Abstract
Kawasaki disease (KD) is an acquired cardiac disease with a high incidence that affects children. KD has various complications, including coronary artery dilation (CAD) and coronary artery aneurysms (CAA). The identification of differentially expressed proteins and the underlying mechanisms may be the key to understanding differences between these KD complications. In the present study, isobaric tags for relative and absolute quantitation were used to identify variations in serum proteins between KD patients with CAD and CAA. In total, 87 (37 upregulated and 50 downregulated) and 65 (33 upregulated and 32 downregulated) significantly differentially-expressed proteins were identified in comparisons between control samples (healthy individuals) and those obtained from patients with KD and with CAD or CAA. Investigation into the underlying biological process revealed that variations between the two complications were associated with the wound healing response, as well as lipoprotein- and cholesterol-associated processes. Important proteins involved in the formation of the wound healing signaling network were identified via enriched biological processes and pathway analysis using ClueGo and ReactomeFIViz software. In the present study, 5 significantly differentially-expressed proteins, including mannose binding lectin 2 (MBL2), complement factor H (CFH), kininogen 1 (KNG1), serpin family C member 1 (SERPINC1) and fibronectin 1 (FN1), were selected and confirmed by western blotting. Analysis indicated that these proteins were associated to immunity, inflammation and metabolism, serving a key role within each module, which has never been reported previously. The present study proposed that MBL2, CFH, KNG1, SERPINC1 and FN1 may be a potentially excellent indicator group for distinguishing the two major KD complications, CAD and CAA.
Keywords: Kawasaki disease, coronary artery dilation, coronary artery aneurysms, potential indicators
Introduction
Kawasaki disease (KD), first described in 1967 by Kawasaki (1), is a type of acute vasculitis with unknown etiology; however, KD is known to induce pathological alterations in medium-sized arteries, particularly coronary arteries. Some patients with KD develop coronary artery lesions and exhibit similar clinical outcomes to those of patients with coronary atherosclerosis (2). Several studies have demonstrated that patients without coronary artery lesions still exhibit a high risk of various diseases, such as atherosclerosis, in adulthood (2–4). KD is a main cause of acquired heart disease in children, predominantly affecting coronary arteries (5).
Without routine therapy, ~25% of patients with KD develop coronary artery dilation or aneurysms (CAD or CAA, respectively) (6). Mild CAD can regress, while CAA and severe CAD are difficult to restore to a normal state. Mueller et al (7) reported that patients with coronary artery sizes of ≤5.0 mm exhibited regression to the normal state, while patients with coronary artery sizes of >5.0 mm exhibited persistence or even increases in size. CAA with a diameter of ≥8.0 mm is strongly associated with the development of coronary artery stenosis and myocardial infarction (7).
These results indicate that KD may exert long-term effects on the coronary artery and cause pathophysiological insults of the cardiovascular system leading to CAD or CAA. To further understand complications in patients with KD, it is important to investigate the mechanisms of CAD and CAA formation, and to characterize the differences among normal coronary arteries, CAA and CAD. Coronary artery specimens are optimal for CAD and CAA studies; however, they are difficult to collect from patients with KD. Thus, blood samples were used in the present study.
Rapid advances in mass spectrometry (MS) for peptide identification and improved protein sequence coverage of complex biological samples (8) has provided highly effective proteomic technologies for biomarker identification and therapeutic target discovery in serum samples (9,10). In order to obtain accurate and reliable results, isobaric tags for relative and absolute quantitation (iTRAQ) and MS were employed for quantitative proteomics analyses, with increased proteome coverage and labelling efficiency to compare proteomes among serum samples from healthy individuals, and patients with KD and CAD or CAA (6). Western blotting was employed to confirm the significantly expressed proteins following bioinformatics analysis. The objective of the present study was to investigate proteomic variations to further our current understanding of the pathogenesis of coronary artery lesions, and provide a basis for the establishment of indicators for these KD complications.
Materials and methods
Preparation of serum samples
Blood samples were collected from 51 children (≤6 years old) who recruited to the present study (Table I). Written informed consent was obtained from the guardians of each participant and the present study was approved by the Ethical Committee of Guangzhou Women and Children's Medical Center [(2013)077; Guangdong, China] based on the Japanese Ministry of Health and Welfare criteria, and the American Heart Association (6,11). A total of 17 age- and sex-matched samples obtained from normal children, served as the controls; individuals were physically examined in Guangzhou Women and Children's Medical Center. The other 34 samples (17 CAA and 17 CAD) were obtained from patients who were diagnosed and excluded of other possible diseases by ≥2 pediatric cardiologists. Blood samples were separated by centrifugation at 1,000 × g for 10 min at 4°C and aliquots of serum were collected and stored at −80°C. Serum samples were processed to remove albumin and immunoglobulin G (IgG) using a ProteoPrep Blue Albumin and IgG Depletion kit (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). The concentrations of these proteins were determined using a Bradford protein assay kit (Bio-Rad Laboratories Inc., Hercules, CA, USA). These protein samples were randomly combined into 17 comparison groups, each of which included 1 control sample, 1 CAA sample and 1 CAD sample. Of the 17 comparison groups, 5 were randomly chosen and pooled together according to each type of sample; 3 pooled samples of the control, CAA and CAD groups were produced, and were subsequently analyzed by iTRAQ (Sciex, Framingham, MA, USA) experiments (Table IA). The other 12 groups out of the total 17 were analyzed via western blotting. Information regarding the groups and diagnosis is presented in Table IB.
Table I.
Clinical indicators of patients with Kawasaki disease.
| Patient | Age (years) | Sex | WBC count (109/1) | N/L | CRP (mg/1) | ALT (U/1) | AST (U/1) | Coronary change (mm) |
|---|---|---|---|---|---|---|---|---|
| A, Patients for iTRAQ experiments | ||||||||
| Patient-1 CAA-1 | 11 months | Female | 11.3 | 0.50 | 13.31 | 50 | 37 | LCA=5.8, RCA=5.7 |
| Patient-2 CAA-2 | 2 years, 5 months | Male | 17.7 | 3.13 | 88.80 | 26 | 18 | LCA=9.7, RCA=7.5 |
| Patient-3 CAA-3 | 1 year, 11 months | Male | 10.5 | 0.88 | 0.21 | 19 | 35 | LCA=12, RCA=8.3 |
| Patient-4 CAA-4 | 1 year, 3 months | Male | 22.8 | 1.20 | 27.37 | 8 | 18 | LCA=5.6, RCA=6.0 |
| Patient-5 CAA-5 | 2 months | Male | 24.8 | 1.75 | 71.51 | 36 | 24 | LCA=4.0, RCA=4.6 |
| Patient-1 CAD-1 | 2 years, 7 months | Male | 23.4 | 5.25 | 211.13 | 7 | 12 | LCA=3.2, RCA=3.7 |
| Patient-2 CAD-2 | 3 years | Male | 14.6 | 11.35 | 9.50 | 10 | 23 | LCA=3.2, RCA=2.8 |
| Patient-3 CAD-3 | 3 years | Male | 11.6 | 4.15 | 45.12 | 14 | 30 | LCA=4.0, RCA=3.7 |
| Patient-4 CAD-4 | 1 year, 1 month | Male | 10.0 | 4.72 | 93.30 | 161 | 32 | LCA=2.9, RCA=3.4 |
| Patient-5 CAD-5 | 3 months | Female | 20.5 | 1.26 | 98.60 | 18 | 25 | LCA=3.5, RCA=3.9 |
| B, Patients for western blotting experiments | ||||||||
| Patient | Age (years) | Sex | WBC count (109/1) | N/L | CRP (mg/1) | ALT (U/1) | AST (U/1) | Coronary change (mm) |
| Patient-6 CAA-6 | 1 year 3 months | Male | 24.3 | 1.71 | 175.60 | 19 | 26 | LCA=4.0, RCA=4.5 |
| Patient-7 CAA-7 | 4 months | Male | 34.7 | 2.95 | 75.05 | 20 | 26 | LCA=4.8, RCA=3.3 |
| Patient-8 CAA-8 | 6 years | Female | 15.5 | 3.77 | 68.57 | 26 | 21 | LCA=6.6, RCA=3.1 |
| Patient-9 CAA-9 | 6 months | Male | 13.31 | 1.09 | 46.06 | 17 | 20 | LCA=3.7, RCA=4.6 |
| Patient-10 CAA-10 | 8 months | Male | 15.6 | 1.60 | 146.70 | 20 | 16 | LCA=3.3, RCA=4.2 |
| Patient-11 CAA-11 | 2 months | Male | 17.0 | 1.62 | 39.90 | 19 | 17 | LCA=4.3, RCA=4.0 |
| Patient-12 CAA-12 | 2 months | Male | 15.4 | 1.85 | 18.18 | 14 | 17 | LCA=3.8, RCA=4.6 m |
| Patient-13 CAA-13 | 3 years | Male | 7.0 | 0.48 | 0.23 | 22 | 31 | LCA=10.4, RCA=9.8 |
| Patient-14 CAA-14 | 1 year, 2 months | Female | 19.6 | 2.82 | 202.37 | 16 | 19 | LCA=4.6, RCA=5.3 |
| Patient-15 CAA-15 | 4 months | Female | 23.1 | 1.42 | 110.05 | 40 | 42 | LCA=3.8, RCA=4.5 |
| Patient-16 CAA-16 | 7 months | Male | 17.5 | 1.67 | 87.36 | 8 | 19 | LCA=5.0, RCA=5.4 |
| Patient-17 CAA-17 | 1 year | Female | 20.8 | 1.76 | 58.00 | 11 | 21 | LCA=5.0, RCA=3.6 |
| Patient-6 CAD-6 | 1 year, 1 month | Male | 12.6 | 0.43 | 73.13 | 35 | 30 | LCA=3.1, RCA=2.8 |
| Patient-7 CAD-7 | 4 months | Male | 13.7 | 2.12 | 113.75 | 27 | 23 | LCA=3.2, RCA=2.0 |
| Patient-8 CAD-8 | 4 years | Male | 19.8 | 7.26 | 59.22 | 73 | 20 | LCA=3.4, RCA=3.1 |
| Patient-9 CAD-9 | 6 months | Male | 21.5 | 5.37 | 71.00 | 50 | 38 | LCA=2.8, RCA=1.9 |
| Patient-10 CAD-10 | 7 months | Male | 12.1 | 0.99 | 78.10 | 14 | 19 | LCA=2.5, RCA=2.0 |
| Patient-11 CAD-11 | 3 months | Female | 9.7 | 0.47 | 15.57 | 84 | 66 | LCA=2.2, RCA=1.5 |
| Patient-12 CAD-12 | 2 months | Male | 24.5 | 12.26 | 209.63 | 24 | 24 | LCA=2.5, RCA=2.1 |
| Patient-13 CAD-13 | 2 years, 9 months | Male | 7.3 | 1.58 | 29.98 | 23 | 23 | LCA=3.4, RCA=2.8 |
| Patient-14 CAD-14 | 7 months | Male | 24.1 | 1.91 | 55.72 | 13 | 20 | LCA=2.7, RCA=2.8 |
| Patient-15 CAD-15 | 7 months | Male | 17.6 | 0.83 | 44.60 | 18 | 19 | LCA=3.0, RCA=2.4 |
| Patient-16 CAD-16 | 4 years | Male | 22.3 | 9.16 | 147.85 | 136 | 39 | LCA=3.0, RCA=2.6 |
| Patient-17 CAD-17 | 1 year, 1 month | Male | 17.5 | 7.79 | 144.10 | 82 | 44 | LCA=3.5, RCA=2.9 |
CAA, coronary artery aneurism; CAD, coronary artery dilation; WBC, white blood cell; N/L, neutrophils/lymphocytes; CRP, C-reactive protein; ALT, alanine transaminase; AST, aspartate aminotransferase; LCA, left coronary artery; RCA, right coronary artery; iTRAQ, isobaric tags for relative and absolute quantitation.
Protein identification and analysis
iTRAQ labeling was performed according to the manufacturer's instructions (Sciex). Briefly, 100 µg of each protein sample was reduced with Tris(2-carboxyethyl)phosphine hydrochloride reducing reagent (Sciex) at 60°C for 1 h, and alkylated with methyl methanethiosulfonate cysteine-blocking reagent (Sciex) for 30 min at room temperature. Then, proteins were digested with 2% trypsin (Promega Corporation, Madison, WI, USA) at 37°C overnight, at a ratio of 1:50 (enzyme-to-substrate). Each sample was labelled separately with the iTRAQ tags (118 tag for control, 113 tag for CAA and 116 tag for CAD), then dehydrated by centrifugation at 6,000 × g for 4 h at 25°C in a vacuum centrifuge.
iTRAQ-labelled samples were firstly diluted to 100 µl with H2O buffer (NH3•H2O, pH=10) prior to high performance liquid chromatography on a Dionex Ultimate 3000 system (Dionex, Sunnyvale, CA, USA) at 25°C on a Gemini-NX 3u C18 110A; 150×2.00 mm Phenomenex column, and Gemini 3u C6-Phenyl 110A; 100×2.0 mm column (all from Phenomenex, Torrance, CA, USA). The flow rate used for reversed-phase column separation was 0.2 ml/min with H2O (mobile phase A) and 80% acetonitrile (mobile phase B) with the following gradient system parameters: 5–10% B for 0–15 min, 15–28% B for 15–48 min, 25–37% B for 48–60 min, 37–95% B for 60–65 min and 95% B for 65–70 min. The elution was monitored by absorbance at 214/280 nm and the fractions were collected every 50 sec; these were pooled for each sample and dehydrated by centrifugation at 6,000 × g for 4 h at 25°C in vacuum centrifugation.
Peptides were separated with mobile phase A (0.1% formic acid), and 5–40% mobile phase B (0.1% formic acid and 80% acetonitrile) for 99 min (0.3 ml/min flow rate). MS analysis was performed on a Q Exactive hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific, Inc., Waltham, MA, USA) with the following parameters: MS spectra were acquired across the scan range of 350–1,800 m/z in high resolution mode (>35,000) and 100 msec was accumulated per spectrum. A maximum of 20 precursors per cycle were chosen for fragmentation, with 120 msec set as the minimum accumulation time for each precursor and dynamic exclusion for 10 sec.
The database searching of MS raw data was conducted against the human protein database by ProteinPilot Software 5.0 (Sciex). The search parameters were set as: ‘Cys alkylation’, ‘Methyl methanethiosulfonate’, ‘Digestion’, ‘Trypsin’, ≤1 missed trypsin cleavage was allowed and the false discovery rate (FDR) was <0.01.
Gene Ontology (GO) term and pathway analysis
The Cytoscape plug-in ClueGO (12) and ReactomeFIViz (13) were applied for the differentially expressed proteins to process the Biological Process GO terms, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (www.genome.jp/kegg/). P<0.05 and FDR <0.01 were selected as threshold values for GO and KEGG pathway enrichment.
Western blot analysis
Serum samples from 36 children (12 control, 12 CAA and 12 CAD) were randomly grouped into 12 sets (for Control, CAA and CAD; details are presented in Table IB) for validation. Proteins from the sera of these patients were extracted and the concentrations were measured using lysis buffer (Beyotime Institute of Biotechnology, Haimen, China) and a Bradford protein assay kit (Bio-Rad Laboratories, Inc.). Following electrophoresis with 20 µg protein per lane via 12–15% SDS-PAGE, the separated proteins were transferred onto polyvinylidene fluoride (PVDF) membranes (Sigma-Aldrich; Merck KGaA) and blocked with 5% skimmed milk for 1 h at room temperature, then incubated with primary antibodies (1:1,000) against: Complement factor H (CFH; cat. no. ab133536), mannose-binding lectin 2 (MBL2; cat. no. ab203303), kininogen 1 (KNG1; cat. no. ab170475), fibronectin (FN1; cat. no. ab2413), antithrombin-III (SERPINC1; cat. no. ab126598) and β-actin (cat. no. ab8227) (all from Abcam, Cambridge, UK) overnight at 4°C. Subsequently, horse radish peroxidase goat anti-rabbit IgG (cat. no. ab6721, 1:2,000; Abcam) was applied to the primary antibody-treated PVDF membranes at room temperature for 2 h. The specific bands on these membranes were visualized using the SuperSignal chemiluminescence system (Promega Corporation).
Results
Characteristics of patients with KD
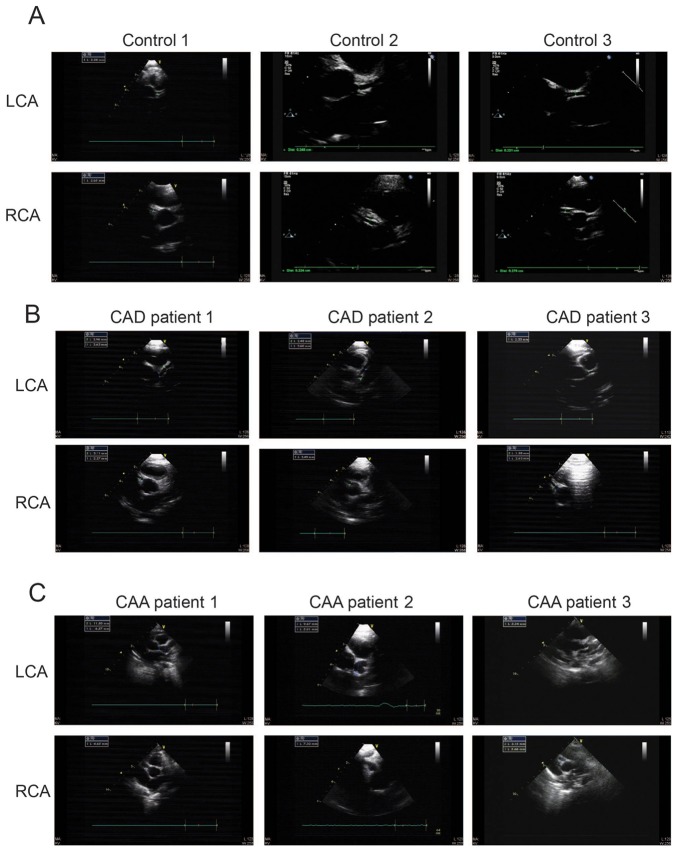
Blood samples were obtained from 51 children (including 17 controls, 17 patients with CAA, and 17 patients with CAD); basic diagnostic information is shown in Table IA and B. The patients with KD were ≤6 years and had clear clinical features; the abnormal degree of coronary alteration revealed that some rare CAA samples were included. Representative echocardiographic images of patients with KD and CAD or CAA, and healthy individuals are presented in Fig. 1. Fig. 1 shows that the diameter of the normal coronary artery was <3 mm and the diameter of CAD was <4 and >3 mm and the diameter of CAA was >5 mm. The maximum diameter of the coronary artery was indicated in the images.
Figure 1.
ECG images of coronary arteries from healthy individuals and patients with Kawasaki disease. Representative ECG images from (A) three children with normal left and right coronary arteries (controls); (B) three patients with CAD; and (C) three patients with CAA. ECG, echocardiography; CAA, coronary artery aneurism; CAD, coronary artery dilation; LCA, left coronary artery; RCA, right coronary artery.
Trial profile and study design
A total of 51 samples were divided into 17 groups, each of which contained one control, one CAD and one CAA sample. Of the 17 groups, 5 were randomly selected and the samples from these groups were pooled based on the sample type for iTRAQ analyses in order to detect significantly differentially expressed proteins. The other 12 groups were analyzed via western blotting. All experiments and validation procedures are summarized in in Fig. 2.
Figure 2.
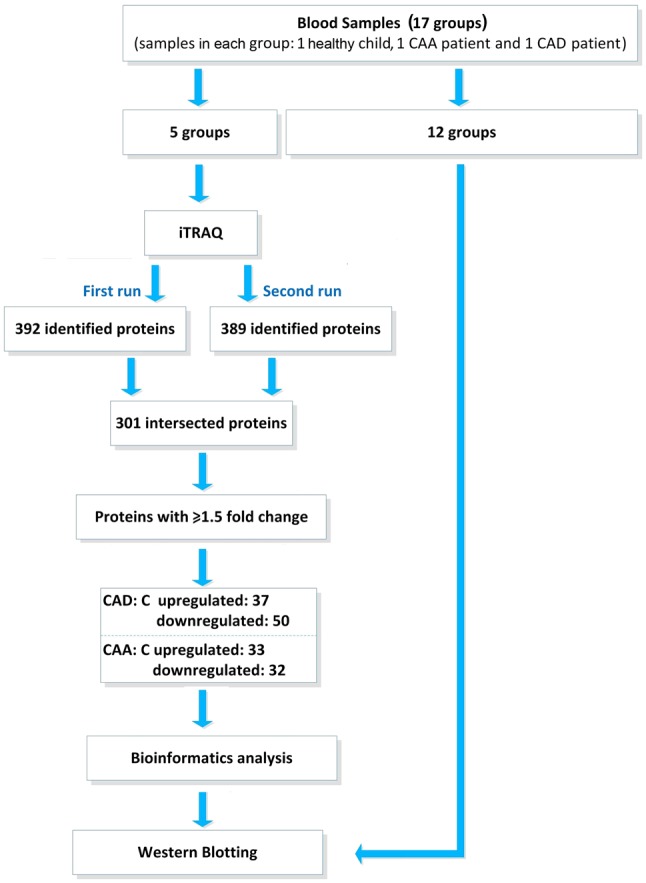
Flow diagram showing the trial profile and study design. CAA, coronary artery aneurism; CAD, coronary artery dilation; iTRAQ, isobaric tags for relative and absolute quantitation.
Identification of differentially expressed proteins in KD patients with CAA or CAD by iTRAQ
Three pooled samples of the control, CAA and CAD groups, were subjected to iTRAQ. As presented in Fig. 2, MS analysis was performed twice using the iTRAQ technique to ensure that reliable results were obtained; 392 and 389 proteins were identified as a result of the two independent experiments. In total, 301 proteins overlapped in the two experiments and were considered the final accurate detected, high-quality proteins for the differential expression analysis. A threshold change of 1.5-fold was established to identify differentially expressed proteins in comparisons between CAD and control samples or CAA and control samples. For comparisons between CAD and control samples, 37 upregulated proteins and 50 downregulated proteins were identified as differentially expressed. For comparisons between CAA and control samples, the numbers of upregulated and downregulated differentially expressed proteins were 33 and 32, respectively (Tables II and III).
Table II.
Identification of differentially expressed proteins between the Kawasaki disease with coronary artery dilation and control groups by isobaric tags for relative and absolute quantitation technology.
| Protein name | Gene name | Accession nos. | 95% CI | Peptides (95%) | Unused ProtScore | Ratio (Con/CAD) |
|---|---|---|---|---|---|---|
| Complement C4-A | C4A | P0C0L4 | 59.46 | 263 | 176.2 | 1.9953 |
| α-1-antitrypsin | SERPINA1 | P01009 | 87.32 | 357 | 147.8 | 3.4041 |
| Haptoglobin | HP | P00738 | 83.25 | 426 | 125.86 | 6.4269 |
| Uncharacterized protein | N/A | B4E1Z4 | 47.47 | 105 | 100.21 | 3.4356 |
| Hemopexin | HPX | P02790 | 74.24 | 155 | 82.36 | 2.3121 |
| α-1-antichymotrypsin | SERPINA3 | P01011 | 67.38 | 119 | 66.07 | 9.0365 |
| α-1-acid glycoprotein 1 | ORM1 | P02763 | 55.22 | 90 | 44.86 | 7.5162 |
| Plasma protease C1 inhibitor | SERPING1 | P05155 | 47.80 | 61 | 44.05 | 3.5318 |
| Complement component C9 | C9 | P02748 | 38.82 | 34 | 40.08 | 9.6383 |
| Inter-α-trypsin inhibitor heavy chain H3 | ITIH3 | Q06033 | 29.10 | 53 | 34.19 | 2.4434 |
| Ig α-1 chain C region | IGHA1 | P01876 | 58.36 | 92 | 33.33 | 2.5119 |
| Leucine-rich α-2-glycoprotein | LRG1 | P02750 | 43.80 | 27 | 23.89 | 4.9204 |
| Hemoglobin subunit β | HBB | P68871 | 88.44 | 41 | 21.25 | 3.0200 |
| Angiotensinogen | AGT | P01019 | 25.77 | 21 | 20.46 | 9.7275 |
| Fibrinogen α chain | FGA | P02671 | 25.17 | 21 | 19.87 | 9.3756 |
| Plastin-2 | LCP1 | P13796 | 21.37 | 11 | 18.50 | 1.6596 |
| Lipopolysaccharide-binding protein | LBP | P18428 | 23.70 | 10 | 18.31 | 3.6983 |
| Fibrinogen γ chain | FGG | C9JEU5 | 33.26 | 11 | 17.56 | 2.5119 |
| α-1-acid glycoprotein 2 | ORM2 | P19652 | 49.75 | 43 | 17.51 | 6.4269 |
| Hemoglobin subunit α | HBA1 | P69905 | 67.61 | 40 | 16.75 | 5.2966 |
| Fibrinogen β chain | FGB | P02675 | 27.09 | 11 | 13.20 | 2.8314 |
| Monocyte differentiation antigen CD14 | CD14 | P08571 | 32.00 | 7 | 12.14 | 2.0512 |
| Serum amyloid A-1 protein | SAA1 | P0DJI8 | 54.92 | 20 | 11.73 | 3.8019 |
| Serum amyloid P-component | APCS | P02743 | 31.39 | 12 | 11.02 | 2.4889 |
| Complement C4-B | C4B | P0C0L5 | 59.12 | 257 | 10.00 | 1.7865 |
| Polymeric immunoglobulin receptor | PIGR | P01833 | 10.34 | 6 | 9.25 | 2.4210 |
| C-reactive protein | CRP | P02741 | 16.52 | 8 | 8.53 | 8.3176 |
| Mannose-binding protein C | MBL2 | P11226 | 16.53 | 4 | 7.33 | 2.5586 |
| Ig ∆ chain C region (fragment) | IGHD | A0A0A0MS09 | 16.05 | 5 | 6.96 | 5.2966 |
| Keratin, type I cytoskeletal 10 | KRT10 | P13645 | 11.82 | 5 | 6.66 | 2.2284 |
| Transgelin-2 (fragment) | TAGLN2 | X6RJP6 | 27.27 | 5 | 6.57 | 3.0479 |
| Protein S100-A9 | S100A9 | P06702 | 30.7 | 3 | 6.39 | 4.9204 |
| α-1-antitrypsin | SERPINA1 | A0A024R6I7 | 87.32 | 348 | 5.19 | 46.1318 |
| Lymphatic vessel endothelial hyaluronic acid receptor 1 | LYVE1 | Q9Y5Y7 | 5.59 | 2 | 4.00 | 2.0701 |
| Protein S100-A8 | S100A8 | P05109 | 23.66 | 2 | 2.14 | 4.4463 |
| Protein IGLV2-11 (fragment) | IGLV2-11 | A0A075B6K3 | 6.723 | 3 | 2 | 5.7544 |
| Resistin | RETN | Q9HD89 | 10.19 | 1 | 2 | 1.8707 |
| Apolipoprotein B-100 | APOB | P04114 | 51.92 | 374 | 399.96 | −2.5586 |
| Serotransferrin | TF | P02787 | 78.08 | 349 | 172.35 | −1.8707 |
| Serum albumin | ALB | P02768 | 74.88 | 271 | 139.74 | −5.2966 |
| Fibronectin | FN1 | P02751 | 36.5 | 101 | 108.76 | −30.4790 |
| Complement factor H | CFH | P08603 | 50.93 | 85 | 92.38 | −2.4660 |
| Complement C5 | C5 | P01031 | 30.91 | 58 | 78.08 | −2.0893 |
| Apolipoprotein A-I | APOA1 | P02647 | 78.28 | 162 | 65.7 | −5.0582 |
| Inter-α-trypsin inhibitor heavy chain H1 | ITIH1 | P19827 | 45.77 | 81 | 56.95 | −2.7040 |
| Plasminogen | PLG | P00747 | 52.59 | 63 | 55.98 | −4.4875 |
| Apolipoprotein A-IV | APOA4 | P06727 | 65.4 | 52 | 45.59 | −1.5996 |
| Afamin | AFM | P43652 | 42.9 | 27 | 37.22 | −2.4889 |
| Heparin cofactor 2 | SERPIND1 | P05546 | 42.48 | 43 | 35.9 | −3.3420 |
| Histidine-rich glycoprotein | HRG | P04196 | 38.67 | 27 | 35.14 | −9.2897 |
| α-2-HS-glycoprotein | AHSG | P02765 | 57.49 | 51 | 34.96 | −1.9231 |
| Apolipoprotein A-II | APOA2 | V9GYM3 | 49.62 | 66 | 30.77 | −5.1051 |
| Serum paraoxonase/arylesterase 1 | PON1 | P27169 | 59.44 | 35 | 30.46 | −2.7040 |
| Apolipoprotein E | APOE | P02649 | 56.78 | 38 | 26.73 | −2.0512 |
| Gelsolin | GSN | P06396 | 25.45 | 24 | 24.19 | −3.8371 |
| Complement component C8 β chain | C8B | F5H7G1 | 19.48 | 14 | 22.71 | −2.5823 |
| Coagulation factor XII | F12 | P00748 | 26.5 | 21 | 21.87 | −2.0893 |
| Coagulation factor XIII B chain | F13B | P05160 | 23.45 | 15 | 21.37 | −1.9231 |
| Pigment epithelium-derived factor | SERPINF1 | P36955 | 30.14 | 12 | 18.6 | −1.9231 |
| Lumican | LUM | P51884 | 41.42 | 17 | 18.42 | −1.5417 |
| β-2-glycoprotein 1 | APOH | P02749 | 43.19 | 16 | 18.2 | −2.1677 |
| N-acetylmuramoyl-L-alanine amidase | PGLYRP2 | Q96PD5 | 38.72 | 17 | 17.6 | −2.3988 |
| Kallistatin | SERPINA4 | P29622 | 30.91 | 13 | 17.58 | −3.1333 |
| Insulin-like growth factor-binding protein complex acid labile subunit | IGFALS | P35858 | 18.18 | 10 | 17.48 | −2.6546 |
| Extracellular matrix protein 1 | ECM1 | Q16610 | 22.04 | 10 | 16.77 | −2.1878 |
| Sex hormone-binding globulin | SHBG | P04278 | 39.55 | 10 | 16.18 | −2.2699 |
| Transthyretin | TTR | P02766 | 72.11 | 27 | 15.31 | −3.0479 |
| Apolipoprotein L1 | APOL1 | O14791 | 34.42 | 11 | 14.3 | −2.3335 |
| Prenylcysteine oxidase 1 | PCYOX1 | Q9UHG3 | 18.02 | 8 | 14 | −3.1623 |
| Retinol binding protein 4, plasma, isoform CRA_b | RBP4 | Q5VY30 | 58.79 | 14 | 11.92 | −3.6983 |
| Phosphatidylinositol-glycan-specific phospholipase D | GPLD1 | P80108 | 10.95 | 8 | 11.54 | −3.6308 |
| Apolipoprotein M | APOM | O95445 | 36.7 | 11 | 10.76 | −3.5318 |
| Tetranectin | CLEC3B | E9PHK0 | 55 | 10 | 10.58 | −2.0893 |
| Protein IGHV3-53 (fragment) | IGHV3-53 | A0A087WSX4 | 60.34 | 21 | 10.43 | −1.9770 |
| Phospholipid transfer protein | PLTP | P55058 | 12.58 | 5 | 10.05 | −1.8030 |
| Apolipoprotein(a) | LPA | P08519 | 11.65 | 7 | 9.86 | −2.8576 |
| Uncharacterized protein | N/A | A0A0G2JPD4 | 71.04 | 150 | 9.19 | −1.9055 |
| Vitamin K-dependent protein C | PROC | E7END6 | 10.1 | 5 | 8 | −1.7378 |
| Cholesteryl ester transfer protein | CETP | P11597 | 10.14 | 6 | 6.52 | −4.9659 |
| Fetuin-B | FETUB | Q9UGM5 | 13.87 | 4 | 6.26 | −2.7040 |
| Apolipoprotein C-III | APOC3 | B0YIW2 | 34.19 | 11 | 6.18 | −2.8840 |
| Uncharacterized protein | N/A | A0A0G2JN06 | 55.05 | 79 | 6.03 | −3.3729 |
| Protein IGHV1-69-2 (fragment) | IGHV1-69-2 | A0A0B4J2H0 | 58.12 | 11 | 6 | −1.7061 |
| Apolipoprotein C-IV | APOC4 | P55056 | 15.75 | 2 | 4 | −2.5823 |
| Apolipoprotein C-I (fragment) | APOC1 | K7ERI9 | 23.38 | 2 | 2.84 | −2.2699 |
| Urea transporter 2 | SLC14A2 | Q15849 | 1.522 | 3 | 2 | −3.4674 |
| Extracellular superoxide dismutase (Cu-Zn) | SOD3 | P08294 | 4.583 | 1 | 1.77 | −1.6144 |
95% CI, percentage of matching amino acids from identified peptides having confidence intervals ≥95%, divided by the total number of amino acids in the sequence; Unused ProtScore, a measure of the protein confidence for a detected protein, calculated from the peptide confidence interval for peptides from spectra that are not already completely used by higher scoring winning proteins. Con, control group; CAD, coronary artery dilation group.
Table III.
Identification of differentially expressed proteins between the Kawasaki disease with coronary artery aneurysm and control groups by isobaric tags for relative and absolute quantitation technology.
| Protein name | Gene name | Accession nos. | 95% CI | Peptides (95%) | Unused ProtScore | Ratio (Con/CAA) |
|---|---|---|---|---|---|---|
| α-1-antitrypsin | SERPINA1 | P01009 | 87.32 | 357 | 147.80 | 1.5996 |
| Haptoglobin | HP | P00738 | 83.25 | 426 | 125.86 | 7.1121 |
| Uncharacterized protein | N/A | B4E1Z4 | 47.47 | 105 | 100.21 | 2.8576 |
| Ig µ chain C region | IGHM | A0A087WYJ9 | 64.94 | 98 | 68.73 | 1.6749 |
| α-1-antichymotrypsin | SERPINA3 | P01011 | 67.38 | 119 | 66.07 | 4.3251 |
| α-1-acid glycoprotein 1 | ORM1 | P02763 | 55.22 | 90 | 44.86 | 6.7920 |
| Complement component C9 | C9 | P02748 | 38.82 | 34 | 40.08 | 6.3096 |
| Pregnancy zone protein | PZP | P20742 | 25.98 | 146 | 24.62 | 5.3951 |
| Leucine-rich α-2-glycoprotein | LRG1 | P02750 | 43.80 | 27 | 23.89 | 3.6983 |
| Angiotensinogen | AGT | P01019 | 25.77 | 21 | 20.46 | 2.2909 |
| Fibrinogen α chain | FGA | P02671 | 25.17 | 21 | 19.87 | 25.3513 |
| Lipopolysaccharide-binding protein | LBP | P18428 | 23.70 | 10 | 18.31 | 4.4463 |
| Fibrinogen γ chain | FGG | C9JEU5 | 33.26 | 11 | 17.56 | 21.2814 |
| α-1-acid glycoprotein 2 | ORM2 | P19652 | 49.75 | 43 | 17.51 | 6.1376 |
| Hemoglobin subunit a | HBA1 | P69905 | 67.61 | 40 | 16.75 | 2.6303 |
| Galectin-3-binding protein | LGALS3BP | Q08380 | 18.63 | 10 | 14.74 | 1.8535 |
| Fibrinogen β chain | FGB | P02675 | 27.09 | 11 | 13.20 | 9.2897 |
| Monocyte differentiation antigen CD14 | CD14 | P08571 | 32.00 | 7 | 12.14 | 1.7219 |
| Serum amyloid A-1 protein | SAA1 | P0DJI8 | 54.92 | 20 | 11.73 | 3.8371 |
| Complement C4-B | C4B | P0C0L5 | 59.12 | 257 | 10.00 | 2.3335 |
| Apolipoprotein(a) | LPA | P08519 | 11.65 | 7 | 9.86 | 3.6644 |
| C-reactive protein | CRP | P02741 | 16.52 | 8 | 8.53 | 3.6644 |
| Hemoglobin subunit γ-2 | HBG2 | P69892 | 43.54 | 6 | 7.70 | 4.0179 |
| Transgelin-2 (fragment) | TAGLN2 | X6RJP6 | 27.27 | 5 | 6.57 | 3.3420 |
| Protein S100-A9 | S100A9 | P06702 | 30.70 | 3 | 6.39 | 5.2966 |
| Protein IGHV1-69-2 (fragment) | IGHV1-69-2 | A0A0B4J2H0 | 58.12 | 11 | 6.00 | 2.4660 |
| α-1-antitrypsin | SERPINA1 | A0A024R6I7 | 87.32 | 348 | 5.19 | 29.1072 |
| Protein IGLV3-19 (fragment) | IGLV3-19 | A0A075B6J8 | 70.54 | 7 | 4.66 | 2.9107 |
| Protein IGLV7-46 (fragment) | IGLV7-46 | A0A075B6I9 | 15.38 | 3 | 3.04 | 1.6596 |
| HCG1782423 (fragment) | IGLV2-18 | A0A075B6J9 | 24.17 | 2 | 2.63 | 3.3113 |
| Protein S100-A8 | S100A8 | P05109 | 23.66 | 2 | 2.14 | 4.8306 |
| Ras-related protein Rap-1b (fragment) | RAP1B | F5H823 | 17.48 | 2 | 2.00 | 2.2699 |
| Serglycin | SRGN | P10124 | 8.23 | 1 | 2.00 | 1.6444 |
| Serotransferrin | TF | P02787 | 78.08 | 349 | 172.35 | −8.5507 |
| Serum albumin | ALB | P02768 | 74.88 | 271 | 139.74 | −3.6983 |
| Fibronectin | FN1 | P02751 | 36.50 | 101 | 108.76 | −3.8726 |
| Apolipoprotein A-I | APOA1 | P02647 | 78.28 | 162 | 65.70 | −3.1623 |
| Inter-α-trypsin inhibitor heavy chain H1 | ITIH1 | P19827 | 45.77 | 81 | 56.95 | −2.0137 |
| Plasminogen | PLG | P00747 | 52.59 | 63 | 55.98 | −2.5119 |
| Kininogen-1 | KNG1 | P01042 | 45.19 | 73 | 54.56 | −2.3121 |
| Apolipoprotein A-IV | APOA4 | P06727 | 65.40 | 52 | 45.59 | −2.7290 |
| Antithrombin-III | SERPINC1 | P01008 | 43.75 | 46 | 44.44 | −2.0893 |
| Afamin | AFM | P43652 | 42.90 | 27 | 37.22 | −1.9953 |
| Histidine-rich glycoprotein | HRG | P04196 | 38.67 | 27 | 35.14 | −2.8054 |
| α-2-HS-glycoprotein | AHSG | P02765 | 57.49 | 51 | 34.96 | −2.2699 |
| Apolipoprotein A-II | APOA2 | V9GYM3 | 49.62 | 66 | 30.77 | −3.3420 |
| Serum paraoxonase/arylesterase 1 | PON1 | P27169 | 59.44 | 35 | 30.46 | −2.0512 |
| Apolipoprotein E | APOE | P02649 | 56.78 | 38 | 26.73 | −1.7378 |
| Gelsolin | GSN | P06396 | 25.45 | 24 | 24.19 | −1.8707 |
| Coagulation factor XII | F12 | P00748 | 26.50 | 21 | 21.87 | −1.8030 |
| Protein AMBP | AMBP | P02760 | 31.25 | 24 | 18.34 | −2.2284 |
| Kallistatin | SERPINA4 | P29622 | 30.91 | 13 | 17.58 | −2.1281 |
| Extracellular matrix protein 1 | ECM1 | Q16610 | 22.04 | 10 | 16.77 | −1.6596 |
| Transthyretin | TTR | P02766 | 72.11 | 27 | 15.31 | −2.8576 |
| Apolipoprotein L1 | APOL1 | O14791 | 34.42 | 11 | 14.30 | −1.8030 |
| Prenylcysteine oxidase 1 | PCYOX1 | Q9UHG3 | 18.02 | 8 | 14.00 | −1.7061 |
| Retinol binding protein 4, plasma, isoform CRA_b | RBP4 | Q5VY30 | 58.79 | 14 | 11.92 | −2.6303 |
| Phosphatidylinositol-glycan-specific phospholipase D | GPLD1 | P80108 | 10.95 | 8 | 11.54 | −1.8880 |
| Apolipoprotein M | APOM | O95445 | 36.70 | 11 | 10.76 | −2.1086 |
| Tetranectin | CLEC3B | E9PHK0 | 55.00 | 10 | 10.58 | −1.8030 |
| Cholinesterase | BCHE | P06276 | 10.96 | 7 | 10.56 | −2.1281 |
| Keratin, type I cytoskeletal 10 | KRT10 | P13645 | 11.82 | 5 | 6.66 | −1.9953 |
| Apolipoprotein C-III | APOC3 | B0YIW2 | 34.19 | 11 | 6.18 | −2.7040 |
| Uncharacterized protein (fragment) | N/A | A0A0C4DH43 | 24.37 | 3 | 4.00 | −2.3768 |
| Urea transporter 2 | SLC14A2 | Q15849 | 1.52 | 3 | 2.00 | −10.0000 |
95% CI, percentage of matching amino acids from identified peptides having confidence intervals ≥95%, divided by the total number of amino acids in the sequence; Unused ProtScore, a measure of the protein confidence for a detected protein, calculated from the peptide confidence interval for peptides from spectra that are not already completely used by higher scoring winning proteins; Con, control group; CAA, coronary artery aneurysm.
Differentially expressed protein categorization and network modelling
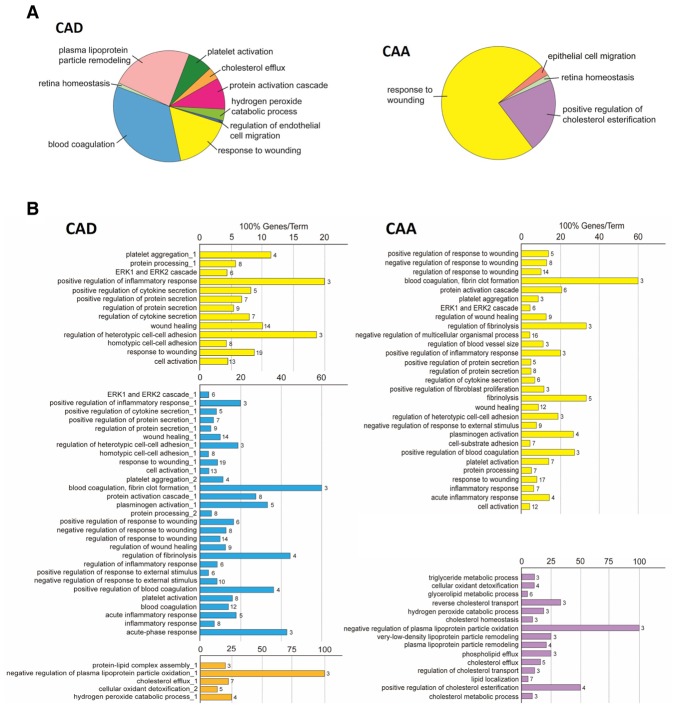
The functions (i.e., biological processes) of these differentially expressed proteins were analyzed using ClueGO, a Cytoscape plug-in that integrates GO terms (12). Pie charts were used to display the differences in the distribution of functional categories for differentially expressed proteins between KD patients with CAD and CAA (Fig. 3A). For the CAD vs. control comparison, 9 enriched processes were observed; however, only 4 processes were enriched in the CAA vs. control comparison. These results are presented as bar charts in Fig. 3B, which use the same color scheme as that of the pie charts. Analysis of the biological processes, as summarized in the two pie charts, notably demonstrated that the wound healing response and lipoproteins are involved in CAD and CAA. In a combined analysis of the corresponding CAA/CAD bar charts and pie charts, the main differentially expressed proteins were involved in the wounding response, and lipoprotein and cholesterol-associated processes, indicating that KD patients with CAA may suffer more harmful attacks in the coronary arteries or surrounding areas.
Figure 3.
Biological process of differentially expressed proteins in KD with CAD and CAA. (A) Pie charts representing the distribution of identified differentially expressed proteins according to their biological process. (B) Bar charts demonstrating the specific processes that correspond to the classification; the same color key that was used in the pie charts has also been applied in these charts. P<0.05 was used as a threshold for gene ontology categories enrichment. KD, Kawasaki disease; CAA, coronary artery aneurism; CAD, coronary artery dilation.
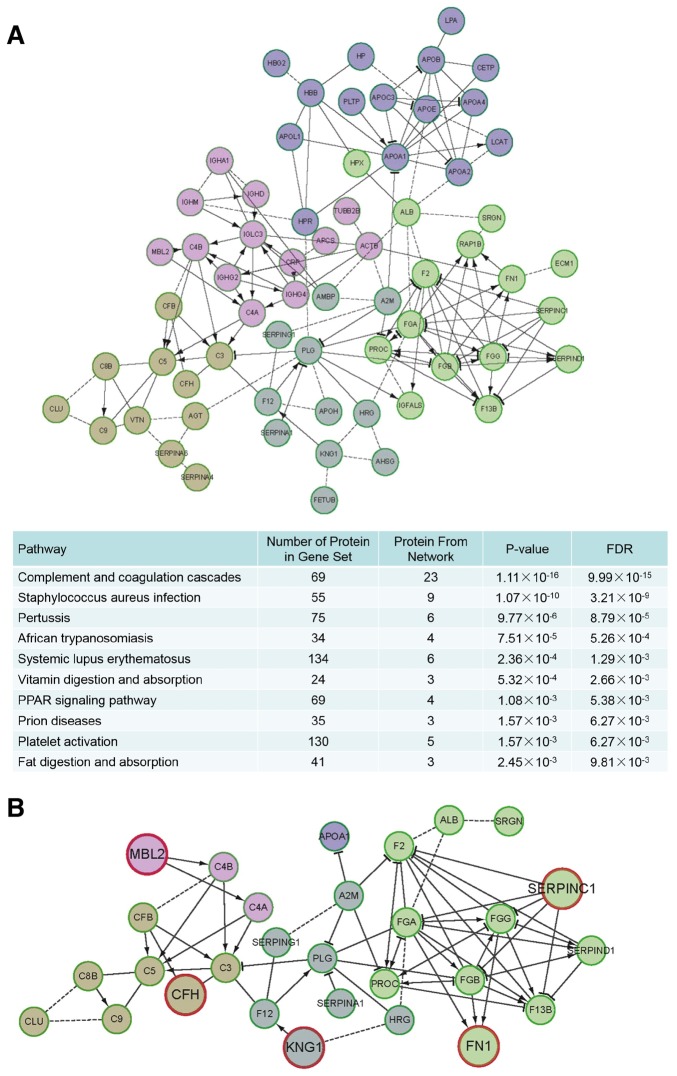
To comprehensively characterize the associations among significantly differentially expressed proteins in CAA and CAD, KEGG pathway enrichment analysis and signaling network modelling were employed using the Cytoscape plug-in ReactomeFIViz (13) (Fig. 4A). The thresholds for pathway enrichment were defined as P<0.05 and FDR <1%. These enriched pathways were associated with bacterial and viral infection (e.g., Staphylococcus aureus infection), inflammatory responses (e.g., complement cascades and platelet activation) and other metabolic processes. Various bacteria and viruses have been reported as infectious agents of KD, such as Staphylococcus aureus (14), which may explain the activation of the inflammatory response pathways. However, KD is a highly-complicated disease, and its etiology and pathology are still unclear. The infection and inflammatory response-associated sub-networks were extracted from the whole differentially expressed protein networks in KD and the sub-network was highly enriched for proteins in infectious and inflammatory pathways. Some of these have demonstrated a high correlation with KD; the expression levels of the fibrinogen γ chain, which belongs to the fibrinogen family, are associated with KD according to a previous study (15). Thus, the sub-network generated was deemed to be associated with KD (Fig. 4B). The network proteins were divided into 5 modules (presented as nodes of different colors in the figure) according to different clusters; proteins that serve a core role in each module and have either not been investigated or have rarely been investigated in previous studies were identified. Specifically, CFH, MBL2, KNG1, FN1 and SERPINC1 were identified as primary candidates involved in CAA and CAD (presented as bigger nodes with red circles). CFH and MBL2 were associated with classical complement proteins (yellow and pink clusters), KNG1 and FN1 were associated with proteins correlated to KD (grey and green clusters), and SERPINC1 is as an inhibitor in a complement protein associated pathway (green cluster). These results clarified the differentially expressed proteins in CAA and CAD samples and the associations among proteins in the network.
Figure 4.
Network of differentially expressed proteins in KD with CAD and CAA. (A) Network based on functionally-enriched KEGG pathways. The proteins in the network were differentially expressed proteins in the present study. (B) Network associated with the variations within patients with KD and CAD/CAA. CFH (yellow cluster), MBL2 (pink cluster), KNG1 (grey cluster), FN1 (green cluster) and SERPINC1 (green cluster) were identified as primary candidates involved in CAA and CAD (presented as bigger nodes with red circles). P<0.05 was a threshold for KEGG pathway enrichment. KD, Kawasaki disease; CAA, coronary artery aneurism; CAD, coronary artery dilation; KEGG, Kyoto Encyclopedia of Genes and Genomes; FN1, fibronectin 1; CFH, complement factor; KNG1, kininogen 1; MBL2, mannose binding lectin 2; SERPINC1, serpin family C member 1; FDR, false discovery rate.
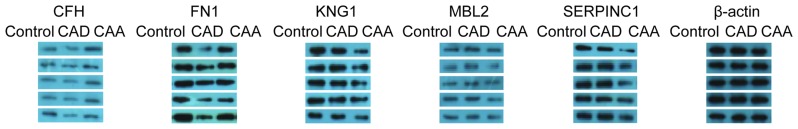
Validation of proteomics results
Alterations in the expression levels of proteins were validated by western blotting using CAD and CAA samples, and normal samples for comparison. Five proteins (CFH, MBL2, KNG1, FN1 and SERPINC1) were validated in 12 groups of samples (each including 1 control, 1 CAD and 1 CAA sample); the results for 5 of 12 validated groups are presented in Fig. 5. The protein expression results for the 5 proteins exhibited high specificity and were consistent with the proteomics results observed in the iTRAQ. In particular, MBL2 and CFH exhibited reduced expression levels within the CAD samples, and the opposite results were obtained for KNG and SERPINC1, in which the expression levels were downregulated within CAA samples. The expression levels of FN1 were downregulated in the CAD and CAA samples when compared with the control samples.
Figure 5.
Protein expression confirmation of MBL2, CFH, KNG1, SERPINC1 and FN1 by western blotting in 5 representative groups comprised of healthy children (control), patients with KD and CAD, and patients with KD and CAA. CAA, coronary artery aneurism; CAD, coronary artery dilation; KD, Kawasaki disease; FN1, fibronectin 1; CFH, complement factor; KNG1, kininogen 1; MBL2, mannose binding lectin 2; SERPINC1, serpin family C member 1.
Discussion
The increased risk of coronary artery complications in patients with KD results in increased morbidity and mortality. The pathological processes of coronary artery lesions include endothelial dysfunction, the destruction of the internal elastic lamina, thrombocytosis and hypercoagulability. CAD and CAA are the main complications of KD, and may cause cardiovascular events (16). It is highly important to determine the mechanisms of CAD/CAA to prevent and treat these complications. In the present study, serum protein level differences between healthy volunteers, patients with CAD and patients with CAA were investigated. The results suggested that several pathways, including those involved in the response to wounding, lipoprotein remodeling, platelet activation and blood coagulation, may differ between healthy volunteers, patients with CAD and patients with CAA.
In the present study, differentially expressed proteins associated with the two coronary artery abnormalities, CAD and CAA, within patients with KD were identified using proteomics methods. A total of 51 samples were divided into 17 groups. Each group contained 1 control sample, 1 CAD sample and 1 CAA sample. Three pooled samples (5 control, 5 CAD and 5 CAA) were generated by combining samples from 5 groups and were used for iTRAQ analyses to detect significantly differentially expressed proteins. The other 12 groups were used for western blot validation. In total, 87 and 65 differentially expressed proteins for the CAD and CAA were obtained, respectively, compared with the control.
The biological processes associated with differentially expressed proteins in CAD and CAA were also analyzed. Variations between the two conditions were identified in the present study; lower representation of proteins associated with the wounding response, lipoprotein modelling, platelet activation and blood coagulation, revealed that patients with KD and CAD may exhibit stress responses associated with coronary artery injury. However, within CAA patients, the representation of proteins involved in the response to wounding and the corresponding cholesterol components increased significantly with respect to control samples, indicating aggravated injury in coronary arteries. All of the identified significant proteins were used to construct a signaling network to clarify the overall pathway associations. The main pathways were enriched for functions in infection and immune associated diseases, inflammation and metabolism. These results are consistent with a previous hypothesis (17,18). Thus, the present study focused on the infection, inflammation and coagulation pathways, which were extracted from the above network as a novel network for further analyses.
As presented in Fig. 3B, a few proteins in this network have been reported in previous studies associated with KD, including complement C3 (C3) (19), apolipoprotein A-I (20), and fibrinogen β chain (21). Accordingly, the focus of the present study was on significant proteins that have not been reported previously and served key roles in each cluster of the network. Thus, the five proteins, CFH, MBL2, KNG1, FN1 and SERPINC1 were selected as candidate proteins associated with CAD and CAA. The western blot results for the five proteins were consistent with those of the iTRAQ analyses. MBL2 and CFH were reduced in KD with CAD compared with controls. KNG1 and SERPINC1 were downregulated in KD with CAA. FN1 was differentially expressed in both diseases; FN1 decreased in CAA and CAD when compared with controls.
CFH and MBL2 were in the complement protein-associated part of the network. Complement proteins, similar to the defense and clearance systems, serve an important role in the innate immune system. Pathogens activate the main complement protein C3 via one of the classical complement proteins, such as lectin, or alternative pathways to form C3b. The process is regulated to avoid excessive C3b production, which damages host cells, or a C3b deficiency, which induces an immune-compromised state via the main regulatory complement factor CFH (22). MBL2 is a macromolecule that contains sugar-binding lectin and collagenous domains, and forms MBL-associated serine proteases to cleave complement proteins C2 and C4. The complex process is also known as the MBL pathway. A reduced level of MBL is associated with increased bacterial or viral infections in children, and MBL is known to serve a dual role in KD. Within children <1-year-old, MBL may be associated with the resistance of infections, and in older children, MBL may be associated with endothelial damage (23). Decreases in CFH and MBL2 were only observed in KD with CAD samples, which suggested a dysfunction in the complement system.
KNG1 and SERPINC1 are located in a coagulation-associated network. To the best of our knowledge, the direct genetic interactions of KNG1 with histidine-proline rich glycoprotein (HRG) and coagulation factor XII have never been reported in previous KD studies. However, some studies have indicated that they may interact with each other in the coagulation pathway. KNG1 and HRG have similar structures and are adjacent to each other on chromosome 3, which suggests that these proteins may exhibit similar functions (24,25). HRG deficiency is associated with enhanced blood coagulation, which indicates that decreased KNG1 is a potential cofactor to enhance coagulation (26). The potent anticoagulant SERPINC1 is a coagulation protease inhibitor and also possesses independent anti-inflammatory properties. Overexpressed SERPINC1 stimulates the expression of Coagulation Factor II and other proteases (27). The protein is mainly associated with the diseases, inherited thrombophilia (28) and SERPINC1 deficiency (29), by affecting the anticoagulation pathways. The downregulation of KNG1 and SERPINC1 observed in KD with CAA samples may increase blood coagulation to enhance thrombosis.
FN1 is a multifunctional glycoprotein that serves in the plasma and extracellular matrix as a soluble dimer, or at the cell surface as a dimer or multimer. It participates in multiple biological processes, including cell adhesion, migration, wound healing, and blood coagulation (30). Both CAD and CAA are characterized by endothelial dysfunction and stress responses to wound healing. Therefore, the decrease in FN1 expression levels in patients with coronary artery lesions may be associated with the aforementioned pathways.
In conclusion, the present study identified five candidate proteins differentially expressed in patients with KD and CAD/CAA: CFH, MBL2, KNG1, FN1 and SERPINC1. These five proteins provide a basis for understanding the differences between CAD and CAA in patients with KD. The alterations in MBL2 and CFH indicated that responses to pathogen infections and the innate immune system may be closely associated with the development of CAD. Decreased levels of KNG1 and SERPINC1 revealed that dysfunctions in coagulation accompanied the development of CAA. Decreased FN1 expression levels in the two conditions indicated that the pathways, including cell adhesion, migration, wound healing and blood coagulation may serve a role in the occurrence of CAD and CAA. The five proteins investigated in the present study may also serve to distinguish CAD and CAA in the early diagnosis of KD.
Acknowledgements
The present study was supported by the Natural Science Fund of China (grant no. 81500275 to H.-L.J), Guangdong Natural Science Foundation (grant no. 2016A030313080 to H.-L.J), Guangdong Natural Science Foundation (grant no. 2016A030310080 to W.-T.L), Fundamental Research Funds for the Central Universities (grant no. 21616305 to W.-T.L), Guangzhou City Scientific Research Project Foundation of Technology and Information Bureau (grant no. 201510010287 to L.Z), the Natural Science Fund of China (81300124 to M.-G.X), Guangdong Medical Science Foundation (grant no. A2016012 to M.-G.X) and Shen Zhen Scientific Plan (grant no. JCYJ20140416141331478 to M.-G.X).
References
- 1.Kawasaki T. Acute febrile mucocutaneous syndrome with lymphoid involvement with specific desquamation of the fingers and toes in children. Allergy. 1967;16:178–222. (In Japanese) [PubMed] [Google Scholar]
- 2.Chen KY, Curtis N, Dahdah N, Kowalski R, Cheung M, Burgner DP. Kawasaki disease and cardiovascular risk: A comprehensive review of subclinical vascular changes in the longer term. Acta Paediatric. 2016;105:752–761. doi: 10.1111/apa.13367. [DOI] [PubMed] [Google Scholar]
- 3.Kato H, Koike S, Yamamoto M, Ito Y, Yano E. Coronary aneurysms in infants and young children with acute febrile mucocutaneous lymph node syndrome. J Pediatr. 1975;86:892–898. doi: 10.1016/S0022-3476(75)80220-4. [DOI] [PubMed] [Google Scholar]
- 4.Uehara R, Belay ED. Epidemiology of Kawasaki disease in Asia, Europe, and the United States. J Epidemiol. 2012;22:79–85. doi: 10.2188/jea.JE20110131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh S, Vignesh P, Burgner D. The epidemiology of Kawasaki disease: A global update. Arch Dis Child. 2015;100:1084–1088. doi: 10.1136/archdischild-2014-307536. [DOI] [PubMed] [Google Scholar]
- 6.Newburger JW, Takahashi M, Gerber MA, Gewitz MH, Tani LY, Burns JC, Shulman ST, Bolger AF, Ferrieri P, Baltimore RS, et al. Diagnosis, treatment, and long-term management of Kawasaki disease: A statement for health professionals from the committee on rheumatic fever, endocarditis and Kawasaki disease, council on cardiovascular disease in the young, American heart association. Circulation. 2004;110:2747–2771. doi: 10.1161/01.CIR.0000145143.19711.78. [DOI] [PubMed] [Google Scholar]
- 7.Mueller F, Knirsch W, Harpes P, Pretre R, Valsangiacomo Buechel E, Kretschmar O. Long-term follow-up of acute changes in coronary artery diameter caused by Kawasaki disease: Risk factors for development of stenotic lesions. Clin Res Cardiol. 2009;98:501–507. doi: 10.1007/s00392-009-0032-2. [DOI] [PubMed] [Google Scholar]
- 8.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 9.Celis JE, Gromov P, Cabezón T, Moreira JM, Ambartsumian N, Sandelin K, Rank F, Gromova I. Proteomic characterization of the interstitial fluid perfusing the breast tumor microenvironment: A novel resource for biomarker and therapeutic target discovery. Mol Cell Proteomics. 2004;3:327–344. doi: 10.1074/mcp.M400009-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Ray S, Reddy PJ, Jain R, Gollapalli K, Moiyadi A, Srivastava S. Proteomic technologies for the identification of disease biomarkers in serum: Advances and challenges ahead. Proteomics. 2011;11:2139–2161. doi: 10.1002/pmic.201000460. [DOI] [PubMed] [Google Scholar]
- 11.Ayusawa M, Sonobe T, Uemura S, Ogawa S, Nakamura Y, Kiyosawa N, Ishii M, Harada K, Kawasaki Disease Research Committee Revision of diagnostic guidelines for Kawasaki disease (the 5th revised edition) Pediatr Int. 2005;47:232–234. doi: 10.1111/j.1442-200x.2005.02033.x. [DOI] [PubMed] [Google Scholar]
- 12.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu G, Dawson E, Duong A, Haw R, Stein L. ReactomeFIViz: A Cytoscape app for pathway and network-based data analysis. F1000Res. 2014;3:146. doi: 10.12688/f1000research.4431.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Komatsu H, Fujisawa T. Kawasaki disease and infection. Nihon Rinsho. 2008;66:278–282. (In Japanese) [PubMed] [Google Scholar]
- 15.Lee KY, Lee HS, Hong JH, Han JW, Lee JS, Whang KT. High-dose intravenous immunoglobulin downregulates the activated levels of inflammatory indices except erythrocyte sedimentation rate in acute stage of Kawasaki Disease. J Trop Pediatr. 2005;51:98–101. doi: 10.1093/tropej/fmh087. [DOI] [PubMed] [Google Scholar]
- 16.Adler AC, Kodavatiganti R. Kawasaki disease and giant coronary artery aneurysms: The role of echocardiography from diagnosis through follow-up. Echocardiography. 2016;33:1245–1250. doi: 10.1111/echo.13271. [DOI] [PubMed] [Google Scholar]
- 17.Greco A, De Virgilio A, Rizzo MI, Tombolini M, Gallo A, Fusconi M, Ruoppolo G, Pagliuca G, Martellucci S, de Vincentiis M. Kawasaki disease: An evolving paradigm. Autoimmun Rev. 2015;14:703–709. doi: 10.1016/j.autrev.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 18.Burns JC, Herzog L, Fabri O, Tremoulet AH, Rodó X, Uehara R, Burgner D, Bainto E, Pierce D, Tyree M, et al. Seasonality of Kawasaki disease: A globalperspective. PLoS One. 2013;8:e74529. doi: 10.1371/journal.pone.0074529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe T. Kidney and urinary tract involvement in Kawasaki disease. Int J Pediatr. 2013;2013:831834. doi: 10.1155/2013/831834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blankier S, McCrindle BW, Ito S, Yeung RS. The role of atorvastatin in regulating the immune response leading to vascular damage in a model of Kawasaki disease. Clin Exp Immunol. 2011;164:193–201. doi: 10.1111/j.1365-2249.2011.04331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoon KL. Update of genetic susceptibility in patients with Kawasaki disease. Korean J Pediatr. 2015;58:84–88. doi: 10.3345/kjp.2015.58.3.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perkins SJ, Fung KW, Khan S. Molecular interactions between complement factor H and its heparin and heparan sulfate ligands. Front Immunol. 2014;5:126. doi: 10.3389/fimmu.2014.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biezeveld MH, Geissler J, Weverling GJ, Kuipers IM, Lam J, Ottenkamp J, Kuijpers TW. Polymorphisms in the mannose-binding lectin gene as determinants of age-defined risk of coronary artery lesions in Kawasaki disease. Arthritis Rheum. 2006;54:369–376. doi: 10.1002/art.21529. [DOI] [PubMed] [Google Scholar]
- 24.Brunel H, Massanet R, Martinez-Perez A, Ziyatdinov A, Martin-Fernandez L, Souto JC, Perera A, Soria JM. The central role of KNG1 gene as a genetic determinant of coagulation pathway-related traits: Exploring metaphenotypes. PLoS One. 2016;11:e0167187. doi: 10.1371/journal.pone.0167187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weng LC, Cushman M, Pankow JS, Basu S, Boerwinkle E, Folsom AR, Tang W. A genetic association study of activated partial thromboplastin time in European Americans and African Americans: The ARIC study. Hum Mol Genet. 2015;24:2401–2408. doi: 10.1093/hmg/ddu732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang W, Schwienbacher C, Lopez LM, Ben-Shlomo Y, Oudot-Mellakh T, Johnson AD, Samani NJ, Basu S, Gögele M, Davies G, et al. Genetic associations for activated partial thromboplastin time and prothrombin time, their gene expression profiles, and risk of coronary artery disease. Am J Hum Genet. 2012;91:152–162. doi: 10.1016/j.ajhg.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iba T, Thachil J. Is antithrombin III for sepsis-associated disseminated intravascular coagulation really ineffective? Intensive Care Med. 2016;42:1193–1194. doi: 10.1007/s00134-016-4288-5. [DOI] [PubMed] [Google Scholar]
- 28.Simioni P, Tormene D, Spiezia L, Tognin G, Rossetto V, Radu C, Prandoni P. Inherited thrombophilia and venous thromboembolism. Semin Thromb Hemost. 2006;32:700–708. doi: 10.1055/s-2006-951298. [DOI] [PubMed] [Google Scholar]
- 29.Cooper PC, Coath F, Daly ME, Makris M. The phenotypic and genetic assessment of antithrombin deficiency. Int J Lab Hematol. 2011;33:227–237. doi: 10.1111/j.1751-553X.2011.01307.x. [DOI] [PubMed] [Google Scholar]
- 30.Ohtsubo H, Okada T, Nozu K, Takaoka Y, Shono A, Asanuma K, Zhang L, Nakanishi K, Taniguchi-Ikeda M, Kaito H, et al. Identification of mutations in FN1 leading to glomerulopathy with fibronectin deposits. Pediatr Nephrol. 2016;31:1459–1467. doi: 10.1007/s00467-016-3368-7. [DOI] [PubMed] [Google Scholar]