Abstract
Gene expression profiles of cutaneous melanoma were analyzed to identify critical genes associated with metastasis. Two gene expression datasets were downloaded from Gene Expression Omnibus (GEO) and another dataset was obtained from The Cancer Genome Atlas (TCGA). Differentially expression genes (DEGs) between metastatic and non-metastatic melanoma were identified by meta-analysis. A protein-protein interaction (PPI) network was constructed for the DEGs using information from BioGRID, HPRD and DIP. Betweenness centrality (BC) was calculated for each node in the network and the top feature genes ranked by BC were selected to construct the support vector machine (SVM) classifier using the training set. The SVM classifier was then validated in another independent dataset. Pathway enrichment analysis was performed for the feature genes using Fisher's exact test. A total of 798 DEGs were identified and a PPI network including 337 nodes and 466 edges was then constructed. Top 110 feature genes ranked by BC were included in the SVM classifier. The prediction accuracies for the three datasets were 96.8, 100 and 94.4%, respectively. A total of 11 KEGG pathways and 13 GO biological pathways were significantly over-represented in the 110 feature genes, including endometrial cancer, regulation of actin cytoskeleton, focal adhesion, ubiquitin mediated proteolysis, regulation of apoptosis and regulation of cell proliferation. A SVM classifier of high prediction accuracy was acquired. Several critical genes implicated in melanoms metastasis were also revealed. These results may advance understanding of the molecular mechanisms underlying metastasis, and also provide potential therapeutic targets.
Keywords: cutaneous melanoma, metastasis, differentially expressed genes, protein-protein interaction interaction, SVM classifier
Introduction
Cutaneous melanoma is the most dangerous type of skin cancer. In the United States, the five-year survival rates is 98% among those with localized disease and 17% among those in whom spread has occurred. Apparently, metastatis is the most important factor affecting prognosis of melanoma patients. Study on melanoma metastasis mechanism would beneficial to the development of novel therapeutic strategies to extend the overall survival of patients.
Certain signaling pathways have been involved in metastasis of cutaneous melanoma, such as integrin alpha V beta 3 signaling (1) and signal transducer and activator of transcription 3 (STAT3) signaling (2). Several indicators and biomarkers associated with metastasis have also been identified in melanoma. LINE1 hypermethylation in peripheral blood of cutaneous melanoma patients is associated with metastasis (3). Increased level of circulating U2 small nuclear RNA fragments indicates metastasis in melanoma patients (4). Chen et al found that Tip60 may regulate melanoma metastasis and could be a potential therapeutic target (5). Thang et al reported that deltex-3-like (DTX3L) stimulates metastasis of melanoma (6). The study by De Semir et al (7) indicated that pleckstrin homology domain-interacting protein (PHIP) is a marker and mediator of melanoma metastasis. Serpin family E member 1 (SERPINE1) expression discriminates site-specific metastasis in human melanoma (8). Galectin-3 expression favors metastasis in murine melanoma (9).
Gene expression profiling is a powerful tool to unveil genes implicated in metastasis of melanoma (10,11). In present study, those gene expression data were combined and more differentially expressed genes (DEGs) were identified via meta-analysis. Feature genes were revealed via support vector machine (SVM) classification. Meanwhile, a SVM classifier was acquired and validated. These findings could advance the understanding about melanoma metastasis and also provide potential therapeutic targets.
Materials and methods
Gene expression data and pre-treatment
Two gene expression datasets (GSE46517 and GSE7553) were retrieved from Gene Expression Omnibus (GEO) with key words ‘cutaneous melanoma’, ‘homo sapiens’ and ‘metastasis’ by the end of May 13th, 2016. Dataset GSE46517 contained 73 metastatic melanoma samples and 31 primary melanoma samples. Dataset GSE7553 contained 40 metastatic melanoma samples and 14 primary melanoma samples. Moreover, gene expression profiles of 481 skin cutaneous melanoma specimens were downloaded from The Cancer Genome Atlas (TCGA, https://tcga-data.nci.nih.gov/tcga) with keyword ‘cutaneous melanoma’, out of which 183 were metastatic melanoma samples, 71 primary melanoma samples and others not specified.
Gene expression dataset GSE7553 was acquired with Affymetrix platform. Background correction and normalization were performed with package oligo (12) of R. Missing values were filled with median method. Backgroud correction was carried out with MSA method. Normalization was achieved with quantiles method. As for dataset GSE46517 from Agilent platform, probes were mapped into genes. For probes corresponding to a same gene, the averaged value was considered as the expression value of the gene. Log2 transformation and normalization with the median were then applied. Level 3 gene expression data from TCGA were normalized with package limma (13) of R.
Screening of differentially expressed genes (DEGs)
Meta-analysis of the three gene expression dataset was performed with package MetaDE (14) of R to identify differentially expressed genes (DEGs). This method firstly tested heterogeneity of gene expression value in various platforms with three statistic parameters: tau2, Q value and Qpval. Then it tested differential expression of genes between different groups with statistic parameters P-value and false positive rate (FDR). To ensure the homogeneity of feature genes, tau2=0, Qpval >0.05 and FDR <0.05 were set as the cut-offs.
Construction of protein-protein interaction (PPI) network
The PPI information was downloaded from Database of Protein, Chemical, and Genetic Interactions (BioGRID, http://thebiogrid.org/) (15), Human Protein Reference Database (HPRD, http://www.hprd.org/) (16) and Database of Interacting Proteins (DIP, http://dip.doe-mbi.ucla.edu/dip/Main.cgi) (17). The protein products of the DEGs were mapped into the whole network and the PPI network for the DEGs was then acquired. The network was then visualized with Cytoscape (18).
Calculation of betweenness centrality (BC)
Feature genes were screened out from the DEGs using betweenness centrality (BC), which reflected hubness of the node in the PPI network. The BC was calculated as follow:
Where σst is the shortest path from s to t; σst(v) is number of the shortest path from s to t passing through node v; BC score is between 0 and 1, and greater BC score indicates higher degree of hubness.
Training and validation of SVM classifier
Gene expresson data from TCGA were chosen as the training set. Genes were ranked based upon BC value and top 10 genes were selected out to train SVM classifier. An increment of 10 genes were added into the classifier until metastatic melanomas could be totally separated from primary melanomas. These DEGs were regarded as feature genes. Gene expression datasets GSE46517 and GSE7553 were used as the validation set. Sensitivity (Se), specificity (Sp), positive predictive value (PPV), negative predictive value (NPV) and area under curve (AUC) were calculated to evaluate the SVM classifier.
Pathway enrichment analysis
Gene ontology (GO) biological pathways and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways related to feature genes were identified using runHyperGO and runHyperKEGG from package EMA of R. The significance was calculated using Fisher's exact test as follow. P-value <0.05 was set as the threshold.
Where N is the total number of gene; M is number of gene in the pathway; K is number of feature gene.
Results
Differentially expressed genes
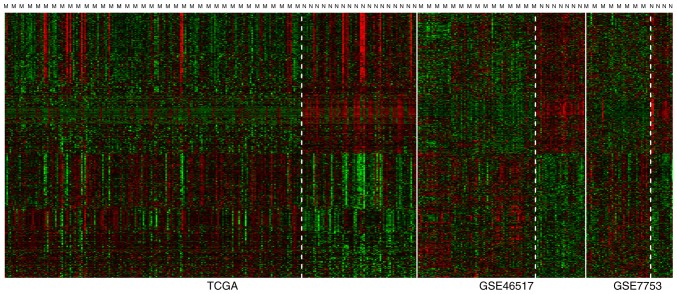
According to the criteria, a total of 798 DEGs were revealed from the three gene expression datasets. Heat map of the expression levels of the 798 DEGs is shown in Fig. 1.
Figure 1.
Heat map of the expression levels of the 798 DEGs. ‘M’ means metastatic cutaneous melanoma; ‘N’ means non-metastatic cutaneous melanoma. X-axis is samples and Y-axis is genes expression level. Green color means higher gene expression level; red color means lower gene expression level. DEGs, differentially expressed genes.
PPI network
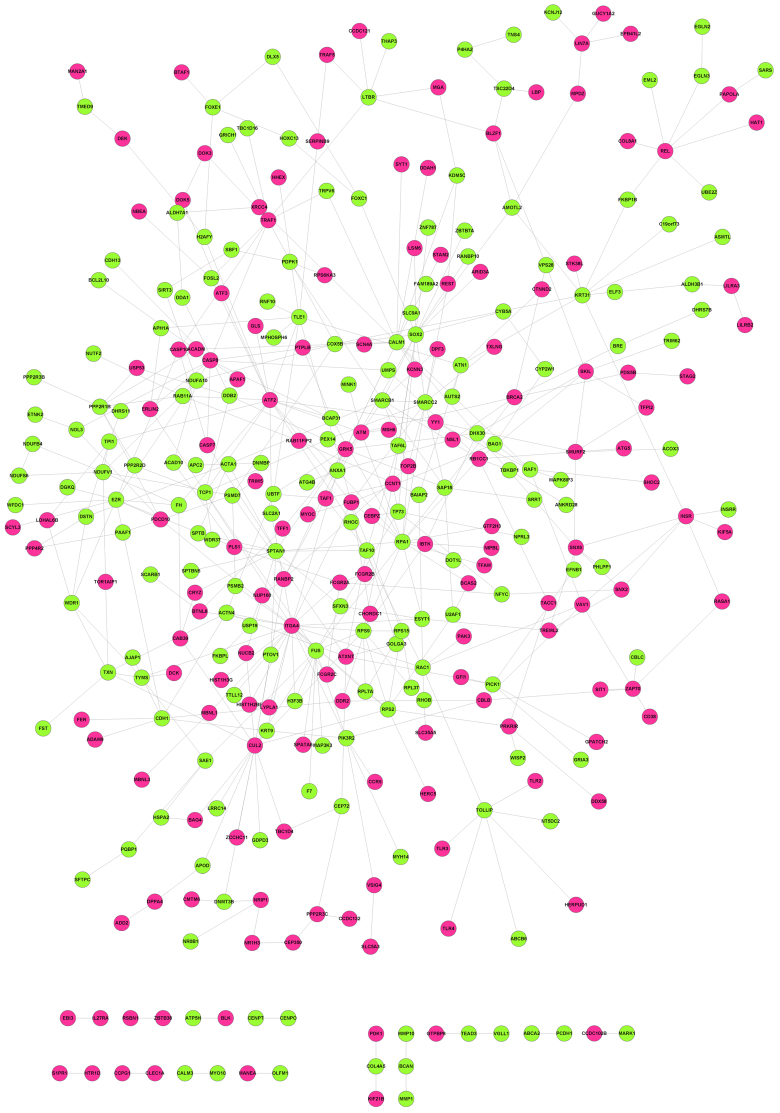
Based upon information from BioGRID, HPRD and DIP, a PPI network containing 337 nodes and 466 edges was obtained (Fig. 2). Distribution of degree is shown in Fig. 3. Most genes (232 genes) showed small degree (Log transformed degree <1) while only 1 gene had Log transformed degree >4. Therefore, like most biological networks, this PPI network exhibited scale-free property. These genes with high degree were hub genes and might play important roles in the development of diseases.
Figure 2.
The protein-protein interaction network of the differentially expressed genes (DEGs). Up-regulated genes in metastatic melanoma are in red, down-regulated genes in metastatic melanoma are in green.
Figure 3.
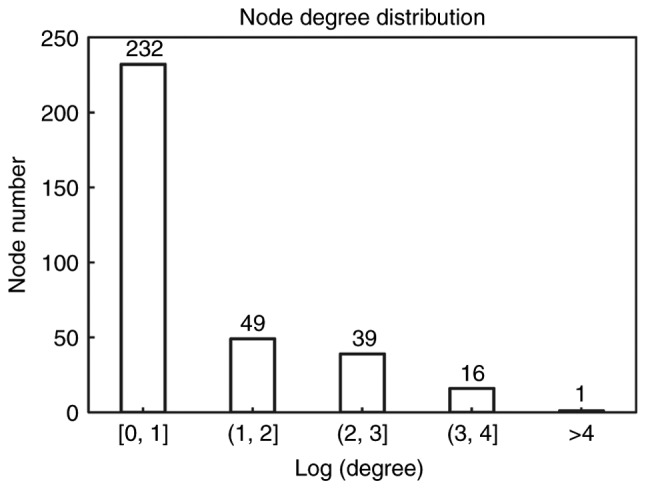
Distribution of degree. X-axis is Log transformed degree and Y-axis is number of node.
SVM classifier
Top 10 DEGs ranked by BC are listed in Table I: Integrin subunit alpha 4 (ITGA4), activating transcription factor 2 (ATF2), SRY-box 2 (SOX2), keratin 31 (KRT31), cullin 2 (CUL2), replication protein A1 (RPA1), calmodulin 1 (CALM1), FUS RNA binding protein (FUS), angiomotin like 2 (AMOTL2) and ras-related C3 botulinum toxin substrate 1 (RAC1).
Table I.
Top 10 genes ranked by betweenness centrality.
| Gene | BC | Exp | Degree | P-value | FDR | Q | Qp | tau2 |
|---|---|---|---|---|---|---|---|---|
| ITGA4 | 2.06×10−1 | 1 | 25 | 1.00×10−20 | 9.22×10−19 | 1.39E+00 | 5.00×10−1 | 0 |
| ATF2 | 1.47×10−1 | 1 | 16 | 2.46×10−6 | 1.78×10−4 | 1.54E+00 | 4.64×10−1 | 0 |
| SOX2 | 1.43×10−1 | 0 | 15 | 3.73×10−4 | 6.85×10−3 | 5.21×10−2 | 9.74×10−1 | 0 |
| KRT31 | 1.36×10−1 | 0 | 10 | 1.00×10−20 | 9.22×10−19 | 4.96×10−1 | 7.80×10−1 | 0 |
| CUL2 | 1.22×10−1 | 1 | 16 | 2.69×10−4 | 5.49×10−3 | 1.19×10−1 | 9.42×10−1 | 0 |
| RPA1 | 1.10×10−1 | 0 | 16 | 4.07×10−3 | 3.69×10−2 | 6.28×10−1 | 7.31×10−1 | 0 |
| CALM1 | 9.78×10−2 | 0 | 13 | 7.63×10−4 | 1.12×10−2 | 8.76×10−1 | 6.45×10−1 | 0 |
| FUS | 9.26×10−2 | 0 | 12 | 6.57×10−6 | 3.27×10−4 | 1.95E+00 | 3.77×10−1 | 0 |
| AMOTL2 | 9.25×10−2 | 0 | 4 | 9.86×10−6 | 4.24×10−4 | 8.40×10−1 | 6.57×10−1 | 0 |
| RAC1 | 8.70×10−2 | 0 | 7 | 2.38×10−3 | 2.54×10−2 | 1.13E+00 | 5.69×10−1 | 0 |
BC, betweenness centrality; Exp, expression; FDR, false positive rate. ‘1’ in column Exp indicates up-regulation in metastatic melanoma compared with non-metastatic melanoma while ‘0’ indicates down-regulation in metastatic melanoma.
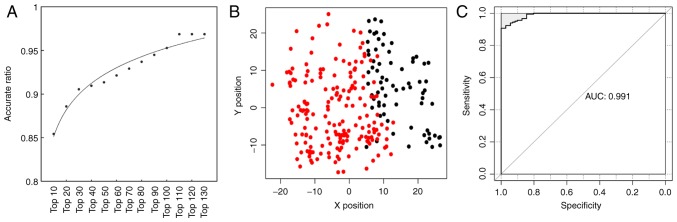
As shown in Fig. 4A, the accuracy increased from 85.4 to 96.8% while more genes were included in the classifier. Details are shown in Table II. The accuracy rate reached 96.8% while top 110 DEGs were included in the classifier, and it didn't increase though more genes were added in. Therefore, the top 110 genes were used to construct the SVM classifier. The scatter plot of prediction result for the training dataset is shown in Fig. 4B and the receiver operating characteristic (ROC) curve is shown in Fig. 4C.
Figure 4.
Accuracy rate with various numbers of differentially expressed genes (A) for the training dataset. Scatter plot showing prediction result of the training dataset (B). Metastatic cutaneous melanoma samples are in red and non-metastatic cutaneous melanoma samples are in black. The receiver operating characteristic (ROC) curve of for the training dataset (C).
Table II.
Prediction results of the SVM classifier in the training dataset with varying numbers of differentially expressed genes.
| SVM classifier | Correct prediction | Wrong prediction | Accuracy rate (%) | Error rate (%) |
|---|---|---|---|---|
| Top10 | 217 | 37 | 85.4 | 14.6 |
| Top20 | 225 | 29 | 88.6 | 11.4 |
| Top30 | 230 | 24 | 90.6 | 9.4 |
| Top40 | 231 | 23 | 90.9 | 9.1 |
| Top50 | 232 | 22 | 91.3 | 8.7 |
| Top60 | 234 | 20 | 92.1 | 7.9 |
| Top70 | 236 | 18 | 92.9 | 7.1 |
| Top80 | 238 | 16 | 93.7 | 6.3 |
| Top90 | 240 | 14 | 94.5 | 5.5 |
| Top100 | 242 | 12 | 95.3 | 4.7 |
| Top110 | 246 | 8 | 96.9 | 3.1 |
| Top120 | 246 | 8 | 96.9 | 3.1 |
| Top130 | 246 | 8 | 96.9 | 3.1 |
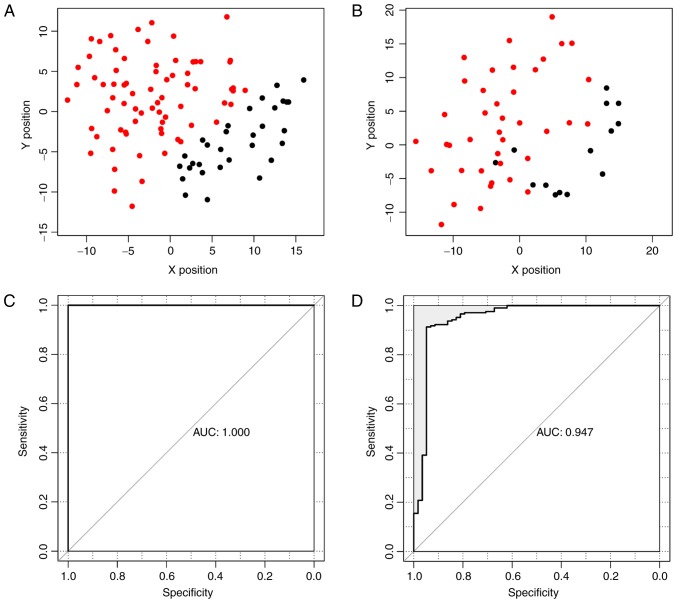
The SVM classifier was validated using dataset GSE46517 and GSE7553. The accuracy rates in dataset GSE46517 (Fig. 5A) and GSE7553 (Fig. 5B) were 100 and 94.4%, respectively. Three non-metastatic melanoma samples from dataset GSE7753 were predicted wrong. The accuracy, sensitivity, specificity, PPV, NPV and AUC are listed in Table III. The receiver operating characteristic (ROC) curves are shown in Fig. 5C and D.
Figure 5.
Scatter plots and ROC curves for dataset GSE46517 (A and C) and GSE7553 (B and D). Metastatic cutaneous melanoma samples are in red and non-metastatic cutaneous melanoma samples are in black. ROC, receiver operating characteristic.
Table III.
Prediction results of the SVM classifier in the 3 independent datasets.
| Dataset | No. of sample | Accuracy | Sensitivity | Specificity | PPV | NPV | AUROC |
|---|---|---|---|---|---|---|---|
| TCGA | 254 | 0.968 | 1.000 | 0.987 | 0.958 | 1.000 | 0.991 |
| GSE46517 | 128 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| GSE7753 | 54 | 0.944 | 1.000 | 0.886 | 0.942 | 1.000 | 0.947 |
PPV, positive predictive value; NPV, negative predictive value; AUROC, area under receiver operating characteristic curve.
Biological pathways
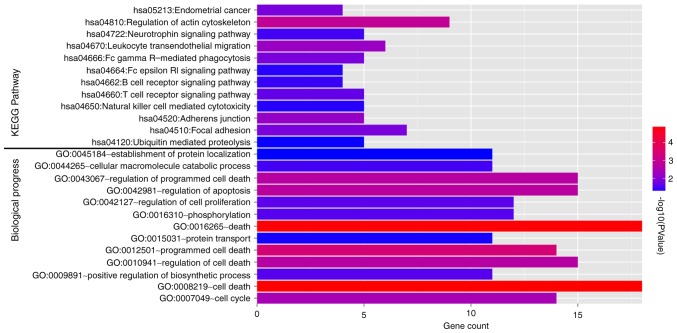
A total of 11 KEGG pathways (Fig. 6, top) and 13 GO biological pathways (Fig. 6, bottom) were identified in the 110 feature genes. Endometrial cancer, regulation of actin cytoskeleton, focal adhesion, ubiquitin mediated proteolysis, regulation of apoptosis, and regulation of cell proliferation were significantly over-represented in the feature genes.
Figure 6.
KEGG pathways and GO biological pathways significantly over-represented in the 110 feature genes. X-axis indicates number of gene and color of the bar indicates-lg (P-value). KEGG, Kyoto Encyclopedia of Genes and Genomes; GO, gene ontology.
Discussion
Three gene expression datasets were downloaded from GEO or TCGA. A total of 798 DEGs were identified in metastatic melanoma compared with non-metastatic melanoma via meta-analysis. A PPI network containing 337 nodes and 466 edges was constructed. BC was calculated for each gene in the network. Top 110 feature genes ranked by BC were included in the SVM classifier. The prediction accuracies for the three datasets were 96.8, 100, and 94.4%, respectively. A total of 11 KEGG pathways and 13 GO biological pathways were significantly over-represented in the 110 feature genes, such as endometrial cancer, regulation of actin cytoskeleton, focal adhesion, ubiquitin mediated proteolysis, regulation of apoptosis, and regulation of cell proliferation. Among the 110 feature genes, several genes have been involved in metastasis of melanoma.
ATF2 is a transcription factor and a member of the leucine zipper family of DNA binding proteins. It confers radiation resistance to human melanoma cells (19). Lau et al further pointed out that transcriptional repression of IFNβ1 by ATF2 confers melanoma resistance to therapy (20). It also promotes melanoma metastasis by suppressing protein fucosylation (21). However, ATF2-derived peptides can inhibit melanoma growth and metastasis (22). Therefore, ATF2 is a critical gene in melanoma metastasis and could be a therapeutic target.
SOX2 is a member of the SRY-related HMG-box (SOX) family of transcription factors involved in the regulation of embryonic development and in the determination of cell fate. It regulates self-renewal and tumorigenicity of human melanoma-initiating cells (23). It also contributes to melanoma cell invasion (24). Downregulation of SOX2 in cancer stem cells suppresses growth and metastasis of lung cancer (25). We speculated that it could be a potential therapeutic target for metastatic melanoma.
RAC1 is a GTPase which belongs to the RAS superfamily of small GTP-binding proteins, which regulate a diverse array of cellular events, including the control of cell growth, cytoskeletal reorganization, and the activation of protein kinases. RAC1 is a critical genes associated with melanoma metastasis. A previous study has reported that a RAC1 mutant (Pro29 to serine, P29S) promotes melanocyte proliferation and migration (26). A recent study indicates that RAC1 P29S regulates PD-L1 expression in melanoma (27). Besides, matrix metallopeptidase 14 modulates melanoma cell dissemination and metastasis through activation of MMP2 and RAC1 (28). Immunity-related GTPase family M member 1 (IRGM1) enhances B16 melanoma cell metastasis through PI3K-RAC1 mediated epithelial mesenchymal transition (29).
Certain genes take parts in metastasis of various cancers and their roles in melanoma metastasis remain to be uncovered. AMOTL2 is related to angiomotin and is a member of the motin protein family. It disrupts apical-basal cell polarity and promotes tumour invasion (30). Raf-1 proto-oncogene (RAF1) activation in gastrointestinal carcinoid cells decreases tumor cell adhesion and thus promotes metastasis (31). Vav guanine nucleotide exchange factor 1 (VAV1) is a member of the VAV gene family. Inhibition of VAV1 can attenuate metastasis of pancreatic cancer (32). Besides, RPA1 is regarded as a useful prognostic indicator in colon cancer (33) and esophageal carcinoma (34). We thought that it might exert prognostic effect in melanoma. Further studies on these genes could expand the knowledge about melanoma metastasis.
In the present study, the biological pathways that were significantly enriched in the metastasis related feature genes in melanoma were screened. To find out the pathways involved in the feature genes in metastatic melanoma, Valsesia et al also conducted a network-guided investigation (35). They connected the genes of somatic copy number alterations in metastatic melanoma with significant biological pathways. The results showed the frequently altered pathways in melanoma, including angiogenesis and melanogenesis, while we found that regulation of apoptosis and regulation of cell proliferation participate in melanoma.
Overall, a multigene SVM predictor of high accuracy was obtained for metastatic melanoma. Several critical genes linked with melanoma metastasis were also revealed. Further researches on the feature genes could improve the understanding about the molecular mechanisms underlying metastasis and provide potential therapeutic targets.
References
- 1.Bai J, Zhang J, Wu J, Shen L, Zeng J, Ding J, Wu Y, Gong Z, Li A, Xu S, et al. JWA regulates melanoma metastasis by integrin alphaVbeta3 signaling. Oncogene. 2010;29:1227–1237. doi: 10.1038/onc.2009.408. [DOI] [PubMed] [Google Scholar]
- 2.Cao HH, Chu JH, Kwan HY, Su T, Yu H, Cheng CY, Fu XQ, Guo H, Li T, Tse AK, et al. Inhibition of the STAT3 signaling pathway contributes to apigenin-mediated anti-metastatic effect in melanoma. Sci Rep. 2016;6:21731. doi: 10.1038/srep21731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Araújo ÉS, Kashiwabara AY, Achatz MI, Moredo LF, De Sá BC, Duprat JP, Rosenberg C, Carraro DM, Krepischi AC. LINE-1 hypermethylation in peripheral blood of cutaneous melanoma patients is associated with metastasis. Melanoma Res. 2015;25:173–177. doi: 10.1097/CMR.0000000000000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuhlmann JD, Wimberger P, Wilsch K, Fluck M, Suter L, Brunner G. Increased level of circulating U2 small nuclear RNA fragments indicates metastasis in melanoma patients. Clin Chem Lab Med. 2015;53:605–611. doi: 10.1515/cclm-2014-1064. [DOI] [PubMed] [Google Scholar]
- 5.Chen G, Cheng Y, Tang Y, Martinka M, Li G. Role of Tip60 in human melanoma cell migration, metastasis, and patient survival. J Invest Dermatol. 2012;132:2632–2641. doi: 10.1038/jid.2012.193. [DOI] [PubMed] [Google Scholar]
- 6.Thang ND, Yajima I, Kumasaka MY, Iida M, Suzuki T, Kato M. Deltex-3-like (DTX3L) stimulates metastasis of melanoma through FAK/PI3K/AKT but not MEK/ERK pathway. Oncotarget. 2015;6:14290–14299. doi: 10.18632/oncotarget.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Semir D, Nosrati M, Bezrookove V, Dar AA, Federman S, Bienvenu G, Venna S, Rangel J, Climent J, Meyer Tamgüney TM, et al. Pleckstrin homology domain-interacting protein (PHIP) as a marker and mediator of melanoma metastasis; Proc Natl Acad Sci USA; 2012; pp. 7067–7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klein RM, Bernstein D, Higgins SP, Higgins CE, Higgins PJ. SERPINE1 expression discriminates site-specific metastasis in human melanoma. Exp Dermatol. 2012;21:551–554. doi: 10.1111/j.1600-0625.2012.01523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Comodo AN, Lacerda Bachi AL, Soares MF, Franco M, de Paulo Castro Teixeira V. Galectin-3 expression favors metastasis in murine melanoma. Adv Biosci Biotechnol. 2013;04:55–62. doi: 10.4236/abb.2013.410A3007. [DOI] [Google Scholar]
- 10.Kabbarah O, Nogueira C, Feng B, Nazarian RM, Bosenberg M, Wu M, Scott KL, Kwong LN, Xiao Y, Cordon-Cardo C, et al. Integrative genome comparison of primary and metastatic melanomas. PLoS One. 2010;5:e10770. doi: 10.1371/journal.pone.0010770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riker AI, Enkemann SA, Fodstad O, Liu S, Ren S, Morris C, Xi Y, Howell P, Metge B, Samant RS, et al. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med Genomics. 2008;1:13. doi: 10.1186/1755-8794-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carvalho B, Bengtsson H, Speed TP, Irizarry RA. Exploration, normalization, and genotype calls of high-density oligonucleotide SNP array data. Biostatistics. 2007;8:485–499. doi: 10.1093/biostatistics/kxl042. [DOI] [PubMed] [Google Scholar]
- 13.Smyth GK. Limma: Linear models for microarray data. Stat Biol Health. 2005:1–420. [Google Scholar]
- 14.Li J, Tseng GC. An adaptively weighted statistic for detecting differential gene expression when combining multiple transcriptomic studies. Ann Appl Stat. 2011;5:994–1019. doi: 10.1214/10-AOAS393. [DOI] [Google Scholar]
- 15.Chatr-Aryamontri A, Breitkreutz BJ, Oughtred R, Boucher L, Heinicke S, Chen D, Stark C, Breitkreutz A, Kolas N, O'Donnell L, et al. The BioGRID interaction database: 2015 update. Nucleic Acids Res. 2015;43(Database issue):D470–DS478. doi: 10.1093/nar/gku1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. Human protein reference database-2009 update. Nucleic Acids Res. 2009;37(Database issue):D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xenarios I, Rice DW, Salwinski L, Baron MK, Marcotte EM, Eisenberg D. DIP: The database of interacting proteins. Nucleic Acids Res. 2000;28:289–291. doi: 10.1093/nar/28.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ronai Z, Yang YM, Fuchs SY, Adler V, Sardana M, Herlyn M. ATF2 confers radiation resistance to human melanoma cells. Oncogene. 1998;16:523–531. doi: 10.1038/sj.onc.1201566. [DOI] [PubMed] [Google Scholar]
- 20.Lau E, Sedy J, Sander C, Shaw MA, Feng Y, Scortegagna M, Claps G, Robinson S, Cheng P, Srivas R, et al. Transcriptional repression of IFNβ1 by ATF2 confers melanoma resistance to therapy. Oncogene. 2015;34:5739–5748. doi: 10.1038/onc.2015.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lau E, Feng Y, Claps G, Fukuda MN, Perlina A, Donn D, Jilaveanu L, Kluger H, Freeze HH, Ronai ZA. The transcription factor ATF2 promotes melanoma metastasis by suppressing protein fucosylation. Sci Signal. 2015;8:ra124. doi: 10.1126/scisignal.aac6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhoumik A, Gangi L, Ronai Z. Inhibition of melanoma growth and metastasis by ATF2-derived peptides. Cancer Res. 2004;64:8222–8230. doi: 10.1158/0008-5472.CAN-04-0714. [DOI] [PubMed] [Google Scholar]
- 23.Santini R, Pietrobono S, Pandolfi S, Montagnani V, D'Amico M, Penachioni JY, Vinci MC, Borgognoni L, Stecca B. SOX2 regulates self-renewal and tumorigenicity of human melanoma-initiating cells. Oncogene. 2014;33:4697–4708. doi: 10.1038/onc.2014.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Girouard SD, Laga AC, Mihm MC, Scolyer RA, Thompson JF, Zhan Q, Widlund HR, Lee CW, Murphy GF. SOX2 contributes to melanoma cell invasion. Lab Invest. 2012;92:362–370. doi: 10.1038/labinvest.2011.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiang R, Liao D, Cheng T, Zhou H, Shi Q, Chuang TS, Markowitz D, Reisfeld RA, Luo Y. Downregulation of transcription factor SOX2 in cancer stem cells suppresses growth and metastasis of lung cancer. Br J Cancer. 2011;104:1410–1417. doi: 10.1038/bjc.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, Cheng E, Davis MJ, Goh G, Choi M, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vu HL, Rosenbaum S, Purwin TJ, Davies MA, Aplin AE. RAC1 P29S regulates PD-L1 expression in melanoma. Pigment Cell Melanoma Res. 2015;28:590–598. doi: 10.1111/pcmr.12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaverdashvili K, Wong P, Ma J, Zhang K, Osman I, Bedogni B. MT1-MMP modulates melanoma cell dissemination and metastasis through activation of MMP2 and RAC1. Pigment Cell Melanoma Res. 2014;27:287–296. doi: 10.1111/pcmr.12201. [DOI] [PubMed] [Google Scholar]
- 29.Tian L, Li L, Xing W, Li R, Pei C, Dong X, Fu Y, Gu C, Guo X, Jia Y, et al. IRGM1 enhances B16 melanoma cell metastasis through PI3K-Rac1 mediated epithelial mesenchymal transition. Sci Rep. 2015;5:12357. doi: 10.1038/srep12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mojallal M, Zheng Y, Hultin S, Audebert S, van Harn T, Johnsson P, Lenander C, Fritz N, Mieth C, Corcoran M, et al. AmotL2 disrupts apical-basal cell polarity and promotes tumour invasion. Nat Commun. 2014;5:4557. doi: 10.1038/ncomms5557. [DOI] [PubMed] [Google Scholar]
- 31.Greenblatt DY, Kunnimalaiyaan M, Chen H. Raf-1 activation in gastrointestinal carcinoid cells decreases tumor cell adhesion. Am J Surg. 2007;193:331–335. doi: 10.1016/j.amjsurg.2006.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Razidlo GL, Magnine C, Sletten AC, Hurley RM, Almada LL, Fernandez-Zapico ME, Ji B, McNiven MA. Targeting pancreatic cancer metastasis by inhibition of Vav1, a driver of tumor cell invasion. Cancer Res. 2015;75:2907–2915. doi: 10.1158/0008-5472.CAN-14-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Givalos N, Gakiopoulou H, Skliri M, Bousboukea K, Konstantinidou AE, Korkolopoulou P, Lelouda M, Kouraklis G, Patsouris E, Karatzas G. Replication protein A is an independent prognostic indicator with potential therapeutic implications in colon cancer. Mod Pathol. 2007;20:159–166. doi: 10.1038/modpathol.3800719. [DOI] [PubMed] [Google Scholar]
- 34.Dahai Y, Sanyuan S, Hong L, Di Z, Chong Z. A relationship between replication protein A and occurrence and prognosis of esophageal carcinoma. Cell Biochem Biophys. 2013;67:175–180. doi: 10.1007/s12013-013-9530-y. [DOI] [PubMed] [Google Scholar]
- 35.Valsesia A, Rimoldi D, Martinet D, Ibberson M, Benaglio P, Quadroni M, Waridel P, Gaillard M, Pidoux M, Rapin B, et al. Network-guided analysis of genes with altered somatic copy number and gene expression reveals pathways commonly perturbed in metastatic melanoma. PLoS One. 2011;6:e18369. doi: 10.1371/journal.pone.0018369. [DOI] [PMC free article] [PubMed] [Google Scholar]