Abstract
MicroRNA (miR)-338-5p has been studied in hepatocellular carcinoma (HCC); however, the diagnostic value and molecular mechanism underlying its actions remains to be elucidated. The present study aimed to validate the diagnostic ability of miR-338-5p and further explore the underlying molecular mechanism. Data from eligible studies, Gene Expression Omnibus (GEO) chips and The Cancer Genome Atlas (TCGA) datasets were gathered in the data mining and the integrated meta-analysis, to evaluate the significance of miR-338-5p in diagnosing HCC comprehensively. The potential target genes of miR-338-5p were achieved from the intersection of the deregulated targets of miR-338-5p from GEO and TCGA in addition to the predicted target genes from 12 online software. A protein-protein-interaction (PPI) network was drawn to illustrate the interaction between target genes and to define the hub genes. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed to investigate the function of the target genes. From the results, miR-338-5p exhibited favorable value in diagnosing HCC. Types of sample and experiment were defined as the possible sources of heterogeneity in meta-analysis. A total of 423 genes were selected as the potential target genes of miR-338-5p, and five genes were defined as the hub genes from the PPI network. The GO and KEGG analyses indicated that the target genes were significantly assembled in the pathways of metabolic process and cell cycle. miR-338-5p may function as a novel diagnostic target for HCC through regulating certain target genes and signaling pathways.
Keywords: miR-338-5p, HCC, GEO, meta-analysis, target genes
Introduction
Hepatocellular carcinoma (HCC) ranked as the 5th most frequent cancer and was also one of the lethal cancers, particularly in People's Republic of China where liver cancer was the most commonly diagnosed cancer and the most prevalent cause of cancer-related deaths followed by lung, stomach, and esophageal cancers (based on the statistics in 2015) (1,2). Because HCC was often diagnosed at an advanced stage, the prognosis of HCC patients was not optimistic (3). Therefore, a better understanding of the pathogenesis of HCC and a novel target for the early screening of HCC might improve the survival of HCC patients (4).
MicroRNAs (miRNAs) are small non-coding RNAs (18–25 nucleotides in length) that regulate the expression of multiple mRNAs at the post-transcriptional level by suppressing the stability and the translation of mRNAs (5,6). The aberrant expressions of miRNAs was observed in various human cancers, and extensive studies suggested that these deregulated miRNAs had the capacity to distinguish malignant tumors of liver, breast, lung, pancreas and leukemia from adjacent non-tumorous tissue (7–12). miR-338-3p and miR-338-5p originate from an intron of the gene encoding apoptosis-associated tyrosine kinase (AATK). Both miR-338-3p and miR-338-5p are co-expressed because they enjoyed the same promoter together (13). miR-338 was the prvious ID of miR-338-3p, which had been reported in variety of diseases (13–16). miR-338* is one of the members of miR-338 family and usually represented as miR-338-5p (17). As a member of the miRNA family, miR-338-5p was reported to be correlated with the carcinogenesis and progression of several human cancers including gastric cancer (18), colorectal cancer (19), and glioblastoma (20). However, there were limited studies on the clinical significance of miR-338-5p in HCC. Chen et al reported the overexpression of miR-338-5p in tumor tissues of the liver and preoperative plasma by miRNA array in Asian patients (21). Whether miR-338-5p is indeed a qualified diagnostic biomarker for HCC and the underlying molecular mechanism of miR-338-5p in HCC remained unclarified.
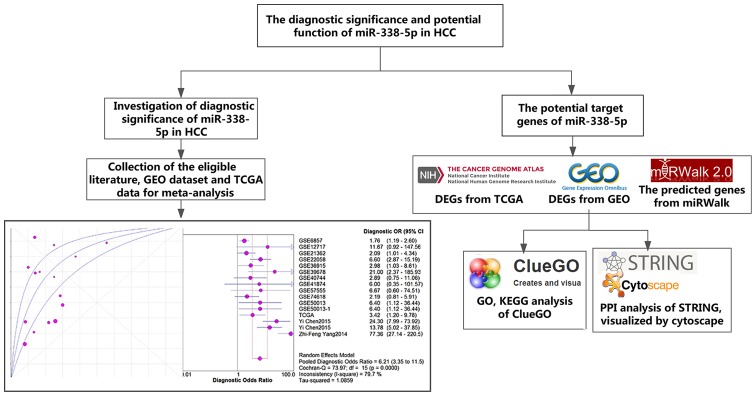
Therefore, this study aimed to investigate the diagnostic significance of miR-338-5p in HCC tissues and the molecular mechanism of miR-338-5p in HCC with a combination of meta-analysis and bioinformatics analysis. Our study confirmed the significance of miR-338-5p for the diagnosis of HCC and might promote the understanding of the molecular mechanism underlying it. The framework of this article was displayed in Fig. 1.
Figure 1.
The framework of this study. The framework described the idea of this study.
Materials and methods
The process of study selection
In order to obtain the comprehensive data of the diagnostic value of miR-338-5p in HCC, a thorough search for the related studies was conducted in Gene Expression Omnibus (GEO) dataset and other database including PubMed, Embase, Cochrane, Web of Science, Sinomed, Chinese VIP, Wanfang database and China National Knowledge Infrastructure (CNKI) until December 15, 2016 with the searching strategies: (miR-338 or miRNA-338 or microRNA-338 or miR338 or miRNA338 or microRNA338 or ‘miR 338’ or ‘miRNA 338’ or ‘microRNA 338’) and (malignan* or cancer or tumor or tumour or neoplas* OR carcinoma) AND (hepatocellular or liver or hepatic or HCC). Studies that meet the following criteria were eligible for the meta-analysis: i) Studies evaluated the expression of miR-338-5p for the diagnosis of HCC; ii) the disease of the patients were validated with golden standard; iii) the number of cases were reported in the study; and iv) the sensitivity and specificity of the diagnostic test were available directly or indirectly from the study. The exclusion criteria of the studies were as follows: i) The content of the studies were irrelevant with HCC; ii) the subjects of experiment were not human beings; iii) there was no sufficient data for researchers to directly acquire or calculate sensitivity or specificity of the diagnostic test; and iv) studies were classified as review, meta-analysis, case study or conference note. Moreover, data of the diagnostic value of miR-338-5p in HCC was also downloaded from the cancer genome atlas (TCGA) (https://cancergenome.nih.gov/).
Data extraction and statistical analysis
The following information and data were extracted from the included studies: The ID no. of each GSE chip, first author, year of publication, country, experiment type, platform of each GSE chip, sample number for the experiment group and control group, tissue types, true positivity (TP), false positivity (FP), false negativity (FN) and true negativity (TN).
MetaDiSc1.4 and STATA12.0 were applied for all the statistical analysis. To explore expression of miR-338-5p, the continuous outcomes of GEO and TCGA datasets were calculated with standard mean difference (SMD). The sensitivity (SEN), specificity (SPE), positive likelihood ratio (PLR), negative likelihood ratio (NLR) and diagnostic odds ratio (DOR) of the included studies were pooled with the bivariate meta-analysis model (22,23). The summary receiver operator characteristic (SROC) curve was plotted according to the sensitivity and specificity from each study. The area under the SROC curve (AUC) calculated from the SROC reflected the capacity of miR-338-5p to differentiate HCC patients from non-cancer patients accurately. An AUC value of 0.5 or 1.0 represents a poor or perfect diagnostic value, respectively (24). Additionally, Q test and I2 statistics were employed to assess the heterogeneity between studies. The random-effects model would be used to pool the results if an I2 value was more than 50% with a P-value <0.10; otherwise, a fixed-effects model would be applied (25,26). To identify the source of heterogeneity, the subgroup analysis was conducted based on the number and features of the included studies. With regard to the publication bias, the Deeks' funnel plot asymmetry test was carried out to detect the publication bias, and P-value <0.05 was indicative of significance.
The target genes of miR-338-5p
The potential target genes of miR-338-5p came from three sources: The differentially expressed genes from GEO, TCGA and the predicted genes from 12 online software (miRWalk, MicroT4, miRanda, mirBridge, miRDB, miRMap, miRNAMap, PicTar2, PITA, RNA22, RNAhybrid and TargetScan). We firstly searched Gene Expression Omnibus (GEO) datasets for deregulated target genes of miR-338-5p from the mRNA profiling data of HCC samples on December 15, 2016. All the GSE chips shared the same platform: GPL570 (Affymetrix Human Genome U133 Plus 2.0 Array). After preliminary screening, 54 studies remained for further selection. Among the 54 studies, Homo sapiens tissue samples instead of cell lines samples were included for further analysis. Finally, 10 HCC datasets (GSE29721, GSE45436, GSE55092, GSE62232, GSE9843, GSE41804, GSE6764, GSE33006, GSE6222 and GSE19665) comprising 431 HCC samples and 198 control samples were chosen for further analysis. Differentially expressed genes (DEGs) between cancer and normal samples of 10 datasets were acquired via GCBI online tool (https://www.gcbi.com.cn/gclib/html/index). Fold-change >1.5, and a P-value <0.05 was set as the threshold for the DEGs. Another database containing high-throughput data: TCGA was also searched. Publicly available miRNA-seq and RNA-seq data of liver HCC was downloaded from the TCGA data portal (December 2016, https://gdc-portal.nci.nih.gov/). Since the TCGA data were a community resource project, additional approval by an ethics committee of our hospital was not mandatory. And the present study adhered to the TCGA publication guidelines and data access policies. From the downloaded data of 377 HCC samples and 50 normal liver samples. R language package DESeq was subsequently used for the calculation of DEGs (Padj <0.05 and the absolute log2 fold-change >1). As for the predicted genes, selection was based on the condition that they were recorded in more than 4 of the 12 prediction software. The selected qualified target genes from the online software and the validated target genes from miRWalk were considered as the potential target genes of miR-338-5p.
The protein-protein-interaction (PPI) network and validation of target genes
To illustrate the interaction between the targets of miR-338-5p, a PPI network was drawn by Cytoskape v.5.3.0. The nodes and edges represented target genes and the interactions between target genes, respectively. Hub genes were identified according to the value of degrees of each node. Protein expression of hub gene was validated by The Human Protein Atlas (HPA), an immunohistochemisty (IHC) database (27). Each antibody in the database has been used for IHC staining of both normal and HCC tissues.
The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of the target genes
The GO and KEGG pathway analysis were performed by the BiNGO and ClueGO plug-in unit in Cytoscape v.3.5.0 for the functional annotation of the target genes. Three GO terms including biological process (BP), cellular component (CC) and molecular function (MF) were utilized to identify the enrichment of target genes. P-value <0.05 was significant.
Results
Eligible studies for the meta-analysis
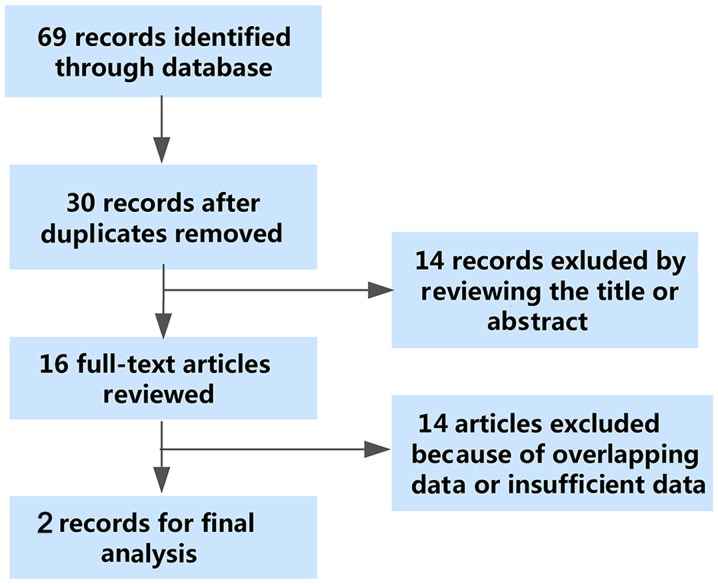
As shown in Fig. 2, the flowchart exhibited the selection and retrieval process of the qualified studies. A total of 69 studies were identified as the initial records, and 30 studies remained after the removal of duplicate records. Then, 14 records were excluded in the preliminary screening of the titles and abstracts of the articles. As a consequence, 16 studies were reviewed in the full text. Among the 16 studies, 14 studies were ineligible due to insufficient data of the diagnostic parameters or duplicate data. Eventually, two studies were enrolled for the meta-analysis. Though two included studies were conducted by the same authors, we failed to validate that the two studies shared the same patient cohorts. Thus, we regard the two studies of Chen et al as two different studies (21,28).
Figure 2.
The flowchart: The detailed process of study selection was recorded in the flowchart. After the preliminary screening and full-text scrutinize, three studies were considered eligible studies for the meta-analysis.
Assessment of the diagnostic value and the integrated meta-analysis
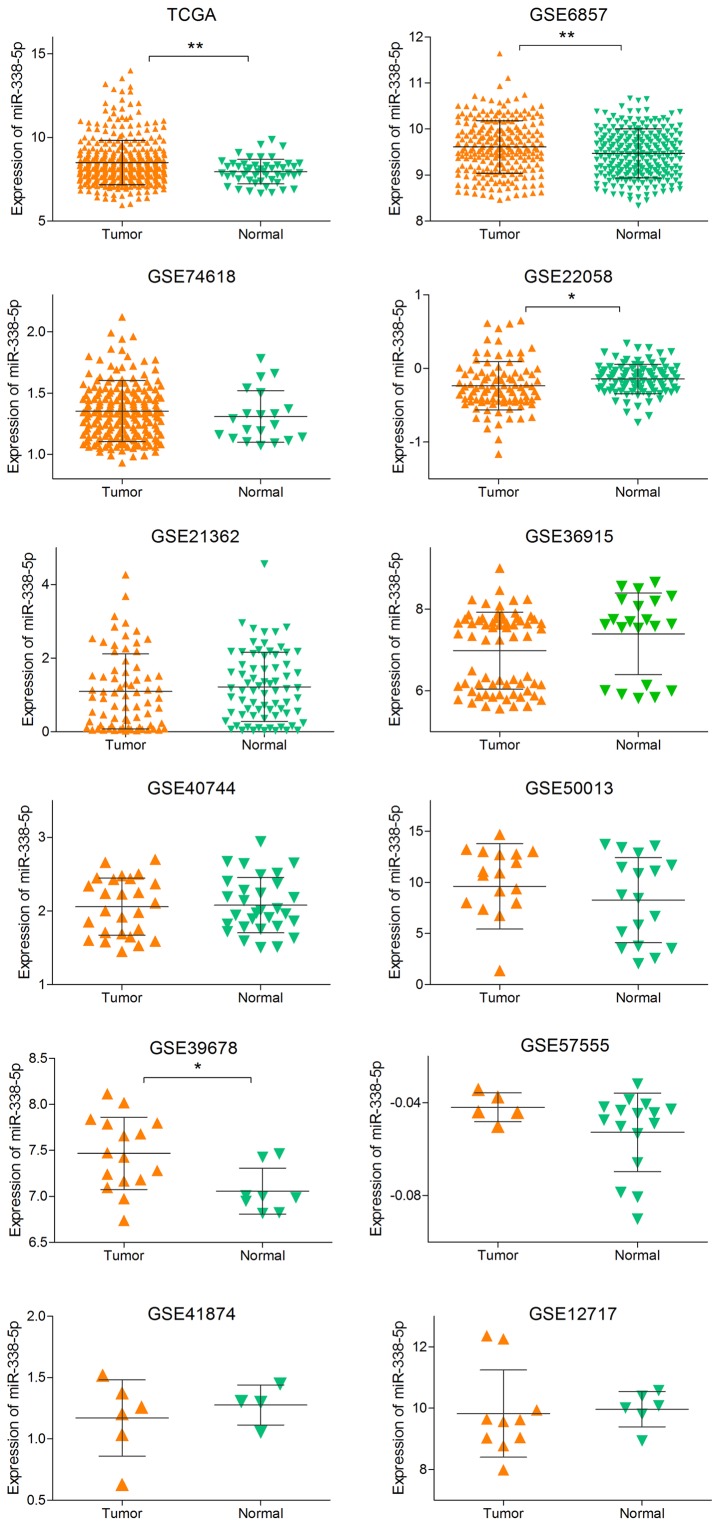
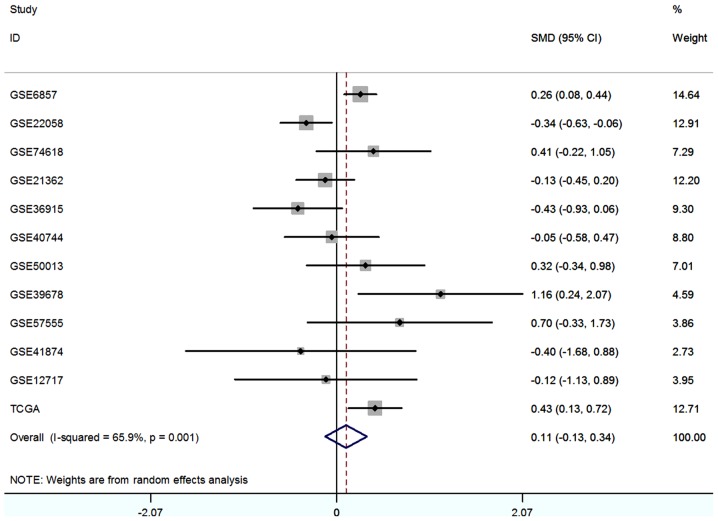
To comprehensively evaluate the diagnostic value of miR-338-5p, we supplemented the literature analysis with GEO data and TCGA data. We searched the GEO dataset with the same searching strategies in literature meta-analysis. Finally, a total of eligible 11 GSE chips were included in our meta-analysis (29–38) (Table I), and the expression level of each study were showed in Fig. 3. With the random-effects model, the forest-plot represented that no significant difference expression was observed between HCC tissue and normal tissue. The pooled SMD (0.11, 95% CI: −0.13, 0.34) was showed in Fig. 4.
Table I.
Basic information and clinical data of the included studies.
| GEO accession | Author, year | Public year | Country | Experiment type | Platform | HCC | Control | Sample type | TP | FP | FN | TN | (Refs.) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GSE6857 | Budhu et al, 2008 | 2008 | USA | Non-coding RNA profiling by array | GPL4700 | 240 | 241 | Tissue | 91 | 62 | 149 | 179 | (29) |
| GSE12717 | Su et al, 2009 | 2008 | USA | Non-coding RNA profiling by array | GPL7274 | 10 | 6 | Tissue | 7 | 1 | 3 | 5 | (30) |
| GSE22058 | Burchard et al, 2010 | 2010 | USA | Non-coding RNA profiling by array | GPL10457 | 96 | 96 | Tissue | 36 | 8 | 60 | 88 | (31) |
| GSE40744 | Diaz et al, 2013 | 2013 | USA | Non-coding RNA profiling by array | GPL14613 | 26 | 19 | Tissue | 8 | 4 | 18 | 26 | (32) |
| GSE50013 | Shen et al, 2013 | 2013 | USA | Non-coding RNA profiling by array | GPL15497 | 18 | 18 | Plasma | 16 | 10 | 2 | 8 | (33) |
| GSE41874 | Morita (unpublished) | 2013 | Japan | Non-coding RNA profiling by array | GPL7722 | 6 | 4 | Tissue | 4 | 1 | 2 | 3 | – |
| GSE57555 | Murakami et al, 2015 | 2015 | Japan | Non-coding RNA profiling by array | GPL16699 | 5 | 16 | Tissue | 4 | 6 | 1 | 10 | (34) |
| GSE74618 | Martinez-Quetglas et al, 2016 | 2016 | Spain | Non-coding RNA profiling by array | GPL14613 | 223 | 20 | Tissue | 131 | 10 | 92 | 10 | (35) |
| GSE36915 | Shih et al, 2012 | 2012 | Taiwan | Non-coding RNA profiling by array | GPL8179 | 68 | 21 | Tissue | 37 | 6 | 31 | 15 | (36) |
| GSE39678 | Noh et al, 2013 | 2013 | South Korea | Non-coding RNA profiling by array | GPL15852 | 16 | 8 | Tissue | 14 | 2 | 2 | 6 | (37) |
| GSE21362 | Sato et al, 2011 | 2011 | Japan | Non-coding RNA profiling by array | GPL10312 | 73 | 73 | Tissue | 27 | 16 | 46 | 57 | (38) |
| TCGA | miRNA-Seq | Illumina | 375 | 50 | Tissue | 156 | 8 | 218 | 42 | ||||
| Literaure | Chen et al, 2015 | 2015 | China | qRT-PCR | 37 | 31 | Plasma | 27 | 0 | 10 | 31 | (21) | |
| Literaure | Chen et al, 2015 | 2015 | China | qRT-PCR | 39 | 25 | Plasma | 35 | 4 | 4 | 21 | (28) |
TP, true positivity; FP, false positivity; FN, false negativity; TN, true negativity.
Figure 3.
The expression level of each study from TCGA and GEO. Scatter plots were created to illustrate the expression of miR-338-5p in normal tissues and HCC for each study from TCGA and GEO. *P<0.05; **P<0.01. TCGA, The Cancer Genome Atlas; GEO, Gene Expression Omnibus; HCC, hepatocellular carcinoma.
Figure 4.
Forest plot of studies calculating SMD of miR-338-5p expression in HCC group compared with control group. SMD, standard mean difference; HCC, hepatocellular carcinoma.
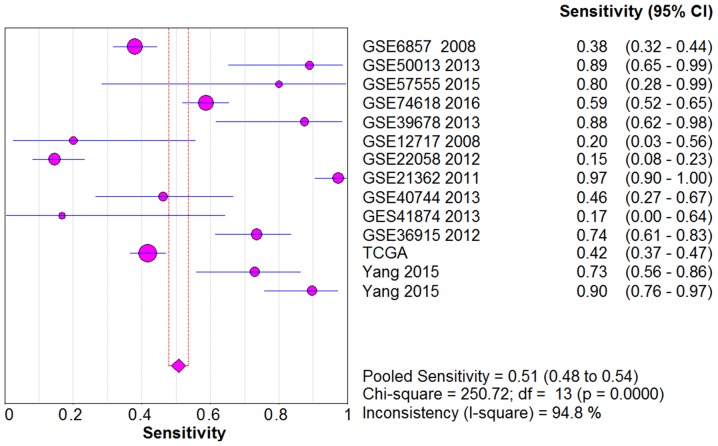
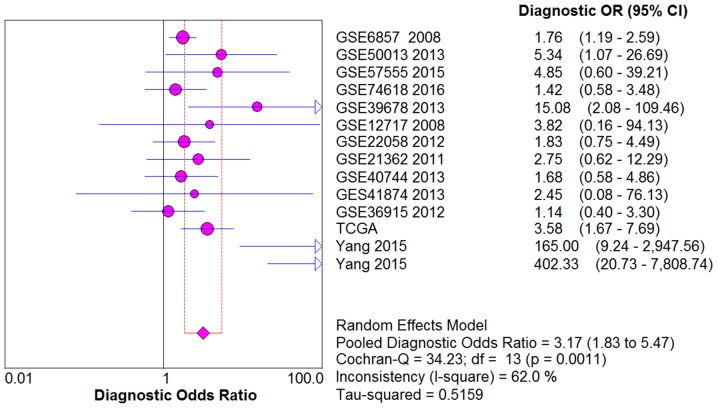
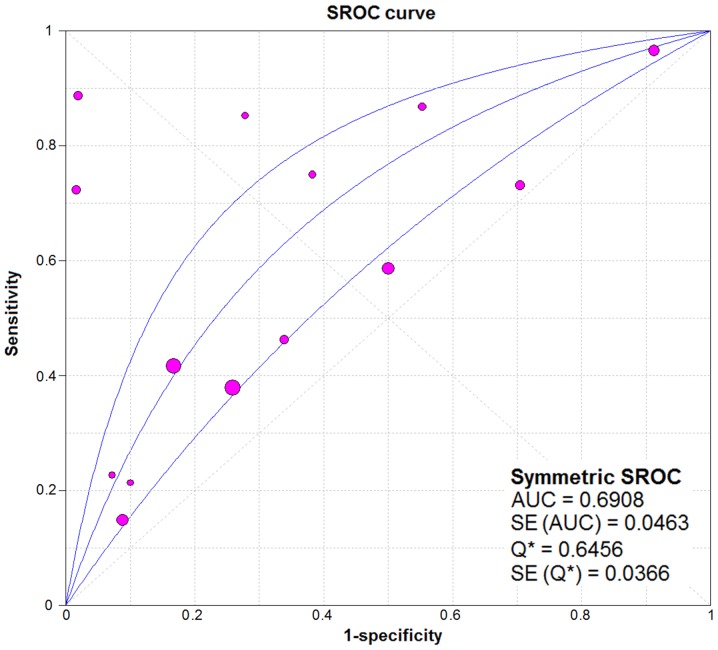
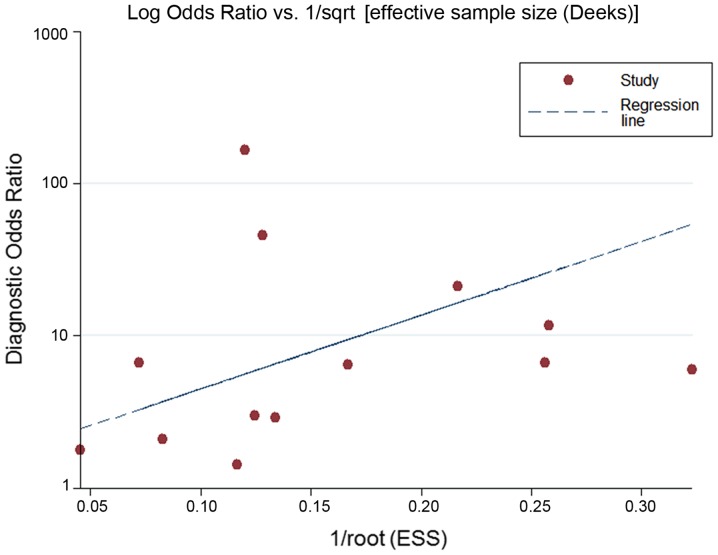
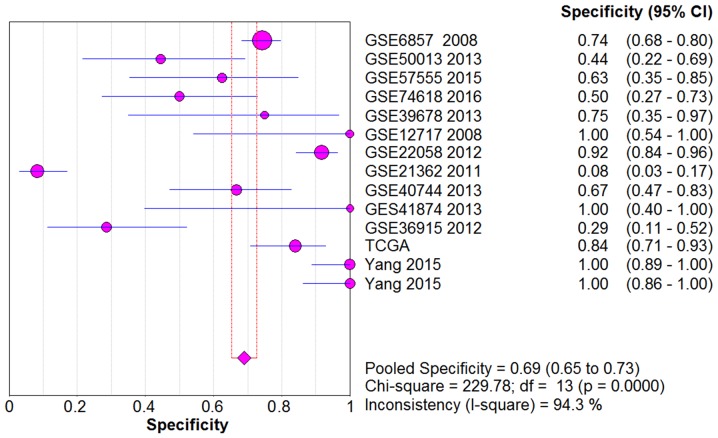
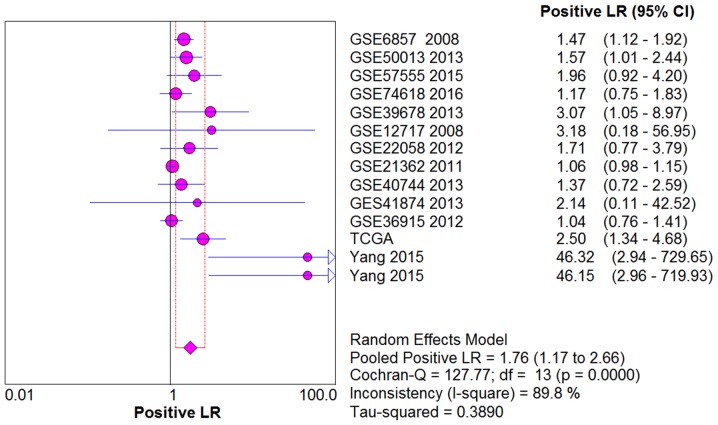
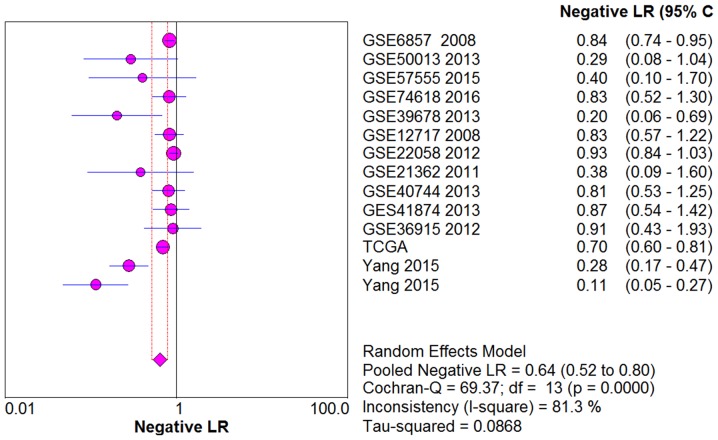
From the chi-square test and I2 test, significant heterogeneity existed in all the pooled effects (SE, SP, PLR, NLR and DOR) between studies (All I2>50%; P<0.05). Therefore, random effects model were employed to estimate the overall SE, SP, PLR, NLR and DOR of all the data. As shown in Figs. 5–9, the SE, SP, PLR, NLR and DOR of all the studies were 0.51 (95% CI: 0.48–0.54), 0.69 (95% CI: 0.65–0.73), 1.76 (95% CI: 1.17–2.66), 0.64 (95% CI: 0.52–0.80) and 3.17 (95% CI: 1.83–5.47). As for the result of SROC, the AUC value of miR-338-5p was 0.691 (Fig. 10). Moreover, the Deeks funnel plot asymmetry test was carried out with Stata 12.0, and no publication bias was detected (P>0.05) (Fig. 11).
Figure 5.
The pooled SE of the integrated meta-analysis. The pooled SE of the integrated meta-analysis was 0.51 (95% CI: 0.48–0.54).
Figure 9.
The pooled DOR of the integrated meta-analysis. The pooled DOR of the integrated meta-analysis was 3.17 (95% CI: 1.83–5.47). DOR, diagnostic odds ratio.
Figure 10.
The pooled SROC of the integrated meta-analysis. The pooled SROC of the integrated meta-analysis was 0.691. SROC, summary receiver operator characteristic.
Figure 11.
The publication bias. The symmetrical Deeks funnel plot indicated no publication bias.
Now that significant heterogeneity existed between the studies, the subgroup analysis was performed to seek the potential sources of heterogeneity. In the subgroup of sample types, the heterogeneity decreased substantially in the pooling estimates of NLR (49.2%) and DOR (5.6%) in the group of tissue. The value of SE, SP, PLR and DOR were obviously higher in the plasma group (0.83, 0.74–0.90; 0.86, 0.77–0.93; 14.02, 0.08–2,395.96; 61.30, 3.61–1,040.31) than in the tissue group (0.48, 0.45–0.51; 0.67, 0.63–0.71; 1.51, 1.08–2.11; 2.05, 1.51–2.77) and the value of NLR was notably lower in the plasma group (0.21, 0.11–0.38) than in the tissue group (0.81, 0.71–0.92).
With regard to the subgroup of experiment, the heterogeneity decreased substantially in the pooling estimate of DOR (10.3%) in the microarray group, and declined heterogeneity of SE (0%), PLR (0%) and DOR (0.0%) were also observed in the group of qRT-PCR. The value of SE, SP, PLR and DOR were obviously higher in the qRT-PCR group (0.82, 0.71–0.90; 1.00, 0.94–1.00; 46.23, 6.60–323.68; 254.42, 32.2–2,010.45) than in the microarray group (0.49, 0.46–0.52; 0.66, 0.62–0.70; 1.51, 1.11–2.06; 2.15, 1.56–2.97) and the value of NLR was notably lower in the qRT-PCR group (0.19, 0.08–0.47) than in the microarray group (0.79, 0.69–0.91). This result confirmed that types of sample and experiment were the possible sources of heterogeneity in this study.
Bioinformatics study of the target genes of miR338-5p
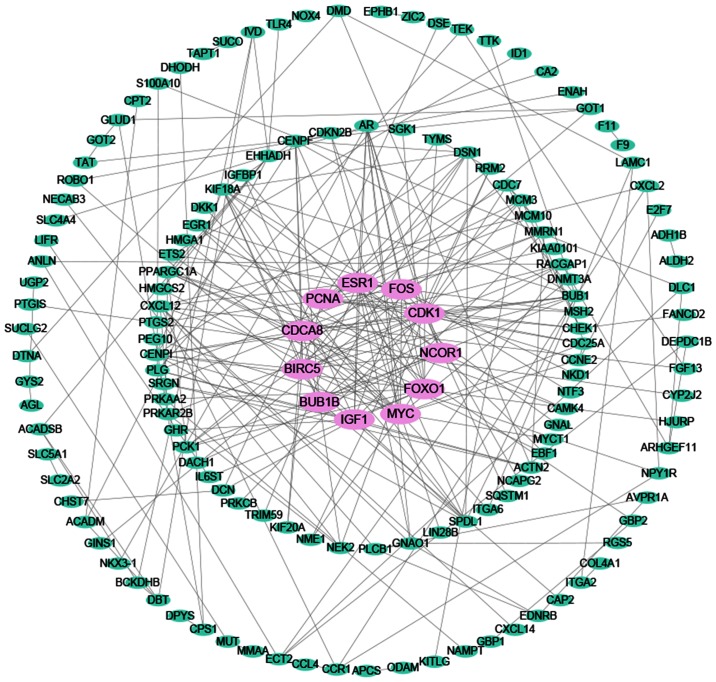
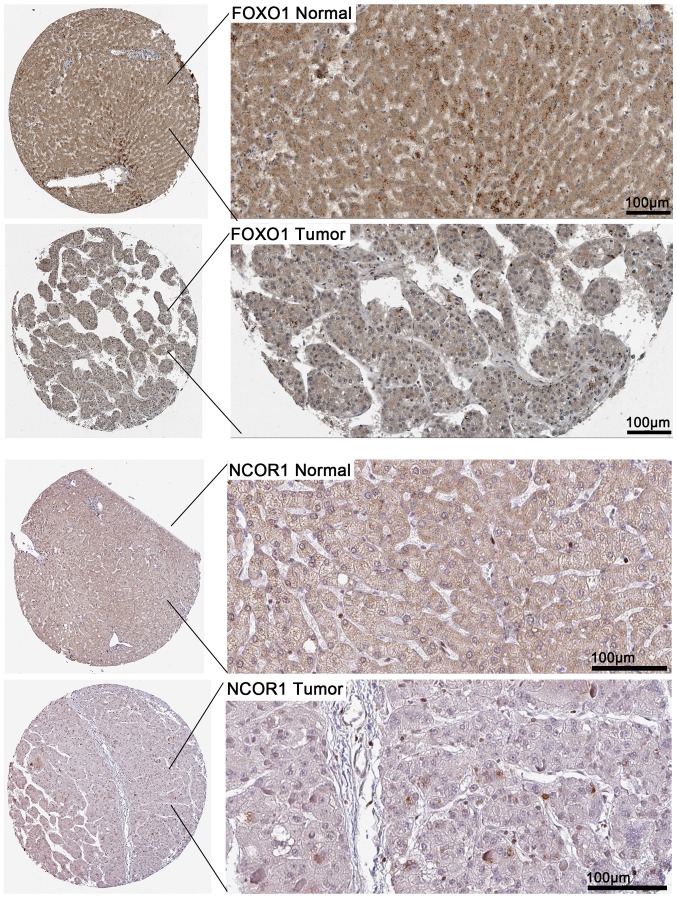
According to the results, a total of 1,698 and 1,798 genes were identified as DEGs targeted by miR-338-5p from TCGA and GEO, respectively. Additionally, a total of 3,610 predicted target genes that appeared in more than four times of the 12 online software were obtained. Taking the intersection of the DEGs from GEO and TCGA as well as the qualified predicted targets genes, we selected 423 genes for the following bioinformatics analyses (Fig. 12). The PPI network shown in Fig. 13 illustrated the interactions between the target genes of miR-338-5p. There were 147 nodes and 248 edges in the network. Hub genes with a degree values of more than 11 including NCOR1, IGF1, FOXO1, FOS, CDCA8, BUB1B, PCNA, ESR1, BIRC5, MYC and CDK1 were emphasized in red while the remaining were colored in green. To verify that these hub genes are targeted by miR-338-5p, we obtained the immunohistochemical staining of several of the hub genes including NCOR1 and FOXO1 in HCC tissues and normal tissues. As shown in Fig. 14, NCOR1 and FOXO1 were found to have medium staining and moderate intensity in cytoplasmic/menbranous of normal tissues, while a lower staining and weaker intensity of these genes were observed in HCC tissues.
Figure 12.
The Venn diagram for the target genes of miR-338-5p. Three circles represented the collected target genes from three different sources (blue for TCGA, yellow for GEO and green for the predicted target genes) There were 1,698, 1,798 and 3,610 potential target genes from TCGA, GEO and online software, respectively. A total of 423 genes were obtained after taking the intersection of the three parts. TCGA, The Cancer Genome Atlas; GEO, Gene Expression Omnibus.
Figure 13.
The PPI network of the target genes of miR-338-5p. A total of 147 nodes and 248 edges constituted the network, from which 11 hub genes colored in red were identified according to the value of degrees. PPI, protein-protein-interaction.
Figure 14.
The immunohistochemical staining of NCOR1 and FOXO1 in normal tissues and HCC tissues. According to the immunohistochemical results of these genes in HPA database, NCOR1 and FOXO1 presented medium staining in normal tissues; while a lower staining of these genes were observed in HCC tissues. HCC, hepatocellular carcinoma; HPA, the Human Protein Atlas.
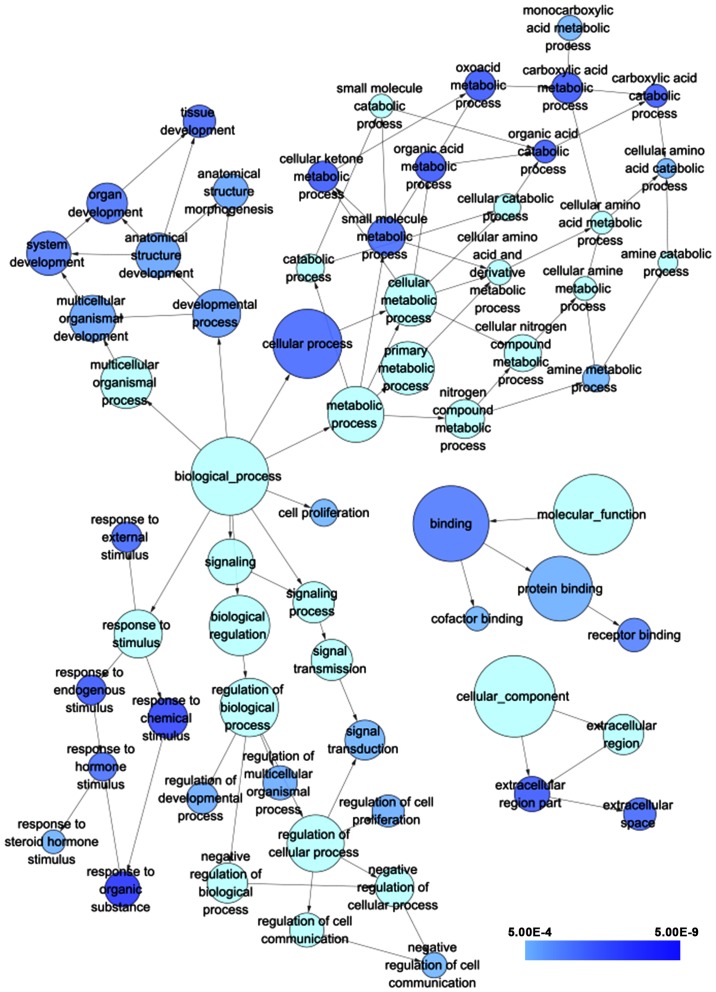
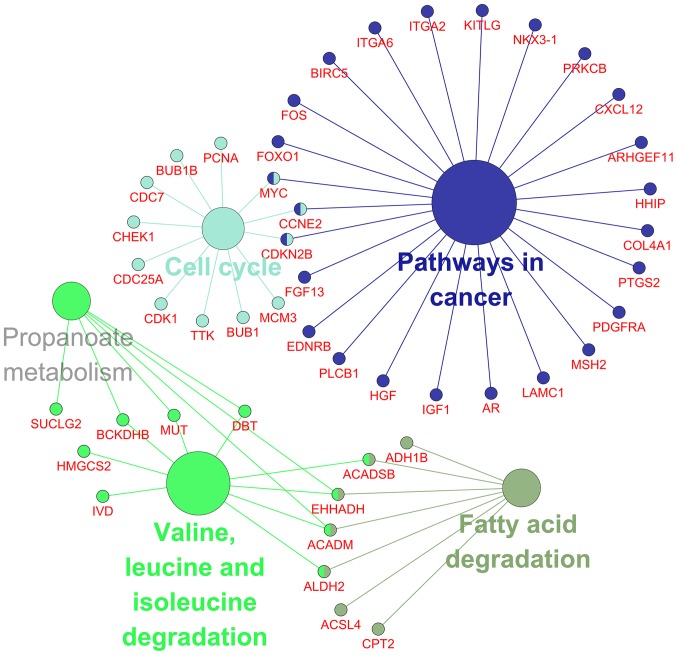
According to the results of GO analysis in cytoskape, the target genes were found to enrich most significantly in the following biological pathways: response to organic substance, response to chemical stimulus and oxoacid metabolic process. As for cellular component and molecular function, target genes mainly assembled in extracellular region part and binding, respectively (Table II; Fig. 15). Moreover, a total of 5 significant pathways were recorded from the KEGG pathway analysis such as valine, leucine and isoleucine degradation, pathways in cancer and cell cycle (Table III; Fig. 16) were the most significant.
Table II.
GO functional annotation of the target genes of miR-338-5p from Cytoskape.
| ID | Category | GO term | P-value | Count |
|---|---|---|---|---|
| GO:0006082 | GO_Biological process | Organic acid metabolic process | 3.87×10−9 | 37 |
| GO:0016054 | GO_Biological process | Organic acid catabolic process | 6.26×10−9 | 16 |
| GO:0008152 | GO_Biological process | Metabolic process | 1.29×10−3 | 159 |
| GO:0009056 | GO_Biological process | Catabolic process | 1.15×10−1 | 28 |
| GO:0007275 | GO_Biological process | Multicellular organismal development | 1.11×10−6 | 102 |
| GO:0048731 | GO_Biological process | System development | 9.71×10−8 | 91 |
| GO:0048519 | GO_Biological process | Negative regulation of biological process | 1.82×10−5 | 72 |
| GO:0048523 | GO_Biological process | Negative regulation of cellular process | 3.81×10−5 | 66 |
| GO:0050789 | GO_Biological process | Regulation of biological process | 3.62×10−4 | 176 |
| GO:0065007 | GO_Biological process | Biological regulation | 6.72×10−5 | 189 |
| GO:0008150 | GO_Biological process | Biological_process | 1.15×10−3 | 336 |
| GO:0032501 | GO_Biological process | Multicellular organismal process | 1.15×10−5 | 133 |
| GO:0044237 | GO_Biological process | Cellular metabolic process | 6.22×10−3 | 132 |
| GO:0034641 | GO_Biological process | Cellular nitrogen compound metabolic process | 2.19×10−2 | 59 |
| GO:0044238 | GO_Biological process | Primary metabolic process | 4.35×10−3 | 140 |
| GO:0050896 | GO_Biological process | Response to stimulus | 4.59×10−5 | 112 |
| GO:0009719 | GO_Biological process | Response to endogenous stimulus | 3.77×10−9 | 34 |
| GO:0050794 | GO_Biological process | Regulation of cellular process | 9.14×10−4 | 166 |
| GO:0023052 | GO_Biological process | Signaling | 5.63×10−5 | 99 |
| GO:0044281 | GO_Biological process | Small molecule metabolic process | 3.29×10−8 | 62 |
| GO:0006519 | GO_Biological process | Cellular amino acid and derivative metabolic process | 5.92×10−5 | 21 |
| GO:0043436 | GO_Biological process | Oxoacid metabolic process | 2.77×10−9 | 37 |
| GO:0019752 | GO_Biological process | Carboxylic acid metabolic process | 2.77×10−9 | 37 |
| GO:0009987 | GO_Biological process | Cellular process | 2.40×10−8 | 258 |
| GO:0046395 | GO_Biological process | Carboxylic acid catabolic process | 6.26×10−9 | 16 |
| GO:0009063 | GO_Biological process | Cellular amino acid catabolic process | 2.78×10−6 | 10 |
| GO:0044248 | GO_Biological process | Cellular catabolic process | 3.54×10−2 | 25 |
| GO:0009725 | GO_Biological process | Response to hormone stimulus | 4.39×10−8 | 30 |
| GO:0048545 | GO_Biological process | Response to steroid hormone stimulus | 2.46×10−6 | 18 |
| GO:0006807 | GO_Biological process | Nitrogen compound metabolic process | 2.95×10−3 | 67 |
| GO:0006520 | GO_Biological process | Cellular amino acid metabolic process | 4.99×10−5 | 16 |
| GO:0009605 | GO_Biological process | Response to external stimulus | 1.81×10−8 | 35 |
| GO:0009308 | GO_Biological process | Amine metabolic process | 4.15×10−6 | 25 |
| GO:0009310 | GO_Biological process | Amine catabolic process | 9.50×10−6 | 10 |
| GO:0032502 | GO_Biological process | Developmental process | 5.43×10−7 | 110 |
| GO:0048856 | GO_Biological process | Anatomical structure development | 8.77×10−7 | 94 |
| GO:0009653 | GO_Biological process | Anatomical structure morphogenesis | 1.19×10−6 | 53 |
| GO:0042221 | GO_Biological process | Response to chemical stimulus | 3.51×10−10 | 70 |
| GO:0010033 | GO_Biological process | Response to organic substance | 4.09×10−11 | 52 |
| GO:0010646 | GO_Biological process | Regulation of cell communication | 1.30×10−5 | 48 |
| GO:0008283 | GO_Biological process | Cell proliferation | 3.68×10−6 | 26 |
| GO:0044282 | GO_Biological process | Small molecule catabolic process | 2.35×10−5 | 18 |
| GO:0023046 | GO_Biological process | Signaling process | 8.53×10−6 | 77 |
| GO:0023060 | GO_Biological process | Signal transmission | 8.53×10−6 | 77 |
| GO:0042127 | GO_Biological process | Regulation of cell proliferation | 1.66×10−6 | 41 |
| GO:0048513 | GO_Biological process | Organ development | 5.37×10−8 | 74 |
| GO:0009888 | GO_Biological process | Tissue development | 2.37×10−8 | 42 |
| GO:0044106 | GO_Biological process | Cellular amine metabolic process | 1.70×10−4 | 18 |
| GO:0032787 | GO_Biological process | Monocarboxylic acid metabolic process | 3.81×10−6 | 21 |
| GO:0007165 | GO_Biological process | Signal transduction | 2.88×10−6 | 71 |
| GO:0010648 | GO_Biological process | Negative regulation of cell communication | 3.44×10−6 | 22 |
| GO:0051239 | GO_Biological process | Regulation of multicellular organismal process | 1.60×10−6 | 48 |
| GO:0042180 | GO_Biological process | Cellular ketone metabolic process | 5.36×10−9 | 37 |
| GO:0050793 | GO_Biological process | Regulation of developmental process | 1.88×10−6 | 39 |
| GO:0005515 | GO_Molecular function | Protein binding | 2.06×10−6 | 223 |
| GO:0003674 | GO_Molecular function | Molecular_function | 4.03×10−4 | 359 |
| GO:0005488 | GO_Molecular function | Binding | 8.63×10−8 | 316 |
| GO:0048037 | GO_Molecular function | Cofactor binding | 3.31×10−6 | 19 |
| GO:0005102 | GO_Molecular function | Receptor binding | 1.45×10−7 | 46 |
| GO:0005575 | GO_Cellular component | Cellular_component | 1.14×10−2 | 370 |
| GO:0005615 | GO_Cellular component | Extracellular space | 1.96×10−8 | 42 |
| GO:0005576 | GO_Cellular component | Extracellular region | 1.10×10−5 | 73 |
| GO:0044421 | GO_Cellular component | Extracellular region part | 3.08×10−9 | 52 |
GO, Gene Ontology.
Figure 15.
The GO terms of target genes of miR-338-5p. The circles represented different GO terms of BP, CC and MF. The darker the color of the circles presents, the greater the significance the term indicated. The relationship between different GO terms were symbolized by arrows. GO, Gene Ontology; BP, biological process; CC, cellular component; MF, molecular function.
Table III.
KEGG pathway analysis of the target genes of miR-338-5p from Cytoskape.
| ID | Name | Category | Term P-value | Count |
|---|---|---|---|---|
| GO:0000280 | Valine, leucine and isoleucine degradation | KEGG | 5.1×10−6 | 9 |
| GO:0005200 | Pathways in cancer | KEGG | 9.5×10−5 | 25 |
| GO:0004110 | Cell cycle | KEGG | 1.4×10−4 | 12 |
| GO:0000071 | Fatty acid degradation | KEGG | 1.7×10−4 | 7 |
| GO:0000640 | Propanoate metabolism | KEGG | 2.0×10−4 | 6 |
KEGG, Kyoto Encyclopedia of Genes and Genomes.
Figure 16.
The network of KEGG pathway analysis. Five large circles in the network represented the corresponding significant pathways from KEGG. Small circles linked by the edges between the large circles symbolized the genes contained in these pathways and the common genes shared by multiple pathways were marked by a combination of colors. KEGG, Kyoto Encyclopedia of Genes and Genomes.
Discussion
Considerable attention has been attracted to miRNAs as promising diagnostic targets for the early screening of human cancers. Prior to our study, several researches have reported some miRNAs had diagnostic value in HCC. A 3-miRNA panel: miR-92-3p, miR-107, and miR-3126-5p discovered by Zhang et al were claimed to distinguish HCC patients in early stage and HCC patients with low-level AFP from their corresponding controls with high accuracy (39). Additionally, some single miRNAs including miR-21 and miR-224 also exhibited prominent diagnostic potential for HCC (40,41) and so far, only one study referred to the diagnostic value of miR-338-5p in HCC with the method of miRNA array. Chen et al (21) reported a moderate ability of miR-338-5p to differentiate HCC from liver cirrhosis with the AUC of 0.799. Furthermore, an extremely strong diagnostic value of miR-338-5p (AUC=0.909) was observed when diagnosing HCC from healthy controls. Despite some advances has been made in exploring the diagnostic capacity of miRNAs for HCC, the diagnostic significance of miR-338-5p in HCC was indefinite, and the relative molecular mechanism has not been elucidated in these studies; therefore our study was the first one to comprehensively assess the diagnostic value of miR-338-5p in HCC with the data from GEO, TCGA and literature as well as to investigate the underlying molecular mechanism through bioinformatics study.
From the meta-analysis result from our collected literature and the integrated meta-analysis, miR-338-5p may serve as a possible diagnostic target for HCC with fair sensitivity and specificity, which enlightened us that miR-338-5p might play an essential role in the occurrence and progression. Previous studies have pointed out that miR-338-5p exerted a tumor suppressive function in a wide range of cancers. In glioblastoma, miR-338-5p was discovered to inhibit the proliferation, invasion and promote apoptosis by targeting EFEMP1 (42); similarly, miR-338-5p significantly attenuated the malignant potential of gastric cancer cells through regulating BMI1 (13). In contrast, miR-338-5p was increased in both blood and tissue of coloreactal cancer (CRC), and represented high area under ROC curve (AUC) of 0.871. The performance of miR-338-5p indicated that it could be a potential biomarker in CRC (19). A similar trend was observed in CRC compared with HCC. Furthermore, according to the subgroup analysis in the integrated meta-analysis, studies with samples from plasma and the method of qRT-PCR were more precise in diagnosing HCC than studies with the controlled conditions, which hinted that the sample type and experiment type may also influence the accuracy of the diagnosis. Although the overall diagnostic ability of miR-338-5p in HCC was the same, which was reflected by the integrated meta-analysis and the meta-analysis from our literature there were still some differences between them. The sensitivity, specificity, diagnostic odds ratio and the area under SROC of the result from the literature meta-analysis were higher than those from the integrated meta-analysis, especially in the evaluation of sensitivity; miR-338-5p showed a poor sensitivity of only 0.51 in the integrated meta-analysis. This might be attributed to the difference in sample type and experiment type as well as the sources of the data. The integrated meta-analysis included GSE datasets from different platforms and TCGA data based on the literature meta-analysis, the samples of which were different. Moreover, due to the limited number of literature, we failed to trace the heterogeneity by carrying out subgroup analysis for our literature meta-analysis. Expanding the sample size was necessary for a more reliable assessment of the diagnostic value of miR-338-5p in HCC.
The results from meta-analysis only provided a superficial hint that miR-338-5p possessed significant diagnostic capacity in HCC and the molecular mechanism underlying it needed further exploration. Thus, we emphasized on the network and functional analysis of the target genes.
We firstly identified the potential target genes of miR-338-5p and further defined the hub genes from PPI network. The 11 hub genes were assumed to correlate closely with miR-338-5p and play essential roles in the miR-338-5p relevant pathogenesis of HCC. Among the hub genes, CDK1 was important protein for the regulation of cell cycles belonging to the cyclin-dependent kinases family (43). The overexpression of CDK1 was detected in various cancers, and a poor prognosis of renal cell carcinoma patients was associated with the high expression of CDK1 and CDK2 (44–48). We hypothesized that CDK1 deregulated by miR-338-5p might promote the deterioration of HCC by affecting the cell cycle of HCC cells. Apart from CDK1, several hub genes such as MYC, BIRC5, IGF1, NCOR1 and FOXO1 participate in the regulation of a wide range of biological processes including cell proliferation, apoptosis and migration (49–57). These genes were reported to be aberrantly expressed in various cancers (58–62) and they were also involved in the malignant progression of HCC (49,50,52,63). It was conceived that miR-338-5p might interact with these molecules through potential signaling pathways to influence the development of HCC. PCNA, a protein that acted as DNA sliding clamp, was found to engage in DNA duplication and repair with its overexpressed in HCC. Further study was necessary to probe into the association between PCNA and miR-338-5p in HCC. In this study, we analyzed 11 hub genes protein expression by HPA database. The result indicated that the expression of FOXO1 and NCOR1 was downregulated in HCC and most likely regulated by miR-338-5p.
GO enrichment analysis was indicative of the possible functions of the target genes in HCC and the results from three GO terms hinted that the target genes were mainly assembled in response to organic substance. Most of the potential functions of the target genes from the GO analysis were accomplished in signaling pathways. Therefore, it is of great importance to investigate the signaling pathways gathered by the target genes of miR-338-5p. From the results of the KEGG pathway analysis, the most significant ten pathways such as pathways in cancer and cell cycle were closely associated with cancer.
Despite the valuable findings acquired from the meta-analysis and bioinformatics study, there were still some limitations in our study. The sample size of our literature was too small for further analysis to identify heterogeneity, which weakened the reliability of our results. Since the samples are from different types, including tissue and plasma, a bias and sensitivity problems might originate from sample types in analysis. Additionally, we only included studies published in Chinese or English, which might cause bias of selection to the meta-analysis. A plausible way to address these issues is to conduct future studies with larger samples and fewer language restrictions to further verify the diagnostic value of miR-338-5p for HCC.
In conclusion, we anticipated that miR-338-5p may serve as a promising diagnostic marker for HCC and miR-338-5p could affect the development of HCC by targeting certain downstream genes and pathways. The future research will be concentrated on validating the target genes of miR-338-5p and its function in the significant signaling pathways mentioned before.
Figure 6.
The pooled SP of the integrated meta-analysis. The pooled SP of the integrated meta-analysis was 0.69 (95% CI: 0.65–0.73).
Figure 7.
The pooled PLR of the integrated meta-analysis. The pooled PLR of the integrated meta-analysis was 1.76 (95% CI: 1.17–2.66). PLR, positive likelihood ratio.
Figure 8.
The pooled NLR of the integrated meta-analysis. The pooled NLR of the integrated meta-analysis was 0.64 (95% CI: 0.52–0.80). NLR, negative likelihood ratio.
References
- 1.Nault JC, De Reyniès A, Villanueva A, Calderaro J, Rebouissou S, Couchy G, Decaens T, Franco D, Imbeaud S, Rousseau F, et al. A hepatocellular carcinoma 5-gene score associated with survival of patients after liver resection. Gastroenterology. 2013;145:176–187. doi: 10.1053/j.gastro.2013.03.051. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Zhu AX. Molecularly targeted therapy for advanced hepatocellular carcinoma in 2012: Current status and future perspectives. Semin Oncol. 2012;39:493–502. doi: 10.1053/j.seminoncol.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 4.Llovet JM, Villanueva A, Lachenmayer A, Finn RS. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol. 2015;12:408–424. doi: 10.1038/nrclinonc.2015.121. [DOI] [PubMed] [Google Scholar]
- 5.Lages E, Ipas H, Guttin A, Nesr H, Berger F, Issartel JP. MicroRNAs: Molecular features and role in cancer. Front Biosci (Landmark Ed) 2012;17:2508–2540. doi: 10.2741/4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neilson JR, Sharp PA. Small RNA regulators of gene expression. Cell. 2008;134:899–902. doi: 10.1016/j.cell.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 7.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets; Proc Natl Acad Sci USA; 2006; pp. 2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, Morgan DL, Postier RG, Brackett DJ, Schmittgen TD. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 10.Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 11.Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias; Proc Natl Acad Sci USA; 2004; pp. 11755–11760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 13.Tong D, Zhao L, He K, Sun H, Cai D, Ni L, Sun R, Chang S, Song T, Huang C, et al. MECP2 promotes the growth of gastric cancer cells by suppressing miR-338-mediated antiproliferative effect. Oncotarget. 2016;7:34845–34859. doi: 10.18632/oncotarget.9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen JS, Liang LL, Xu HX, Chen F, Shen SL, Chen W, Chen LZ, Su Q, Zhang LJ, Bi J, et al. miR-338-3p inhibits epithelial-mesenchymal transition and metastasis in hepatocellular carcinoma cells. Oncotarget. 2016;8:71418–71429. doi: 10.18632/oncotarget.10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen X, Wei L, Zhao S. miR-338 inhibits the metastasis of lung cancer by targeting integrin β3. Oncol Rep. 2016;36:1467–1474. doi: 10.3892/or.2016.4928. [DOI] [PubMed] [Google Scholar]
- 16.Weng HL, Wang MJ. Effects of microRNA-338-3p on morphine-induced apoptosis and its underlying mechanisms. Mol Med Rep. 2016;14:2085–2092. doi: 10.3892/mmr.2016.5506. [DOI] [PubMed] [Google Scholar]
- 17.Zhuang Y, Dai J, Wang Y, Zhang H, Li X, Wang C, Cao M, Liu Y, Cai H, Zhang D, Wang Y. miR-338* suppresses fibrotic pathogenesis in pulmonary fibrosis through targeting LPA1. Am J Transl Res. 2016;8:3197–3205. [PMC free article] [PubMed] [Google Scholar]
- 18.Xing Z, Yu L, Li X, Su X. Anticancer bioactive peptide-3 inhibits human gastric cancer growth by targeting miR-338-5p. Cell Biosci. 2016;6:53. doi: 10.1186/s13578-016-0112-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yong FL, Law CW, Wang CW. Potentiality of a triple microRNA classifier: miR-193a-3p, miR-23a and miR-338-5p for early detection of colorectal cancer. BMC Cancer. 2013;13:280. doi: 10.1186/1471-2407-13-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Besse A, Sana J, Lakomy R, Kren L, Fadrus P, Smrcka M, Hermanova M, Jancalek R, Reguli S, Lipina R, et al. miR-338-5p sensitizes glioblastoma cells to radiation through regulation of genes involved in DNA damage response. Tumour Biol. 2016;37:7719–7727. doi: 10.1007/s13277-015-4654-x. [DOI] [PubMed] [Google Scholar]
- 21.Chen Y, Chen J, Liu Y, Li S, Huang P. Plasma miR-15b-5p, miR-338-5p, and miR-764 as biomarkers for hepatocellular carcinoma. Med Sci Monit. 2015;21:1864–1871. doi: 10.12659/MSM.893082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol. 2005;58:882–893. doi: 10.1016/j.jclinepi.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 23.Glas AS, Lijmer JG, Prins MH, Bonsel GJ, Bossuyt PM. The diagnostic odds ratio: A single indicator of test performance. J Clin Epidemiol. 2003;56:1129–1135. doi: 10.1016/S0895-4356(03)00177-X. [DOI] [PubMed] [Google Scholar]
- 24.Harbord RM, Deeks JJ, Egger M, Whiting P, Sterne JA. A unification of models for meta-analysis of diagnostic accuracy studies. Biostatistics. 2007;8:239–251. doi: 10.1093/biostatistics/kxl004. [DOI] [PubMed] [Google Scholar]
- 25.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson D, White IR, Thompson SG. Extending DerSimonian and Laird's methodology to perform multivariate random effects meta-analyses. Stat Med. 2010;29:1282–1297. doi: 10.1002/sim.3602. [DOI] [PubMed] [Google Scholar]
- 27.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, et al. Towards a knowledge-based Human protein atlas. Nat Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y, Huang P, Chen J, Liu Y, Li SL, Wang Z, Song D. Plasma circulating miR-338-5p, miR-21-5p and miR-15b-5p are potential biomarkers for screening hepatocellular carcinoma. J Third Mil Med Univ. 2015;37:1720–1726. (In Chinese) [Google Scholar]
- 29.Budhu A, Jia HL, Forgues M, Liu CG, Goldstein D, Lam A, Zanetti KA, Ye QH, Qin LX, Croce CM, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- 30.Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, Zhuang SM. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135–1142. doi: 10.1158/0008-5472.CAN-08-2886. [DOI] [PubMed] [Google Scholar]
- 31.Burchard J, Zhang C, Liu AM, Poon RT, Lee NP, Wong KF, Sham PC, Lam BY, Ferguson MD, Tokiwa G, et al. microRNA-122 as a regulator of mitochondrial metabolic gene network in hepatocellular carcinoma. Mol Syst Biol. 2010;6:402. doi: 10.1038/msb.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diaz G, Melis M, Tice A, Kleiner DE, Mishra L, Zamboni F, Farci P. Identification of microRNAs specifically expressed in hepatitis C virus-associated hepatocellular carcinoma. Int J Cancer. 2013;133:816–824. doi: 10.1002/ijc.28075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen J, Wang A, Wang Q, Gurvich I, Siegel AB, Remotti H, Santella RM. Exploration of genome-wide circulating microRNA in hepatocellular carcinoma: miR-483-5p as a potential biomarker. Cancer Epidemiol Biomarkers Prev. 2013;22:2364–2373. doi: 10.1158/1055-9965.EPI-13-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murakami Y, Kubo S, Tamori A, Itami S, Kawamura E, Iwaisako K, Ikeda K, Kawada N, Ochiya T, Taguchi YH. Comprehensive analysis of transcriptome and metabolome analysis in Intrahepatic cholangiocarcinoma and hepatocellular carcinoma. Sci Rep. 2015;5:16294. doi: 10.1038/srep16294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinez-Quetglas I, Pinyol R, Dauch D, Torrecilla S, Tovar V, Moeini A, Alsinet C, Portela A, Rodriguez-Carunchio L, Solé M, et al. IGF2 is up-regulated by epigenetic mechanisms in hepatocellular carcinomas and is an actionable oncogene product in experimental models. Gastroenterology. 2016;151:1192–1205. doi: 10.1053/j.gastro.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 36.Shih TC, Tien YJ, Wen CJ, Yeh TS, Yu MC, Huang CH, Lee YS, Yen TC, Hsieh SY. MicroRNA-214 downregulation contributes to tumor angiogenesis by inducing secretion of the hepatoma-derived growth factor in human hepatoma. J Hepatol. 2012;57:584–591. doi: 10.1016/j.jhep.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 37.Noh JH, Chang YG, Kim MG, Jung KH, Kim JK, Bae HJ, Eun JW, Shen Q, Kim SJ, Kwon SH, et al. miR-145 functions as a tumor suppressor by directly targeting histone deacetylase 2 in liver cancer. Cancer Lett. 2013;335:455–462. doi: 10.1016/j.canlet.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 38.Sato F, Hatano E, Kitamura K, Myomoto A, Fujiwara T, Takizawa S, Tsuchiya S, Tsujimoto G, Uemoto S, Shimizu K. MicroRNA profile predicts recurrence after resection in patients with hepatocellular carcinoma within the Milan Criteria. PLoS One. 2011;6:e16435. doi: 10.1371/journal.pone.0016435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Li T, Qiu Y, Zhang T, Guo P, Ma X, Wei Q, Han L. Serum microRNA panel for early diagnosis of the onset of hepatocellular carcinoma. Medicine (Baltimore) 2017;96:e5642. doi: 10.1097/MD.0000000000005642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okajima W, Komatsu S, Ichikawa D, Miyamae M, Kawaguchi T, Hirajima S, Ohashi T, Imamura T, Kiuchi J, Arita T, et al. Circulating microRNA profiles in plasma: Identification of miR-224 as a novel diagnostic biomarker in hepatocellular carcinoma independent of hepatic function. Oncotarget. 2016;7:53820–53836. doi: 10.18632/oncotarget.10781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan SR, Liu ZJ, Yu S, Bao YX. Investigation of the value of miR-21 in the diagnosis of early stage HCC and its prognosis: A meta-analysis. Genet Mol Res. 2015;14:11573–11586. doi: 10.4238/2015.September.28.9. [DOI] [PubMed] [Google Scholar]
- 42.Lei D, Zhang F, Yao D, Xiong N, Jiang X, Zhao H. miR-338-5p suppresses proliferation, migration, invasion, and promote apoptosis of glioblastoma cells by directly targeting EFEMP1. Biomed Pharmacother. 2017;89:957–965. doi: 10.1016/j.biopha.2017.01.137. [DOI] [PubMed] [Google Scholar]
- 43.Yang W, Cho H, Shin HY, Chung JY, Kang ES, Lee EJ, Kim JH. Accumulation of cytoplasmic Cdk1 is associated with cancer growth and survival rate in epithelial ovarian cancer. Oncotarget. 2016;7:49481–49497. doi: 10.18632/oncotarget.10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsaur I, Makarević J, Hudak L, Juengel E, Kurosch M, Wiesner C, Bartsch G, Harder S, Haferkamp A, Blaheta RA. The cdk1-cyclin B complex is involved in everolimus triggered resistance in the PC3 prostate cancer cell line. Cancer Lett. 2011;313:84–90. doi: 10.1016/j.canlet.2011.08.026. [DOI] [PubMed] [Google Scholar]
- 45.Sung WW, Lin YM, Wu PR, Yen HH, Lai HW, Su TC, Huang RH, Wen CK, Chen CY, Chen CJ, Yeh KT. High nuclear/cytoplasmic ratio of Cdk1 expression predicts poor prognosis in colorectal cancer patients. BMC Cancer. 2014;14:951. doi: 10.1186/1471-2407-14-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Willder JM, Heng SJ, McCall P, Adams CE, Tannahill C, Fyffe G, Seywright M, Horgan PG, Leung HY, Underwood MA, Edwards J. Androgen receptor phosphorylation at serine 515 by Cdk1 predicts biochemical relapse in prostate cancer patients. Br J Cancer. 2013;108:139–148. doi: 10.1038/bjc.2012.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Banerjee SK, Weston AP, Zoubine MN, Campbell DR, Cherian R. Expression of cdc2 and cyclin B1 in Helicobacter pylori-associated gastric MALT and MALT lymphoma: Relationship to cell death, proliferation, and transformation. Am J Pathol. 2000;156:217–225. doi: 10.1016/S0002-9440(10)64722-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hongo F, Takaha N, Oishi M, Ueda T, Nakamura T, Naitoh Y, Naya Y, Kamoi K, Okihara K, Matsushima T, et al. CDK1 and CDK2 activity is a strong predictor of renal cell carcinoma recurrence. Urol Oncol. 2014;32:1240–1246. doi: 10.1016/j.urolonc.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 49.Tian J, Hu X, Gao W, Zhang J, Chen M, Zhang X, Ma J, Yuan H. Identification a novel tumor-suppressive hsa-miR-599 regulates cells proliferation, migration and invasion by targeting oncogenic MYC in hepatocellular carcinoma. Am J Transl Res. 2016;8:2575–2584. [PMC free article] [PubMed] [Google Scholar]
- 50.Cao L, Li C, Shen S, Yan Y, Ji W, Wang J, Qian H, Jiang X, Li Z, Wu M, et al. OCT4 increases BIRC5 and CCND1 expression and promotes cancer progression in hepatocellular carcinoma. BMC Cancer. 2013;13:82. doi: 10.1186/1471-2407-13-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Le Coz V, Zhu C, Devocelle A, Vazquez A, Boucheix C, Azzi S, Gallerne C, Eid P, Lecourt S, Giron-Michel J. IGF-1 contributes to the expansion of melanoma-initiating cells through an epithelial-mesenchymal transition process. Oncotarget. 2016;7:82511–82527. doi: 10.18632/oncotarget.12733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang XW, Shen GZ, Cao LQ, Jiang XF, Peng HP, Shen G, Chen D, Xue P. MicroRNA-1269 promotes proliferation in human hepatocellular carcinoma via downregulation of FOXO1. BMC Cancer. 2014;14:909. doi: 10.1186/1471-2407-14-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ko YS, Cho SJ, Park J, Kim Y, Choi YJ, Pyo JS, Jang BG, Park JW, Kim WH, Lee BL. Loss of FOXO1 promotes gastric tumour growth and metastasis through upregulation of human epidermal growth factor receptor 2/neu expression. Br J Cancer. 2015;113:1186–1196. doi: 10.1038/bjc.2015.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ren JW, Li ZJ, Tu C. miR-135 post-transcriptionally regulates FOXO1 expression and promotes cell proliferation in human malignant melanoma cells. Int J Clin Exp Pathol. 2015;8:6356–6366. [PMC free article] [PubMed] [Google Scholar]
- 55.Martinez-Iglesias OA, Alonso-Merino E, Gómez-Rey S, Velasco-Martín JP, Martín Orozco R, Luengo E, García Martín R, Ibáñez de Cáceres I, Fernández AF, Fraga MF, et al. Autoregulatory loop of nuclear corepressor 1 expression controls invasion, tumor growth, and metastasis; Proc Natl Acad Sci USA; 2016; pp. E328–E337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu JJ, Wu YX, Zhao FJ, Xia SJ. miR-96 promotes cell proliferation and clonogenicity by down-regulating of FOXO1 in prostate cancer cells. Med Oncol. 2014;31:910. doi: 10.1007/s12032-014-0910-y. [DOI] [PubMed] [Google Scholar]
- 57.Zhang B, Gui LS, Zhao XL, Zhu LL, Li QW. FOXO1 is a tumor suppressor in cervical cancer. Genet Mol Res. 2015;14:6605–6616. doi: 10.4238/2015.June.18.3. [DOI] [PubMed] [Google Scholar]
- 58.Li K, Chen MK, Situ J, Huang WT, Su ZL, He D, Gao X. Role of co-expression of c-Myc, EZH2 and p27 in prognosis of prostate cancer patients after surgery. Chin Med J (Engl) 2013;126:82–87. [PubMed] [Google Scholar]
- 59.Liu Z, Jiang Y, Hou Y, Hu Y, Cao X, Tao Y, Xu C, Liu S, Wang S, Wang L, et al. The IκB family member Bcl-3 stabilizes c-Myc in colorectal cancer. J Mol Cell Biol. 2013;5:280–282. doi: 10.1093/jmcb/mjt020. [DOI] [PubMed] [Google Scholar]
- 60.Duffy MJ, O'Donovan N, Brennan DJ, Gallagher WM, Ryan BM. Survivin: A promising tumor biomarker. Cancer Lett. 2007;249:49–60. doi: 10.1016/j.canlet.2006.12.020. [DOI] [PubMed] [Google Scholar]
- 61.Grimberg A. Mechanisms by which IGF-I may promote cancer. Cancer Biol Ther. 2003;2:630–635. doi: 10.4161/cbt.2.6.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xie L, Ushmorov A, Leithäuser F, Guan H, Steidl C, Färbinger J, Pelzer C, Vogel MJ, Maier HJ, Gascoyne RD, et al. FOXO1 is a tumor suppressor in classical Hodgkin lymphoma. Blood. 2012;119:3503–3511. doi: 10.1182/blood-2011-09-381905. [DOI] [PubMed] [Google Scholar]
- 63.Chun YS, Huang M, Rink L, Von Mehren M. Expression levels of insulin-like growth factors and receptors in hepatocellular carcinoma: A retrospective study. World J Surg Oncol. 2014;12:231. doi: 10.1186/1477-7819-12-231. [DOI] [PMC free article] [PubMed] [Google Scholar]