Abstract
Novel biomarker discovery plays a crucial role in providing more sensitive and specific disease detection. Unfortunately many low-abundance biomarkers that exist in biological fluids cannot be easily detected with mass spectrometry or immunoassays because they are present in very low concentration, are labile, and are often masked by high-abundance proteins such as albumin or immunoglobulin. Bait containing poly(N-isopropylacrylamide) (NIPAm) based nanoparticles are able to overcome these physiological barriers. In one step they are able to capture, concentrate and preserve biomarkers from body fluids. Low-molecular weight analytes enter the core of the nanoparticle and are captured by different organic chemical dyes, which act as high affinity protein baits. The nanoparticles are able to concentrate the proteins of interest by several orders of magnitude. This concentration factor is sufficient to increase the protein level such that the proteins are within the detection limit of current mass spectrometers, western blotting, and immunoassays. Nanoparticles can be incubated with a plethora of biological fluids and they are able to greatly enrich the concentration of low-molecular weight proteins and peptides while excluding albumin and other high-molecular weight proteins. Our data show that a 10,000 fold amplification in the concentration of a particular analyte can be achieved, enabling mass spectrometry and immunoassays to detect previously undetectable biomarkers.
Keywords: Bioengineering, Issue 90, biomarker, hydrogel, low abundance, mass spectrometry, nanoparticle, plasma, protein, urine
Introduction
Despite the completion of the human genome sequencing, significant progress has not been made in identifying biomarkers predictive of early stage disease, or that correlate with therapeutic outcome, or prognosis1. One reason for this lack of progress is that many potentially significant biomarkers exist at a concentration below the detection limit of conventional mass spectrometry and other biomarker discovery platforms. Mass spectrometry (MS) and Multiple Reaction Monitoring (MRM) have a detection sensitivity typically greater than 50 ng/ml while the majority of the analytes measured by immunoassays in a clinical laboratory fall in the range between 50 pg/ml and 10 ng/ml. This means that many biomarkers, particularly in the early stage of a disease cannot be detected by conventional MS and MRM2. In addition the presence of high-abundance proteins such as albumin and immunoglobulin in complex biological fluids often mask by billion-fold excess low-abundance, low molecular weight proteins and peptides3, 4. For this reason several sample preparatory steps are required prior to mass spectrometry sequencing and identification. One such preparatory step employs the depletion of high-abundance proteins with commercially available depletion columns5-8. Unfortunately this step leads to the reduction of the yield of candidate biomarkers because they are often non-covalently associated with carrier proteins that are being removed. Another challenge is represented by the stability of candidate biomarkers ex-vivo once the samples are collected. Proteins are subject to degradation by endogenous or exogenous proteases9. Hydrogel nanoparticles can transcend these critical challenges by amplifying the putative biomarker concentration to a level within the range of the assay, while protecting the protein from degradation10-13.
It’s important to note that LMW proteins in blood are a mixture of small intact proteins as well as fragments of large proteins. Tissue-derived proteins larger than 60 kDa are too large to passively enter the blood stream through the vascular basement membrane, but they can be represented in blood as peptides or protein fragments14. Our goal is to measure novel circulating biomarkers that can be candidates for early detection of disease, patient stratification for therapy, and monitoring the response to therapy. Our nanoparticles are created to selectively exclude high abundance immunoglobulins and albumin, while simultaneously capturing smaller proteins and peptides and concentrating them up to 100-fold depending on the starting volume.
Our group identified a set of small organic dyes which can successfully act as high affinity molecular baits for proteins and peptides. Protein-dye binding is thought to be due to a combination of hydrophobic and electrostatic interactions. The aromatic rings on the dye interleave with proteins via hydrophobic pockets on the protein surface11.
The baits, depending on their chemistry, show a particular affinity for selected classes of analytes. The baits compete with the carrier proteins, such as albumin, for the proteins or peptides. The low-molecular weight proteins/peptides become trapped in the particle. High-molecular weight proteins such as albumin and immunoglobulin are prevented from entering the particle because of the sieving capability due to the restrictive pore of the hydrogel11(Figure 1).
Hydrogel nanoparticles are synthesized by precipitation polymerization initiated by ammonium persulfate11. N-isopropylacrylamide (NIPAm), co-monomers of acrylic acid (AAc) and allylamine (AA) and cross-linker N,N’-Methylenebisacrylamide (BIS) are allowed to react at 70 °C for 6 hr in dilute conditions11, 13. The high protein binding affinity of poly(N-isopropylacrylamide-co-acrylic acid) (poly(NIPAm-co-AAc) nanoparticlesis achieved by covalently incorporating amino-containing dyes (i.e., sulfonatedanthraquinonetriazine dyes) into the nanoparticles through an amidation reaction performed in aqueous or organic solvents depending on the hydrophilic/hydrophobic characteristics of the dyes11, 13. Nucleophilic substitution of the amine groups in the nanoparticle with the chloride atom of an anthraquinonetriazine dye is utilized to create dye-containing poly(NIPAm-co-Allylamine) (AA) nanoparticles11, 12. A two-step polymerization process is utilized to create hydrogel nanoparticles containing an outer shell of vinylsulfonic acid (VSA)11, 13.
Hydrogel nanoparticles can be applied to various biological fluids, including whole blood, plasma, serum, cerebrospinal fluid, sweat, and urine. In one step, in solution, the nanoparticles perform a rapid (within minutes) sequestration and concentration of low molecular weight analytes10, 11, 13, 15-18. Proteins are subsequently eluted from the nanoparticles and detected using western blotting19-21, mass spectrometry10, 11, 13, 15, 18, 22, 23, immunoassays/ELISA10, 11, 15, 18, or reverse phase protein microarray16, 24 assays. Nanoparticles functionalized with chemical bait, and presenting a core or core shell architecture, capture and concentrate proteins based on the bait/shell physicochemical properties. Different dyes incorporated into the nanoparticles will therefore capture different subsets of proteins with varying efficiency based on the dye affinity, pH of the solution, and the presence/absence of competing high-abundant proteins13. Furthermore, the quantity of nanoparticles in relation to the volume of the solution will affect the protein yield from the nanoparticles. These aspects of hydrogel nanoparticle harvesting are demonstrated using three different nanoparticle baits for harvesting proteins from plasma samples which contain high amounts of protein, and from urine samples which typically do not contain large amounts of protein. In this protocol we demonstrate harvesting and concentrating tumor necrosis factor alpha (TNFα) from plasma samples using poly(NIPAm-co-AAc), Poly(NIPAm/dye), and core- shell nanoparticles (Poly(NIPAm-co-VSA)). Poly(NIPAm/dye) nanoparticles are shown to concentrate Mycobacterium species antigen that was added to human urine samples, to mimic Mycobacterium tuberculosis infected individuals.
Protocol
Human plasma and urine was collected from healthy volunteer donors, with written informed consent, following George Mason University Institutional Review Board approved protocols. Donors were equally distributed between Caucasian males and females between the ages of 25 and 42. Samples were analyzed individually and were not pooled.
1. Nanoparticle Processing of Serum or Plasma Samples
Potential low abundant biomarkers in plasma are captured, in solution, with hydrogel nanoparticles. The particles are added to the plasma, incubated, separated by centrifugation, washed, and the captured proteins are eluted. The eluted proteins are dried under nitrogen flow for downstream mass spectrometry sequencing and identification.
Dilute 500 µl of serum 1:2 with 50 mM TrisHCl pH 7 (500 µl serum + 500 µl TrisHCl) in a microcentrifuge tube.
Add 500 µL of poly(NIPAm/AAc) core nanoparticles. Incubate for 15 min at RT.
Spin the sample at 16,100 x g, 25 °C for 10 min in a centrifuge equipped with a fixed-angle rotor. Remove and discard the supernatant.
Add 500 µl sodium thiocyanate (25 mM) to the pellet. Resuspend the nanoparticles by vigorously pipetting up and down multiple times.
Spin the sample at 16,100 x g, 25 °C for 10 min in a centrifuge with a fixed angle rotor. Remove and discard the supernatant.
Add 500 µl of MilliQ water to the nanoparticle pellet. Resuspend the nanoparticles by vigorously pipetting up and down multiple times.
Spin the sample at 16,100 x g, 25 °C for 10 min in a centrifuge with a fixed angle rotor. Remove and discard the supernatant.
Prepare fresh elution buffer: Add 700 µl acetonitrile to 300 µl ammonium hydroxide in a clean tube. Caution: Ammonium hydroxide is corrosive. Use with appropriate ventilation. Note: The elution buffer must be prepared immediately before use. Do not store the elution buffer.
Add 300 µl of elution buffer to the nanoparticle pellet. Resuspend the nanoparticles by vigorously pipetting up and down multiple times. Incubate for 15 min at RT.
Spin the sample at 16,100 x g, 25 °C for 10 min in a centrifuge with a fixed angle rotor. Remove and save the eluate in a clean, labeled microcentrifuge tube.
Repeat steps 1.9 – 1.10. Combine the two eluates into one microcentrifuge tube.
Dry the eluates under nitrogen flow in a nitrogen evaporator manifold at 42.1 °C, with air flow set to 8 (Figure 2F).
Store the dried eluate at RT for O/N storage, or at -20 °C for long term storage, prior to mass spectrometry, western blotting, or ELISA assays (optional).
2. Nanoparticle Processing of Urine Samples
Normal urine contains less than 30 mg/dl protein and less than 1+ blood. However, many diseases/conditions may alter the normal levels of urine protein and blood. To aid in determining the optimal volume of nanoparticles to add to the urine sample, a urinalysis is performed prior to nanoparticle harvesting. Urine biomarkers may exist in extremely low concentrations, which may require optimizing the ratio of nanoparticles to urine volume. This procedure describes nanoparticle harvesting of urine samples for downstream western blot analysis.
Collect urine samples in a clean, dry plastic cup. A 22 ml minimum volume is required. Store urine specimens at -80 °C until ready to analyze.
Thaw the frozen urine at RT or at 4 °C O/N. Mix briefly on a vortex mixer. Pour at least 22 ml of urine into a 50 ml conical bottom polypropylene tube.
Spin the urine in a centrifuge with a swing-out rotor at 3,700 x g for 15 min. Decant the urine, without disturbing the pellet, into a clean 50 ml conical bottom polypropylene tube. Discard the pellet.
- Perform urinalysis using a multi-analyte “urine dipstick” reagent strip. Note: store the reagent strips in a tightly closed container away from direct sunlight and humidity17, 18.
- Lay the reagent strip, face-up, on a clean, dry paper towel.
- Use a disposable pipette to aspirate 1 ml of the urine prepared in step 2.3.
- Set a timer for 2 min but do not start it. Quickly dispense 1 - 2 drops of urine on each test pad of the reagent strip. Immediately start the timer. Note: Do not submerge the reagent strip directly in the urine container. The dye/chemical indicators in the reagent strip can potentially leach out of the reagent strip into the urine container.
- At the time indicated on the reagent strip container, record the qualitative results for the various analytes on the reagent strip by comparing the color of the individual reagent strips to the corresponding color-coded indicators on the reagent container. Typical normal urine results are: (normal or negative) for blood, protein, leukocytes, nitrite, glucose, ketone, bilirubin, and urobilinogen; Urine pH (5.5 - 7.0); specific gravity (1.001 - 1.020). Note: High protein levels in a urine specimen may compete with the protein of interest for binding sites on the nanoparticles. To maximize protein harvesting with nanoparticles, the nanoparticle volume/urine volume ratio can be adjusted to either limit competition from high abundance proteins, or can be optimized to harvest proteins that exist in very low concentrations. If the urine protein value is 1+ or greater, add 2x volumes of nanoparticles in step 2.5 below. If the analyte of interest is a very low abundant protein, it may be necessary to increase the volume of nanoparticles up to 2 ml (Figure 3).
Transfer 20 ml of the clarified urine from step 2.3 into a clean 50 ml conical bottom polypropylene tube. Do not disturb any debris/pellet that may be in the bottom of the urine tube. Add 200 µl of nanoparticles to the 20 ml urine sample. Mix briefly on a vortex mixer. Note: If the urine protein value is 1+ or greater, add 400 µl of nanoparticles to 20 ml urine.
Incubate the urine/nanoparticle mixture for 30 min at RT, without rocking/mixing.
Spin the urine/nanoparticle suspension in a centrifuge equipped with a swing-out rotor at 3,700 x g for 10 min. Note: If a pellet is not visible following centrifugation, spin the samples for an additional 2 – 7 min.
Remove and discard the supernatant. Add 500 µl MilliQ water to the nanoparticle pellet.
Resuspend the nanoparticles by vigorously pipetting up and down multiple times. Transfer the nanoparticle solution to a clean 1.5 ml microcentrifuge tube.
Spin the nanoparticles in a centrifuge equipped with a fixed-angle rotor at 16,100 x g for 10 min. Note: If a pellet is not visible following centrifugation, spin the samples for an additional 2 – 7 min.
Remove and discard the supernatant.
Repeat steps 2.8 and 2.11 (twice) with 200 µl 18 MilliQ water to wash the nanoparticles.
Prepare fresh elution buffer: Add 970 µl acetonitrile to 30 µl ammonium hydroxide in a clean tube. Caution: Ammonium hydroxide is corrosive. Use with appropriate ventilation. Note: The elution buffer must be prepared immediately before use. Do not store the elution buffer.
Add 20 μl elution buffer to the nanoparticle pellet. Resuspend the nanoparticles by vigorously pipetting up and down multiple times. Incubate for 15 min at RT.
Spin the nanoparticle samples at 16,100 x g for 10 min. Remove and save the supernatant in a clean, 1.5 ml microcentrifuge tube. Do not disrupt the nanoparticle pellet. Discard the pellet in the biohazard trash.
Place the samples in a rack in a chemical fume hood. Open the caps and incubate at RT for 30 min. Alternatively, place the samples under nitrogen flow at 40 °C until dry (1 - 2 hr).
Add 15 µl Tris-glycine SDS sample buffer (2x) to the samples. Heat at 100°C in a dry heat block for 5 min with the caps open or until the volume left in the tube is no more than 20 µl. Note: do not use a boiling water bath. The humidity from the water bath will prevent evaporation of the buffer.
Place the cap on the tubes. Remove the tubes from the heat block. The sample can be stored at -80 °C or used immediately for downstream western blot analysis.
Representative Results
Hydrogel Nanoparticle Size and Uniformity
Poly(NIPAm-AAc) particles have been produced with extremely high yield and reproducibility between and within batches. The particles have very good colloidal stability at RT during the time required for capture, storage, and elution of proteins (at least 48 hr), and nanoparticle precipitation has not been observed (Figure 1)11. The colloidal stability may be very important for rapid protein/peptide uptake by the nanoparticles.
Potential Low-abundance Biomarkers Harvested from Plasma
The incorporation of the dye bait drives the uptake of molecules in solution and assures that captured molecules are preserved from degradation13, 15, 18, 25. For this reason, protease inhibitors are not needed during sample pre-processing. This attribute of the nanoparticles makes them suitable as a preservation technology for sample collection in the field and for shipment of biological fluids at RT.
The bait molecule can be incorporated in the hydrogel particle by copolymerization or covalent binding to functional groups present in the nanoparticle. As an example, poly(NIPAm-co-AAc) nanoparticles were constructed with amino containing dyes via zero length crosslinking amidation reactions, while nanoparticles with an outer shell containing vinylsulfonic acid (VSA) copolymer were created by a second polymerization reaction10-13. Different types of nanoparticles can be produced by incorporating a different chemical dye within the nanoparticle and thus enhance harvesting of specific classes of proteins. An important concept of our technology is that different baits can show a preferential affinity for different proteins because the dye-protein interaction is dependent mainly on a combination of hydrophobic interactions, 3-D interactions, and electrostatic forces. We have observed that some analytes can be captured with a high affinity by chemically different dyes because of the complexity of the binding forces. See Tamburro et al. for an extensive explanation and examples of this principle13. For example, four different types of capture bait (nanoparticles) were used to harvest TNFα from plasma using 200 µl of particles and 200 µl plasma for each type of nanoparticle. In all cases after treatment with the nanoparticles, TNFα was not detectable in the supernatant by SDS-PAGE with silver staining, whereas TNFα was detectable in the nanoparticle eluate (Figure 1C). (Lane 1 = recombinant TNFα (MW 17,000 Da), Lanes 2 & 3, 4 & 5, 6 & 7 and 8 & 9 represent results for four different types of nanoparticle baits respectively; C = control, S = supernatant, P = nanoparticle eluate).
Dose-response for Different Types of N anoparticles
In order to perform a calibration curve, and thus assess yield and precision, experiments need to be performed with spiked-in proteins of known concentration. Published results for a variety of body fluids and analytes demonstrate how the nanoparticle pre-processing maintains linearity of the assay and establishes a calibration curve while enhancing the effective limit of detection10, 16, 26. We have also published studies of between-laboratory precision using the nanoparticles to conduct multi-reaction monitoring mass spectrometry with enhanced sensitivity27.
To compare IL-17 harvesting efficiency by different types of nanoparticle, recombinant IL-17 (17.5 kDa) was added to plasma samples to which either poly(NIPAm-co-AAc) or poly(NIPAm-co-VSA) nanoparticles were subsequently added. Two sets of four serial dilutions of 100 ng of recombinant IL-17 were prepared in 100 µl plasma (100 ng/100 µl, 50 ng/100 µl, 25 ng/100 µl and 12.5 ng/100 µl). poly(NIPAm-co-AAc) or poly(NIPAm-co-VSA) particles were added to the respective plasma samples in a 1:1 (v/v) particle:plasma ratio. A spectrophotometric total protein assay was performed on the supernatant samples. Western blotting with anti-rabbit polyclonal IL-17 was performed on 20 μg of plasma supernatant (S) after nanoparticle harvesting and the nanoparticle eluates (P) (Figure 4). Both nanoparticle types adequately harvested and concentrated IL-17 at each dilution. The additional bands on the western blot represent human serum albumin and immunoglobulin that cross-react with the secondary antibody. (AAc particles: Lanes 1 & 2 = 1 ng/µl IL-17, Lanes 3 & 4 = 0.50 ng/µl, Lanes 5 & 6 = 0.25 ng/µl, Lanes 7 & 8 = 0.125 ng/µl, Lane 9 = IL-17; VSA particles: Lanes 1 & 2 = 1 ng/µl IL-17, Lanes 3 & 4 = 0.50 ng/µl, Lanes 5 & 6 = 0.25 ng/µl, Lanes 7 & 8 = 0.125 ng/µl).
Detection of Potentially Diagnostic Proteins in Urine
Human urine samples, obtained from healthy volunteer donors with written informed consent, were spiked with a recombinant Mycobacterium species antigen Early Secretory Target Mycobacterium Tuberculosis protein (ESAT-6) to mimic Mycobacterium tuberculosis infected individuals. 1 µg each of ESAT-6 (15 kDa) and IL-2 (15.5 kDa) were added to 1 ml aliquots of human urine collected from healthy volunteer donors. Urinalysis was performed on the samples with a urine reagent strip to determine the optimal ratio of nanoparticles to urine, which was based on the presence/absence of urinary proteins. Poly(NIPAm/dye) nanoparticles were used to harvest proteins from treated and untreated urine samples. One-dimensional gel electrophoresis of the nanoparticle eluates was performed followed by silver staining. Samples in U4 and U6 lanes represent nanoparticle eluates from ESAT-6 and IL-2 treated urine samples, clearly showing prominent bands at 15 kDa (Figure 5). Mass spectrometry of the eluates identified ESAT-6 (UniProt accession POA567, 84.0 coverage, 9 peptides) and IL-2 (UniProt accession P60568, 38.56 coverage, 3 peptides). Urine without nanoparticle harvesting is shown in the lane labeled “Raw”.
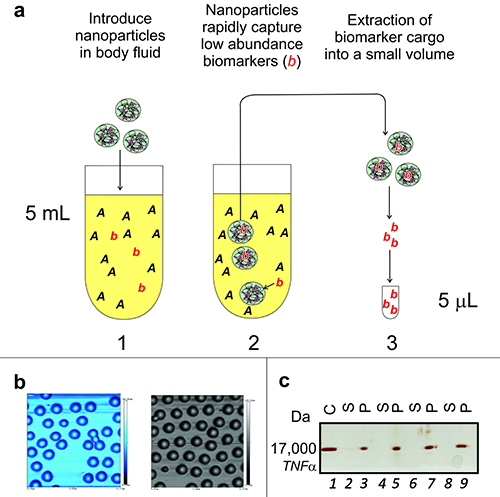 Figure 1. Nanoparticles harvest and concentrate low abundance proteins
from complex biological fluids. (A) Workflow for harvesting
proteins. Total processing time is approximately 1.5 hr. Proteins in solution are
concentrated from blood, serum, plasma, urine, sweat, saliva, or other body fluids
(1,000-fold concentration depicted). (B) Batch to batch comparison
showing the uniform size of nanoparticles. Atomic Force Microscopy on mica shows
uniformity of size (0.7 µm diameter) and absence of clumping in two batches of
nanoparticles. (C) 1-D gel electrophoresis, with subsequent silver
staining, of Tumor Necrosis Factor alpha (TNFα) sequestered from plasma. Please click here to view a larger version of this figure.
Figure 1. Nanoparticles harvest and concentrate low abundance proteins
from complex biological fluids. (A) Workflow for harvesting
proteins. Total processing time is approximately 1.5 hr. Proteins in solution are
concentrated from blood, serum, plasma, urine, sweat, saliva, or other body fluids
(1,000-fold concentration depicted). (B) Batch to batch comparison
showing the uniform size of nanoparticles. Atomic Force Microscopy on mica shows
uniformity of size (0.7 µm diameter) and absence of clumping in two batches of
nanoparticles. (C) 1-D gel electrophoresis, with subsequent silver
staining, of Tumor Necrosis Factor alpha (TNFα) sequestered from plasma. Please click here to view a larger version of this figure.
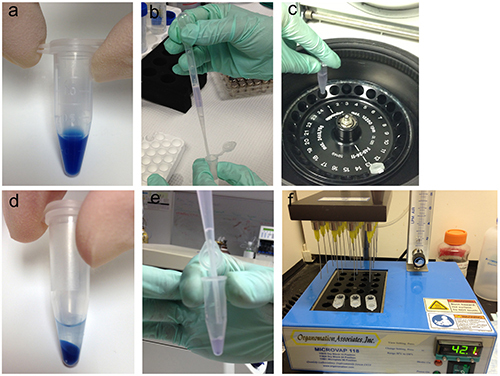 Figure 2. Nanoparticle harvesting procedure from plasma/serum for
downstream mass spectrometry analysis. (A) Nanoparticles
remain suspended in solution (Poly(NIPAm/dye) shown). (B) Nanoparticles
are added to the diluted plasma sample. (C) The nanoparticle-plasma
suspension is spun in a centrifuge to separate the nanoparticles from the
supernatant. (D) Post centrifugation, the nanoparticles form a distinct
pellet in the bottom of the tube. (E) After washing the nanoparticles,
the eluate containing the harvested proteins is removed and saved for downstream
analysis. (F) The eluate is dried under nitrogen flow in an evaporator
at 42.1 °C, air flow 8, for 1 - 2 hr prior to mass spectrometry. Please click here to view a larger version of this figure.
Figure 2. Nanoparticle harvesting procedure from plasma/serum for
downstream mass spectrometry analysis. (A) Nanoparticles
remain suspended in solution (Poly(NIPAm/dye) shown). (B) Nanoparticles
are added to the diluted plasma sample. (C) The nanoparticle-plasma
suspension is spun in a centrifuge to separate the nanoparticles from the
supernatant. (D) Post centrifugation, the nanoparticles form a distinct
pellet in the bottom of the tube. (E) After washing the nanoparticles,
the eluate containing the harvested proteins is removed and saved for downstream
analysis. (F) The eluate is dried under nitrogen flow in an evaporator
at 42.1 °C, air flow 8, for 1 - 2 hr prior to mass spectrometry. Please click here to view a larger version of this figure.
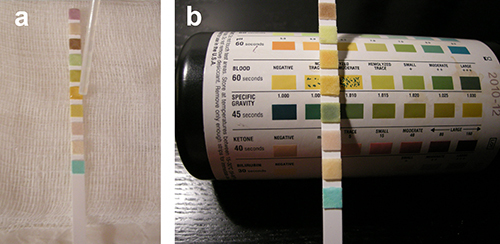 Figure 3. Urinalysis using multi-analyte reagent strips for specimen
quality control. (A) Each pad on the reagent strip is impregnated with
chemicals, enzymes, and/or indicator dyes which react with a different urine analyte
(e.g., glucose, bilirubin, ketone, specific gravity, blood, pH,
protein, urobilinogen, nitrite, and/or leukocyte esterase). Urine is clarified by
centrifugation. A drop of urine is added to each pad on the reagent strip.
(B) The color of each pad is compared visually to the color blocks
on the container, at the specified time. Please click here to view a larger version of this figure.
Figure 3. Urinalysis using multi-analyte reagent strips for specimen
quality control. (A) Each pad on the reagent strip is impregnated with
chemicals, enzymes, and/or indicator dyes which react with a different urine analyte
(e.g., glucose, bilirubin, ketone, specific gravity, blood, pH,
protein, urobilinogen, nitrite, and/or leukocyte esterase). Urine is clarified by
centrifugation. A drop of urine is added to each pad on the reagent strip.
(B) The color of each pad is compared visually to the color blocks
on the container, at the specified time. Please click here to view a larger version of this figure.
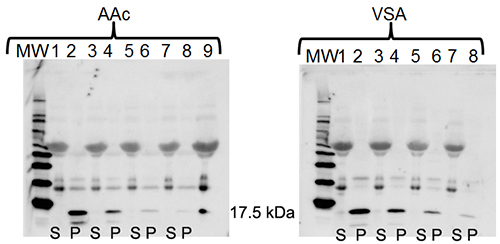 Figure 4. Harvesting IL-17 from plasma using
core (AAc) or core-shell (VSA) nanoparticles.1-D gel
electrophoresis and western blotting with anti-IL-17 demonstrates the ability of
Acrylic Acid (AAc) and VSA core shell nanoparticles to harvest various
concentrations of IL-17 from plasma. (S = supernatant after nanoparticle harvesting,
P = nanoparticle eluate. AAc particles: Lanes 1 & 2 = 1 ng/µl IL-17, Lanes 3
& 4 = 0.50 ng/µl, Lanes 5 & 6 = 0.25ng/µl, Lanes 7 & 8 = 0.125 ng/µl,
Lane 9 = IL-17; VSA particles: Lanes 1 & 2 = 1 ng/µl IL-17, Lanes 3 & 4 =
0.50 ng/µl, Lanes 5 & 6 = 0.25 ng/µl, Lanes 7 & 8 = 0.125 ng/µl) Please click here to view a larger version of this figure.
Figure 4. Harvesting IL-17 from plasma using
core (AAc) or core-shell (VSA) nanoparticles.1-D gel
electrophoresis and western blotting with anti-IL-17 demonstrates the ability of
Acrylic Acid (AAc) and VSA core shell nanoparticles to harvest various
concentrations of IL-17 from plasma. (S = supernatant after nanoparticle harvesting,
P = nanoparticle eluate. AAc particles: Lanes 1 & 2 = 1 ng/µl IL-17, Lanes 3
& 4 = 0.50 ng/µl, Lanes 5 & 6 = 0.25ng/µl, Lanes 7 & 8 = 0.125 ng/µl,
Lane 9 = IL-17; VSA particles: Lanes 1 & 2 = 1 ng/µl IL-17, Lanes 3 & 4 =
0.50 ng/µl, Lanes 5 & 6 = 0.25 ng/µl, Lanes 7 & 8 = 0.125 ng/µl) Please click here to view a larger version of this figure.
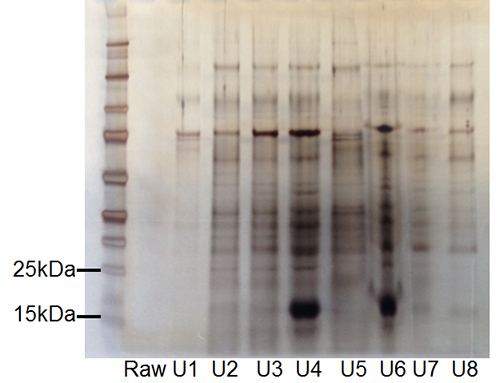 Figure 5. Recovery of
Mycobacterium tuberculosis
antigen and cytokine IL-2 from urine using hydrogel
nanoparticles.Silver staining following 1-D gel
electrophoresis of urine samples. Samples in lanes U4 and U6 represent eluates from
urine samples containing recombinant ESAT-6 and IL-2, clearly showing prominent
bands at 15 kDa. Mass spectrometry of the eluates identified ESAT-6 (UniProt
accession POA567, 84.0 coverage, 9 peptides) and IL-2 (UniProt accession P60568,
38.56 coverage, 3 peptides). (RAW=urine without nanoparticle harvesting; U1, U2, U3,
U5, U7, U8 = urine lacking recombinant ESAT-6 and IL-2 following nanoparticle
harvesting). Please click here to view a larger version of this figure.
Figure 5. Recovery of
Mycobacterium tuberculosis
antigen and cytokine IL-2 from urine using hydrogel
nanoparticles.Silver staining following 1-D gel
electrophoresis of urine samples. Samples in lanes U4 and U6 represent eluates from
urine samples containing recombinant ESAT-6 and IL-2, clearly showing prominent
bands at 15 kDa. Mass spectrometry of the eluates identified ESAT-6 (UniProt
accession POA567, 84.0 coverage, 9 peptides) and IL-2 (UniProt accession P60568,
38.56 coverage, 3 peptides). (RAW=urine without nanoparticle harvesting; U1, U2, U3,
U5, U7, U8 = urine lacking recombinant ESAT-6 and IL-2 following nanoparticle
harvesting). Please click here to view a larger version of this figure.
| Protein name | GI protein number | Concentration in serum / plasma [ng/ml] | Reference |
| abl-interactor 1 isoform a | 61743942 | Unknown | |
| AMP-activated protein kinase gamma2 subunit isoform a | 33186925 | Unknown | |
| Cas-Br-M (murine) ecotropic retroviral transforming sequence | 52426745 | Unknown | |
| chemokine (C-C motif) ligand 18 (pulmonary and activation-regulated) | 4506831 | 30 | 34 |
| chemokine (C-C motif) ligand 5 | 22538814 | 1.5 | 31 |
| chondroadherin | 153251229 | Unknown | |
| chromosome 16 open reading frame 80 | 8392875 | Unknown | |
| Consortin | 213021160 | Unknown | |
| defensin, alpha 4, corticostatin | 4503303 | Unknown | |
| EF-hand domain family, member D2 | 20149675 | Unknown | |
| glycoprotein Ib (platelet), beta polypeptide | 4504073 | Unknown | |
| guanine nucleotide binding protein (G protein), q polypeptide | 40254462 | Unknown | |
| Heparanase | 148746204 | Unknown | |
| insulin-like growth factor 2 (somatomedin A) | 189083846 | 100 | 30 |
| lacritin | 15187164 | Unknown | |
| leukocyte cell-derived chemotaxin 2 | 59806345 | 200,000 | 33 |
| lipocalin 2 (oncogene 24p3) | 38455402 | 0.05 | 28 |
| Monoglyceride lipase, isoform CRA_b | 6005786 | Unknown | |
| N-ethylmaleimide-sensitive factor attachment protein, alpha | 47933379 | Unknown | |
| one cut homeobox 2 | 119220564 | Unknown | |
| outer dense fiber of sperm tails 4 | 171184433 | Unknown | |
| palate, lung and nasal epithelium associated | 18765705 | Unknown | |
| peptidoglycan recognition protein 2 | 156616294 | Unknown | |
| platelet-derived growth factor alpha polypeptide | 77695917 | 4 | 29 |
| Protein CASC3 | 15721939 | Unknown | |
| RAB27B, member RAS oncogene family | 5729997 | Unknown | |
| Ras association (RalGDS/AF-6) domain family 6 isoform a | 29789443 | Unknown | |
| ras-related GTP-binding protein RAB10 | 33695095 | Unknown | |
| ring finger protein 166 | 30520320 | Unknown | |
| serum deprivation response (phosphatidylserine binding protein) | 4759082 | Unknown | |
| solute carrier family 33 (acetyl-CoA transporter), member 1 | 4757708 | Unknown | |
| ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 isoform a | 27765091 | 15 | 32 |
| thymosin-like 3 | 34013530 | Unknown | |
| transducin-like enhancer of split 3 (E(sp135) homolog, Drosophila) | 157384982 | Unknown | |
| transglutaminase 1 | 4507475 | Unknown | |
| WD repeat domain 1 | 9257257 | Unknown | |
| WD repeat domain 91 | 222080092 | Unknown | |
| xin actin-binding repeat containing 2 | 119372317 | Unknown |
Table 1. Example low abundance proteins harvested by nanoparticles from serum/plasma. (PeptideAtlas peptidome mass spectrometry)28-34. Adapted with permissionfrom reference 13(Tamburro, D. et al.Multi-functional core-shell nanoparticles: Discovery of previously invisible biomarkers. J Am Chem Soc, 133, 19178-19188, (2011).) Copyright 2011 American Chemical Society.
Discussion
Clinical Relevance
A serum or plasma sample is thought to contain low-abundance circulating proteins and peptides which can provide a rich source of information regarding the state of the organism as a whole. Despite the promise of serum proteomics, there are three fundamental and serious physiologic barriers thwarting biomarker discovery and translation to clinical benefit10, 11, 16, 25.
1. Important diagnostic biomarkers may exist in extremely low-abundance (concentration) in blood. Early stage diseased tissue, such as pre-metastatic cancer lesions, may constitute less than a few mm3. Biomarkers shed into the circulation from such a small tissue volume will become highly diluted in the entire blood volume. Relevant analytes may exist below the detection limits of mass spectrometry and conventional immunoassays.
2. Low abundance biomarkers/proteins are masked by the presence of high abundant proteins such as albumin and immunoglobulin, which represent up to 90% of the plasma proteome14.
3. Proteins and peptides are susceptible to degradation by endogenous and exogenous proteinases following venipuncture and sample transport/storage. Protein degradation can lead to false positive or false negative results35.
Hydrogel nanoparticles were developed as porous, buoyant, polymers containing anthraquinonetriazine dyes and/or vinylsulfonic acid shells for protein and peptide harvesting and preservation in body fluids11, 13. Our group synthesized and tested hydrogel nanoparticles functionalized with a plethora of organic dyes (i.e., sulfonated and non-sulfonatedanthraquinonetriazine dyes) and showed the preferential affinities for several protein analytes11, 13. We also established protocols for biomarker discovery that use hydrogel nanoparticles with lymph, saliva, cerebrospinal fluid, sweat, urine, plasma, blood, or serum specimens11, 13, 23, 24, 26.
Optimizing the nanoparticle harvesting parameters prior to harvesting proteins from precious clinical/research specimens is necessary for ideal results. The amount of protein in the samples should be quantified to determine the optimum ratio of nanoparticles to sample volume. For analytes of unknown concentration, a dose-response curve using recombinant proteins provides information regarding the limits of detection. If there is adequate sample, various types of nanoparticles can be used to determine the ideal nanoparticle for the analyte and specimen.
Urinalysis prior to nanoparticle harvesting provides a urine sample quality control check. The affinity of the nanoparticle dye bait depends on the isoelectric point of the protein of interest and the pH of the surrounding medium. Urine pH between 5.5 and 7.0 is optimal for protein harvesting with the poly(NIPAm/dye) nanoparticles. The presence of hemolyzed or intact red blood cells (1+ blood on the reagent strip) does not interfere with nanoparticle harvesting.
In this protocol we focused on the application of the hydrogel nanoparticles to harvest proteins from plasma and urine. Future applications could comprise other body fluids such as vitreous of the eye, or synovial fluid. Potential in vivo applications can be envisioned for the future in which nanoparticles are injected into diseased tissue to collect biomolecules. Future work will also be focused on the development of new devices for the capture and preservation of biomarkers, such as a nanoparticle-based skin patch for the analysis of the skin transudate proteome.
Disclosures
Benjamin Espina is an employee of Ceres Nanosciences, Inc. that produces reagents used in this Article. Lance Liotta, Alessandra Luchini and Virginia Espina hold patents (US patent 7,935,518 and/or 8,497,137) on the nanoparticle technology used in this Article. As university employees they are entitled to receive royalty from these patents per university policy. Lance Liotta and Alessandra Luchini are shareholders in Ceres Nanosciences and serve on the Scientific Advisory Board.
Acknowledgments
Michael Henry, Dublin City University, kindly assisted with the data collection and analysis shown in Figure 5. This work was supported partially by (1) George Mason University, (2) the Italian IstitutoSuperiore di Sanita’ in the framework of the Italy/USA cooperation agreement between the U.S. Department of Health and Human Services, George Mason University, and the Italian Ministry of Public Health, (3) NIH, IMAT program grants 1R21CA137706-01 and 1R33CA173359-01 to LAL, and (4) Ceres Nanosciences, Inc.
References
- Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422(6928):198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- Gerszten RE, et al. Challenges in translating plasma proteomics from bench to bedside: update from the NHLBI Clinical Proteomics Programs. Am J Physiol Lung Cell Mol Physiol. 2008;295(1):L16–L22. doi: 10.1152/ajplung.00044.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrell K, et al. Analysis of low-abundance, low-molecular-weight serum proteins using mass spectrometry. J Biomol Tech. 2004;15(4):238–248. [PMC free article] [PubMed] [Google Scholar]
- Petricoin EF, Belluco C, Araujo RP, Liotta LA. The blood peptidome: a higher dimension of information content for cancer biomarker discovery. Nat Rev Cancer. 2006;6(12):961–967. doi: 10.1038/nrc2011. [DOI] [PubMed] [Google Scholar]
- Camerini S, Polci ML, Liotta LA, Petricoin EF, Zhou W. A method for the selective isolation and enrichment of carrier protein-bound low-molecular weight proteins and peptides in the blood. Proteomics Clin Appl. 2007;1(2):176–184. doi: 10.1002/prca.200600618. [DOI] [PubMed] [Google Scholar]
- Geho D, et al. Fractionation of serum components using nanoporous substrates. Bioconjug Chem. 2006;17(3):654–661. doi: 10.1021/bc0503364. [DOI] [PubMed] [Google Scholar]
- Sennels L, et al. Proteomic analysis of human blood serum using peptide library beads. J Proteome Res. 2007;6(10):4055–4062. doi: 10.1021/pr070339l. [DOI] [PubMed] [Google Scholar]
- Zheng X, Baker H, Hancock WS. Analysis of the low molecular weight serum peptidome using ultrafiltration and a hybrid ion trap-Fourier transform mass spectrometer. J Chromatogr A. 2006;1120(1-2):173–184. doi: 10.1016/j.chroma.2006.01.098. [DOI] [PubMed] [Google Scholar]
- Marshall J, et al. Processing of serum proteins underlies the mass spectral fingerprinting of myocardial infarction. J Proteome Res. 2003;2(4):361–372. doi: 10.1021/pr030003l. [DOI] [PubMed] [Google Scholar]
- Fredolini C, et al. Concentration and Preservation of Very Low Abundance Biomarkers in Urine, such as Human Growth Hormone (hGH), by Cibacron Blue F3G-A Loaded Hydrogel Particles. Nano Res. 2008;1(6):502–518. doi: 10.1007/s12274-008-8054-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchini A, et al. Smart hydrogel particles: biomarker harvesting: one-step affinity purification, size exclusion, and protection against degradation. Nano Lett. 2008;8(1):350–361. doi: 10.1021/nl072174l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patanarut A, et al. Synthesis and characterization of hydrogel particles containing Cibacron Blue F3G-A. Colloids Surf A Physicochem Eng Asp. 2010;362(1-3):8–19. doi: 10.1016/j.colsurfa.2010.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamburro D, et al. Multifunctional core-shell nanoparticles: discovery of previously invisible biomarkers. J Am Chem Soc. 2011;133(47):19178–19188. doi: 10.1021/ja207515j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1(11):845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- Fredolini C, et al. Nanoparticle technology: amplifying the effective sensitivity of biomarker detection to create a urine test for hGH. Drug Test Anal. 2009;1(9-10):447–454. doi: 10.1002/dta.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo C, et al. Core-shell hydrogel particles harvest, concentrate and preserve labile low abundance biomarkers. PLoS One. 2009;4(3):e4763. doi: 10.1371/journal.pone.0004763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchini A, et al. Nanoparticle technology: addressing the fundamental roadblocks to protein biomarker discovery. Curr Mol Med. 2010;10(2):133–141. doi: 10.2174/156652410790963268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchini A, et al. Application of Analyte Harvesting Nanoparticle Technology to the Measurement of Urinary HGH in Healthy Individuals. J Sports Med Doping Stud. 2012;2(6) doi: 10.4172/2161-0673.1000e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eslami A, Lujan J, Western blotting: sample preparation to detection. J Vis Exp. 2010. [DOI] [PMC free article] [PubMed]
- Gallagher S, Chakavarti D. Immunoblot analysis. J Vis Exp. 2008. [DOI] [PMC free article] [PubMed]
- Penna A, Cahalan M. Western Blotting using the Invitrogen NuPage Novex Bis Tris minigels. J Vis Exp. 2007. p. 264. [DOI] [PMC free article] [PubMed]
- Bosch J, et al. Analysis of urinary human growth hormone (hGH) using hydrogel nanoparticles and isoform differential immunoassays after short recombinant hGH treatment: preliminary results. J Pharm Biomed Anal. 2013;85:194–197. doi: 10.1016/j.jpba.2013.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredolini C, et al. Investigation of the ovarian and prostate cancer peptidome for candidate early detection markers using a novel nanoparticle biomarker capture technology. AAPS J. 2010;12(4):504–518. doi: 10.1208/s12248-010-9211-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo C, et al. A novel biomarker harvesting nanotechnology identifies Bak as a candidate melanoma biomarker in serum. Exp Dermatol. 2010;20(1):29–34. doi: 10.1111/j.1600-0625.2010.01187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchini A, Longo C, Espina V, Petricoin EF, 3rd, Liotta LA. Nanoparticle technology: Addressing the fundamental roadblocks to protein biomarker discovery. J Mater Chem. 2009;19(29):5071–5077. doi: 10.1039/b822264a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas TA, et al. The use of hydrogel microparticles to sequester and concentrate bacterial antigens in a urine test for Lyme disease. Biomaterials. 2010;32(4):1157–1166. doi: 10.1016/j.biomaterials.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash A, et al. Interlaboratory reproducibility of selective reaction monitoring assays using multiple upfront analyte enrichment strategies. J Proteome Res. 2012;11(8):3986–3995. doi: 10.1021/pr300014s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KM, et al. Implication of lipocalin-2 and visfatin levels in patients with coronary heart disease. Eur J Endocrinol. 2008;158(2):203–207. doi: 10.1530/EJE-07-0633. [DOI] [PubMed] [Google Scholar]
- Hughes AD, Clunn GF, Refson J, Demoliou-Mason C. Platelet-derived growth factor (PDGF): actions and mechanisms in vascular smooth muscle. Gen Pharmacol. 1996;27(7):1079–1089. doi: 10.1016/s0306-3623(96)00060-2. [DOI] [PubMed] [Google Scholar]
- Izycki T, et al. Serum levels of IGF-I and IGF-II in patients with lung cancer during chemotherapy. Exp Oncol. 2004;26(4):316–319. [PubMed] [Google Scholar]
- Jalosinski M, Karolczak K, Mazurek A, Glabinski A. The effects of methylprednisolone and mitoxantrone on CCL5-induced migration of lymphocytes in multiple sclerosis. Acta Neurol Scand. 2008;118(2):120–125. doi: 10.1111/j.1600-0404.2008.00998.x. [DOI] [PubMed] [Google Scholar]
- Kitazume S, et al. Molecular insights into beta-galactoside alpha2,6-sialyltransferase secretion in vivo. Glycobiology. 2009;19(5):479–487. doi: 10.1093/glycob/cwp003. [DOI] [PubMed] [Google Scholar]
- Saito T, et al. Increase in hepatic NKT cells in leukocyte cell-derived chemotaxin 2-deficient mice contributes to severe concanavalin A-induced hepatitis. J Immunol. 2004;173(1):579–585. doi: 10.4049/jimmunol.173.1.579. [DOI] [PubMed] [Google Scholar]
- Struyf S, et al. PARC/CCL18 is a plasma CC chemokine with increased levels in childhood acute lymphoblastic leukemia. Am J Pathol. 2003;163(5):2065–2075. doi: 10.1016/S0002-9440(10)63564-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael IP, et al. Human tissue kallikrein 5 is a member of a proteolytic cascade pathway involved in seminal clot liquefaction and potentially in prostate cancer progression. J Biol Chem. 2006;281(18):12743–12750. doi: 10.1074/jbc.M600326200. [DOI] [PubMed] [Google Scholar]


