Abstract
Gene expression variation is a major contributor to phenotypic diversity within species and is thought to play an important role in adaptation. However, examples of adaptive regulatory polymorphism are rare, especially those that have been characterized at both the molecular genetic level and the organismal level. In this study, we perform a functional analysis of the Drosophila melanogaster CG9509 enhancer, a cis-regulatory element that shows evidence of adaptive evolution in populations outside the species’ ancestral range in sub-Saharan Africa. Using site-directed mutagenesis and transgenic reporter gene assays, we determined that 3 single nucleotide polymorphisms are responsible for the difference in CG9509 expression that is observed between sub-Saharan African and cosmopolitan populations. Interestingly, while 2 of these variants appear to have been the targets of a selective sweep outside of sub-Saharan Africa, the variant with the largest effect on expression remains polymorphic in cosmopolitan populations, suggesting it may be subject to a different mode of selection. To elucidate the function of CG9509, we performed a series of functional and tolerance assays on flies in which CG9509 expression was disrupted. We found that CG9509 plays a role in larval growth and influences adult body and wing size, as well as wing loading. Furthermore, variation in several of these traits was associated with variation within the CG9509 enhancer. The effect on growth appears to result from a modulation of active ecdysone levels and expression of growth factors. Taken together, our findings suggest that selection acted on 3 sites within the CG9509 enhancer to increase CG9509 expression and, as a result, reduce wing loading as D. melanogaster expanded out of sub-Saharan Africa.
Author summary
Much of the phenotypic variation that is observed within species is thought to be caused by variation in gene expression. Variants within cis-regulatory elements, which affect the expression of nearby genes within the same DNA strand, are thought to be an abundant resource upon which natural selection can act. Understanding the functional consequences of adaptive cis-regulatory changes is important, as it can help elucidate the mechanisms underlying phenotypic evolution in general and provide insight into the development and maintenance of biodiversity. However, functional analyses of these types of changes remain rare. Here we present a functional analysis of an adaptively evolving enhancer element of a D. melanogaster gene called CG9509, of previously unknown function. We show that 3 single nucleotide polymorphisms located within the enhancer of this gene are responsible for an increase in CG9509 expression in cosmopolitan populations (outside of south and central Africa) relative to sub-Saharan populations, which include ancestral populations. We further show that CG9509 is involved in the regulation of growth rate and body size determination and propose that the CG9509 enhancer underwent positive selection to reduce wing loading as the species expanded out of sub-Saharan Africa.
Introduction
Gene expression variation is extensive both within and between species and is believed to underlie much of the phenotypic diversity observed among species, as well as among populations of the same species [1–2]. Furthermore, expression variation is thought to provide an abundant source of material for adaptation, as alterations in gene expression are more easily fine-tuned on a temporal and tissue-specific scale than changes in protein structure [3–4]. In particular, cis-regulatory elements, which are adjacent to genes and directly affect their expression, are thought to be frequent targets of adaptive evolution [2–6]. Despite this prediction, examples of adaptive cis-regulatory changes remain comparatively rare, although the number of such examples continues to grow [7–16]. The discrepancy between the predicted abundance and actual instances of identified adaptive cis-regulatory divergence is likely in part due to the difficulty in detecting regulatory adaptation, as well as in determining its effect on an organismal phenotype that may be the target of selection. Even in some of the best-studied species, the function of many genes remains unknown, and alterations in those with known functions often have pleiotropic effects, making it difficult to determine the link between an expression change and an adaptive organismal phenotype. As more instances of adaptive cis-regulatory evolution are uncovered, it is important to identify the genetic and molecular mechanisms that underlie them, which can help to further our understanding of the mechanisms of phenotypic evolution and shed light on the origins of biodiversity [17]. However, studies performing in-depth functional analyses of individual adaptively evolving cis-regulatory elements remain even more rare than those documenting adaptive cis-regulatory divergence (e.g., [18–21]).
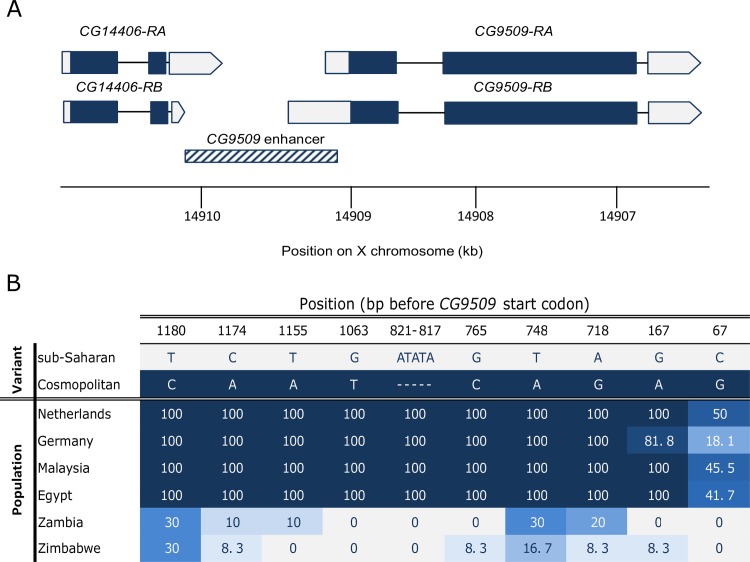
Transcriptomic methods have proven effective at identifying putatively adaptive alterations in gene expression within and between species [22–28]. CG9509 is a gene initially identified as a candidate for adaptive cis-regulatory divergence through one such study that compared expression between a derived, European and an ancestral, sub-Saharan African (henceforth sub-Saharan) population of D. melanogaster [24]. Until now, the function of CG9509 has remained unknown, although it has been predicted to have oxidoreductase activity [29] and/or play a role in ecdysteroid metabolism [30]. Adult CG9509 expression was found to be 2–3-fold higher in the European population than in the sub-Saharan population (Fig 1A) [12,24], and this expression difference extends to other cosmopolitan (here defined as populations outside of south and central Africa) and sub-Saharan populations [31]. Transgenic reporter gene experiments revealed that variation within a 1.2-kb cis-regulatory element upstream of the gene (referred to here as the CG9509 enhancer, Fig 2A) can account fully for the expression divergence and shows evidence of a selective sweep in cosmopolitan populations [12,31]. This suggests that positive selection acted on the CG9509 enhancer to increase CG9509 expression after D. melanogaster’s expansion out of sub-Saharan Africa, which is estimated to have occurred approximately 15,000 years ago [32–34], but before the separation of European and Asian populations approximately 2,500–5,000 years ago [31,35]. Within the CG9509 enhancer, there are 9 single nucleotide polymorphisms (SNPs) and 1 insertion/deletion (indel) polymorphism (Fig 2B) that show large frequency differences between the populations and are candidates for the target(s) of selection responsible for the expression divergence. In all cases, the cosmopolitan variant is inferred to be the derived state, while the sub-Saharan variant is inferred to be ancestral [31].
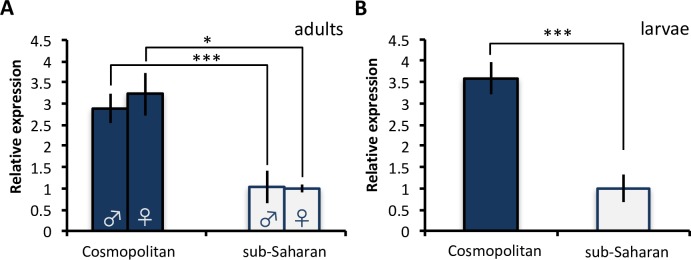
Fig 1. CG9509 expression in cosmopolitan and sub-Saharan African D. melanogaster.
Relative expression levels as determined by quantitative reverse transcription PCR (qRT-PCR). Shown are (A) adult males (data from [31]) and females (data from [12]) and (B) late wandering third instar larvae (N = 21–35 isofemale strains per type with 2 biological replicates per strain). Blue bars represent cosmopolitan flies, and white bars represent sub-Saharan flies. Underlying data can be found in S1 Data. Error bars show the standard error of the mean. Differences between populations were assessed with a t test. *P < 0.05, ***P < 0.005.
Fig 2. The CG9509 gene region.
(A) Schematic of the CG9509 gene and enhancer. Blue boxes represent exons, and white boxes represent untranslated regions, with the pointed ends indicating the direction of transcription. RNA sequencing (RNA-seq) data [28] indicate that the CG9509-RA transcript is more abundant, accounting for >99% of all transcripts. The CG9509 enhancer is indicated by a hatched box. (B) Single nucleotide polymorphisms (SNPs) and indels in the CG9509 enhancer with >10% frequency in cosmopolitan populations. Cosmopolitan sequence variants are indicated in blue, and sub-Saharan variants are indicated in white, with lighter shades of blue indicating a mixture of both variants in the population. For each population, the observed frequency of the cosmopolitan variant (in percent) is shown [31].
In this study, we use site-directed mutagenesis and transgenic reporter genes to determine the effect of individual SNP and indel variants within the CG9509 enhancer on gene expression. We also use RNA interference (RNAi) and a newly discovered CG9509 hypomorph allele to reveal some of CG9509’s previously unknown biological functions. We find that 3 SNPs within the CG9509 enhancer contribute to the expression divergence seen between cosmopolitan and sub-Saharan alleles. Interestingly, 2 SNPs that have a small effect on expression are fixed in cosmopolitan populations and appear to have been the targets of a selective sweep, while the SNP with the largest effect on expression is at intermediate frequency in cosmopolitan populations, suggesting that another type of selection may be acting on this site. We also show that CG9509 expression influences larval growth and thus plays a role in determining adult body size and wing loading (i.e., the ratio of body mass to wing area). Furthermore, the genetic variants influencing CG9509 expression are associated with variation in these phenotypic traits. Our results suggest that selection on the 3 SNPs within the CG9509 enhancer occurred in order to reduce wing loading outside of sub-Saharan Africa.
Results
Larval CG9509 expression divergence
Previous studies focused solely on adult CG9509 expression variation [12,31]. To determine if the adult expression pattern is established earlier in development, we surveyed larval CG9509 expression. Because D. melanogaster developmental gene expression is highly dynamic with a high transcriptional turnover, even among stages that are only a few hours apart [36], we focused our expression analysis on a well-established larval stage in order to ensure that any observed expression divergence is due to population divergence rather than developmental stage variation. To this end, we surveyed CG9509 expression in late wandering third instar larvae of 3 cosmopolitan populations (the Netherlands, Egypt, and Malaysia) and 2 sub-Saharan populations (Zimbabwe and Zambia). Similar to adults (Fig 1A), larval CG9509 expression in cosmopolitan populations was significantly higher than in sub-Saharan populations by 3–5.5-fold (t test, P < 5 × 10−4, Fig 1B and S1 Fig).
Functional analysis of sequence variants in the CG9509 enhancer
To determine which variants in the CG9509 enhancer (Fig 2B) contribute to the expression divergence between cosmopolitan and sub-Saharan D. melanogaster (Fig 1 and S1 Fig), we created a series of transgenic reporter gene constructs that were introduced into the D. melanogaster genome. Because these reporter gene constructs were tested in a common genomic background, our results should be free from the confounding effects of trans-acting factors. Briefly, a cosmopolitan and a sub-Saharan enhancer allele were cloned in front of a LacZ reporter gene, and 6 sites of interest at positions 1174, 1155, 1063, 821–817, 765, and 67 (Fig 2B) were mutated individually and in various combinations (Fig 3). The tested sites were chosen as those at which the derived variant was fixed in all cosmopolitan populations, but at a frequency ≤10% in sub-Saharan populations. Position 67, at which the derived variant is at intermediate frequency in cosmopolitan populations but absent from sub-Saharan populations (Fig 2B), was also tested because it had previously been associated with CG9509 expression variation [31,37]. Mutations were first introduced into the cosmopolitan enhancer allele, changing the nucleotide(s) to the ancestral (sub-Saharan) state. Sites found to have an effect on expression in the cosmopolitan background were then mutated to the cosmopolitan state in the sub-Saharan background. Adult and larval expression driven by the cosmopolitan enhancer was 3–4-fold higher than that driven by the sub-Saharan enhancer (Fig 3), which is in line with the expression divergence found in natural populations (Fig 1 and S1 Fig). Thus, the CG9509 enhancer can account for nearly all of the expression divergence between cosmopolitan and sub-Saharan larvae and adults.
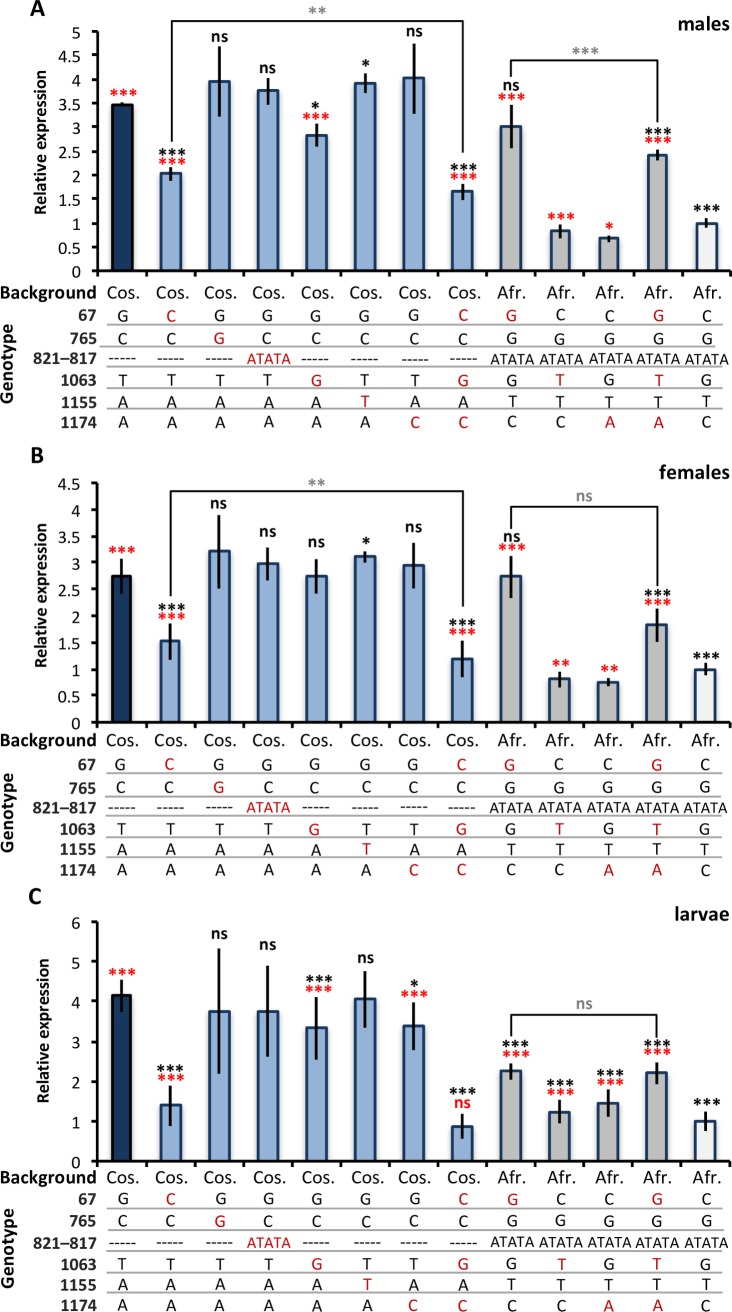
Fig 3. Transgenic reporter gene expression.
Reporter gene expression in (A) males, (B) females, and (C) larvae (N = 4–8 per strain and sex or stage). Expression of the LacZ reporter gene was measured with a β-galactosidase enzymatic assay. Blue bars indicate expression driven by a wild-type cosmopolitan (Cos.) enhancer, while white bars show expression driven by a wild-type sub-Saharan African (Afr.) enhancer. Light blue bars indicate expression driven by the enhancer after mutations were introduced into a cosmopolitan background. Dark gray bars show expression driven by the enhancer after mutations were introduced in a sub-Saharan African background. “Genotype” indicates the nucleotides at positions 1174, 1155, 1063, 821–817, 765, and 67 before the CG9509 start codon, respectively. Mutated sites are shown in red. Underlying data can be found in S1 Data. Significance was assessed using a t test, and a Bonferroni multiple test correction was applied. Significance is represented by black asterisks for comparisons to the original cosmopolitan enhancer, red asterisks for the original sub-Saharan enhancer, and gray asterisks for comparisons between mutated enhancers. ns, not significant; *P < 0.05, **P < 0.01, ***P < 0.005.
One SNP accounts for most of the expression divergence in adults
We determined that the SNP at position 67 (Fig 2B) accounts for the majority of adult expression divergence between cosmopolitan and sub-Saharan populations. The sub-Saharan “C” variant of this SNP is at intermediate frequency in cosmopolitan populations and, when introduced into the cosmopolitan enhancer background, leads to an approximately 45% reduction in expression (Fig 3A and 3B). However, expression was still significantly higher than that driven by the sub-Saharan enhancer (Fig 3A and 3B). When the cosmopolitan “G” variant was introduced into the sub-Saharan background, expression increased 3-fold (Fig 3A and 3B), resulting in expression equivalent to cosmopolitan levels (Fig 3A and 3B). Thus, while the derived variant “G” is sufficient to produce adult cosmopolitan expression in a sub-Saharan background, background-specific epistatic effects, presumably due to other site(s) within the cosmopolitan enhancer, appear to prevent a complete return to sub-Saharan expression when the “C” variant is introduced into a cosmopolitan background.
To better understand how the variant at position 67 affects adult expression, we performed an ANOVA using sex, background, the variant at position 67, and the interaction between the variant at position 67 and background, sex, and the other tested sites within the CG9509 enhancer. We found that background, sex, and the variant at position 67 had a highly significant effect on expression (P < 0.001, S1 Table); however, the interaction between the variant at position 67 and background was not significant (P > 0.1, S1 Table). On the other hand, the interaction between the variant at position 67 and sex (P < 0.005, S1 Table), as well as 4 of the 5 other tested variants, was significant (P < 0.05 for each, S1 Table), suggesting that (1) our inability to recreate sub-Saharan expression in a cosmopolitan background is likely due to epistatic effects with other sites in the CG9509 enhancer haplotype rather than the genomic background as a whole and (2) epistatic interactions between sites in regulatory regions may be common. Indeed, we detected several sex-, background-, and/or stage-specific effects in our reporter gene assays (Fig 3). Next, we calculated partial eta squared (η2p), which is a standardized measure of effect size in which effect sizes on the magnitude of approximately 0.01, 0.06, and 0.14 are generally considered to represent small, medium, and large effect sizes, respectively [38]. All of the tested factors had a large effect on expression (η2p > 0.12, S1 Table). The largest effect was for the variant at position 67 (η2p = 0.975, S1 Table), followed by sex (η2p = 0.896, S1 Table) and background (η2p = 0.772, S1 Table), suggesting that the variant at position 67 is the strongest predictor of adult CG9509 expression.
Three SNPs contribute to the expression divergence in larvae
We identified 3 SNPs in the CG9509 enhancer at positions 67, 1063, and 1174 (Fig 2B) that can account for the majority of the expression divergence between cosmopolitan and sub-Saharan African larvae. Of the 3, the SNP at position 67, which accounts for the majority of adult expression divergence, has the largest individual effect on expression (Fig 3). Introduction of the cosmopolitan “G” variant into a sub-Saharan background increased expression by 2.25-fold (Fig 3C), while introduction of the sub-Saharan “C” variant into the cosmopolitan background reduced expression by 2-fold (Fig 3C). At position 1063, introducing the cosmopolitan “T” variant into the sub-Saharan background increased expression by 25% (Fig 3C), while introducing a sub-Saharan “G” variant into the cosmopolitan background reduced expression by 20% (Fig 3C). Similarly, at position 1174, introducing a sub-Saharan “C” variant into the cosmopolitan background reduced expression by 20% (Fig 3C), while introducing a cosmopolitan “A” variant into the sub-Saharan background increased expression by 50% (Fig 3C). When the sub-Saharan variants at all 3 positions were introduced together in the cosmopolitan background, expression was reduced to levels equivalent to sub-Saharan expression (Fig 3C). When the cosmopolitan variants at all 3 positions were introduced together in the sub-Saharan background, expression increased 2.2-fold; however, it remained 50% lower than cosmopolitan expression (Fig 3C). Furthermore, when all 3 cosmopolitan variants were present in the sub-Saharan background, expression did not differ from when only the cosmopolitan “G” variant at position 67 was introduced (t test, P = 1, Fig 3C). Thus, background-specific epistatic effects, presumably caused by interactions with other sub-Saharan variants within the CG9509 enhancer, appear to prevent expression from reaching the full cosmopolitan level in a sub-Saharan background. Our expression analyses corroborate these findings. When larval CG9509 expression in each population is partitioned according to the variant at position 67, the “G” variant makes a large contribution to the expression divergence observed between cosmopolitan and sub-Saharan larvae but cannot account for all of it (S1 Fig).
Functional analysis of CG9509
In order to elucidate the function of CG9509 and the effects of its expression on organismal phenotypes, we performed a series of functional and tolerance assays on flies in which CG9509 expression was disrupted. For this, we used both a newly identified hypomorph allele and RNAi. The hypomorph allele (CG9509del) was discovered as a spontaneous mutation in an isofemale line derived from Munich, Germany. It contains a frameshift-causing deletion within the CG9509 coding region and shows greatly reduced levels of CG9509 mRNA (S2 Fig). As a control, flies homozygous for CG9509del were compared to wild-type isofemale lines derived from the same population at the same time. RNAi knockdown of CG9509 expression was achieved by crossing a ubiquitous Act5C-GAL4 driver line to a transgenic line containing an RNAi hairpin construct specific to CG9509 (RNAi-CG9509) and flanked by a yeast upstream activating sequence (UAS). As a control, CG9509 knockdown flies were compared to flies of the host strain from which they were derived (UAS-).
Effect of CG9509 expression on cold, ethanol, and insecticide tolerance
Flies with disrupted CG9509 expression were tested for cold, ethanol, and insecticide (DDT and malathion) tolerance. If CG9509 expression plays a role in tolerance to cold temperatures or any of the tested compounds, we expect that a decrease in CG9509 expression should lead to a decrease in tolerance. CG9509 expression was not positively associated with tolerance to cold or the tested compounds and, unexpectedly, was negatively associated with insecticide and female cold tolerance (Table 1, S3 Fig).
Table 1. Effect of CG9509 knockdown in adult tolerance assays.
| Tolerance assay | Effecta | P valueb |
|---|---|---|
| DDT | + | 0.013 |
| Malathion | + | 2.84 × 10−5 |
| Cold (females) | + | 0.009 |
| Cold (males) | 0 | 0.248 |
| Ethanol | 0 | 0.733 |
aDirectional effect of CG9509 knockdown on tolerance.
bP values were obtained using generalized linear models (glms) with a quasibinomial distribution using sex, fly strain, and compound concentration as factors or, for cold tolerance, a t test.
Effect of CG9509 expression on developmental timing, larval growth rate, adult body size, and wing loading
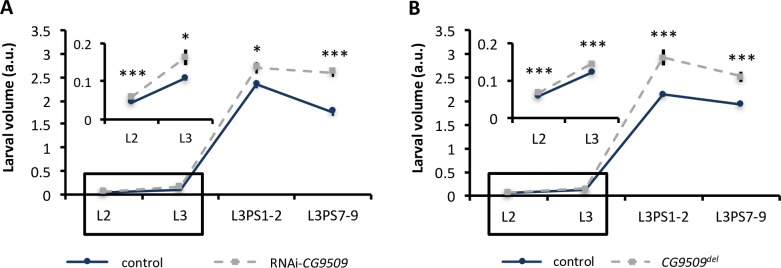
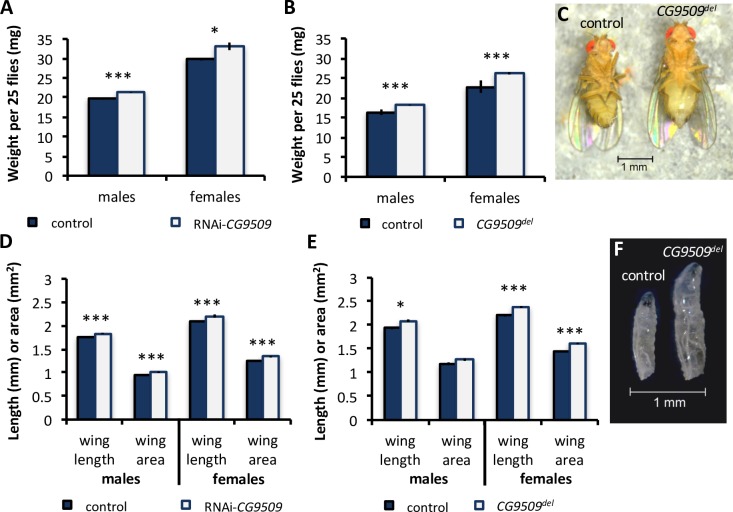
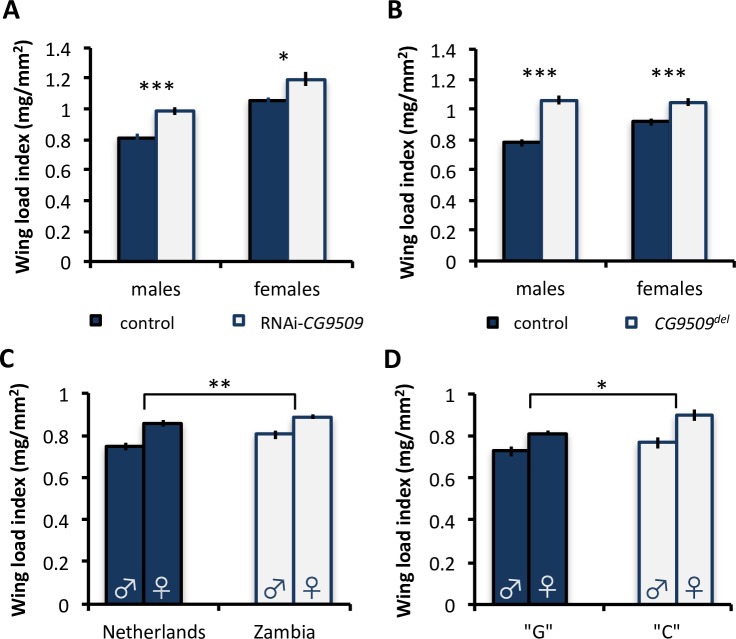
RNAi-knockdown and CG9509del larvae were measured for larval growth rate and developmental timing (as measured by duration of the larval stage and wandering stage). CG9509 expression had no effect on developmental timing (t test, P > 0.42, S4 Fig). In contrast, disruption of CG9509 expression was associated with increased larval growth rate (Fig 4), and CG9509 knockdown and CG9509del larvae were significantly larger than control larvae at all stages (Figs 4 and 5F). We further examined adult body size, as measured by body weight and wing size (length and area), in RNAi-knockdown and CG9509del flies. Disrupting CG9509 expression increased body weight by 8%–15% (Fig 5A and 5B). Similarly, disruption of CG9509 expression increased wing size by 4%–11% (Fig 5D and 5E), which was significant for all comparisons, except male wing area in CG9509del flies. Next, we examined wing loading, which is the relationship between mass and wing area [39] and is dependent upon relative body size. To this end, we measured the wing load index (wet weight/right wing area) in CG9509del and RNAi-knockdown flies. Disruption of CG9509 expression increased wing loading in both males and females by 10%–35% (Fig 6A and 6B). The fact that similar results were obtained independently for the CG9509del and RNAi-knockdown flies, which have different genetic backgrounds, suggests that the effect is a direct cause of losing CG9509 expression and not an artifact of off-target gene silencing or mutations at other loci.
Fig 4. Effect of CG9509 expression on larval growth rate.
Larval volume in arbitrary units (a.u.) in (A) control (blue lines; N = 15–19 per stage) and RNAi-CG9509 (gray, hatched lines; N = 15–20 per stage) larvae and (B) control (blue lines; N = 60 per stage) and CG9509del (gray, hatched lines; N = 15 per stage) larvae. Four larval stages were examined: second instar larvae (L2) after the first-to-second instar larval molt, third instar larvae (L3) after the second-to-third instar larval molt, early wandering third instar larvae (L3PS1-2), and late wandering third instar larvae (L3PS7-9). Insets show the boxed region on a larger scale. Underlying data can be found in S1 Data. Error bars represent the standard error of the mean. Significance was assessed via a t test for each larval stage, and a Bonferroni multiple test correction was applied. *P < 0.05, ***P < 0.005.
Fig 5. Effect of CG9509 expression on body size.
(A, B) Body weight per 25 flies in (A) control (blue; N = 5–6 per sex) and RNAi-CG9509 (white; N = 4 per sex) flies and (B) control (blue; N = 16 per sex) and CG9509del (white; N = 6–7 per sex) flies. (D, E) Wing length and wing area in (D) control (blue) and RNAi-CG9509 (white) flies (N = 15 per line and sex) and (E) control (blue; N = 20 per sex) and CG9509del (white; N = 9–13 per sex) flies. (C, F) Pictured are control (left) and CG9509del (C) females and (F) third instar larvae after the second-to-third instar molt. Underlying data can be found in S1 Data. Error bars represent the standard error of the mean. Significance was assessed via t test. *P < 0.05, ***P < 0.005.
Fig 6. Effect of CG9509 expression on wing loading.
Wing load index in (A) control (blue; N = 10–15 per sex) and RNAi-CG9509 (white; N = 10 per sex) flies, (B) control (blue; N = 20 per sex) and CG9509del (white; N = 14–15 per sex) flies, (C) flies from a Dutch (blue; N = 12 isofemale strains with 5 replicates each per sex) and a Zambian (white; N = 10 strains with 5 replicates each per sex) population, and (D) flies from the Dutch population partitioned according to the sequence variant at position 67. The high-expression, cosmopolitan “G” variant (6 strains with 5 replicates each per sex) is shown in blue, and the low-expression, sub-Saharan “C” variant (6 strains with 5 replicates each per sex) is shown in white. Underlying data can be found in S1 Data. Error bars represent the standard error of the mean. (A,B) Significance was assessed via a t test. (C,D) Significance was assessed using an ANOVA with sex, isofemale line, and population or variant at position 67 as factors. *P < 0.05, **P < 0.01, ***P < 0.005.
Adult body size and wing loading in a sub-Saharan and a cosmopolitan population
If selection favored a more active CG9509 enhancer in cosmopolitan populations because of its negative effect on adult body size and/or wing loading, we would expect to find reduced adult body size and/or wing loading in cosmopolitan populations relative to sub-Saharan populations. To test this, we examined a sub-Saharan (Zambia) and a cosmopolitan (the Netherlands) population to determine if CG9509 expression and enhancer variation could be associated with between-population differences in adult body size and/or wing loading. Contrary to the expectation, wing area and length were approximately 2%–10% reduced in the Zambian population in comparison to the Dutch population; however, these differences were not significant (ANOVA, P > 0.08 for both, S5 Fig). For adult body weight, there was a significant difference that also ran contrary to the expectation, with a significant reduction in the sub-Saharan population in comparison to the Dutch population (P < 10−5, S5 Fig). Wing loading, on the other hand, was significantly lower (approximately 3%–7.5%, P = 0.007, Fig 6C) in the Dutch population, which was in line with the expectation. However, it should be noted that these populations differ at more than just the CG9509 locus; thus, at least some of the phenotypic differences observed between these populations could be due to correlated changes in trans-acting or other cis-acting factors.
Effect of CG9509 expression on active ecdysone levels and the expression of 2 known growth regulators
CG9509 has been predicted to be involved in metabolism of the steroid hormone ecdysone [30], pulses of which act as temporal signals during D. melanogaster development [40,41]. To estimate active ecdysone levels in early and late wandering third instar larvae, we measured expression of E74B, which is directly activated by ecdysone and often used as a readout of active ecdysone levels [42,43], in RNAi-knockdown and CG9509del larvae. It must be noted, however, that because E74B expression is an indirect measure of active ecdysone levels, it may not correspond perfectly with actual ecdysone titers. When CG9509 expression was disrupted, E74B expression decreased by approximately 50% in both early and late wandering third instar larvae (Fig 7A and 7B, S6 Fig). During larval development, ecdysone and insulin signaling interact to influence growth [44,45]. Integral to this interaction are the growth regulators forkhead box, sub-group O (dFOXO), and Myc (dMyc). We examined dMyc and dFOXO expression in early and late wandering third instar RNAi-knockdown and CG9509del larvae, as these genes are known to have developmental stage-specific effects [40,44]. Disrupting CG9509 expression resulted in a 40%–50% decrease in both dMyc and dFOXO expression in late wandering third instar larvae (Fig 7D and S6 Fig). Disrupting CG9509 expression also decreased dMyc and dFOXO expression in early wandering third instar larvae; however, we could only detect this decrease in CG9509del larvae (Fig 7C and S6 Fig). This discrepancy could be due to the decreased efficiency of CG9509 RNAi-knockdown in this stage (76% versus >97% in all other examined stages). Thus, the association of CG9509 expression with expression of these 2 important growth regulators occurs in at least 1 larval stage.
Fig 7. Effect of CG9509 expression on active ecdysone levels and dMyc and dFOXO expression.
(A, B) Levels of active ecdysone, approximated by relative E74B expression as measured by quantitative reverse transcription PCR (qRT-PCR) in (A) early and (B) late wandering third instar larvae in control (blue; N = 8 per stage) and CG9509del (white; N = 4 per stage) flies. (C, D) Relative gene expression of dMyc and dFOXO in control (blue; N = 8 per stage) and CG9509del (white; N = 4 per stage) (C) early wandering third instar larvae and (D) late wandering third instar larvae. Expression is shown relative to the control for each stage. Underlying data can be found in S1 Data and S2 Data. Error bars represent the standard error of the mean. Significance was assessed via a t test. *P < 0.05, **P < 0.01, ***P < 0.005.
Association of CG9509 enhancer variants with body size and wing-loading variation
To determine if there was an association between SNP variation in the CG9509 enhancer and variation in either body size or wing loading, we performed further analyses in the Dutch population, as well as in the reconstituted homozygotes, hemizygotes, and/or heterozygotes of the F2 generation from reciprocal crosses of individuals from either the Dutch or a Rwandan population (S1 Text). The genetic background of these reconstituted F2 individuals represents a mixture of the 2 original parental genomes, which allows us to better disentangle the effects of SNP variation within the CG9509 enhancer from the effects of other variants in the genome (S1 Text), including trans-acting variants. Thus, association of CG9509 enhancer SNP variants with body size or wing-loading variation within this shared background should represent true associations, rather than spurious associations caused by linked variation elsewhere in the genome.
Effect of the SNP at position 67 on body size and wing loading
To determine if body size and/or wing-loading variation was associated with the SNP at position 67, we examined adult body weight, wing size, and wing loading in the cosmopolitan population from the Netherlands. The high-expression "G" variant at this position is at intermediate frequency in this population (Fig 2B). Wing loading, body weight, and wing size (as measured by length and area) in flies with the high CG9509 expression “G” variant were reduced by approximately 3%–10.5% in comparison to those with the ancestral “C” variant (ANOVA, P < 0.025 in all cases, Fig 6D and S5 Fig). These differences are driven by a shift in the phenotype distribution of “C” versus “G” variants rather than by a single isofemale line or measurement (S7 Fig). To further characterize the magnitude of the effect of carrying a “G” versus a “C” at position 67 on these phenotypes, we calculated partial eta squared (η2p), which is a standardized measure of effect size in which effect sizes on the magnitude of approximately 0.01, 0.06, and 0.14 are generally considered to represent small, medium, and large effect sizes, respectively [38]. In this cosmopolitan population, the variant at position 67 has a moderate (wing loading η2p = 0.046) to large (wing area η2p = 0.311, wing length η2p = 0.487) effect on these traits. To exclude the possibility that these associations are caused by variation at a linked site, we calculated the degree of linkage disequilibrium, r2 [46], between position 67 and all SNPs, excluding singletons, within a 50-kb region centered around position 67 in this population. We did not detect any significant association between the variant at position 67 and any other SNP in this region (Fisher’s exact test, P > 0.06 for all comparisons, S5 Table). However, it should be noted that our analysis does not take into account potential variation in trans-acting factors that may also contribute to body size and wing-loading variation in this population.
To further confirm the association of body size and wing-loading variation with sequence variation at position 67, we measured body weight, wing size (length and area), and wing load index in the F2 offspring of a set of reciprocal crosses between a “G” variant and a “C” variant isofemale line from the Dutch population. Briefly, we performed reciprocal crosses between the “C” and “G” variant isofemale lines and then crossed the F1 offspring within each cross. In the F2 generation, we measured the phenotype and genotyped flies using a PCR- and restriction enzyme-based assay. Presence of the high-expression “G” variant was associated with reduced body weight, wing area, wing length, and wing loading, with the magnitude of reduction generally increasing with the number of “G” variant alleles (Table 2). This reduction was significant for body weight, wing length, and wing area (ANOVA, P < 0.015 for each comparison) and represents a medium-sized effect of the SNP variant at position 67 (body weight η2p = 0.063, wing length η2p = 0.102, wing area η2p = 0.070) on these 3 traits in a cosmopolitan background (Table 2). On the other hand, this reduction was nonsignificant for wing loading (P = 0.317), corresponding to only a small-sized (η2p = 0.011) effect of the SNP variant at position 67 on this trait in a cosmopolitan background (Table 2).
Table 2. Effect of single nucleotide polymorphism (SNP) variation at position 67 on adult body size and wing loading in F2 offspring of reciprocal crosses between a “G” variant and a “C” variant isofemale line from a Dutch population.
| SNP 67a | Sexb | Weight (mg) ± SEM | Wing area (mm2) ± SEM | Wing length (mm) ± SEM | Wing load index (mg/mm2) ± SEM |
|---|---|---|---|---|---|
| G/G | F | 1.534 ± 0.036 | 1.277 ± 0.020 | 2.129 ± 0.020 | 1.184 ± 0.025 |
| G/C | F | 1.560 ± 0.034 | 1.288 ± 0.014 | 2.157 ± 0.014 | 1.214 ± 0.027 |
| C/C | F | 1.565 ± 0.035 | 1.280 ± 0.016 | 2.153 ± 0.011 | 1.224 ± 0.024 |
| G | M | 1.204 ± 0.026 | 0.977 ± 0.013 | 1.833 ± 0.012 | 1.245 ± 0.026 |
| C | M | 1.324 ± 0.034 | 1.046 ± 0.019 | 1.904 ± 0.018 | 1.269 ± 0.026 |
| P valuec | 0.012 | 0.011 | 0.002 | 0.317 | |
| η2pd | 0.063 | 0.07 | 0.102 | 0.011 |
aNucleotide at position 67 in the CG9509 enhancer. Males are hemizygous and therefore carry only 1 variant.
bM, male; F, female
cSignificance was assessed using an ANOVA with sex, cross, and variant at the position of interest as factors. The effect of the SNP variant was assumed to be additive, with cosmopolitan homo- and hemizygotes assigned a value of 2, sub-Saharan homo- and hemizygotes assigned a value of 0, and heterozygotes assigned a value of 1.
dEffect size of the SNP at position 67 on the surveyed trait
Effect of the SNPs at positions 1063 and 1174 on body size and wing loading
The cosmopolitan variants at positions 1063 and 1174 are fixed in the surveyed cosmopolitan populations and absent or at too low of a frequency in the surveyed sub-Saharan populations (Fig 2B) to be able to detect an association between these SNPs and body size or wing-loading variation within 1 of the surveyed populations. For this reason, we utilized 3 isofemale lines isolated from a Rwandan population, where the cosmopolitan SNPs are at moderate frequency (9.5%–16%) [47]. Due to the low frequency and linkage of these SNPs in the Rwandan population, the effects of these positions were surveyed together using 3 isofemale lines that differed along a block spanning positions 1174, 1155, 1063, and 821–817 but were otherwise identical at the other positions of interest within the CG9509 enhancer (Fig 2B, S1 Text). While we are unable to separate the effects of the SNP variants at these 4 positions, we have shown that the variants at positions 1155 and 821–817 do not contribute to CG9509 expression divergence (Fig 3).
Two sets of reciprocal crosses between 2 isofemale lines with a cosmopolitan “A” and “T” at positions 1174 and 1063, respectively, (hereafter designated “AT” variant) and an isofemale line with a sub-Saharan “C” and “G” at these positions (hereafter designated “CG” variant” (S1 Text) were performed, and body weight, wing size (length and area), and wing load index were measured in the F2 offspring. Briefly, we performed separate reciprocal crosses between the 2 “AT” variant isofemale lines and the “CG” variant isofemale line and then crossed the F1 offspring within each cross. Next, in the F2 generation, we measured the phenotype in homo- and hemizygotes and genotyped flies using sequencing. Presence of the cosmopolitan “AT” variant was associated with reduced body weight, wing area, wing length, and wing loading (Table 3). This reduction was significant for body weight and wing area (ANOVA, P < 0.007 for both comparisons) and moderately significant for wing length (ANOVA, P = 0.052), corresponding to a moderate (wing length η2p = 0.0315) or medium-sized (body weight η2p = 0.069, wing area η2p = 0.061) effect of the variants at positions 1174 and 1063 on these 3 traits in this population (Table 3). On the other hand, this reduction was not significant for wing loading (P = 0.128), corresponding to only a small-sized (η2p = 0.020) effect of variants at positions 1174 and 1063 on this trait in this population (Table 3).
Table 3. Effect of single nucleotide polymorphism (SNP) variation at positions 1174 and 1063 on adult body size and wing loading in F2 offspring of reciprocal crosses between 2 cosmopolitan “AT” variant isofemale lines and a sub-Saharan “CG” variant line from a Rwandan population.
| SNPs 1174 and 1063a | Sexb | Weight (mg) ± SEM | Wing area (mm2) ± SEM | Wing length (mm) ± SEM | Wing load index (mg/mm2) ± SEM |
|---|---|---|---|---|---|
| AT/AT | F | 1.549 ± 0.028 | 1.360 ± 0.015 | 2.219 ± 0.015 | 1.142 ± 0.024 |
| CG/CG | F | 1.617 ± 0.033 | 1.394 ± 0.014 | 2.256 ± 0.014 | 1.163 ± 0.023 |
| AT | M | 1.154 ± 0.027 | 1.016 ± 0.013 | 1.905 ± 0.013 | 1.140 ± 0.029 |
| CG | M | 1.234 ± 0.020 | 1.066 ± 0.008 | 1.931 ± 0.009 | 1.164 ± 0.022 |
| P valuec | 0.003 | 0.0071 | 0.052 | 0.128 | |
| η2pd | 0.069 | 0.061 | 0.032 | 0.012 |
aNucleotide at positions 1174 and 1063, respectively, in the CG9509 enhancer. Males are hemizygous and therefore carry only 1 variant.
bM, male; F, female
cSignificance was assessed using an ANOVA with sex, cross, and variant at the position of interest as factors. The effect of the SNP variant was assumed to be additive, with cosmopolitan homo- and hemizygotes assigned a value of 2, sub-Saharan homo- and hemizygotes assigned a value of 0, and heterozygotes assigned a value of 1.
dEffect size of positions 1174 and 1063 on the surveyed trait
Discussion
We have shown that the between-population expression divergence of CG9509 occurs in both larvae and adults (Fig 1) and is driven by nucleotide polymorphism within the CG9509 enhancer (Fig 3). We identified 3 SNPs that can account for the majority of the expression divergence, 2 of which (at positions 1174 and 1063, Fig 2B) only affect expression in larvae and have a relatively small effect on larval expression (Fig 3C). The third SNP (position 67, Fig 2B) accounts for the majority of the expression divergence in both adults and larvae (Fig 3). We propose that selection on the CG9509 enhancer occurred in 2 phases. First, the derived variants at positions 1174 and 1063 were the targets of the previously identified selective sweep [12]. These variants are fixed in cosmopolitan populations but absent or at low frequency in ancestral, sub-Saharan African populations (Fig 2B). Thus, this sweep likely occurred during or shortly after D. melanogaster’s expansion out of Africa but before the separation of European and Asian populations. A single haplotype spans positions 1174 and 1063 in cosmopolitan populations, suggesting that both derived variants were fixed by a single selective sweep. The derived variants at these positions are also present at low frequency in central African populations [47], suggesting that selection acted on standing variation. In a second step, we propose that the large-effect derived variant at position 67 arose as a new mutation on the selected haplotype and rose to intermediate frequency more recently in cosmopolitan populations. Consistent with this view, the derived variant is absent from sub-Saharan and central African populations (Fig 2B) [47]. It has been proposed that advantageous regulatory mutations with large effects are likely to display overdominance and, thus, remain polymorphic within populations [48]. The large effect of the derived variant at position 67 on CG9509 expression (Fig 3 and S1 Fig) and its intermediate frequency in cosmopolitan populations (Fig 2B) are consistent with this model. However, other causes for its maintenance at intermediate frequency, such as sexual antagonism, temporally varying selection, or the interaction of alleles at multiple loci, are also possible [48–51].
Until now, CG9509’s function and, therefore, the organismal phenotype(s) affected by variation in its expression have remained unknown. Here, we used RNAi-mediated knockdown of CG9509 expression and a newly identified CG9509 hypomorph allele to show that increased CG9509 expression is associated with reduced wing loading (Fig 6). Wing loading in a Dutch population was associated with polymorphism at position 67 (Fig 6D), and we also found that flies from Zambia had greater wing loading than those from the Netherlands (Fig 6C). When we surveyed wing loading in the F2 offspring of crosses between fly strains containing either cosmopolitan or sub-Saharan CG9509 enhancer variants affecting expression, wing loading varied in the expected direction (Tables 2 and 3), but this variation was small (approximately 1%–3%) and nonsignificant (Tables 2 and 3). Some of this discrepancy between our findings in Dutch and Zambian populations versus F2 offspring may be due to trans-acting variation present among the inbred lines from different populations. This variation should be greatly reduced in the F2 offspring, which share a more homogenous trans environment after a generation of recombination. However, we did detect significant associations between these variants and both body weight and wing size (Tables 2 and 3). Our inability to detect significant associations with wing loading may be a result of this trait being a ratio of 2 measurements, which increases trait variance and reduces statistical power. It is also possible that the effect size is too small to be detected as significant in our experimental design. Previous studies have shown that even small changes in flight load can lead to differences in flight performance [52–53], especially at low temperatures [53], and therefore impact fitness.
Consistent with our findings, several studies have documented clinal variation in wing loading among Drosophila populations across multiple continents [39,53–56], with reduced wing loading at higher latitudes, and this cline is thought to be maintained by selection. In Drosophila, and indeed all flying animals, relative size is important for flight aerodynamics. Furthermore, previous microarray comparisons of gene expression have found overexpression of muscle-related genes (including flight muscle components) in flies from Zimbabwe relative to those from the Netherlands [24,25], suggesting that flies in the ancestral range require a greater investment in flight muscle. This suggests that improved flight ability may have been an important adaptation as D. melanogaster expanded its species range, and selection on the CG9509 enhancer likely favored the reduction in wing loading conferred by increased larval CG9509 expression as D. melanogaster expanded out of Africa. Drosophila wing beat frequency and power output decrease as temperature decreases, resulting in reduced flight ability at cooler temperatures [53], and previous studies suggest that reduced wing loading may help counteract this effect [39,57–58]. Thus, improved flight ability may represent an adaptation to lower temperatures in the derived species range; however, general improvement of flight ability could also be adaptive. The energy conserved by improved flight ability could be used for other processes or aid in survival when resources are scarce. Improved flight ability might also aid in predator evasion or dispersion, which may have helped facilitate D. melanogaster’s expansion to new territories. However, it is important to note that, although selection for reduced wing loading represents a plausible scenario for adaptive regulatory evolution at the CG9509 locus, we cannot rule out the possibility that selection acted on an unobserved, pleiotropic trait associated with variation in the CG9509 enhancer.
We have shown that increased CG9509 expression is associated with reduced larval growth (Fig 4) and adult body size (Fig 5) and were able to associate body size variation with CG9509 enhancer sequence variation (Tables 2 and 3), with the high-expression, cosmopolitan variants associated with decreased body size. We also showed that weight is reduced in a sub-Saharan population in comparison to a Dutch population (S5 Fig). However, these results are contrary to expectations if selection acted on the CG9509 enhancer to reduce body size in cosmopolitan populations but in line with well-documented, latitudinal body size clines that are thought to be maintained by selection [55,59]. We additionally showed that increased CG9509 expression is associated with increased levels of the maturation hormone ecdysone (Fig 7A and 7B, S6 Fig). Most likely, the increased active ecdysone levels result in the reduced larval growth rate and a subsequently smaller body size, since the antagonistic interaction of ecdysone with insulin signaling is known to suppress larval growth [43,60], which in turn reduces adult body size. However, the mechanism through which CG9509 expression adjusts larval growth to reduce wing loading remains unknown. The effect on wing loading (Fig 6) is at least in part due to CG9509’s effect on active ecdysone levels (Fig 7A and 7B, S6 Fig), as ecdysone plays a key role in regulating proportional growth and coordinating the growth of individual organs with each other as well as with the entire body [61].
CG9509 is expressed in the larval fat body [62], which acts as a coordinator of larval growth [40,41,44]. Ecdysone signaling specifically in the fat body antagonizes insulin signaling in part via down-regulation of the positive growth regulator dMyc and translocation of the negative growth regulator dFOXO to the nucleus, where it activates the expression of target genes [44,45,63]. When we knocked down CG9509 expression, we found a decrease in both dMyc and dFOXO expression in late wandering third instar larvae (Fig 7D and S6 Fig), which is the stage during which the peak of the final and largest larval ecdysone titer occurs, signaling the onset of pupariation [40,41]. We detected a similar decrease in the early wandering third instar larval stage, which coincides with another, smaller ecdysone peak [40,41], but only in CG9509del larvae (Fig 7D). The reduction in dFOXO transcript expression is interesting, as it is dFOXO protein localization that suppresses growth [44,45,63]. However, a study documenting insulin/TOR network transcriptional variation found strong covariance for dFOXO transcript abundance and the expression of dFOXO-affected genes [64]. Thus, the expression of genes downstream of dFOXO may also be affected. The reduction in dMyc expression (Fig 7D) is counterintuitive, since its up-regulation in the fat body is expected during CG9509 expression knockdown. However, the effect of ecdysone on dMyc expression is both stage- and tissue-specific [40]; thus, the expression decrease is likely in another tissue and may represent a part of the mechanism through which CG9509 expression adjusts proportional growth to affect wing loading. Interestingly, previous studies have documented negative correlations of the expression of growth-associated genes with stress tolerance [65,66], which we also found for CG9509 expression (Table 1). However, this correlation could simply be a by-product of body size, which has been shown to correlate with stress tolerance in Drosophila [67].
We identified 3 SNPs that account for the majority of CG9509 expression divergence observed between cosmopolitan and sub-Saharan D. melanogaster (Figs 1 and 3). Indeed, when we mutated these SNPs, we were able to recover 100% of this expression divergence in the cosmopolitan background (Fig 3), although we also found evidence that unidentified SNPs in the CG9509 enhancer have epistatic effects on expression in the sub-Saharan background in larvae and the cosmopolitan background in adults (Fig 3). However, these epistatic effects are small relative to the magnitude of expression divergence that could be attributed to the 3 SNPs of major effect. Furthermore, the context-dependent nature of these effects makes it unlikely that they have been targets of positive selection, which acts most efficiently on additive genetic variation [68]. While we assume that the identified SNPs exert their effects on gene expression through interactions with trans-acting factors, the specific trans-acting factors that are involved remain unknown. To identify potential transcription factors that might interact with the identified SNPs, we scanned representative cosmopolitan and sub-Saharan CG9509 enhancer sequences for predicted transcription factor binding sites (TFBSs) [69]. All of the identified SNPs overlapped with at least 1 predicted TFBS, and for each SNP, differential binding (absence or a lower binding score in 1 sequence) was predicted for 2–8 TFBS matrix models (S2 Table). The majority of the identified transcription factors are known to be involved in developmental regulation and morphogenesis, including several forkhead box factors, the Iroquois complex genes, hairy, Distal-less, slow border cells, and twist [70–74]. Several, such as fork head and the Broad-Complex [75,76], are also known to be involved in insulin and/or ecdysone signaling.
It is important to elucidate both the mechanisms behind and the selective forces driving the adaptive divergence of cis-regulatory elements, as these examples help us to understand the genetic basis of phenotypic evolution, which can give further insights about biodiversity. Our results provide evidence that in cosmopolitan populations of D. melanogaster, positive selection has acted on 3 SNPs within the CG9509 enhancer to increase CG9509 expression and thereby reduce wing loading. While 2 of these SNPs appear to be the targets of a completed selective sweep, the third, which has the largest effect on CG9509 expression, has been maintained at intermediate frequency, suggesting that it has been subject to another mode of selection. Using natural variation, a mutant allele, and RNAi, we provide the first experimental evidence of CG9509’s function. We show that its expression influences larval and adult body size, as well as the ratio of wing-to-body size. We propose that the reduced wing loading conferred by elevated CG9509 expression represents an adaptation to improve flight ability as D. melanogaster expanded out of Africa. Because of the remarkable body size increase seen in CG9509del larvae and adults, we propose that the gene be named fezzik (fiz) after the giant character in The Princess Bride.
Materials and methods
D. melanogaster lines
All flies were maintained as inbred, isofemale lines under standard conditions (22°C, 14 hours light:10 hours dark cycle, cornmeal-molasses medium). The phiX-86Fb stock [77], containing a mapped attP site on the third chromosome (cytological position: 3R 86F), was obtained from the Bloomington Stock Center (Indiana, United States) and used for phiC31 site-specific integration.
Population samples
Expression of CG9509 was surveyed in isofemale lines derived from the following locations: Leiden, the Netherlands (12 lines); Kuala Lumpur, Malaysia (11 lines); Cairo, Egypt (12 lines); Siavonga, Zambia (11 lines); and Lake Kariba, Zimbabwe (11 lines). Body size and wing-loading assays were performed in the Dutch and Zambian populations. To survey the effect of CG9509 SNP variants on body size and wing-loading variation, a series of reciprocal crosses and subsequent body size and wing-loading assays were performed using 2 Dutch lines and 3 lines (RG11N, RG25, and RG28) from a population in Gikongoro, Rwanda [47]. All of the surveyed populations, with the exception of Rwanda, were used in a previous study of adult expression and sequence variation associated with the CG9509 enhancer region [31]. The Zimbabwean and Dutch populations were also used in a previous study of sequence and expression variation associated with the CG9509 enhancer region [12] as well as genome-wide expression studies [24–26,28].
Crosses to test the association between CG9509 enhancer SNPs and phenotype
Body size and wing-loading assays were performed on the F2 offspring of 3 pairs of reciprocal crosses. Reciprocal crosses of 30–40 females and 15–20 males were performed for each pair. Forty to fifty F1 progeny were allowed to randomly mate, and phenotypes were measured in the F2 generation. Adults for phenotype measurements were staged as described in S1 Text. Crosses were performed using 2 Dutch lines with either a “C” or a “G” variant at position 67 but identical at other sites of interest in the CG9509 enhancer (Fig 2B) and 3 Rwandan lines (RG11N, RG25, and RG38) [47] containing either cosmopolitan or sub-Saharan variants at positions 1174, 1155, 1063, and 821–817 (Fig 2B, S1 Text). Flies were genotyped using either a PCR followed by restriction enzyme digestion for the Dutch crosses or sequencing for the Rwandan crosses (S1 Text).
CG9509 hypomorph and knockdown lines
CG9509, E74B, dFOXO, and dMyc expression analyses as well as body size, larval growth rate, developmental timing, wing-loading, and tolerance assays were performed on flies in which the open reading frame (ORF) of the CG9509 gene was disrupted and/or in which CG9509 expression was knocked down by RNAi. We discovered a mutant CG9509 allele as a naturally occurring variant in an isofemale line from a Munich population. In this line (CG9509del), a deletion introduces a frameshift that leads to a premature stop codon 232 amino acids into the CG9509 ORF (S2 Fig). Our quantitative reverse transcription PCR (qRT-PCR) assay (see below) was able to detect the expression of CG9509 mRNA in this line, but only at very low levels (S2 Fig), suggesting that it is degraded by the nonsense-mediated decay pathway. Given the disrupted ORF and the very low expression, we assume that CG9509 function is greatly reduced in this line. For this reason, we refer to it as a hypomorph allele. As a control, 4 lines from the same Munich population, showing representative CG9509 expression for the population, were used. We further confirmed that the observed phenotypes are likely driven specifically by the CG9509 hypomorph allele rather than variation located elsewhere in the genome (S1 Text).
The knockdown of CG9509 expression was achieved using an RNAi construct under the control of the yeast GAL4/UAS system. A D. melanogaster line producing a hairpin RNA complementary to CG9509 mRNA under the control of a UAS (RNAi-CG9509, transformant ID: 107089) as well as a line containing an empty vector at the same genomic location (UAS-, transformant ID: 60100), which we used as a control, were obtained from the Vienna Drosophila Resource Center (Vienna, Austria) [78]. The RNAi-CG9509 and UAS- lines were crossed to an Act5C-GAL4/Cyo driver line, and the progeny were used in subsequent body size, larval growth rate, developmental timing, wing-loading, and tolerance assays, as well as in expression analyses. Using qRT-PCR, CG9509 expression knockdown efficiency was estimated to be 98.6% for adult females, 98.9% for adult males, 76.0% for early wandering third instar larvae, and 97.6% for late wandering third instar larvae.
Expression analysis
In adults, CG9509 expression is highly enriched in the Malpighian tubule, while in larvae, it is enriched in the Malpighian tubule and fat body [62]; therefore, we surveyed expression in whole flies and larvae. Total RNA was extracted from 3–5 adult males (aged 4–6 days) or 1–3 early or late third instar wandering larvae, and a DNAse I digestion was performed using the MasterPure RNA Purification Kit (Epicentre; Madison, Wisconsin, US). Two biological replicates were performed for each line and/or stage. Using random hexamer primers and Superscript III reverse transcriptase (Invitrogen; Carlsbad, California, US), 3 μg total RNA for each replicate was reverse transcribed following the manufacturer’s protocol. TaqMan Gene Expression Assays (Invitrogen; Carlsbad, California, US) were then performed on the resulting cDNA using probes specific to CG9509 (Dm01838873_g1), dFOXO (Dm02140207_g1), dMyc (Dm01843706_m1), and/or E74B (Dm01793592_m1) as well as a probe specific to the ribosomal protein gene RpL32 (Dm02151827_g1), which was used as an endogenous control. The ΔΔCt method was used to calculate normalized gene expression [79]. Briefly, for each biological replicate, the average threshold cycle (Ct) of 2 technical replicates was measured, and ΔCt was calculated as the mean Ct difference between the probe of interest and the RpL32 probe. The fold-change difference in expression relative to the Zimbabwe population for population comparisons or the control lines for CG0509 hypomorph and knockdown comparisons was then calculated as 2–(ΔCtX–ΔCtY), where ΔCtX is the mean ΔCt value for each biological replicate of the line of interest and ΔCtY is the mean ΔCt value of either the Zimbabwe or control lines. Significance was assessed with a t test. When more than 3 comparisons were made using the same data, a Bonferroni multiple test correction was applied.
Transgenic reporter gene assays
The CG9509 enhancer region, spanning coordinates 14,909,008–14,910,193 of the X chromosome (release 6), was PCR-amplified from 2 cosmopolitan strains and 1 sub-Saharan strain as described in [12] and cloned into the pCR2.1-TOPO vector (Invitrogen; Carlsbad, California, US). The effects of 6 sub-Saharan sequence variants (positions 67, 765, 821–817, 1063, 1155, and 1174; Fig 2B) in the cosmopolitan background were examined. The sub-Saharan African variants were introduced into the cosmopolitan sequence using either standard cloning techniques or site-directed mutagenesis (S1 Text) [80]. For sites shown to affect reporter gene expression, the cosmopolitan variants were introduced into the sub-Saharan enhancer, and constructs with all contributing sites were generated in both a cosmopolitan and sub-Saharan background using site-directed mutagenesis (S1 Text) for a total of 13 reporter gene constructs. The original and the mutated enhancer sequences were confirmed via sequencing (S1 Text). The Escherichia coli LacZ coding region was then inserted downstream of the CG9509 enhancer sequence, and both were introduced into the pattB integration vector [77] using standard cloning techniques (S1 Text). The pattB vectors containing the CG9509 enhancer and the LacZ reporter gene were microinjected into early-stage embryos of the phiX-86Fb (attP site at cytological band 86F) strain [77], which contains a stable source of phiC31 integrase on the X chromosome. After microinjection, surviving flies were crossed to a white- strain to remove the integrase source, and stable lines homozygous for each of the constructs were established. A subset of the microinjections was performed by Rainbow Transgenic Flies (Camarillo, CA, US).
In adults, CG9509 expression is highly enriched in the Malpighian tubule, while in larvae, it is enriched in the Malpighian tubule and fat body [62], and adult reporter gene in whole flies has been shown to be a good proxy for Malpighian tubule expression [12]; therefore, we surveyed reporter gene expression in whole flies and larvae. For each reporter gene construct, β-galactosidase activity was measured in groups of 15 adult 4–6-day-old males or females or 8 late wandering third instar larvae. Soluble proteins were extracted, and a β-galactosidase activity assay was performed as described in [81] with the following modifications: flies or larvae were frozen with liquid nitrogen and homogenized before the addition of 200 μl of the 0.1 M Tris-HCl, 1 mM EDTA, and 7 mM 2-mercaptoethanol buffer (pH 7.5). β-galactosidase activity was measured spectrophotometrically by following the change in absorbance at 420 nm at 37°C. Four to eight biological replicates were performed per stage or sex. Significance was assessed using a t test, and a Bonferroni multiple test correction was applied for each stage and sex. To better understand the effect of position 67 on reporter gene expression in adults, an ANOVA using sex, background, the variant at position 67, and the interaction between the variant at position 67 and background, sex, and the other tested sites within the CG9509 enhancer was performed.
Body size assays
Weight
The wet weight of flies was measured in groups of 25 males or females or, for F2 offspring, single flies. Groups of flies were lightly anesthetized with CO2 and placed in preweighed 1.5 mL Eppendorf tubes on ice for 5 minutes before being weighed on a Mettler H51 scale (d = 0.01 mg, error = 0.05 mg). The weight of 25 flies was then calculated as the weight of 25 flies and tube minus the weight of the tube. For each line and sex, 4 replicates were performed for population comparisons, 5–7 replicates were performed for the CG9509del and RNAi-knockdown lines, and 4–5 replicates were performed for all control lines. For F2 offspring, 13–35 individual flies were weighed for each genotype and sex. Significance was assessed using a t test for CG9509del and RNAi comparisons. For population comparisons, significance was assessed using an ANOVA with sex, isofemale line, and population or variant at position 67 as factors. For F2 offspring comparisons, significance was assessed using an ANOVA with sex, cross, and SNP variant(s) as factors. The effect of the SNP variant(s) was assumed to be additive, with cosmopolitan homo- and hemizygotes assigned a value of 2, sub-Saharan homo- and hemizygotes assigned a value of 0, and heterozygotes assigned a value of 1.
Wing size
For each fly, the right wing (or the left wing if the right wing was damaged) was dissected in isopropanol, mounted in Euparal (Carl Roth; Karlsruhe, Germany), and allowed to dry at least 1 week before being photographed. Wings were photographed using a Nikon D5100 camera and compound microscope. Images were analyzed in ImageJ [82]. A piece of millimeter paper was included in all images for scale. Wing length was measured in a straight line from the humeral-costal break to the third longitudinal vein, and wing area was estimated as previously described [55]. For each line and sex, wing size was measured for 5 flies for population comparisons and 10–15 biological replicates per sex for RNAi-CG9509/Act5C-GAL4, UAS-/Act5C-GAL4, and CG9509del and control lines. For F2 offspring, 11–35 wings were measured for each genotype and sex. Significance was assessed using a t test for CG9509del and RNAi comparisons. For population comparisons, significance was assessed using an ANOVA with sex, isofemale line, and population or variant at position 67 as factors. For F2 offspring comparisons, significance was assessed using an ANOVA with sex, cross, and SNP variant(s) as factors. The effect of the SNP variant(s) was assumed to be additive, with cosmopolitan homo- and hemizygotes assigned a value of 2, sub-Saharan homo- and hemizygotes assigned a value of 0, and heterozygotes assigned a value of 1.
Wing-loading assays
Wing load index was calculated as the wet weight of a fly divided by the area of its right wing. Flies were lightly anesthetized with CO2 and placed in preweighed 1.5 mL Eppendorf tubes on ice for 5 minutes before being weighed on a Mettler H51 scale (d = 0.01 mg, error = 0.05 mg). The weight of a fly was then calculated as the weight of the fly and tube minus the weight of the tube. For each fly, the right wing (or the left wing if the right wing was damaged) was then dissected, and the wing area was estimated as described above. For each line and sex, wing loading was measured for 5 flies for population comparisons and 10–15 flies for RNAi-CG9509/Act5C-GAL4 and CG9509del lines as well as their respective control lines. For F2 offspring, wing loading was measured for 11–35 flies for each genotype and sex. Significance was assessed using a t test for CG9509del and RNAi comparisons. For population comparisons, significance was assessed using an ANOVA with sex, isofemale line, and population or variant at position 67 as factors. For F2 offspring comparisons, significance was assessed using an ANOVA with sex, cross, and SNP variant(s) as factors. The effect of the SNP variant(s) was assumed to be additive, with cosmopolitan homo- and hemizygotes assigned a value of 2, sub-Saharan homo- and hemizygotes assigned a value of 0, and heterozygotes assigned a value of 1.
Larval growth assays
To assess larval growth rate, larval volume was measured in the following stages: second instar approximately 48 hours after egg laying (AEL), early third instar (72 hours AEL), early wandering third instar (110 hours AEL), and late wandering third instar (116 hours AEL). Larvae were staged as described in S1 Text. Before imaging, larvae were placed on ice for at least 5 minutes. Larvae were photographed using a Nikon D5100 camera and a compound microscope, and images were analyzed in ImageJ [82]. A piece of millimeter paper was included in all images for scale. Larval volume was calculated as 4/3π(L/2)2(d/2), where L = length and d = diameter [60]. For each stage and line, larval volume was measured in 15–20 larvae for RNAi-CG9509 /Act5C-GAL4 and CG9509del lines as well as their respective controls. Significance at each larval stage was assessed using a t test, and a Bonferroni multiple test correction was applied.
Developmental timing assays
As a measure of developmental timing, the time from the first instar larval stage to pupariation and the duration of the wandering stage were measured. As described in S1 Text, flies were allowed to lay eggs for 12 hours, and first instar larvae were collected. Larvae were transferred in groups of 50 to cornmeal-molasses medium and allowed to mature. In order to measure the duration of the larval stage (L1 to pupariation), pupariation was recorded every 2 hours for 25–110 larvae per line. In order to measure the duration of the wandering stage, larvae were screened for onset of wandering behavior every hour and transferred individually to a petri dish containing moistened filter paper. Pupariation was recorded every hour for 10–50 larvae per line. Both assays were performed at 25°C to prevent fluctuations in developmental timing due to temperature.
Tolerance assays
DDT, malathion, ethanol, and cold tolerance assays were performed using RNAi-CG9509/Act5C-GAL4 and UAS-/Act5C-GAL4 flies. For DDT, malathion, and ethanol tolerance assays, for each line, sex, and concentration, 6–8 tolerance chambers with 20 flies each were exposed to 4 concentrations of a compound, and mortality was measured as the number of flies dead or unable to move after 30 minutes (malathion), 2 hours (DDT), or 48 hours (ethanol). For ethanol tolerance assays, tolerance chambers consisted of a plastic vial (diameter = 25 mm, height = 95 mm) with compressed cotton at the bottom containing 2.5 ml ethanol solution supplemented with 5% sucrose and sealed with a cork. For DDT and malathion assays, tolerance chambers consisted of glass vials (h = 5 cm, r = 1.65 cm) in which 200 μl of DDT (Dr. Ehrenstorfer; Augsburg, Germany) or malathion (Dr. Ehrenstorfer; Augsburg, Germany) diluted in acetone was swirled until the acetone dried; the vials were allowed to dry an additional hour before addition of flies and were sealed with compressed cotton soaked in 5% sucrose solution. For all assays, 2–3 control chambers containing only 5% sucrose solution were also tested. The data for each assay were fit to a generalized linear model using concentration, line, and sex as factors (unless sex was not significant, in which case it was removed from the model) and a quasibinomial distribution using the glm function in R [83]. For cold tolerance assays, for each line and sex, 25 groups of 5 flies were exposed to an ice water bath for 5 hours, and the time in minutes until each fly had recovered from chill coma (able to stand upright again) was recorded. The mean recovery time for each vial was calculated, and a t test was applied to assess significance.
Supporting information
(XLSX)
(XLSX)
Relative expression in late wandering third instar larvae in (A) the Netherlands (Net.), Malaysia (Mal.), Egypt (Egy.), Zimbabwe (Zim.), and Zambia (Zam.) populations (N = 10–12 isofemale strains per population with 2 biological replicates per strain). Blue bars represent cosmopolitan populations, and white bars represent sub-Saharan populations. (B) Relative expression in each population represented according to the variant at position 67. The high-expression, cosmopolitan “G” variant is shown in blue, and the low-expression, sub-Saharan “C” variant is shown in white. For simplicity, Zambian and Zimbabwean expression are presented together as sub-Saharan African (Afr.) expression. Underlying data can be found in S1 Data. Error bars indicate the standard error of the mean. Differences between populations were tested by a t test, and a Bonferroni multiple test correction was applied. ns, not significant; ●P < 0.10, *P < 0.05, **P < 0.01, ***P < 0.005.
(PDF)
(A) Relative expression of CG9509 in the CG9509del line as determined by quantitative reverse transcription PCR (qRT-PCR). For comparison, the average expression of the population in which the CG9509del line was discovered (Munich) is shown. Underlying data can be found in S1 Data. Error bars represent the standard error of the mean. (B) DNA sequence alignment spanning the deletion within the CG9509del coding region. Sequences of flies from the CG9509del source population in Munich (MU) are shown for comparison. (C) Amino acid alignment spanning the frameshift within the CG9509del coding region. Sequences of flies from the CG9509del source population are shown for comparison (MU). The asterisk indicates a stop codon.
(PDF)
Adult (A) DDT, (B) malathion, (C) ethanol (N = 6–8 replicates per line, sex, and concentration), and (D) cold tolerance assay results (N = 25 per line and sex) for control (blue lines or bars) and RNAi-CG9509 (gray hatched lines or white bars) flies. Underlying data can be found in S2 Data. In panels A–C, significance was assessed using a generalized linear model with a quasibinomial distribution. In panel D, significance was assessed using a t test. ns, not significant; *P < 0.05, **P < 0.01, ***P < 0.005.
(PDF)
(A, B) Duration of larval stage in (A) control (blue; N = 25) and RNAi-CG9509 (white; N = 51) flies and (B) control (blue; N = 242) and CG9509del (white; N = 107) flies. (C, D) Duration of wandering stage in (C) control (blue; N = 16) and RNAi-CG9509 (white; N = 40) flies and (D) CG9509del (white; N = 10) and control (blue; N = 40) flies. Underlying data can be found in S2 Data. Error bars represent the standard deviation. The knockdown and hypomorph lines were not significantly different from their respective control lines for either stage (t test; P > 0.4 for all comparisons).
(PDF)
(A) Body weight per 25 flies (N = 10–12 isofemale lines per population with 4 replicates per sex), (C) wing length (N = 10–12 isofemale lines per population with 4 replicates per sex), and (E) wing area in a Dutch (blue bars) and a Zambian (white bars) population (N = 10–12 isofemale lines per population with 4 replicates per sex). (B) Body weight per 25 flies (N = 6 isofemale lines per variant with 4 replicates per sex), (D) wing length (N = 6 isofemale lines per variant with 4 replicates per sex), and (F) wing area in a Dutch population separated according to the variant at position 67 (N = 6 isofemale lines per variant with 4 replicates per sex). The derived, high-expression “G” is shown in blue, and the ancestral, low-expression “C” variant is shown in white. Error bars indicate the standard error of the mean. Underlying data can be found in S2 Data. Significance was assessed with an ANOVA using sex, isofemale line, and population or the variant at position 67 as factors (shown in black). Significance was additionally assessed in both populations simultaneously with population and the variant at position 67 included as factors (shown in red). ns, not significant; ●0.05 < P < 0.10, *P < 0.05, **P < 0.01, ***P < 0.005.
(PDF)
(A, B) Levels of active ecdysone, approximated by relative E74B expression as measured by quantitative reverse transcription PCR (qRT-PCR) in (A) early and (B) late wandering third instar larvae in control (blue) and RNAi-CG9509 (white) flies (N = 9–10 per line). (C, D) Relative gene expression of dMyc and dFOXO in control (blue) and RNAi-CG9509 (white) (C) early wandering third instar larvae and (D) late wandering third instar larvae (N = 9–10 per line). Expression is shown relative to the control for each stage. Underlying data can be found in S2 Data. Error bars represent the standard error of the mean. Significance was assessed via a t test. *P < 0.05, **P < 0.01, ***P < 0.005.
(PDF)
Distribution of (A) wing loading, (C) wing area, (E) wing length, and (G) body weight in a Dutch (blue) and a Zambian (yellow) population. Distribution of (B) wing loading, (D) wing area, (F) wing length, and (H) body weight in a Dutch population separated according to the variant at position 67 (6 isofemale lines per variant). The derived, high-expression “G” is shown in blue, and the ancestral, low-expression “C” variant is shown in yellow.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(DOCX)
Acknowledgments
We thank Hilde Lainer, Hedwig Gebhart, Simone Lange, Annabella Königer, and Eliza Argyridou for technical assistance in the lab. We also thank Lisa Marie Keitel, Sandra Petrone Mendoza, Asya Martirosyan, and Keshika Ravichandran who helped at various stages of this project.
Abbreviations
- indel
insertion/deletion
- qRT-PCR
quantitative reverse transcription PCR
- RNAi
RNA interference
- SNP
single nucleotide polymorphism
- TFBS
transcription factor binding site
- UAS
upstream activating sequence
Data Availability
All sequence files are available from the GenBank database (accession numbers MG195568 – MG195570, HF913659.1, HF913664.1, HF913717.1). Dutch whole genome sequence data are available from http://evol.bio.lmu.de/downloads.
Funding Statement
Deutsche Forschungsgemeinschaft http://www.dfg.de/en/ (grant number PA 903/5). Received by JP. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Deutsche Forschungsgemeinschaft http://www.dfg.de/en/ (grant number PA 903/8-1). Received by JP. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975; 188(4184): 107–16. [DOI] [PubMed] [Google Scholar]
- 2.Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, et al. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol. 2003; 20(9): 1377–419. doi: 10.1093/molbev/msg140 [DOI] [PubMed] [Google Scholar]
- 3.Carroll SB. Endless forms: The evolution of gene regulation and morphological diversity. Cell. 2000; 101(6): 577–80. [DOI] [PubMed] [Google Scholar]
- 4.Carroll SB. Evo-Devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell. 2008; 134(1): 25–36. doi: 10.1016/j.cell.2008.06.030 [DOI] [PubMed] [Google Scholar]
- 5.Prud'homme B, Gompel N, Carroll SB. Emerging principles of regulatory evolution. Proc Natl Acad Sci U S A. 2007; 104(suppl 1): 8605–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007; 8(3): 206–16. doi: 10.1038/nrg2063 [DOI] [PubMed] [Google Scholar]
- 7.Ingram CJE, Mulcare CA, Itan Y, Thomas MG, Swallow DM. Lactose digestion and the evolutionary genetics of lactase persistence. Human Genetics. 2008; 124(6): 579–91. doi: 10.1007/s00439-008-0593-6 [DOI] [PubMed] [Google Scholar]
- 8.González J, Macpherson JM, Petrov DA. A recent adaptive transposable element insertion near highly conserved developmental loci in Drosophila melanogaster. Mol Biol Evol. 2009; 26(9): 1949–61. doi: 10.1093/molbev/msp107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daborn PJ, Yen JL, Bogwitz MR, Le Goff G, Feil E, Jeffers S, et al. A Single P450 Allele Associated with Insecticide Resistance in Drosophila. Science. 2002; 297(5590): 2253–6. doi: 10.1126/science.1074170 [DOI] [PubMed] [Google Scholar]
- 10.Fraser HB, Levy S, Chavan A, Shah HB, Perez JC, Zhou Y, et al. Polygenic cis-regulatory adaptation in the evolution of yeast pathogenicity. Genome Res. 2012; 22(10): 1930–9. doi: 10.1101/gr.134080.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sucena É, Stern DL. Divergence of larval morphology between Drosophila sechellia and its sibling species caused by cis-regulatory evolution of ovo/shaven-baby. Proc Natl Acad Sci U S A. 2000; 97(9): 4530–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saminadin-Peter SS, Kemkemer C, Pavlidis P, Parsch J. Selective sweep of a cis-regulatory sequence in a non-African population of Drosophila melanogaster. Mol Biol Evol. 2012; 29(4): 1167–74. doi: 10.1093/molbev/msr284 [DOI] [PubMed] [Google Scholar]
- 13.Guio L, Barrón MG, González J. The transposable element Bari-Jheh mediates oxidative stress response in Drosophila. Mol Ecol. 2014; 23(8): 2020–30. doi: 10.1111/mec.12711 [DOI] [PubMed] [Google Scholar]
- 14.Mateo L, Ullastres A, González J. A transposable element insertion confers xenobiotic resistance in Drosophila. PLoS Genet. 2014; 10(8): e1004560 doi: 10.1371/journal.pgen.1004560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koshikawa S, Giorgianni MW, Vaccaro K, Kassner VA, Yoder JH, Werner T, et al. Gain of cis-regulatory activities underlies novel domains of wingless gene expression in Drosophila. Proc Natl Acad Sci U S A. 2015; 112(24): 7524–9. doi: 10.1073/pnas.1509022112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Catalán A, Glaser-Schmitt A, Argyridou E, Duchen P, Parsch J. An indel polymorphism in the MtnA 3' untranslated region is associated with gene expression variation and local adaptation in Drosophila melanogaster. PLoS Genet. 2016; 12(4): e1005987 doi: 10.1371/journal.pgen.1005987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wittkopp PJ, Kalay G. cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet. 2011; 13(1): 59–69. [DOI] [PubMed] [Google Scholar]
- 18.Gompel N, Prud'homme B, Wittkopp PJ, Kassner VA, Carroll SB. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. 2005; 433(7025): 481–7. doi: 10.1038/nature03235 [DOI] [PubMed] [Google Scholar]
- 19.Prud'homme B, Gompel N, Rokas A, Kassner VA, Williams TM, Yeh SD, et al. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature. 2006; 440(7087): 1050–3. doi: 10.1038/nature04597 [DOI] [PubMed] [Google Scholar]
- 20.Kwasnieski JC, Mogno I, Myers CA, Corbo JC, Cohen BA. Complex effects of nucleotide variants in a mammalian cis-regulatory element. Proc Natl Acad Sci U S A. 2012; 109(47): 19498–503. doi: 10.1073/pnas.1210678109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang J, Zhou Y, Hu X, Lam L, Henry C, Green EM, et al. The molecular mechanism of a cis-regulatory adaptation in yeast. PLoS Genet. 2013; 9(9): e1003813 doi: 10.1371/journal.pgen.1003813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cooper TF, Rozen DE, Lenski RE. Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc Natl Acad Sci U S A. 2003; 100(3): 1072–7. doi: 10.1073/pnas.0334340100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holloway AK, Lawniczak MK, Mezey JG, Begun DJ, Jones CD. Adaptive gene expression divergence inferred from population genomics. PLoS Genet. 2007; 3(10): 2007–13. doi: 10.1371/journal.pgen.0030187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hutter S, Saminadin-Peter SS, Stephan W, Parsch J. Gene expression variation in African and European populations of Drosophila melanogaster. Genome Biol. 2008; 9(1): R12 doi: 10.1186/gb-2008-9-1-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Müller L, Hutter S, Stamboliyska R, Saminadin-Peter SS, Stephan W, Parsch J. Population transcriptomics of Drosophila melanogaster females. BMC Genomics. 2011; 12: 81 doi: 10.1186/1471-2164-12-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Catalán A, Hutter S, Parsch J. Population and sex differences in Drosophila melanogaster brain gene expression. BMC Genomics. 2012; 13: 654 doi: 10.1186/1471-2164-13-654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schoville SD, Barreto FS, Moy GW, Wolff A, Burton RS. Investigating the molecular basis of local adaptation to thermal stress: population differences in gene expression across the transcriptome of the copepod Tigriopus californicus. BMC Evol Biol. 2012; 12: 170 doi: 10.1186/1471-2148-12-170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huylmans AK, Parsch J. Population- and sex-biased gene expression in the excretion organs of Drosophila melanogaster. G3 (Bethesda). 2014; 4(12): 2307–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McQuilton P, St Pierre SE, Thurmond J, Consortium F. FlyBase 101—the basics of navigating FlyBase. Nucleic Acids Res. 2012; 40(Database issue): D706–14. doi: 10.1093/nar/gkr1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iida K, Cox-Foster DL, Yang X, Ko WY, Cavener DR. Expansion and evolution of insect GMC oxidoreductases. BMC Evol Biol. 2007; 7: 75 doi: 10.1186/1471-2148-7-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glaser-Schmitt A, Catalán A, Parsch J. Adaptive divergence of a transcriptional enhancer between populations of Drosophila melanogaster. Philos Trans R Soc Lond B Biol Sci. 2013; 368(1632): 20130024 doi: 10.1098/rstb.2013.0024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glinka S, Ometto L, Mousset S, Stephan W, De Lorenzo D. Demography and natural selection have shaped genetic variation in Drosophila melanogaster: a multi-locus approach. Genetics. 2003; 165(3): 1269–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ometto L, Glinka S, De Lorenzo D, Stephan W. Inferring the effects of demography and selection on Drosophila melanogaster populations from a chromosome-wide scan of DNA variation. Mol Biol Evol. 2005; 22(10): 2119–30. doi: 10.1093/molbev/msi207 [DOI] [PubMed] [Google Scholar]
- 34.Li H, Stephan W. Inferring the demographic history and rate of adaptive substitution in Drosophila. PLoS Genet. 2006; 2(10): e166 doi: 10.1371/journal.pgen.0020166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laurent SJ, Werzner A, Excoffier L, Stephan W. Approximate Bayesian analysis of Drosophila melanogaster polymorphism data reveals a recent colonization of Southeast Asia. Mol Biol Evol. 2011; 28(7): 2041–51. doi: 10.1093/molbev/msr031 [DOI] [PubMed] [Google Scholar]
- 36.White KP, Rifkin SA, Hurban P, Hogness DS. Microarray analysis of Drosophila development during metamorphosis. Science. 1999; 286(5447): 2179–84. [DOI] [PubMed] [Google Scholar]
- 37.Massouras A, Waszak SM, Albarca-Aguilera M, Hens K, Holcombe W, Ayroles JF, et al. Genomic variation and its impact on gene expression in Drosophila melanogaster. PLoS Genet. 2012; 8(11): e1003055 doi: 10.1371/journal.pgen.1003055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen J. Statistical power analysis for the behavioral sciences. 2nd ed. Hillsdale, N.J.: L. Erlbaum Associates; 1988. [Google Scholar]
- 39.Gilchrist GW, Huey RB. Plastic and genetic variation in wing loading as a function of temperature within and among parallel clines in Drosophila subobscura. Integr Comp Biol. 2004; 44(6): 461–70. doi: 10.1093/icb/44.6.461 [DOI] [PubMed] [Google Scholar]
- 40.Quinn L, Lin J, Cranna N, Lee AJE, Mitchell N, Hannan R. Steroid hormones in Drosophila: How ecdysone coordinates developmental signalling with cell growth and division In: Abduljabbar PH, editor. Steroids—Basic Science. online: InTech; 2012. [Google Scholar]
- 41.Mirth CK, Riddiford LM. Size assessment and growth control: how adult size is determined in insects. Bioessays. 2007; 29(4): 344–55. doi: 10.1002/bies.20552 [DOI] [PubMed] [Google Scholar]
- 42.Karim FD, Thummel CS. Ecdysone coordinates the timing and amounts of E74A and E74B transcription in Drosophila. Genes Dev. 1991; 5(6): 1067–79. [DOI] [PubMed] [Google Scholar]
- 43.Caldwell PE, Walkiewicz M, Stern M. Ras activity in the Drosophila prothoracic gland regulates body size and developmental rate via ecdysone release. Curr Biol. 2005; 15(20): 1785–95. doi: 10.1016/j.cub.2005.09.011 [DOI] [PubMed] [Google Scholar]
- 44.Tennessen JM, Thummel CS. Coordinating growth and maturation—insights from Drosophila. Curr Biol. 2011; 21(18): R750–7. doi: 10.1016/j.cub.2011.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamanaka N, Rewitz KF, O'Connor MB. Ecdysone control of developmental transitions: lessons from Drosophila research. Annu Rev Entomol. 2013; 58: 497–516 doi: 10.1146/annurev-ento-120811-153608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lewontin RC. The interaction of selection and linkage. II. Optimum models. Genetics. 1964; 50: 757–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pool JE, Corbett-Detig RB, Sugino RP, Stevens KA, Cardeno CM, Crepeau MW, et al. Population genomics of sub-Saharan Drosophila melanogaster: African diversity and non-African admixture. PLoS Genet. 2012; 8(12): e1003080 doi: 10.1371/journal.pgen.1003080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sellis D, Callahan BJ, Petrov DA, Messer PW. Heterozygote advantage as a natural consequence of adaptation in diploids. Proc Natl Acad Sci U S A. 2011; 108(51): 20666–71. doi: 10.1073/pnas.1114573108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pavlidis P, Metzler D, Stephan W. Selective sweeps in multilocus models of quantitative traits. Genetics. 2012; 192(1): 225–39. doi: 10.1534/genetics.112.142547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ellegren H, Parsch J. The evolution of sex-biased genes and sex-biased gene expression. Nat Rev Genet. 2007; 8(9): 689–98. doi: 10.1038/nrg2167 [DOI] [PubMed] [Google Scholar]
- 51.Bergland AO, Behrman EL, O'Brien KR, Schmidt PS, Petrov DA. Genomic evidence of rapid and stable adaptive oscillations over seasonal time scales in Drosophila. PLoS Genet. 2014; 10(11): e1004775 doi: 10.1371/journal.pgen.1004775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lehmann FO, Dickinson MH. The changes in power requirements and muscle efficiency during elevated force production in the fruit fly Drosophila melanogaster. J Exp Biol. 1997; 200(Pt 7): 1133–43. [DOI] [PubMed] [Google Scholar]
- 53.Lehmann FO. Ambient temperature affects free-flight performance in the fruit fly Drosophila melanogaster. J Comp Physiol B. 1999; 169(3): 165–71. [DOI] [PubMed] [Google Scholar]
- 54.Azevedo RB, James AC, McCabe J, Partridge L. Latitudinal variation of wing: thorax size ratio and wing-aspect ratio in Drosophila melanogaster. Evolution. 1998; 1353–62. doi: 10.1111/j.1558-5646.1998.tb02017.x [DOI] [PubMed] [Google Scholar]
- 55.Gilchrist AS, Partridge L. A comparison of the genetic basis of wing size divergence in three parallel body size clines of Drosophila melanogaster. Genetics. 1999; 153(4): 1775–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhan V, Parkash R, Aggarwal DD. Effects of body-size variation on flight-related traits in latitudinal populations of Drosophila melanogaster. J Genet. 2014. April 1; 93(1): 103–12. [DOI] [PubMed] [Google Scholar]
- 57.Stalker HD. Chromosome studies in wild populations of Drosophila melanogaster. II. Relationship of inversion frequencies to latitude, season, wing-loading and flight activity. Genetics. 1980; 95(1): 211–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frazier MR, Harrison JF, Kirkton SD, Roberts SP. Cold rearing improves cold-flight performance in Drosophila via changes in wing morphology. J Exp Biol. 2008; 211(Pt 13): 2116–22. doi: 10.1242/jeb.019422 [DOI] [PubMed] [Google Scholar]
- 59.James AC, Azevedo RB, Partridge L. Cellular basis and developmental timing in a size cline of Drosophila melanogaster. Genetics. 1995; 140(2): 659–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Colombani J, Bianchini L, Layalle S, Pondeville E, Dauphin-Villemant C, Antoniewski C, et al. Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science. 2005; 310(5748): 667–70. doi: 10.1126/science.1119432 [DOI] [PubMed] [Google Scholar]
- 61.Mirth CK, Shingleton AW. Integrating body and organ size in Drosophila: recent advances and outstanding problems. Front Endocrinol (Lausanne). 2012; 3: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007; 39(6): 715–20. doi: 10.1038/ng2049 [DOI] [PubMed] [Google Scholar]
- 63.Delanoue R, Slaidina M, Léopold P. The steroid hormone ecdysone controls systemic growth by repressing dMyc function in Drosophila fat cells. Dev Cell. 2010; 18(6): 1012–21. doi: 10.1016/j.devcel.2010.05.007 [DOI] [PubMed] [Google Scholar]
- 64.Nuzhdin SV, Brisson JA, Pickering A, Wayne ML, Harshman LG, McIntyre LM. Natural genetic variation in transcriptome reflects network structure inferred with major effect mutations: insulin/TOR and associated phenotypes in Drosophila melanogaster. BMC Genomics. 2009; 10(1): 124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Broughton SJ, Piper MD, Ikeya T, Bass TM, Jacobson J, Driege Y, et al. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl Acad Sci U S A. 2005; 102(8): 3105–10. doi: 10.1073/pnas.0405775102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Simon AF, Shih C, Mack A, Benzer S. Steroid control of longevity in Drosophila melanogaster. Science. 2003; 299(5611): 1407–10. doi: 10.1126/science.1080539 [DOI] [PubMed] [Google Scholar]
- 67.Gibbs AG, Matzkin LM. Evolution of water balance in the genus Drosophila. J Exp Biol. 2001; 204(Pt 13): 2331–8. [DOI] [PubMed] [Google Scholar]
- 68.Hartl DL, Clark AG. Principles of population genetics. 3rd ed. Sunderland, Mass: Sinauer Associates; 1997. [Google Scholar]
- 69.Mathelier A, Fornes O, Arenillas DJ, Chen CY, Denay G, Lee J, et al. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2016; 44(D1): D110–5. doi: 10.1093/nar/gkv1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carlsson P, Mahlapuu M. Forkhead transcription factors: key players in development and metabolism. Dev Biol. 2002; 250(1): 1–23. [DOI] [PubMed] [Google Scholar]
- 71.Carroll SB, Whyte JS. The role of the hairy gene during Drosophila morphogenesis: stripes in imaginal discs. Genes & Development. 1989; 3: 905–16. [Google Scholar]
- 72.Mann RS, Morata G. The developmental and molecular biology of genes that subdivide the body of Drosophila. Annu Rev Cell Dev Biol. 2000; 16: 243–71. doi: 10.1146/annurev.cellbio.16.1.243 [DOI] [PubMed] [Google Scholar]
- 73.Panganiban G, Rubenstein JL. Developmental functions of the Distal-less/Dlx homeobox genes. Development. 2002; 129(19): 4371–86. [DOI] [PubMed] [Google Scholar]
- 74.Hombría JC, Brown S. The fertile field of Drosophila Jak/STAT signalling. Curr Biol. 2002; 12(16): R569–75. [DOI] [PubMed] [Google Scholar]
- 75.Liu Y, Lehmann M. Genes and biological processes controlled by the Drosophila FOXA orthologue Fork head. Insect Mol Biol. 2008; 17(2): 91–101. doi: 10.1111/j.1365-2583.2007.00785.x [DOI] [PubMed] [Google Scholar]
- 76.Karim FD, Guild GM, Thummel CS. The Drosophila Broad-Complex plays a key role in controlling ecdysone-regulated gene expression at the onset of metamorphosis. Development. 1993; 118(3): 977–88. [DOI] [PubMed] [Google Scholar]
- 77.Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific φC31 integrases. Proc Natl Acad Sci U S A. 2007; 104(9): 3312–7. doi: 10.1073/pnas.0611511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007; 448(7150): 151–6. doi: 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- 79.Pfaffl MW. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001; 29(9): e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zheng L, Baumann U, Reymond J-L. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004; 32(14): e115 doi: 10.1093/nar/gnh110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hense W, Baines JF, Parsch J. X chromosome inactivation during Drosophila spermatogenesis. PLoS Biol. 2007; 5(10): e273 doi: 10.1371/journal.pbio.0050273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012; 9(7): 671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2015. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
Relative expression in late wandering third instar larvae in (A) the Netherlands (Net.), Malaysia (Mal.), Egypt (Egy.), Zimbabwe (Zim.), and Zambia (Zam.) populations (N = 10–12 isofemale strains per population with 2 biological replicates per strain). Blue bars represent cosmopolitan populations, and white bars represent sub-Saharan populations. (B) Relative expression in each population represented according to the variant at position 67. The high-expression, cosmopolitan “G” variant is shown in blue, and the low-expression, sub-Saharan “C” variant is shown in white. For simplicity, Zambian and Zimbabwean expression are presented together as sub-Saharan African (Afr.) expression. Underlying data can be found in S1 Data. Error bars indicate the standard error of the mean. Differences between populations were tested by a t test, and a Bonferroni multiple test correction was applied. ns, not significant; ●P < 0.10, *P < 0.05, **P < 0.01, ***P < 0.005.
(PDF)
(A) Relative expression of CG9509 in the CG9509del line as determined by quantitative reverse transcription PCR (qRT-PCR). For comparison, the average expression of the population in which the CG9509del line was discovered (Munich) is shown. Underlying data can be found in S1 Data. Error bars represent the standard error of the mean. (B) DNA sequence alignment spanning the deletion within the CG9509del coding region. Sequences of flies from the CG9509del source population in Munich (MU) are shown for comparison. (C) Amino acid alignment spanning the frameshift within the CG9509del coding region. Sequences of flies from the CG9509del source population are shown for comparison (MU). The asterisk indicates a stop codon.
(PDF)
Adult (A) DDT, (B) malathion, (C) ethanol (N = 6–8 replicates per line, sex, and concentration), and (D) cold tolerance assay results (N = 25 per line and sex) for control (blue lines or bars) and RNAi-CG9509 (gray hatched lines or white bars) flies. Underlying data can be found in S2 Data. In panels A–C, significance was assessed using a generalized linear model with a quasibinomial distribution. In panel D, significance was assessed using a t test. ns, not significant; *P < 0.05, **P < 0.01, ***P < 0.005.
(PDF)
(A, B) Duration of larval stage in (A) control (blue; N = 25) and RNAi-CG9509 (white; N = 51) flies and (B) control (blue; N = 242) and CG9509del (white; N = 107) flies. (C, D) Duration of wandering stage in (C) control (blue; N = 16) and RNAi-CG9509 (white; N = 40) flies and (D) CG9509del (white; N = 10) and control (blue; N = 40) flies. Underlying data can be found in S2 Data. Error bars represent the standard deviation. The knockdown and hypomorph lines were not significantly different from their respective control lines for either stage (t test; P > 0.4 for all comparisons).
(PDF)
(A) Body weight per 25 flies (N = 10–12 isofemale lines per population with 4 replicates per sex), (C) wing length (N = 10–12 isofemale lines per population with 4 replicates per sex), and (E) wing area in a Dutch (blue bars) and a Zambian (white bars) population (N = 10–12 isofemale lines per population with 4 replicates per sex). (B) Body weight per 25 flies (N = 6 isofemale lines per variant with 4 replicates per sex), (D) wing length (N = 6 isofemale lines per variant with 4 replicates per sex), and (F) wing area in a Dutch population separated according to the variant at position 67 (N = 6 isofemale lines per variant with 4 replicates per sex). The derived, high-expression “G” is shown in blue, and the ancestral, low-expression “C” variant is shown in white. Error bars indicate the standard error of the mean. Underlying data can be found in S2 Data. Significance was assessed with an ANOVA using sex, isofemale line, and population or the variant at position 67 as factors (shown in black). Significance was additionally assessed in both populations simultaneously with population and the variant at position 67 included as factors (shown in red). ns, not significant; ●0.05 < P < 0.10, *P < 0.05, **P < 0.01, ***P < 0.005.
(PDF)
(A, B) Levels of active ecdysone, approximated by relative E74B expression as measured by quantitative reverse transcription PCR (qRT-PCR) in (A) early and (B) late wandering third instar larvae in control (blue) and RNAi-CG9509 (white) flies (N = 9–10 per line). (C, D) Relative gene expression of dMyc and dFOXO in control (blue) and RNAi-CG9509 (white) (C) early wandering third instar larvae and (D) late wandering third instar larvae (N = 9–10 per line). Expression is shown relative to the control for each stage. Underlying data can be found in S2 Data. Error bars represent the standard error of the mean. Significance was assessed via a t test. *P < 0.05, **P < 0.01, ***P < 0.005.
(PDF)
Distribution of (A) wing loading, (C) wing area, (E) wing length, and (G) body weight in a Dutch (blue) and a Zambian (yellow) population. Distribution of (B) wing loading, (D) wing area, (F) wing length, and (H) body weight in a Dutch population separated according to the variant at position 67 (6 isofemale lines per variant). The derived, high-expression “G” is shown in blue, and the ancestral, low-expression “C” variant is shown in yellow.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(DOCX)
Data Availability Statement
All sequence files are available from the GenBank database (accession numbers MG195568 – MG195570, HF913659.1, HF913664.1, HF913717.1). Dutch whole genome sequence data are available from http://evol.bio.lmu.de/downloads.