U4 5′ SL (lanes 1, 2, 3, 4, 5, 6). NHP2L1 protein was added to the assay (lanes 2, 3, 4, 5, 6). Increasing concentrations of topotecan were added to the reactions (lanes 3 [0.2μM], 4 [2μM], 5 [20μM], 6 [200μM]). The shifted band (lane 2) indicates that NHP2L1 binds to Biotin-U4 5′SL. (C) The degree of displacement was determined by measuring by densitometry of the shifted band using ImageJ and the results were expressed as a percentage of the NHP2L1 U4 complex without topotecan.
Characterization of small molecules interfering with NHP2L1 and U4 interaction
To confirm our findings, we used an orthogonal assay to test the inhibition of the NHP2L1-U4 interaction by topotecan. Electrophoretic Mobility Shift Assay (EMSA) documented that NHP2L1 binds to U4 5′SL, as shown by the shift in the EMSA, and documented greater inhibition with increasing concentrations of topotecan, confirming results obtained by TR-FRET (Fig. 2B, C). To determine how topotecan interferes with the NHP2L1-U4 5′SL interaction, we determined whether topotecan binds to U4 5′ SL and (or) to NHP2L1 protein. Surface plasmon resonance with immobilized NHP2L1 or U4 showed that topotecan binds to U4 in a time-dependent manner (Fig. 3A), with markedly less binding to NHP2L1 (Fig. 3B and Suppl. Table S3). Further, we used WaterLogsy (1D NMR) and 1H-15N TROSY NMR (2D NMR) methods to monitor the binding of topotecan to U4 and NHP2L1. The binding of topotecan to U4 5′ SL was confirmed by 1D NMR as several peaks shifted upon adding topotecan to U4 (Suppl. Fig. S4). The 2D 1H-15N spectrum of NHP2L1 shows well dispersed peaks (Suppl. Fig. S5A) comparable to the previously published NMR spectrum of NHP2L16, 18, 24. The assignments were further confirmed with 3D data collected using 13C, 15N-labeled sample. Titrations of topotecan did not change the spectrum, suggesting that topotecan does not bind to NHP2L1 even at 1:20 molar ratio (Suppl. Fig. S5A). There was an overall decrease in the intensity of the peaks as a function of topotecan concentration as shown by the 1D slices (Suppl. Fig. S5B). To confirm whether topotecan interferes with NHP2-U4 interaction, we used 2D 1H-15N NMR spectroscopy. We first titrated U4 5′ S L into a solution of 15N labelled NHP2L1 and measured 1H-15N spectrum of the complex. Upon binding of U4 5′ S L to NHP2L1, the NMR signals of several residues were shifted (Fig. 3C, 3D; blue contours). The addition of high concentration of topotecan to NHP2L1-U4 complex (1:1:20) partially restores the free NHP2L1 signals (Fig. 3C, 3D, 3E; magenta). These findings confirm that topotecan displaces NHP2L1 from U4 only at high concentrations. The 1D spectrum of this complex also showed additional resonances for topotecan only with U4 – NHP2L1 complex, suggesting topotecan is able to directly bind to the NHP2L1-U4 complex (Suppl. Fig. S6A, S6B).
Figure 3.
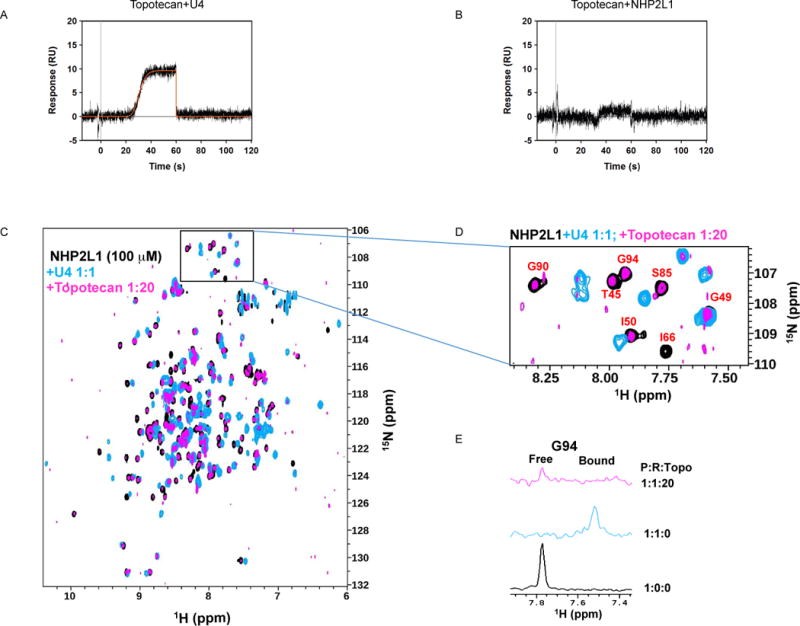
NMR and equilibrium analysis of topotecan binding to NHP2L1 protein or to U4 5′ SL by surface plasmon resonance (SPR). Topotecan was injected at a maximum concentration of 50μM using the OneStep gradient injection mode over surfaces of U4 5′ SL (A) or NHP2L1 protein (B). The data (black line) were globally fit to a 1:1 equilibrium affinity binding model (orange line). (C) Overlay of 2D 1H-15N TROSY spectra of 100μM 15N-labelled NHP2L1 in the absence (black) and presence (blue) of 100μM U4 5′ SL. The NMR signal of specific NHP2L1 residues changes upon addition of U4 5′ SL, indicating an interaction. Topotecan was added to NHP2L1-U4 complex as shown in magenta (2mM topotecan). (D) Enlarged view of selected residues of the 2D 1H, 15N TROSY NMR spectra that exhibited displacement of NHP2L1 and U4 by topotecan. (E) 1D slice through the residue G94 peak marked from the three different spectra indicating the displacement of U4 from NHP2L1 by topotecan.
Inhibition of NHP2L1 and U4 interaction and splicing
Because the interaction of NHP2L1 and U4 is required for splicing, we assessed the effect of topotecan on pre-mRNA splicing, using a previously reported splicing reporter vector, pTN2320. This construct contains reporter genes for β-galactosidase and luciferase separated by an exon-intron-exon cassette. Although β-galactosidase is constitutively expressed, luciferase is only expressed when splicing occurs. The ratio of luciferase to β–galactosidase activities is used to quantify splicing efficiency. These data shown that topotecan significantly decreased splicing efficiency (p<0.001) without changing significantly β–galactosidase activities (p=0.4) (Fig. 4A). Overexpression of NHP2L1 (Fig. 4B) reduced significantly the effect of topotecan on splicing (Fig. 4C) (p=0.04), consistent with topotecan inhibiting splicing by interfering with the NHP2L1 U4 interaction.
Figure 4.
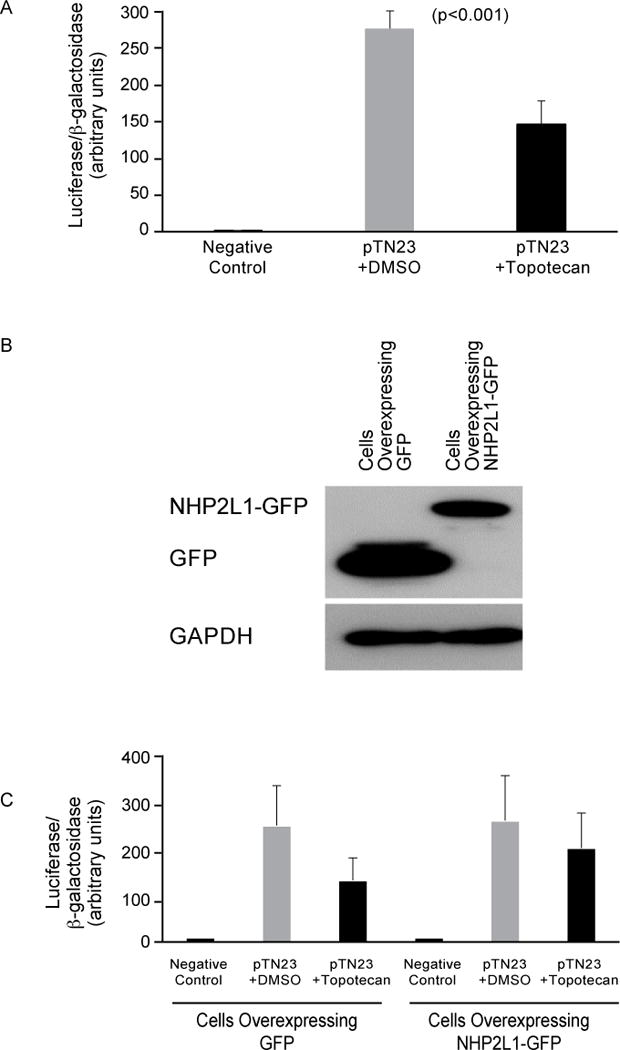
Inhibition of NHP2L1-U4 interaction and splicing. (A) HEK 293 cells were transfected with the pTN23 vector or not (negative control) and the cells were treated with topotecan (250nM) or with DMSO. After 24 hours, the luciferase and β-galactosidase activities were determined. The ratio of luciferase to β-galactosidase activities are shown. Error bars represent the standard deviation of three independent experiments and each experiment was performed in duplicate. Student’s t-test was used. (B) HEK293 cells were transfected with plasmid expressing GFP or NHP2L1-GFP. Expression of GFP and NHP2L1-GFP proteins were detected by western blot using an anti-GFP antibody. GAPDH was used as internal control. (C) The HEK 293 cells overexpressing GFP or NHP2L1-GFP were transfected with the pTN23 vector or not (negative control) and cells were treated with topotecan (250nM) or with DMSO. After 24 hours the luciferase and β-galactosidase activities were determined. The ratio of luciferase to β-galactosidase activities are shown. Error bars represent the standard deviation of three independent experiments and each experiment was performed in duplicate.
Discussion
Protein-nucleic acid interactions are essential for multiple cellular processes and small molecule disruptors of these interactions could be powerful tools for investigation of these biological processes and could have potential as therapeutics25.
In the present study, we have successfully developed a TR-FRET assay to assess the interaction between NHP2L1 protein and U4 (RNA), two key players in splicing. This assay confirms the specific and high binding affinity of NHP2L1 to U4 as previously shown3, 6, 21 and provides a tool for discovery of small molecules that inhibits this process.
The identification of compounds that perturb protein-nucleic acid interactions is extremely challenging25 and there is a paucity of reports of small molecules inhibitors of any protein-DNA26 or protein-RNA interactions27.
By using a high-throughput TR-FRET screen, we have identified for the first time several small-molecules inhibitors that disrupt the complex formed by NHP2L1 and U4. These small molecules are chemically diverse, reflecting the overall diversity of the compound library, a good predictor for library performance28-30. Moreover the presence of several distinct chemical small molecules among our top hits suggest there may be different mechanisms by which they interfere with NHP2L1 and U4. It is possible that some could bind to the protein and or the RNA with different interaction sites, as previously shown in other protein and RNA interaction27. It will be important for future experiments to determine precisely how the different small molecules disrupt the interaction between NHP2L1 and U4 and if they inhibit a broader set of nucleic acid protein interaction. We have focused on our top hit topotecan and our results show that it binds to the U4 (RNA), consistent with previous findings that very few compounds target the protein in an RNA-protein complex31. Topotecan and related camptothecin derivatives are widely used anticancer agents that exert their pharmacologic effects primarily by inhibiting topoisomerase I32-33. Topoisomerase interacts with DNA whereas NHP2L1 interacts with RNA, and the mechanism by which topotecan and camptothecin derivatives inhibit Topoisomerase I likely differs from the way they interfere with NHP2L1 and U4. Indeed topotecan does bind to DNA alone at high concentrations but not to topoisomerase alone34, rather it stabilizes the transient covalent topoisomerase I – DNA complex35. Our new findings show that topotecan binds to the NHP2L1-U4 complex (as shown by NMR) and binds U4 alone, but it disrupts this interaction instead of stabilizing it. The other difference is while only the 10-OH camptothecin derivatives, namely SN38, topotecan, and N-demethyl-topotecan are able to disrupt the interaction of U4 and NHP2L1, camptothecin and its derivative, irinotecan, which are known to inhibit topoisomerase I34, 36-39 were less effective at disrupting the NHP2L1 and U4 interaction. Attempts to cocrystallize topotecan and NHP2L1-U4 did not produce crystals, precluding further structural studies at this time.
The current study has limitations including that we used only the pTN23 splicing reporter vector and not endogenous RNA to show the effects of topotecan on splicing. Also further in-vivo studies are needed in order to determine the concentrations of topotecan and of the other small molecules that are required to alter the in-vivo function of NHP2L1-U4 and by extension splicing and to determine the impact of this inhibition on the known and potentially new therapeutic effects of these small molecules.
In summary we have identified the first documented inhibitors of the interaction between NHP2L1 and U4, providing new insights into how topotecan interferes with this interaction. Taken together, our data reveal new functions of known drugs, which could facilitate the development of therapeutics that modify splicing and thereby gene function.
Supplementary Material
Acknowledgments
We thank Darcie Miller, Sivaraja Vaithiyalingam, Amanda Nourse, Duane Currier for their technical assistance. We thank Elizabeth Stevens for preparation of the figures. We thank Ian C. Eperon (University of Leicester) for the pTN23 plasmid. We thank the shared research resources of the St. Jude Children’s Research Hospital Comprehensive Cancer Center. The study was supported in part by National Institutes of Health Grants R37 CA36401 [W.E.E. and M.V.R.], U01 GM92666 [M.V.R. and W.E.E.], P50 GM115279 [W.E.E. and M.V.R.] Comprehensive Cancer Center grant CA21765, from the National Cancer Institute, and by the American Lebanese Syrian Associated Charities (ALSAC).
Footnotes
Declaration of Conflicting Interests
The authors declare no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Lee SC, Abdel-Wahab O. Therapeutic targeting of splicing in cancer. Nature medicine. 2016;22(9):976–86. doi: 10.1038/nm.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maslon MM, Heras SR, Bellora N, et al. The translational landscape of the splicing factor SRSF1 and its role in mitosis. eLife. 2014:e02028. doi: 10.7554/eLife.02028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vidovic I, Nottrott S, Hartmuth K, et al. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Molecular cell. 2000;6(6):1331–42. doi: 10.1016/s1097-2765(00)00131-3. [DOI] [PubMed] [Google Scholar]
- 4.Saito H, Fujiwara T, Shin S, et al. Cloning and mapping of a human novel cDNA (NHP2L1) that encodes a protein highly homologous to yeast nuclear protein NHP2. Cytogenetics and cell genetics. 1996;72(2-3):191–3. doi: 10.1159/000134186. [DOI] [PubMed] [Google Scholar]
- 5.Leung A K, Lamond AI. In vivo analysis of NHPX reveals a novel nucleolar localization pathway involving a transient accumulation in splicing speckles. The Journal of cell biology. 2002;157(4):615–29. doi: 10.1083/jcb.200201120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu S, Li P, Dybkov O, et al. Binding of the human Prp31 Nop domain to a composite RNA-protein platform in U4 snRNP. Science (New York, NY) 2007;316(5821):115–20. doi: 10.1126/science.1137924. [DOI] [PubMed] [Google Scholar]
- 7.Falb M, Amata I, Gabel F, et al. Structure of the K-turn U4 RNA: a combined NMR and SANS study. Nucleic acids research. 2010;38(18):6274–85. doi: 10.1093/nar/gkq380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vrijens K, Lin W, Cui J, et al. Identification of small molecule activators of BMP signaling. PloS one. 2013;8(3):e59045. doi: 10.1371/journal.pone.0059045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leonardi R, Zhang YM, Yun MK, et al. Modulation of pantothenate kinase 3 activity by small molecules that interact with the substrate/allosteric regulatory domain. Chemistry & biology. 2010;17(8):892–902. doi: 10.1016/j.chembiol.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walters BJ, Lin W, Diao S, et al. High-throughput screening reveals alsterpaullone, 2-cyanoethyl as a potent p27Kip1 transcriptional inhibitor. PloS one. 2014;9(3):e91173. doi: 10.1371/journal.pone.0091173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu DD, Lin W, Chen T, et al. Development of time resolved fluorescence resonance energy transfer-based assay for FXR antagonist discovery. Bioorganic & medicinal chemistry. 2013;21(14):4266–78. doi: 10.1016/j.bmc.2013.04.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bemis GW, Murcko MA. The properties of known drugs. 1. Molecular frameworks. Journal of medicinal chemistry. 1996;39(15):2887–93. doi: 10.1021/jm9602928. [DOI] [PubMed] [Google Scholar]
- 13.Schuffenhauer A, Ertl P, Roggo S, et al. The scaffold tree–visualization of the scaffold universe by hierarchical scaffold classification. Journal of chemical information and modeling. 2007;47(1):47–58. doi: 10.1021/ci600338x. [DOI] [PubMed] [Google Scholar]
- 14.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papalia G, Myszka D. Exploring minimal biotinylation conditions for biosensor analysis using capture chips. Analytical biochemistry. 2010;403(1-2):30–5. doi: 10.1016/j.ab.2010.03.044. [DOI] [PubMed] [Google Scholar]
- 16.Quinn JG. Modeling Taylor dispersion injections: determination of kinetic/affinity interaction constants and diffusion coefficients in label-free biosensing. Analytical biochemistry. 2012;421(2):391–400. doi: 10.1016/j.ab.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 17.Quinn JG. Evaluation of Taylor dispersion injections: determining kinetic/affinity interaction constants and diffusion coefficients in label-free biosensing. Analytical biochemistry. 2012;421(2):401–10. doi: 10.1016/j.ab.2011.11.023. [DOI] [PubMed] [Google Scholar]
- 18.Soss SE, Flynn PF. NMR assignment of the human spliceosomal 15.5K protein. Journal of biomolecular NMR. 2007;38(2):175. doi: 10.1007/s10858-006-9100-3. [DOI] [PubMed] [Google Scholar]
- 19.Keller R. The computer aided resonance assignment tutorial. CANTINA Verlag; Goldau, Switerland: 2004. [Google Scholar]
- 20.Nasim MT, Chowdhury HM, Eperon IC. A double reporter assay for detecting changes in the ratio of spliced unspliced mRNA in mammalian cells. Nucleic acids research. 2002;30(20):e109. doi: 10.1093/nar/gnf108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nottrott S, Hartmuth K, Fabrizio P, et al. Functional interaction of a novel 15.5kD [U4/U6.U5] tri-snRNP protein with the 5′ stem-loop of U4 snRNA. The EMBO journal. 1999;18(21):6119–33. doi: 10.1093/emboj/18.21.6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emami-Nemini A, Roux T, Leblay M, et al. Time-resolved fluorescence ligand binding for G protein-coupled receptors. Nature protocols. 2013;8(7):1307–20. doi: 10.1038/nprot.2013.073. [DOI] [PubMed] [Google Scholar]
- 23.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation Validation of High Throughput Screening Assays. Journal of biomolecular screening. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 24.Soss SE, Flynn PF. Functional implications for a prototypical K-turn binding protein from structural dynamical studies of 15.5K. Biochemistry. 2007;46(51):14979–86. doi: 10.1021/bi701254q. [DOI] [PubMed] [Google Scholar]
- 25.Chan LL, Pineda M, Heeres JT, et al. A general method for discovering inhibitors of protein-DNA interactions using photonic crystal biosensors. ACS chemical biology. 2008;3(7):437–48. doi: 10.1021/cb800057j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neher TM, Shuck SC, Liu JY, et al. Identification of novel small molecule inhibitors of the XPA protein using in silico based screening. ACS chemical biology. 2010;5(10):953–65. doi: 10.1021/cb1000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mei HY, Cui M, Heldsinger A, et al. Inhibitors of protein-RNA complexation that target the RNA: specific recognition of human immunodeficiency virus type 1 TAR RNA by small organic molecules. Biochemistry. 1998;37(40):14204–12. doi: 10.1021/bi981308u. [DOI] [PubMed] [Google Scholar]
- 28.Franzini RM, Neri D, Scheuermann J. DNA-encoded chemical libraries: advancing beyond conventional small-molecule libraries. Accounts of chemical research. 2014;47(4):1247–55. doi: 10.1021/ar400284t. [DOI] [PubMed] [Google Scholar]
- 29.Petrone PM, Wassermann AM, Lounkine E, et al. Biodiversity of small molecules–a new perspective in screening set selection. Drug discovery today. 2013;18(13-14):674–80. doi: 10.1016/j.drudis.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Nissink JW, Schmitt S, Blackburn S, et al. Stratified high-throughput screening sets enable flexible screening strategies from a single plated collection. Journal of biomolecular screening. 2014;19(3):369–78. doi: 10.1177/1087057113498933. [DOI] [PubMed] [Google Scholar]
- 31.Childs-Disney JL, Stepniak-Konieczna E, Tran T, et al. Induction and reversal of myotonic dystrophy type 1 pre-mRNA splicing defects by small molecules. Nature communications. 2013;4:2044. doi: 10.1038/ncomms3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsiang YH, Liu LF. Identification of mammalian DNA topoisomerase I as an intracellular target of the anticancer drug camptothecin. Cancer research. 1988;48(7):1722–6. [PubMed] [Google Scholar]
- 33.Herben VM, ten Bokkel Huinink WW, Beijnen JH. Clinical pharmacokinetics of topotecan. Clinical pharmacokinetics. 1996;31(2):85–102. doi: 10.2165/00003088-199631020-00001. [DOI] [PubMed] [Google Scholar]
- 34.Thomas CJ, Rahier NJ, Hecht SM. Camptothecin: current perspectives. Bioorganic & medicinal chemistry. 2004;12(7):1585–604. doi: 10.1016/j.bmc.2003.11.036. [DOI] [PubMed] [Google Scholar]
- 35.Staker BL, Hjerrild K, Feese MD, et al. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(24):15387–92. doi: 10.1073/pnas.242259599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szafranska AE, Hitchman TS, Cox RJ, et al. Kinetic and mechanistic analysis of the malonyl CoA:ACP transacylase from Streptomyces coelicolor indicates a single catalytically competent serine nucleophile at the active site. Biochemistry. 2002;41(5):1421–7. doi: 10.1021/bi012001p. [DOI] [PubMed] [Google Scholar]
- 37.Laco GS. Evaluation of two models for human topoisomerase I interaction with dsDNA and camptothecin derivatives. PloS one. 2011;6(8):e24314. doi: 10.1371/journal.pone.0024314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li QY, Zu YG, Shi RZ, et al. Review camptothecin: current perspectives. Current medicinal chemistry. 2006;13(17):2021–39. doi: 10.2174/092986706777585004. [DOI] [PubMed] [Google Scholar]
- 39.Tanizawa A, Kohn KW, Kohlhagen G, et al. Differential stabilization of eukaryotic DNA topoisomerase I cleavable complexes by camptothecin derivatives. Biochemistry. 1995;34(21):7200–6. doi: 10.1021/bi00021a035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


