Abstract
Androgens have been recognized to be primary causative agents of prostate cancer. Following binding to the androgen receptor (AR), androgens serve important roles in the carcinogenesis of prostate cancers. ARs serve an important role during all stages of prostate cancer, and inhibiting their function may help to slow prostate cancer growth. In the present study, the AR gene was targeted in androgen-positive prostate cancer cells using the clustered regularly interspaced short palindromic repeats-associated protein (CRISPR/Cas) system. A total of three different single-guide RNAs (sgRNAs) were designed according to the three different target sites in the AR gene. The optimal sgRNA with a specific target effect was effectively screened to cleave the AR gene in androgen-positive prostate cancer cell lines, and to suppress the growth of androgen-sensitive prostate cancer in vitro. The AR-sgRNA-guided CRISPR/Cas system was able to disrupt the AR at specific sites and inhibit the growth of androgen-sensitive prostate cancer cells; further studies demonstrated that the decreased cell proliferation was due to cellular apoptosis. The results of the present study suggested that the CRISPR/Cas system may be a useful therapeutic strategy for the treatment of prostate cancer.
Keywords: clustered regularly interspersed short palindromic repeats-associated protein system, androgen receptor, LNCaP cells, prostate cancer, gene therapy
Introduction
Prostate cancer is one of the most malignant types of tumour in developed countries, and prostate cancer-associated mortality has recently increased. According to statistics from the American Cancer Society in 2016 (1), >1.8 million new cases of prostate cancer occur each year. In addition, >26,000 patients die from prostate cancer per year, accounting for 27% of the total number of patients. Due to diverse dietary habits and more advanced medical equipment, early and small prostate cancer lesions are increasingly detected. The appropriate treatment for prostate cancer depends on the disease stage. Radical prostatectomy is the primary choice for the treatment of early stage prostate cancer. However, endocrine therapy may be more effective for the treatment of prostate tumours that have progressed to an advanced stage.
Gene therapy is a novel treatment approach that is primarily in an experimental stage. The androgen-androgen receptor (AR) signalling pathway serves an important role during all stages of prostate cancer. Following binding to the AR in prostate epithelial cells, androgens translocate to the cell nucleus. Once in the nucleus, androgens bind to androgen response elements upstream of the target genes, which leads to DNA transcription, promotes the abnormal proliferation of prostate epithelial cells and enables carcinogenesis (2). During the androgen-sensitive stage of prostate cancer, the blockade or removal of androgens prevents the binding of androgens to the AR, which may delay the development of prostate cancer. However, at 18 months, prostate cancer becomes androgen-independent. Therefore, the tumour growth may not be inhibited using androgen suppressors. However, numerous studies have demonstrated that the AR retains a role in androgen-independent prostate cancer via AR gene mutation and gene amplification, and acts together with co-modulating factors to stimulate the abnormal activation of other signalling pathways (3–5). Therefore, the development of prostate cancer may be suppressed by inhibiting AR.
The clustered regularly interspaced short palindromic repeats-associated protein (CRISPR/Cas) system has recently become an extensively used gene-editing technology (6,7). Compared with the previous zinc-finger nuclease (ZFN) and transcription activator-like effector nuclease (TALEN) technologies, the CRISPR/Cas system is more widely used in basic and in certain clinical studies (8). A number of researchers have begun to apply the CRISPR/Cas system in studies investigating inherited genetic diseases, and certain studies have achieved relevant results (9–13). For example, Kawamura et al (14) established homeobox protein NANOG (NANOG)- and NANOGP8-knockout DU145 cell lines, which exhibited a significantly decreased malignant potential compared with control cells.
In the present study, a site-specific CRISPR/Cas system was designed to cleave the AR gene in androgen-positive prostate cancer cell lines and decrease the growth of androgen-dependent prostate cancer in vitro. Stable LNCaP cell lines harbouring Cas9 and single-guide RNAs (sgRNAs) were constructed to knock out AR, and the knockout activity and efficiency of CRISPR was investigated. The results of the present study demonstrated that the treatment with CRISPR/Cas significantly inhibited the growth of LNCaP cells. These results suggested that the CRISPR/Cas system may be a potential therapeutic strategy for the treatment of prostate cancer.
Materials and methods
Cell culture
The human LNCaP cell line was purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and preserved in the Laboratory of the Second Affiliated Hospital of Soochow University. All cells were cultured in RPMI 1640 medium (HyClone; GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 100 U/ml penicillin, 100 µg/ml streptomycin and 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The cell culture plates were maintained in humidified incubators at 37°C with 5% CO2.
Construction of AR-Cas9-sgRNA stable cell lines
Lentiviral vectors (LVs) harbouring Cas9 and sgRNA (Shanghai GeneChem Co., Ltd., Shanghai, China) were constructed for transfection of the LNCaP cells. According to the manufacturer's protocol, the LVs harbouring Cas9 were transfected into LNCaP cells using polybrene (Shanghai GeneChem Co., Ltd.), and the LNCaP-Cas9 stable cell lines were isolated. In addition, three different sgRNAs were designed according to the three different target sites in the AR gene and packaged into the LVs to form LV-sgRNA-AR230, LV-sgRNA-AR231 and LV-sgRNA-AR232. Sequencing was performed using the Sanger method (Shanghai GeneChem Co., Ltd.) to confirm the sequence of the three sgRNAs. The sequences of the sgRNAs were as follows: sgRNA-AR230, CATTTCCGAAGACGACAAGA; sgRNA-AR231, TGTCCAGCACACACTACACC; and sgRNA-AR232, GCACTATTGATAAATTCCGA. The three LV-sgRNA-ARs were transfected into the LNCaP cells to eventually generate the LV-Cas9-sgRNA-AR stable cell lines, and 48 h following LV transfection, subsequent experiments were performed. The LV transfection altered the growth of the LNCaP cells to a certain degree.
Polymerase chain reaction (PCR) detection of the mutation
The LV-Cas9-sgRNA-AR-infected and LV-Cas9-control-infected LNCaP cells (LNCaP-Cas9-sgRNA-AR and LNCaP-Cas9-control) were collected into Eppendorf tubes for the detection of the mutations. The mutations were detected using the Knockout and Mutation Detection kit (Shanghai GeneChem Co., Ltd.), which may efficiently recognize and detect specific double-stranded DNA mutations. DNA was extracted from the samples using a TIANamp Genomic DNA kit (Tiangen Biotech Co., Ltd., Beijing, China). The specific sequences of the three primers were as follows: sgRNA-230 sense, 5′-AAGACTGGGGGTATGATCACC-3′ and sgRNA-230 antisense, 5′-AGGCCAGTATCATTAAGTCCC-3′; sgRNA-231 sense, 5′-ATCCAAGGATATGCTAGGTTGG-3′ and sgRNA-231 antisense, 5′-GAGACTTGTAACAATCCCTCTC-3′; sgRNA-232 sense, 5′-AAGACTGGGGGTATGATCACC-3′ and sgRNA-232 antisense, 5′-AGGCCAGTATCATTAAGTCCC-3′.
The PCR reactions were performed according to the manufacturer's protocol, using the following cycling conditions: 35 cycles of degradation at 95°C for 20 sec, annealing at 55°C for 20 sec, and extension at 72°C for 30 sec. The DNA product was digested using a T7E1 enzyme digestion reaction system at 45°C for 20 min, and a stop buffer was added to terminate the reaction. The samples were stained with GelRed dye (Biotium, Inc., Freemont, CA, USA), and detected following separation using 2% agarose gel electrophoresis at 105 V for 30 min.
Cell proliferation assay
A Cell Counting kit (CCK)-8 assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was used to assess the proliferation of the transfected cells. The cells were seeded at an initial density of 5×104 cells/ml in 96-well plates (5 wells/group) and incubated for 24, 48, 72, 96 and 120 h post-transfection with LV-sgRNA-AR230. For the analysis, 10 µl CCK-8 reagent was added to each well, and the plates were incubated for 2 h at 37°C in a 5% CO2 incubator. The optical density at 450 nm was then measured. All experiments were performed in triplicate, and the mean results were calculated.
Measurement of cellular apoptosis and the cell cycle
Cellular apoptosis was detected using an Annexin V-allophycocyanin (APC)/7-amino-actinomycin D (7-ADD) kit (MultiSciences Biotech Co., Ltd., Hangzhou, China). 7-AAD is a nucleic-acid dye that is superior to propidium iodide (PI) for multicoloured fluorescence analyses. In the present study, flow cytometry was used to analyse cellular apoptosis. The LNCaP-Ca9-sgRNA-AR230 and LNCaP-Cas9-control cell samples were trypsinized and centrifuged at 300 × g for 5 min at room temperature. Following two washes with PBS buffer at 300 × g for 5 min at room temperature, the cells were re-suspended with 500 µl 1X binding buffer at a concentration of 1–3×106 cells/ml. Subsequently, 5 µl Annexin V-APC and 10 µl 7-AAD was added to the cells and incubated at room temperature in the dark for 15 min. Using a CytoFLEX flow cytometer (Beckman Coulter, Inc., Brea, CA, USA), the cells were divided into four regions (Q1-Q4). Region Q1 was representative of mechanical error; region Q2 was representative of late apoptotic or necrotic cells; region Q3 was representative of early apoptotic cells; and region Q4 was representative of living cells. Data analysis was performed using FlowJo software (version 7.6.1; BD Biosciences, Franklin Lakes, NJ, USA).
For the cell cycle assay, the LNCaP-Cas9-sgRNA-AR230 and LNCaP-Cas9-control cell samples were washed twice with 1 ml cold PBS buffer, and the cells were fixed in 70% ethanol for 12–24 h at −20°C. The cells were washed with cold PBS buffer and stained with 500 µl PI at 37°C in the dark for 30 min. The analyses were performed using a CytoFLEX flow cytometer (Beckman Coulter, Inc.). The percentage of cellular apoptosis was calculated according to the percentage of the sub-G1 peak in the cell cycle profile following the PI staining (15).
Statistical analysis
All data are presented as the mean ± standard deviation. The statistical analyses were performed using SPSS 19.0 software (IBM Corp., Armonk, NY, USA). Significant differences between the groups were analysed using one-way analysis of variance followed by a Newman-Keuls post hoc test and t-tests. P<0.05 was considered to indicate a statistically significant difference.
Results
Construction of sgRNAs targeting the AR gene
A total of three AR targets were selected in the present study. All three sgRNAs were expressed using the following lentiviral preparations: LV-sgRNA-AR230, LV-sgRNA-AR231, and LV-sgRNA-AR232. These viral packaged sgRNAs were confirmed by sequencing, as presented in Fig. 1.
Figure 1.
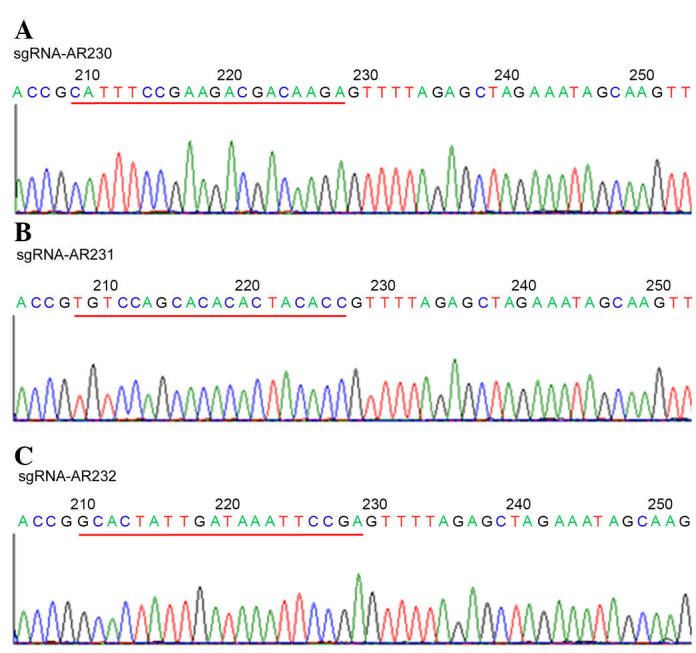
Sequencing diagrams of three sgRNAs. (A) sgRNA-AR230, CATTTCCGAAGACGACAAGA; (B) sgRNA-AR231, TGTCCAGCACACACTACACC; (C) sgRNA-AR232, GCACTATTGATAAATTCCGA. The red underlined sections refer to sequences of the three sgRNAs. sgRNA, single-guide RNA.
Confirmation of gene editing activity by PCR detection of mutations following viral infection
The viral preparations were used to infect LNCaP cells that permanently express Cas9. The cells were harvested 7 days subsequent to the viral infection for DNA extraction and mutation analysis. As presented in Fig. 2A, the expected sizes of the PCR-amplified products were obtained for the three sgRNA-associated CRISPR/Cas9 preparations as follows: 983, 702 and 983 bp for sgRNA-AR230, sgRNA-AR231 and sgRNA-AR232, respectively.
Figure 2.
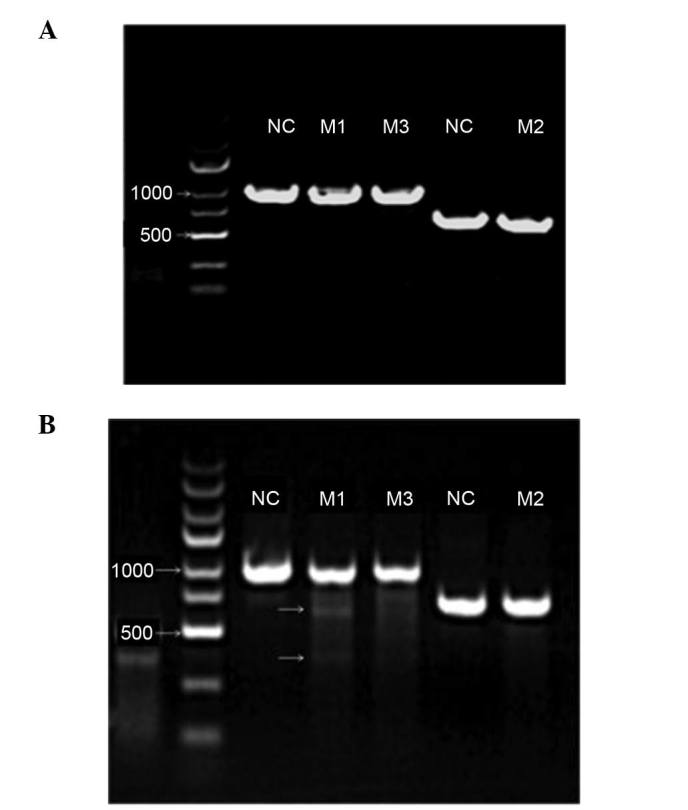
Enzymatic electrophoresis of three target sites determined using T7E1 endonuclease. (A) Images of agarose gel demonstrating that the correct fragments of the three different androgen receptor target sites relevant to LV-230, LV-231 and LV-232 were amplified. Lanes M1, M2, and M3 exhibit the amplified products at the expected sizes (LV230, 983 bp; LV231, 702 bp; and LV-232, 983 bp). The two NC lanes exhibit the corresponding products in the NC groups. The same primers were used in lanes M1 and M3 and, therefore, the NC was shared. (B) Agarose gel image exhibiting the mismatch-associated short fragments in the M1 lane, indicating the gene-editing effect of the single-guide RNA-associated clustered regularly interspaced short palindromic repeats-associated protein system. No such obvious gene editing effect was detected in the M2 and M3 lanes. NC, negative control.
These three viral preparations exhibited different gene editing effects in the LNCaP cells. Using T7E1 endonuclease (Fig. 2B), gene editing was observed in the cells infected with the virus expressing sgRNA-AR230, and not in the cells infected with the other two viruses. Therefore, the sgRNA-AR230 viral preparation was used in the following cell proliferation and apoptosis analyses.
Effects of AR-knockout on tumour cell proliferation
Since androgens may promote prostate cancer cell proliferation via the AR, the present study assessed whether prostate cancer cell proliferation was affected by knocking out AR. At 24 h subsequent to the viral infection, green fluorescent protein was observed in ~80% of the LNCaP cells. The CCK-8 assays demonstrated that the cells in the control group (LNCaP-Cas9-control) proliferated significantly more rapidly compared with those in the experimental group (LNCaP-Cas9-sgRNA-AR230). Notably, the partial knockout of AR using the CRISPR/Cas9 system suppressed cell proliferation, and the most significant alterations were observed at 48–72 h. As presented in Fig. 3, the difference in cell proliferation between the two groups was significant (P<0.05). Fig. 4 illustrates the cell proliferation in the experimental and control groups 24 and 72 h post-transfection. The cell proliferation rate in the experimental group was decreased compared with that in the control group 72 h post-transfection.
Figure 3.
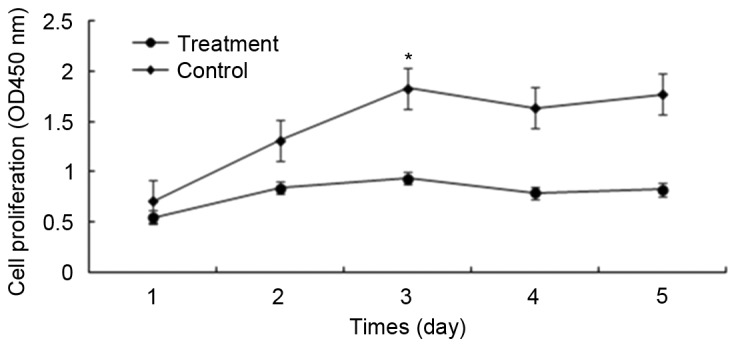
Proliferation of LNCaP prostate cancer cells. The Cell Counting kit-8 assay demonstrated that knocking down the androgen receptor using the clustered regularly interspaced short palindromic repeats-associated protein system inhibited prostate cancer cell proliferation. Treatment, LNCaP-Cas9-single-guide RNA-AR230; Control, LNCaP-Cas9-control. OD, optical density.
Figure 4.
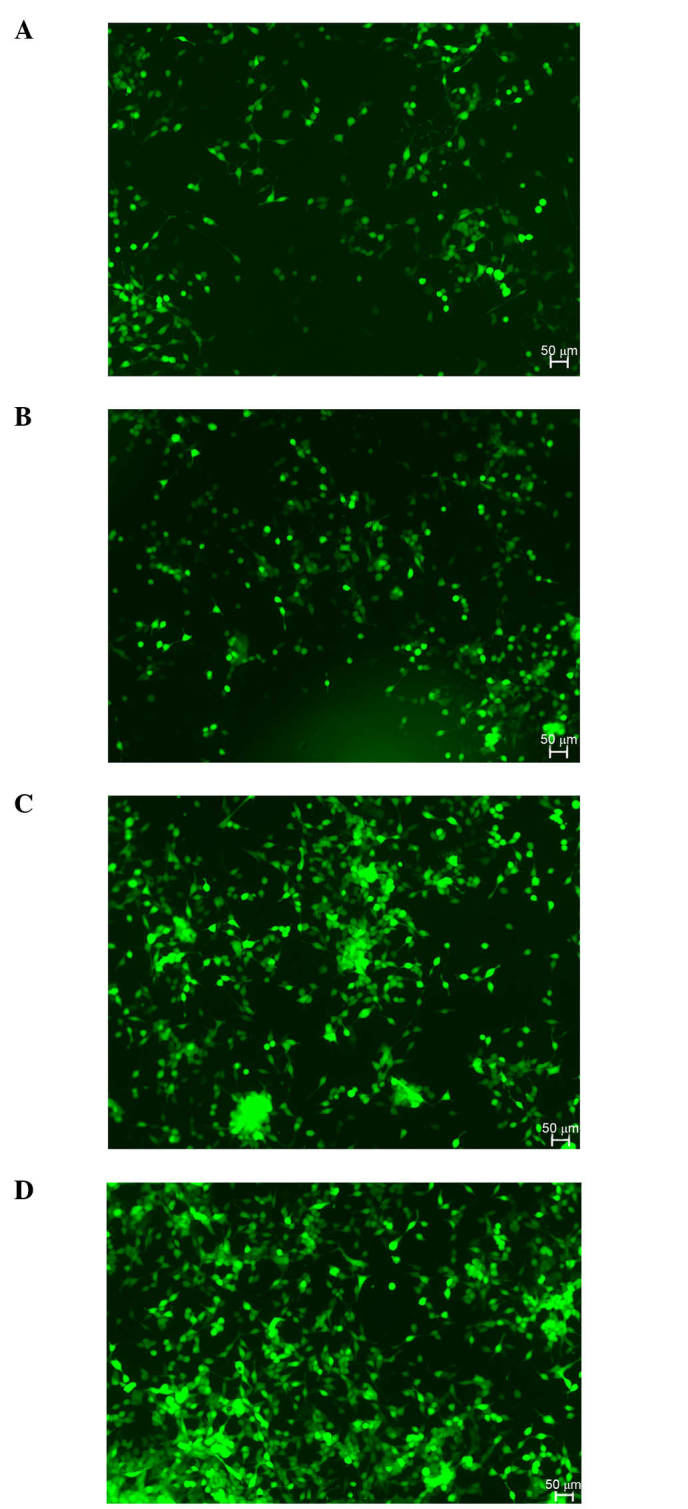
LNCaP cell proliferation 24 and 72 h post-transfection, demonstrated using green fluorescence microscopy. The images illustrate LNCaP cell growth status 24 h subsequent to the (A) LV-Cas9-sgRNA-AR230 and (B) LV-Cas9-control virus transfection; and LNCaP cell growth status 72 h following the (C) LV-Cas9-sgRNA-AR230 and (D) LV-Cas9-control virus transfection. sgRNA, single-guide RNA.
Effects of AR knockdown on tumour cell apoptosis
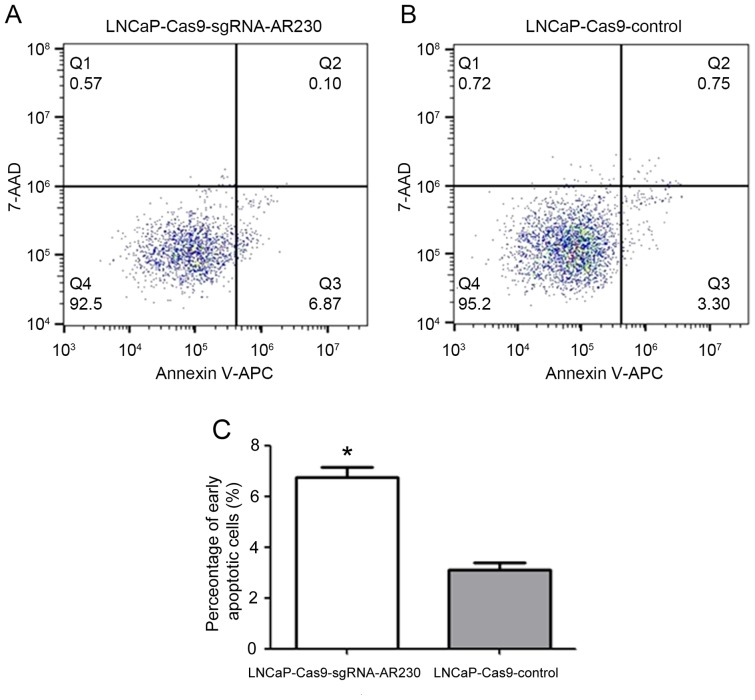
Cellular apoptosis was analysed by flow cytometry as presented in Fig. 5. To investigate whether the decreased cell proliferation was due to cellular apoptosis, flow cytometry was used to evaluate the effect of AR knockdown on cellular apoptosis. It was observed that the proportion of early apoptosis (region Q3) in the LNCaP-Cas9-sgRNA-AR 230-treated cells was increased compared with that induced by the LNCaP-Cas9-control treatment (6.87% vs. 3.30%). The difference between the two groups was statistically significant (P<0.05) according to the results of a two-tailed Student's t-test.
Figure 5.
Analysis of cellular apoptosis using flow cytometry. Virus-infected LNCaP cells were stained using Annexin V-APC and 7-AAD. Apoptotic cells and necrotic cells are presented for (A) experimental and (B) control cells. Cells were divided into four regions (Q1-Q4). Region Q1 is representative of mechanical error; region Q2 is representative of late apoptotic and necrotic cells; region Q3 is representative of early apoptotic cells; and region Q4 is representative of living cells. (C) The percentage of early apoptotic cells in the experimental group (LNCaP-Cas9-sgRNA-AR230) and the control group (LNCaP-Cas9-control; *P<0.05). 7-AAD, 7-amino-actinomycin D; APC, allophycocyanin.
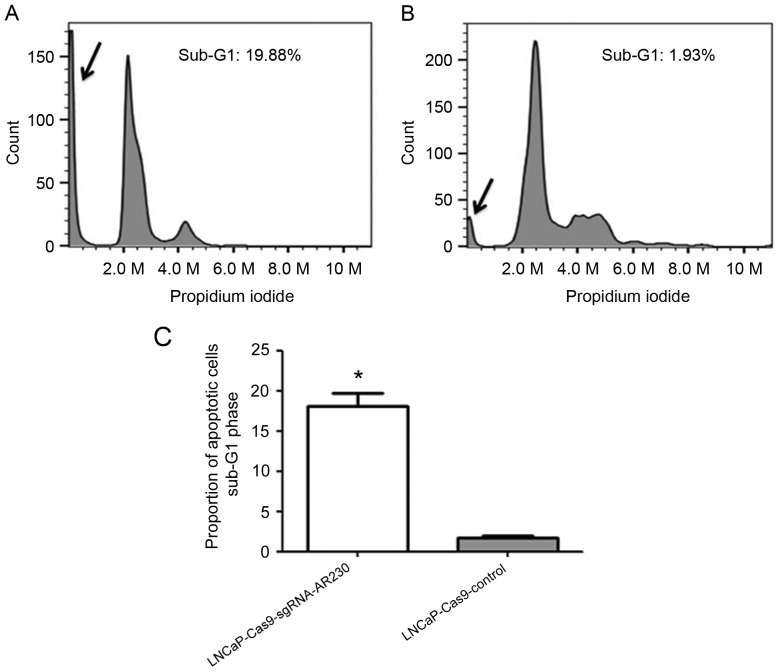
The sub-G1 peak in the cell cycle profile was detected in the two groups, as presented in Fig. 6. The proportion of sub-G1 phase cells was 19.88% in the LNCaP-Cas9-sgRNA-AR230 group and 1.93% in the LNCaP-Cas9-control group, (Fig. 6). A significant increase in the number of cells in the sub-G1 phase was observed in the LNCaP-Cas9-sgRNA-AR230 group compared with that in the LNCaP-Cas9-control group (P<0.05).
Figure 6.
Analysis of the cell cycle using flow cytometry. The DNA profile was analysed using propidium iodide staining and flow cytometry. The percentage of cellular apoptosis is calculated using the percentage of the sub-G1 peak. (A) The LNCaP-Cas9-sgRNA-AR230 cells exhibited high numbers of cells in the sub-G1 peak. (B) The LNCaP-Cas9-control cell samples displayed a low sub-G1 peak. Arrows, sub-G1 peak. (C) The proportions of cells in apoptotic sub-G1 phase from the experimental group (LNCaP-Cas9-sgRNA-AR230) and the control group (LNCaP-Cas9-control; *P<0.05).
Discussion
The A-AR signalling pathway serves a significant role during all stages of prostate cancer. During the early stages, resecting the prostate gland may yield a positive therapeutic outcome. During the middle stages of prostate carcinogenesis, decreasing the levels of androgen via surgery or anti-androgenic drugs may be effective. Following a certain period (~18 months), prostate cancer frequently becomes insensitive to androgens and continues to grow under androgen-depleted conditions. Nevertheless, the AR continues to serve important roles during the androgen refractory stage of prostate cancer via multiple mechanisms, including gene mutations, gene amplification and the abnormal activation of other relevant signalling pathways (16,17). Consequently, editing the expression of ARs may be a reliable treatment for prostate cancer.
The present study partially knocked out the AR using gene-editing technology and observed its effects on the proliferation and apoptosis of LNCaP cancer cells. The CRISPR/Cas9 system was used to successfully edit the AR gene. The partial AR knockout significantly inhibited the growth of LNCaP cells by inhibiting cell proliferation, and the blockade was implemented by promoting cellular apoptosis. According to the cellular apoptosis and cell cycle analysis using flow cytometry, apoptosis was significantly increased in the experimental group compared with that in the control group. This observation is consistent with previous studies (18,19).
The CRISPR/Cas9 system has recently become a hot topic in the field of gene editing. Compared with ZFN and TALEN technologies, the CRISPR/Cas9 system has advantages in operability and repeatability (20,21). The CRISPR/Cas9 system has been widely used in studies investigating various tumours, including cervical cancer, liver cancer, lymphoma and prostate cancer. The present study used the CRISPR/Cas9 system to edit the AR gene, leading to a partial knockout in androgen-dependent LNCaP cells. Stable LNCaP cells containing sgRNA and Cas9 were constructed. These cells represented a solid foundation for further studies. The influence of AR gene editing by CRISPR/Cas9 was observed in the LNCaP cells, although a statistically significant difference was not noted.
However, the present study has certain limitations. The scope of this study was limited to androgen-dependent prostate cancer cells due to their hormone dependence. In future studies, androgen-independent prostate cancer cells may be used to further analyse the CRISPR/Cas9 system. The present study confirmed that CRISPR/Cas9 was able to edit the AR gene, although the quantitative accuracy of the knockout of the AR gene was not investigated. In addition, the CRISPR/Cas9 system has off-target effects. Therefore, future studies may investigate methods to avoid these off-target effects.
In conclusion, the CRISPR/Cas9 system was able to edit the expression of AR and restrain the growth of androgen-dependent prostate cancer cells in vitro, suggesting the potential of the CRISPR/Cas9 system in future cancer therapy.
Acknowledgements
The authors were financially supported by the Suzhou Science and Technology Bureau Development Plan (grant no. SYS201475), the Jiangsu Provincial Special Program of Clinical Medical Science (grant no. BL2014040), the Suzhou Clinical Special Disease Diagnosis and Treatment Program (grant no. LCZX201406), the Suzhou Science and Technology Development Plan (grant no. SS201534), and the Suzhou Science and Technology Development Plan Guidance Project (grant no. SYSD2015091). This project additionally received the Second Affiliated Hospital of Soochow University Preponderant Clinic Discipline Group project funding (grant no. XKQ2015009).
References
- 1.DeSantis CE, Siegel RL, Sauer AG, Miller KD, Fedewa SA, Alcaraz KI, Jemal A. Cancer statistics for African Americans, 2016: Progress and opportunities in reducing racial disparities. CA Cancer J Clin. 2016;66:290–308. doi: 10.3322/caac.21340. [DOI] [PubMed] [Google Scholar]
- 2.Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 3.Saraon P, Jarvi K, Diamandis EP. Molecular alterations during progression of prostate cancer to androgen independence. Clin Chem. 2011;57:1366–1375. doi: 10.1373/clinchem.2011.165977. [DOI] [PubMed] [Google Scholar]
- 4.Yuan X, Balk SP. Mechanisms mediating androgen receptor reactivation after castration. Urol Oncol. 2009;27:36–41. doi: 10.1016/j.urolonc.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen MB, Rokhlin OW. Mechanisms of prostate cancer cell survival after inhibition of AR expression. J Cell Biochem. 2009;106:363–371. doi: 10.1002/jcb.22022. [DOI] [PubMed] [Google Scholar]
- 6.Horii T, Hatada I. Genome engineering using the CRISPR/Cas system. World J Med Genet. 2014;4:69–76. doi: 10.5496/wjmg.v4.i3.69. [DOI] [Google Scholar]
- 7.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pu J, Frescas D, Zhang B, Feng J. Utilization of TALEN and CRISPR/Cas9 technologies for gene targeting and modification. Exp Biol Med (Maywood) 2015;240:1065–1070. doi: 10.1177/1535370215584932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seeger C, Sohn JA. Targeting hepatitis B virus with CRISPR/Cas9. Mol Ther Nucleic Acids. 2014;3:e216. doi: 10.1038/mtna.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu XJ, Qi X, Zheng DH, Ji LJ. Modeling cancer processes with CRISPR-Cas9. Trends Biotechnol. 2015;33:317–319. doi: 10.1016/j.tibtech.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 11.Tang H, Shrager JB. CRISPR/Cas-mediated genome editing to treat EGFR-mutant lung cancer: A personalized molecular surgical therapy. EMBO Mol Med. 2016;8:83–85. doi: 10.15252/emmm.201506006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, Watanabe T, Kanai T, Sato T. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med. 2015;21:256–262. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- 13.Wang G, Zhao N, Berkhout B, Das AT. CRISPR-Cas9 can inhibit HIV-1 replication but NHEJ repair facilitates virus escape. Mol Ther. 2016;24:522–526. doi: 10.1038/mt.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawamura N, Nimura K, Nagano H, Yamaguchi S, Nonomura N, Kaneda Y. CRISPR/Cas9-mediated gene knockout of NANOG and NANOGP8 decreases the malignant potential of prostate cancer cells. Oncotarget. 2015;6:22361–22374. doi: 10.18632/oncotarget.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang HT, Jun WU, Zhang HF, Zhu QF. Efflux of potassium ion is an important reason of HL-60 cells apoptosis induced by tachyplesin. Acta Pharmacol Sin. 2006;27:1367–1374. doi: 10.1111/j.1745-7254.2006.00377.x. [DOI] [PubMed] [Google Scholar]
- 16.Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocr Rev. 2004;25:276–308. doi: 10.1210/er.2002-0032. [DOI] [PubMed] [Google Scholar]
- 17.Shore N. Management of early-stage prostate cancer. Am J Manag Care. 2014;20(12 Suppl):S260–S272. [PubMed] [Google Scholar]
- 18.Yu L, Wang X, Zhu D, Ding W, Wang L, Zhang C, Jiang X, Shen H, Liao S, Ma D, et al. Disruption of human papillomavirus 16 E6 gene by clustered regularly interspaced short palindromic repeat/Cas system in human cervical cancer cells. Onco Targets Ther. 2014;8:37–44. doi: 10.2147/OTT.S64092. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Hu Z, Yu L, Zhu D, Ding W, Wang X, Zhang C, Wang L, Jiang X, Shen H, He D, et al. Disruption of HPV16-E7 by CRISPR/Cas system induces apoptosis and growth inhibition in HPV16 positive human cervical cancer cells. Biomed Res Int 2014. 2014:612823. doi: 10.1155/2014/612823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding Q, Regan SN, Xia Y, Oostrom LA, Cowan CA, Musunuru K. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell. 2013;12:393–394. doi: 10.1016/j.stem.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pattanayak V, Guilinger JP, Liu DR. Determining the specificities of TALENs, Cas9, and other genome editing enzymes. Methods Enzymol. 2014;546:47–78. doi: 10.1016/B978-0-12-801185-0.00003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]