Abstract
Pancreatic cancer is one of the most fatal types of cancer and is associated with a dismal prognosis. Gemcitabine-based chemotherapy is clinically used for the treatment of advanced pancreatic cancer. However, many forms of pancreatic cancer have acquired resistance to gemcitabine. In order to prevent patients from suffering from the side effects of chemotherapy and to have the chance to receive more effective intervention, assessment of whether the patient pancreatic cancer cells are resistant to gemcitabine before clinical practice is crucial. Recently, patient-derived xenograft (PDX) models have been regarded as a practical approach for preclinical drug resistance test. In the present study, we harvested tumor specimens from 28 pancreatic cancer patients to establish PDX models. The tumor formation rate of the xenografts was 100%, several of which could be re-implanted in nude mice for more than 10 passages. Primary cells were further obtained from the PDX xenografts to determine their morphological features and evaluate their proliferation rate, migration capacity and angiopoietic ability. In addition, the sensitivities of the primary cells and PDX xenografts to gemcitabine were correlated with each other. When compared to the gemcitabine-sensitive cells, the gemcitabine-resistant cells had a higher level of MCF2L expression, suggesting that MCF2L plays an important role in gemcitabine resistance.
Keywords: pancreatic cancer, primary cell, PDX model, gemcitabine, chemoresistance, MCF2L
Introduction
Pancreatic cancer is a devastating disease. It is attributed to obscure causes and generally displays a dismal prognosis. Particularly, it remains intractable owing to the fact that most types of pancreatic cancer are resistant to traditional chemotherapies. Pancreatic ductal adenocarcinoma (PDAC) is by far the most common tumor type of the exocrine pancreas, accounting for 85 to 90% of all pancreatic tumors (1). Although research on PDAC has been carried out for half a century, its 5-year survival rate (approximately 15-5%) remains the lowest as compared to other digestive tract tumors (2). Only 20% of these patients have the chance to undergo curative operation. More than 80% of the patients are diagnosed with PDAC at later stages, for whom chemotherapy is the only choice (3). Gemcitabine is one of the few FDA-approved agents used in the treatment of advanced pancreatic cancer, and inhibits tumor growth by replacing cytidine during DNA replication, thus blocking the biosynthesis of deoxyribonucleotide by inactivating ribonucleotide reductase (4). Unfortunately, this therapy typically prolongs the survival of fewer than 20% of the patients by only a few months. Some of the patients become gemcitabine-resistant and succumb to progressive disease after several courses, while the vast majority of the patients have to bear the side effects with little clinical remission after the treatment of gemcitabine (5). Thus, evaluation of the susceptibility of pancreatic cancer patients to gemcitabine before clinical practice is crucial. Furthermore, unraveling of the molecular mechanisms of gemcitabine resistance may improve the efficacy of treatment.
Nude mice and commercial cell lines are used for the research of the malignant biological characteristics of pancreatic cancer, but immunodeficient nude mice have a poor track record for predicting the response to therapy in clinical pancreatic cancer patients. In addition, long culture duration makes commercial cells prone to genetic drift (6,7). Compared to other tumor cell lines, there are only 15 commercial PDAC cell lines. The classical cell lines, such as Capan-1, Aspc-1, CFPAC, are derived from ascites, lymph node metastases and other peritoneal metastases (8,9). As far as we know, the primary tumor and their metastatic derivatives usually undergo gene mutation, thus it is open to discussion regarding whether the conclusion we have acquired from metastasis-derived cells could be applied to primary tumors. Thus, primary cells, which can imitate the epigenetic phenotypes, are recommended to obtain a greater phenotypic heterogeneity as compared to the disposable cell lines, and can upgrade the pancreatic cancer cell line family. Few primary models are available due to the limited amounts of clinical samples. On the other hand, when rapid-growing fibroblasts overgrow or tumor cells suffer bacterial contamination, the establishment of primary cells fails. In this regard, patient-derived xenograft (PDX) models have been utilized in order to overcome these difficulties. Moreover, primary cells can be isolated from these xenograft-passaged tumors. Both PDX models and primary cells assumedly play important roles in the study of pancreatic cancer chemoresistance.
The mechanisms of the chemoresistance of pancreatic cancer have been proposed as follows (10–12). The microenvironment of PDAC has two marked features: lack of vessels and rich in dense stroma. Lack of vessel leads to failure of drug delivery. The stroma of PDAC is a dynamic environment filled with various components, including pancreatic stellate cells, matrix metalloproteinases, and growth factors. The interaction between microenvironmental components and tumor cells could impact the features of tumor cells themselves, such as the chemoresistance. Previous studies on the internal biological processes underlying drug resistance have recognized the role of traditional drug-resistance related genes, such as MDR1, LRP and RRM1, in PDAC chemoresistance. A recent study identified many new protein (e.g., MCF2L, VMP1, and MAML3) that may lead to gemcitabine resistance through the differenial expression spectrum between gemcitabine-resistance strains and -sensitive strains (13). In the present study, our preliminary screening results suggest that gemcitabine-resistant pancreatic cancer cells exhibit a higher level of MCF2 transforming sequence-like protein (MCF2L) mRNA, and therefore we chose MCF2 to interrogate its role in the resistance of the primary cells to gemcitabine. The goal of this study was to establish and validate preclinical in vivo and in vitro models for screening of the patients who are unable to respond to gemcitabine. In addition, we attempted to identify the molecules involved in gemcitabine resistance, which may serve as novel therapeutic targets for PDAC.
Materials and methods
Establishment of pancreatic cancer PDX (patient-derived xenograft) models
The human study protocol for the present study was reviewed and approved by the Research Ethics Committee of Ruijin Hospital, Shanghai Jiaotong University School of Medicine. All of the eligible patients had been informed of the essentials of this study, and written consent was obtained before enrollment of each patient into this study. Surgically resected primary tumor tissues from the patients with primary pancreatic cancer were harvested and then separately placed in a sterile culture dish. After dissection and removal of the necrotic areas, fatty tissues, blood clots and connective tissues with forceps and scissors, each tumor specimen was washed with 1% antibiotics-containing Dulbecco's phosphate buffered saline (DPBS) twice and subsequently transferred into another new dish where it was finely trimmed into a 20–30 mm3 fragment. Immediately following this process, the tumor sample was implanted subcutaneously, by using an 18-gauge trocar, in the fore and/or hind bilateral flanks of 6- to 8-week-old female BALB/c nude mice (Shanghai SLAC Laboratory Animal Co., Ltd.). The general health of the mice was monitored daily and growth of the tumor xenograft was monitored twice a week. Once the first generation of xenografts (designed as P0) was established (when the tumor size reached 500–800 mm3), serial implantations in BALB/c nude mice were performed to expand the xenograft tumors (i.e. P1, P2, P3, and beyond). Tumor size was measured periodically using a digital caliper (Cal Pro, Sylvac, Switzerland), and tumor volume was calculated as 0.5 × length × width2.
Establishment of pancreatic primary cancer cells
Tumor samples were resected from the xenograft mouse model and then placed in a sterile culture dish. After dissection and removal of the necrotic areas, fatty tissues, blood clots and connective tissues with forceps and scissors, the tumor specimens were washed twice with DPBS that contains 100 U/ml penicillin and 0.1 mg/ml streptomycin. The samples were transferred into a new dish where they were finely minced on ice with sterile scissors. Approximately 15–20 ml of 1X Accumax solution was added to carry out enzymatic dissociation of the cells from the primary tumor tissues by vigorous pipetting. Following this process, the resultant sample suspension was equally distributed between two 50-ml centrifuge tubes and incubated at 37°C for 1–2 h in a shaking water bath. Then, the dissociated cells were re-suspended in 35–40 ml of 1X DPBS (for each 50 ml centrifuge tube) by pipetting. The resultant suspension was subjected to filtration using 70-µm cell strainers placed on two 50-ml centrifuge tubes, and the filtrates were centrifuged at 1200 rpm for 5 min at room temperature. The supernatants were aspirated and then washed with antibiotic-containing DPBS twice. Finally, the primary cells were harvested and cultured with serum-containing medium, which was changed every second or third day. The cells were passaged using trypsin/EDTA (Beyotime Biotechnology) when reaching sub-confluence. Images of the cultures were captured with Olympus 1681 (Olympus, Hamburg, Germany).
Immunofluorocytochemical profiling
The expression of cytokeratin-8 (CK-8), epithelial cell adhesion molecule (EpCAM) and pancreas/duodenum homeobox protein-1 (PDX-1) in the isolated primary cells was evaluated by immunofluorocytochemistry. For immunofluorescence labeling, one drop of cell suspension at a low density was dripped on the cover glass pretreated with poly-L-lysine, and then subjected to incubation at 37°C for 2–4 h, which can make the cells fully adherent. After washing with PBS, the cells were fixed with 4% paraformaldehyde for 10 min at room temperature and then permeabilized with 2 mg/l to 0.03 mg/l of Triton X-100 in PBS. The nonspecific binding sites were blocked by incubating with 10 mg/l bovine serum albumin (BSA) in PBS for 45 min at room temperature. Slides were then incubated overnight at 4°C with the following primary Abs: anti-CK-8 (1:500, Abcam), anti-PDX1 (1:500, Abcam), anti-EpCAM (1:500, Santa Cruz Biotechnology), and the negative control was incubated with 1X PBS. After incubation, the slides were washed with PBS and further incubated with the donkey anti-mouse, donkey anti-goat, donkey anti-rabbit secondary Ab (dilution ratio 1:200, Life Technologies) for 2 h at room temperature. After extensive washes, the slides were mounted in glycerine (Beyotime) and observed using a confocal laser scanning microscope (Radiance Plus; Bio-Rad Laboratories, Milan, Italy).
Exome library preparation and sequencing
The genomic DNA of primary cells was sequenced by next generation sequencing (NGS) to uncover the exome mutations of KRAS and TP53. A total of 150 ng to 1 µg of DNA extracted from primary cells by MALBAC (multiple annealing and looping-based amplification cycles) was sheared into fragments around 175 bp using the Covaris system (Covaris). The sheared DNA was purified with Agencourt AMPure XP SPRI beads (Beckman Coulter). The DNA was blunted with 5′-phosphorylated ends using the NEB Quick Blunting kit and ligated to truncated PE P7 adaptors and barcoded P5 adaptors using NEBNext Quick Ligation Module. After cleanup with Agencourt AMPure XP SPRI beads and nick fill-in with Bst polymerase large fragment (New England Biolabs), the DNA fragments with adaptors were enriched by PCR. A total amount of 500 ng of DNA pooled from four barcoded libraries was used for hybridization and posthybridization amplification following the manufacturer's protocol (SureSelectXT Target Enrichment System for Illumina Paired-End Sequencing Library, version 1.3.1, February 2012, pp37-60; Agilent Technologies). The posthybridization amplification product was quality checked and sequenced with Illumina HiSEq. 2000/2500 2X 100-bp paired-end (PE) reads.
CCK-8 assay
Cells (5×104) were seeded in 200 µl aliquots into 96-well plates. At the indicated time points, 20 µl CCK-8 solution (Dojindo, Tokyo, Japan) was added to the cells, and the plates were incubated at 37°C in 5% CO2 humidified chamber for an additional 2 h. The absorbance at 450 nm was measured to determine the number of viable cells in each well. All experiments were performed in quintuplicate.
In vitro migration assay
To quantify the in vitro motility of the 7 cell isolates, 8 µm Transwell cell culture chambers (Corning Costar, Milan, Italy) were used. For migration assay, the 7 cell isolates were starved in medium containing 1% FBS (fetal bovine serum) for 24 h before the assay. A cell suspension (200 µl; 2×105 cells/ml) in RPMI-1640 with 1% FBS was added to the upper chambers, whereas 600 µl of RPMI-1640 with 10% FBS was placed in the lower wells. Thereafter, the cells were incubated at 37°C in 5% CO2 humidified chamber for 24 and 48 h. The number of cells that migrated through the filters and seeded to the lower chamber was photographed at each interval. Migration experiments were conducted in quintuplicate.
Endothelial tube formation assay
Briefly, each well of 96-well culture plates was coated with 100 µl Matrigel (BD Biosciences, Franklin Lakes, NJ, USA), which was left to solidify at 37°C for 2 h. Human umbilical vein endothelial cells (HUVECs) were re-suspended in the supernatants obtained from each primary cell. Cell resuspension solution (200 µl) containing 5×104 HUVECs were then added to the solidified Matrigel. After 3 h of incubation at 37°C with 5% CO2, the tubule numbers were assessed under a light microscope.
Western blot analysis of EMT markers
For western blot analysis, cells at confluence were washed twice with PBS and harvested by trypsinization. Cells were lysed in RIPA supplemented with cocktail and PMSF (Solarbio), and the protein concentration was determined using a Bradford assay (Bio-Rad Laboratories). Total protein extract (50 µg) of each sample was separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and electroblotted on polyvinylidene fluoride membranes (Merk Millipore). The transferred membranes were probed with specific primary Ab at 1:1000 dilutions: anti-E-cadherin, anti-N-cadherin, anti-vimentin, anti-catenin, anti-claudin. Anti-GAPDH at a dilution of 1:1000 (Cell Signaling Technology) was used as an internal control. After 3 washes, anti-mouse and anti-rabbit horseradish peroxidase-conjugated secondary Ab (1:5000, Cell Signaling Technology) were added to the membranes. Bands were visualized by incubating the membranes with enhanced chemiluminescence reagent (Thermo Fisher Scientific) and exposing the membranes to X-ray film.
Real-time qPCR analysis
Cells, harvested by trypsinization, were subjected to RNA extraction using the RNeasy kit (Qiagen, Milan, Italy). Two microliters of total RNA (1 µg) was used for cDNA synthesis by using reverse transcription (RT) reaction (ReverTra Ace-α-™, Toyobo, Osaka, Japan). cDNAs was subjected to real-time quantitative PCR analysis by using specific sets of primers designed for the MCF2L gene (5′-AAGCCCGGTTATCACCTTCC-3′ and 5′-GGAGGTCCATTTGTCCCGT-3′). GAPDH gene (5′-GGAGCGAGATCCCTCCAAAAT-3′ and 5′-GGCTGTTGTCATACTTCTCATGG-3′) was used as an internal control.
In vivo efficacy study
We chose the most gemcitabine sensitive cell line PC-07-0049 and the most resistant cell line PC-07-0037 to conduct animal tests. Gemcitabine was purchased from Melonepharma Co., Ltd. (Dalian, China) and was formulated in distilled water for the in vivo study. Tumor tissues were cut into small fragments of ~30 mm3 under sterile conditions. BALB/c nude mice were implanted subcutaneously with a tumor fragment by using a trocar. When the average tumor size reached 150–200 mm3, tumor size-matched mice were randomly assigned to two groups with 5 mice in each group. The tumor-bearing mice were administered gemcitabine at 30 mg/kg (i.p., thrice per week), or vehicle (250 µl, i.p., thrice per week) for two weeks. Tumor volumes and body weights were measured using calipers thrice a week.
Statistical analysis
The difference in tumor volumes between treatment groups was analyzed for significance using one-way ANOVA followed by Dunnett's test. The difference in expression profile of the MCF2L mRNA between different PDX model-derived cells was analyzed for significance using the Student's t-test. The statistical analyses were performed using the IBM SPSS Statistics 19.0 software. P<0.05 was considered to indicate a statistically significant difference.
Results
Patient information, PDX modeling, and primary pancreatic cell isolation
According to the method mentioned above, we harvested 28 tumor specimens, which were derived from patients clinically diagnosed with primary pancreatic cancer but without known or suspected peritoneal metastases, to establish the PDX (patient-derived xenograft) models. It should be specified that the patients who were selected for this study had neither been subjected to adjuvant chemotherapy/radiotherapy nor undergone radical pancreaticoduodenectomy. As anticipated, we acquired 28 PDX models after the initial inoculation; the P0 tumor formation rate in this regard was 100%. However, merely 18 of the 28 tumor specimens had the capacity to expand the xenograft numbers in the PDX mice over 10 successive generations; the remainder failed to re-establish tumor xenografts after the second passage. As to these expandable PDX models, the average time it took to form xenografts with a size of 500 mm3 was 36±7.8 days. As indicated in Table I, postoperative pathological examination of the donors showed that 15 patients had pancreatic ductal adenocarcinoma (of which 8 were identified in the head of the pancreas and 7 were found in its tail). The other 3 patients were confirmed to suffer from pancreatic duct mucus adenoma, duodenal papillary adenocarcinoma, and pancreatic adenosquamous carcinoma, respectively. The clinical information (i.e. demographic) of the 18 patients is shown in Table II. In addition, we further performed isolation of the primary cells from 7 of the 18 xenografts, which were able to be expanded for more than 10 generations in the PDX mice. Of the 7 primary cell isolates, PC-07-0038 became inert after the 19th passage, which suggested the occurrence of cell growth arrest. Accordingly, 6 constantly growing primary cell isolates, namely PC-07-0001, PC-07-0015, PC-07-0034, PC-07-0037, PC-07-0045, and PC-07-0049, were selected for the later drug sensitivity assessment.
Table I.
Pathologic examination of the patients whose tumor specimens enabled the establishment of PDX models and the time required for P0 tumor formation.
| PDX model | Pathological type | Time spent on P0 tumor formationa (days) |
|---|---|---|
| PC-07-0001 | PDAC arising from the head of the pancreas | 48±7 |
| PC-07-0015 | PDAC arising from the head of the pancreas | 27±9 |
| PC-07-0019 | Duodenal papillary adenocarcinoma | 20±3 |
| PC-07-0020 | PDAC arising from the head of the pancreas | 104±14 |
| PC-07-0021 | PDAC arising from the head of the pancreas | 62±9 |
| PC-07-0022 | PDAC arising from the head of the pancreas | 34±4 |
| PC-07-0023 | PDAC arising from the body and tail of the pancreas | 27±4 |
| PC-07-0034 | PDAC arising from the body and tail of the pancreas | 36±4 |
| PC-07-0035 | PDAC arising from the head of the pancreas | 12±2 |
| PC-07-0037 | PDAC arising from the body and tail of the pancreas | 35±3 |
| PC-07-0038 | PDAC arising from the head of the pancreas | 28±6 |
| PC-07-0040 | PDAC arising from the head of the pancreas | 34±10 |
| PC-07-0041 | PDAC arising from the head of the pancreas | 33±7 |
| PC-07-0043 | Pancreatic intraductal papillary mucinous carcinoma | 27±12 |
| PC-07-0045 | PDAC arising from the body and tail of the pancreas | 40±10 |
| PC-07-0048 | Pancreatic adenosquamous carcinoma | 35±11 |
| PC-07-0049 | PDAC arising from the head of the pancreas | 29±4 |
| PC-07-0053 | PDAC arising from the head of the pancreas | 23±3 |
‘Time spent on P0 tumor formation’ represents the length of duration allowed for xenograft tumors to grow to 500 mm3 after subcutaneous inoculation. PDAC, pancreatic ductal adenocarcinoma.
Table II.
Demographic characteristics of the patients included in the study.
| PDX model | Sex | Age | Diabetesa | CA199b |
|---|---|---|---|---|
| PC-07-0001 | F | 55 | N | >>1000 |
| PC-07-0015 | M | 78 | Y | 25.5 |
| PC-07-0019 | M | 76 | Y | 109 |
| PC-07-0020 | M | 65 | Y | >>1000 |
| PC-07-0021 | F | 78 | N | 2.7 |
| PC-07-0022 | F | 71 | Y | 5.6 |
| PC-07-0023 | M | 70 | Y | >>1000 |
| PC-07-0034 | M | 62 | N | 790 |
| PC-07-0035 | M | 67 | Y | 399 |
| PC-07-0037 | F | 59 | Y | >>1000 |
| PC-07-0038 | F | 82 | Y | >>1000 |
| PC-07-0040 | F | 63 | N | >>1000 |
| PC-07-0041 | M | 69 | N | 360 |
| PC-07-0043 | F | 74 | N | 21 |
| PC-07-0045 | M | 65 | Y | >>1000 |
| PC-07-0048 | M | 69 | N | 2.6 |
| PC-07-0049 | F | 47 | N | 39 |
| PC-07-0053 | F | 70 | N | >>1000 |
Patients with/without complications of diabetes.
The normal value of CA199 ranges between 0 and 37 U/ml.
Size, shape, arrangement and growth of the cultured primary cells
All of the isolated cells were shown to have the capability of completely attaching to the bottom of the culture dish. Cell culture medium testing showed that RPMI-1640 supplemented with 10% FBS provided the optimum growth conditions for the primary cell isolates FC-07-0001, FC-07-0015, FC-07-0034, and FC-07-0049. By contrast, the isolates FC-07-0037, FC-07-0038 and FC-07-0045 displayed best growth rates when cultivated in DMEM/F12 supplemented with 2, 5, and 10% FBS, respectively. The average volume of FC-07-0001, FC-7-0038, and FC-07-0049 was comparatively larger than that of FC-07-0015, FC-07-0034, FC-07-0037, and FC-07-0045 (data not shown). The morphologic features of the isolated cells are specified in Table III. As for the cell expandability, the primary cells isolated from the PDX models PC-07-0015, PC-07-0034, PC-07-0037, PC-07-0045, and PC-07-0049 could still maintain an active state (i.e. being capable of growing well) when they were multiplied over 20 passages in culture. As indicated above, cells isolated from the PDX model PC-07-0038 were shown to go into an inert state when they had undergone 19 passages.
Table III.
Morphologic features of the cells isolated from xenografts of the PDX models.
| Primary cells from the PDX model | Shape | Arrangement | Other notes |
|---|---|---|---|
| PC-07-0001 | Long quadrilateral or triangular | Being scattered disorderly | |
| PC-07-0015 | Ovoid or round | Being fused into irregular sheets | |
| PC-07-0034 | Ovoid or round | Without clear boundary; being fused into a monolayer | |
| PC-07-0037 | A variety of shapes | Being fused into round sheets | Small nuclei |
| PC-07-0038 | Circular or polygonal | Being scattered disorderly | Large nuclei; many granules in the cytoplasm |
| PC-07-0045 | Round | Being fused into round or irregular ball; well-defined contour | Large nuclei; some are multinucleated |
| PC-07-0049 | Polygonal | Being scattered disorderly | Large volume; small nuclei |
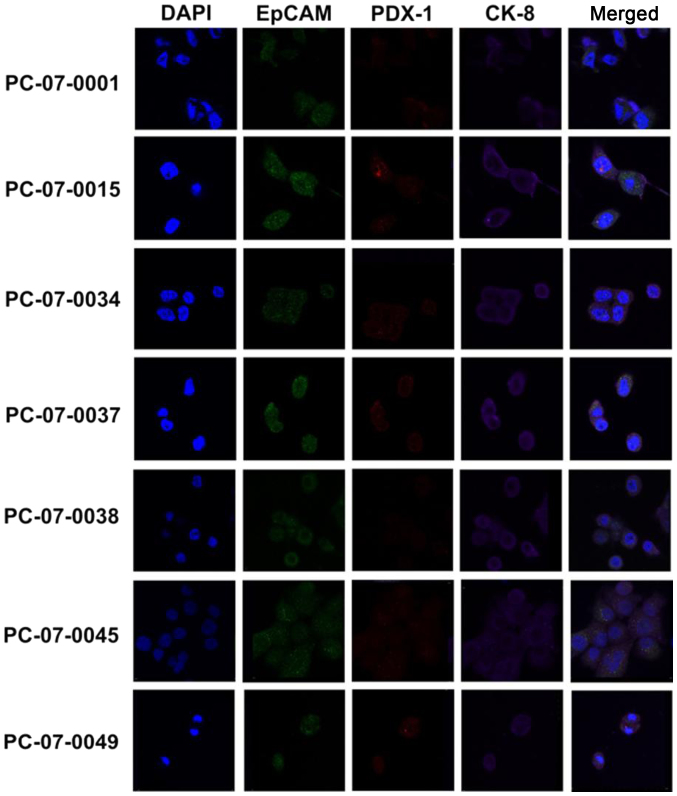
All the primary cell isolates express CK-8, EpCAM and PDX-1
The expression of CK-8, EpCAM, and PDX-1 in the primary cells isolated from the 7 PDX models was evaluated by immunofluorocytochemical assay. As demonstrated in Fig. 1, all 7 primary cell isolates were shown to express CK-8, EpCAM and PDX-1. Notably, the merged images revealed that CK-8 (purple fluorescence), EpCAM (green fluorescence), and PDX-1 (red fluorescence) were located in the cytosol, plasma membrane, and nucleus/cytosol, respectively.
Figure 1.
Immunofluorocytochemical analysis of the expression of CK-8, EpCAM, and PDX-1 in primary cells respectively isolated from the 7 PDX models. DAPI (blue fluorescence) specifically stains nuclei of individual primary cell isolates. Green fluorescence denotes expression of EpCAM; red fluorescence denotes PDX-1; and purple fluorescence denotes CK-8.
Isolated primary cells display high frequency of KRAS and TP53 mutations
By analyzing the whole-exome sequencing data of the 7 primary cells, we found that all of them contained several mutations in the KRAS oncogene and TP53 tumor-uppressor gene. The mutation frequency of KRAS was 100% (7/7), while that of TP53 was 71.4% (5/7). In all 7 primary isolates, KRAS was mutated at codon 12. PC-07-0015, PC-07-0034, PC-07-0045 and PC-07-0049 had a G to R transition (GGT-GRT); PC-07-0001 showed a G to A transversion (GGT-GAT); and PC-07-0037 and PC-07-0038 had a G to D transition (GGT-GDT). TP53 inactivating mutations were found in 5 out of 7 cell isolates (Table IV).
Table IV.
Mutation loci of the key oncogene KRAS and tumor-suppressor gene TP53 in the isolated primary cells.
| Primary cells | TP53 | KRAS |
|---|---|---|
| PC-07-0001 | R248Q, R116Q, R209Q | G12A |
| PC-07-0015 | C238R, C199R, C79R, C106R | G12R |
| PC-07-0034 | G12R | |
| PC-07-0037 | H179R, H140R, H20R, H47R | G12D |
| PC-07-0038 | R175H, R136H, R16H, R43H | G12D |
| PC-07-0045 | R175H, R136H, R16H, R43H | G12R |
| PC-07-0049 | G12R |
Proliferation, motility and angiopoietic ability of isolated primary cells
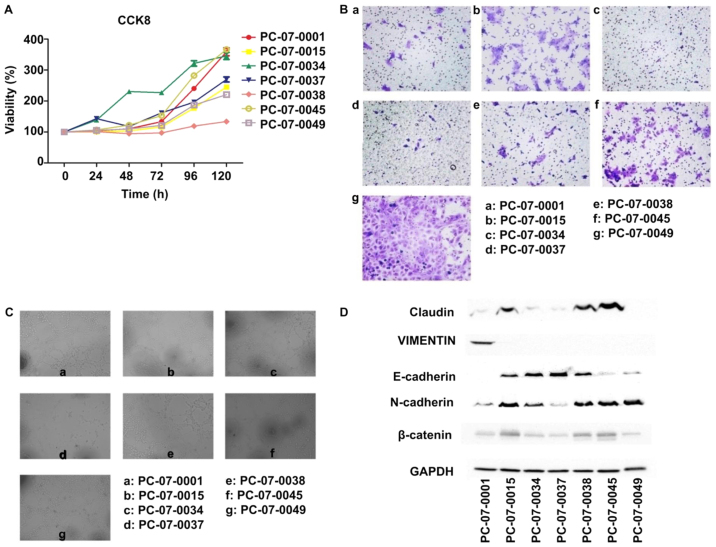
The results of the CCK-8 assay showed that the relative proliferation rates of the 7 cells were different. PC-07-0034 demonstrated the fastest growth rate, with a doubling time of approximately 36 h, whereas PC-073-0038 exhibited the slowest growth rate, with a doubling time of 145 h (Fig. 2A). The Transwell assay showed that the number of PC-07-0049 cells passing through the basement membrane was the highest, which indicated that the motility of PC-07-0049 was the most advanced in all 7 cells, followed by PC-07-0045 and PC-07-0015. In contrast, there were few PC-07-0034 and PC-07-0037 cells passing through the membrane indicating that the migration rates of PC-07-0034 and PC-07-0037 were the slowest (Fig. 2B). In the endothelial tube formation assay, following treatment with the culture media of PC-07-0001, PC-07-0038 and PC-07-0049, the HUVECs were capable of forming integrated tubes; on the contrary, the culture media of PC-07-0037 and PC-07-0045 were unable to induce endothelial tube formation. Imperfect tubes were found in HUVECs treated with culture media of PC-07-0015 and PC-07-0034 (Fig. 2C).
Figure 2.
Proliferation rates (A), migration capacities (B), angiopoietic abilities (C) of the isolated primary cells and (D) expression of EMT markers in these cells.
Expression of EMT markers in the isolated primary cells
Results of the western blot analysis demonstrated that PC-07-0045 and PC-07-0049 expressed the lowest level of E-cadherin and the highest level of N-cadherin (Fig. 2D). However, the results of PC-07-0037 were reversed. Claudin and β-catenin were most highly expressed in PC-07-0015, PC-07-0038 and PC-07-0045. The expression of vimentin was solely found in PC-07-0001.
Viability of the isolated primary cells in response to gemcitabine
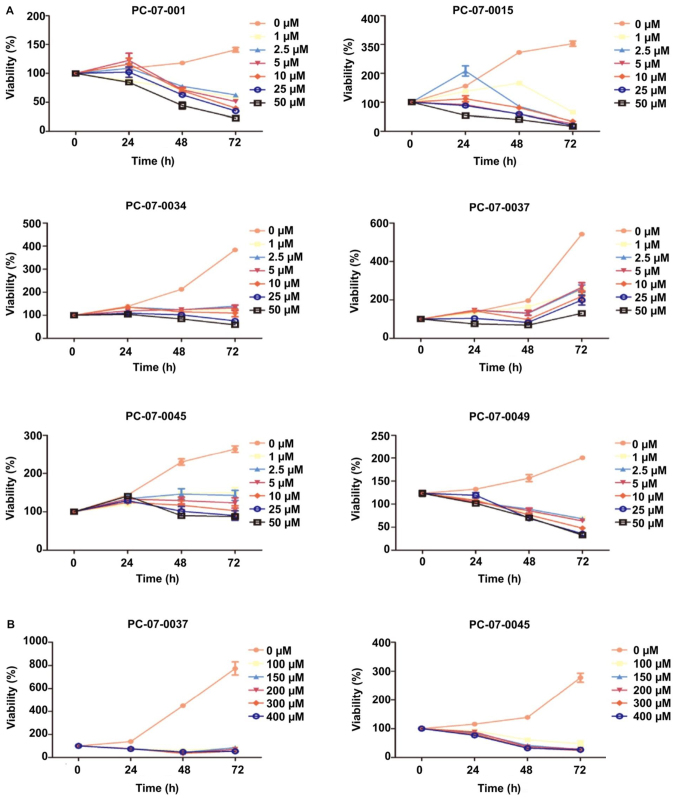
The dose response of each primary cell to gemcitabine was evaluated by the CCK-8 assay. The 7 isolated primary cells were treated with the following concentrations of gemcitabine: 0, 2.5, 5, 10, 20, and 50 µM. The IC50 values of the cells derived from PC-07-0001, PC-07-0015 and PC-07-0049 were 4.332, 1.534 and 9.697 µM, respectively. As for the cells from PC-07-0034, PC-07-0037 and PC-07-0045, the IC50 values were 55.18, 409.6 and 62.87 µM respectively. Nevertheless, PC-07-0037 and PC-07-0045 cells subjected to treatment with 50 µM gemcitabine were still alive even at 72 h post-treatment, suggesting they were less sensitive to gemcitabine (Fig. 3A). When subjected to higher doses (100, 150, 200, 300 and 400 µM) of gemcitabine, their vulnerabilities significantly increased (Fig. 3B). As a consequence, in view of their vulnerabilities to lower and higher doses of gemcitabine, these cells could be divided into the sensitive groups (PC-07-0001, PC-07-0015, and PC-07-0049) and the resistant groups (PC-07-0034, PC-07-0037, and PC-07-0045). The IC50 values of the two groups were significantly different (P<0.05).
Figure 3.
Dose-dependent decrease in cell viability of the isolated primary cells to gemcitabine. (A) Except for PC-07-0037 and PC-07-0045, most isolated primary cells were vulnerable to lower doses of gemcitabine. (B) Cells derived from the PDX models PC-07-0037 and PC-07-0045 were sensitive to higher doses of gemcitabine.
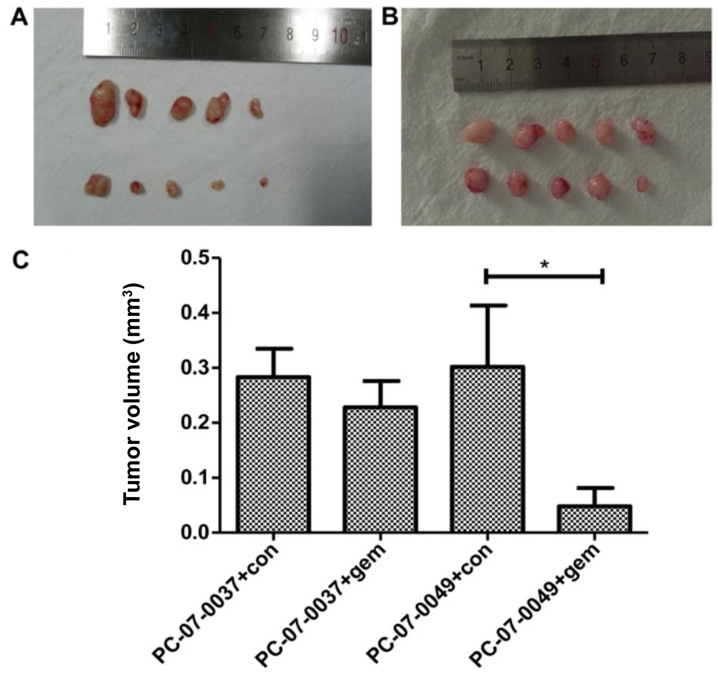
The PC-07-0049 and PC-07-0037 PDX mice respond differently to gemcitabine
At 2-weeks post-treatment, the volume of the tumor in the PC-07-0049 PDX mice (1,579.87±274.8 mm3) was significantly smaller than that of the control group (3,318.63±549.47 mm3) (P<0.05), which indicates that gemcitabine significantly inhibited the growth of inoculated sensitive pancreatic cancer cells in mice (Fig. 4A and C). However, as for the resistant xenograft (i.e., PC-07-0037 implantation), there was no significant difference between the gemcitabine treatment and control groups (Fig. 4B and C).
Figure 4.
Differential inhibitory effects of gemcitabine on the tumor volume of sensitive and resistant xenografts in PDX mice. (A) The sensitive xenograft (PC-07-0049); (B) the resistant xenograft (PC-07-0037); (C) the quantification data (*P<0.05).
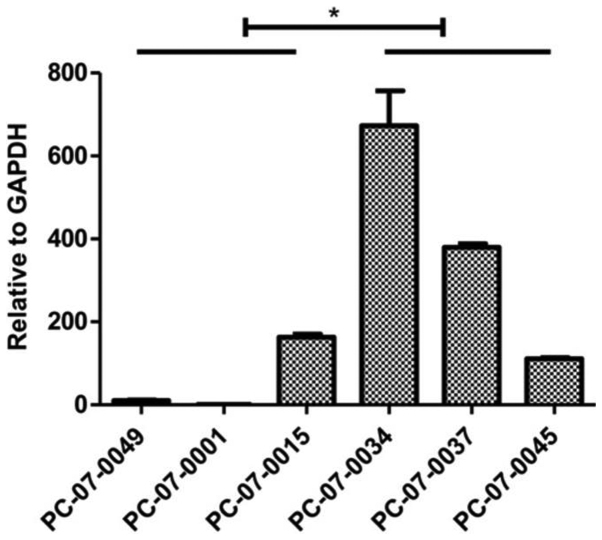
mRNA of MCF2L is highly expressed in the chemoresistant groups. The results of the real-time qPCR assay indicated that expression levels of MCF2 transforming sequence-like protein (MCF2L) mRNA were significantly higher in the resistant cells, as compared to that noted in the sensitive cells (P<0.05) (Fig. 5).
Figure 5.
Expression levels of MCF2L mRNA in the isolated primary cells. Compared to the gemcitabine-sensitive cells, the gemcitabine-resistant cells PC-07-0034, PC-07-0037, and PC-07-0045 had comparatively higher expression levels of MCF2L (*P<0.05).
Discussion
To achieve better outcomes of gemcitabine treatment, it is important to evaluate the therapeutic effects of gemcitabine on pancreatic cancer patients before clinical practice. In general, research associated with drug sensitivity testing is inseparable from the research tools in vitro. Immortalized cells provide a substitute for a limited number of tissue specimens, and therefore becomes an important part of the in vitro experimental platform (14). However, there are only 15 pancreatic cancer cell lines commonly used for testing (15). Many classical cell lines are not derived from primary tumors, but from metastatic tissues instead. Whether the experimental conclusions from metastasis-derived cell lines can be used to explain the mechanisms involved in primary tumor formation is still questionable, as accumulation of gene mutations occurs while primary tumors become metastatic. Furthermore, the commercial cells are prone to genetic drift after long-term culture and may not represent patient characteristics (16–18), which indicates that their derived xenograft models could not accurately predict drug response during clinical therapy. While the PDX (patient-derived xenograft) models are more advantageous for testing drug susceptibility, the primary cells isolated from the PDX models are therefore capable of evaluating drug responses in vitro. Primary cells have been successfully established from pancreatic ductal adenocarcinoma, neuroendocrine tumor, biliary papilloma and normal tissue by means of direct adherent culture methods or trypsin enzyme digesting method (19–22).
To validate whether the cells we harvested through the above-mentioned are exactly tumor cells of human pancreatic ductal adenocarcinoma, we assessed the expression of pancreatic ductal-specific marker PDX-1, pancreatic tumor marker CK8 and epithelial-specific marker EpCAM. After the body becomes mature, PDX-1 specifically expresses in the pancreas as a transcription factor and is considered to be a master gene that controls pancreatic endocrine cell development (23). It contributes to the development of the pancreas and participates in the differentiation of pancreas islet cells. PDX-1 is expressed in pancreatic ductal epithelial cells, β cells that secrete insulin, and delta cells that release somatostatin (24). A series of cytoskeleton markers, for example, cytokeratin, could be used to identify whether the human pancreatic cancer cells were of ductal epithelial origin (25). Cytokeratin, belonging to the cytoskeleton protein family, is one of the intermediate neurofilament protein expressed by epithelial cells. It plays an important role in maintaining epithelial cell morphology. CK-8 is an alkaline cell keratin and is mainly expressed in the cytoplasm of epithelial malignant cells. As a characteristic marker of epithelial cells, CK-8 could become a new target for cancer treatment. High expression of CK-8 protein could alter the epithelial phenotype and promote malignant transformation of cells (26,27). Epithelial cell adhesion molecule (EpCAM) is a calcium-independent epithelial cell adhesion molecule, and it is also a novel tumor marker identified in recent years. EpCAM is widely expressed in lung cancer, colorectal cancer, pancreatic cancer and thyroid cancer, but is seldom expressed in normal epithelium. In this regard, we considered it a cancer cell marker of isolated primary cells (28,29). In our study, we found that all of the 7 cells expressed CK-8, EpCAM and PDX-1. CK-8 was shown to be located in the cytoplasm, EPCAM in the plasma membrane, and PDX-1 in the nucleus and cytoplasm. Thus, we confirmed that all the isolated cells were pancreatic ductal epithelial tumor cells.
We found that morphologic and phenotypic features of the primary cells varied between different PDX models, which may be attributed to the genetic and epigenetic diversities between individual patients. Such diversities conduce to the difference in their clinical manifestations and differential drug sensitivity. We analyzed the proliferation rate, migration capacity, ability of blood vessel formation, and genomic characteristics of the primary cells derived from the different PDX models. Similar to the development of all epithelial tumors, our study also indicated that activation of oncogenes and inactivation of tumor-suppressor genes, due to genetic mutations, account for the occurrence of pancreatic cancer (30,31). By analyzing the exome sequencing data of the 7 primary cells, we found that all the primary cells contained several mutations in the well-known oncogene KRAS and tumor-suppressor gene TP53, and their mutation frequencies are consistent with published sequencing data (32,33). KRAS mutation was found in all 7 cell isolates and were all mutated at codon 12. Of the 7 cells, 4 (PC-07-0015, PC-07-0034, PC-07-0045 and PC-07-0049) had a G to R transition (GGT-GRT). PC-07-0001 showed a G to A transversion (GGT-GAT), whereas PC-07-0037 and PC-07-0038 had a G to D transition (GGT-GDT). TP53 inactivating mutations were found in 5 out of 7 cell lines, which was also consistent with a previous report described as follows. Dong et al reported that most P53 mutations are located in one of 4 missense mutation hot spots, including a.a.129-146, a.a.171-179, a.a.234-260 and a.a.270-287 (34,35).
In our study, all the cells had 3–4 mutation loci, 8 of which were located at the common loci reported in the previous study. The CCK-8 assay indicated that the proliferation rate of the 7 cells were different. PC-07-0034 grew the fastest, with a doubling time of around 36 h. By contrast, the doubling time of PC-07-0038 was 145 h. The proliferation rates of the remaining cells just fell in between. The digestive passage proportion of cells also reflects its growth rate. PC-07-0034 should be passaged into 3 or 4 culture dishes. Cell confluence (more than 90%) will be achieved in 24 h if they are passaged into two dishes. We also found several differences in the migration ability of these isolated cells. Transwell assay showed that the number of PC-07-0049 cells migrating across the basement membrane was the largest, which indicated that the motility of PC-07-0049 was the highest among all 7 cells. In contrast, there were few PC-07-0037 cells that accessed across the membrane, indicating that the migration capacity of PC-07-0037 was weak. An important mechanism of tumor invasion is the occurrence of epithelial-mesenchymal transition (EMT). Our western blot data demonstrated that PC-07-0045 and PC-07-0049 expressed the highest levels of E-cadherin and the lowest level of N-cadherin. However, PC-07-0037 displayed opposite results. The differential motility of the 3 cells may be explained by levels of EMT, which are affected by the expression of the two epithelial cadherins.
Vimentin is the most important intermediate filament in ectomesenchymal cells, and it is absent in normal epithelial cells. Its expression has been noted to be upregulated during tumor transformation (36,37). The expression of vimentin was only found in PC-07-0001, indicating that the motility of PC-07-0001 may be associated with vimentin. β-catenin and E-cadherin have been shown to be colocalized in intercellular adhesion connection (38,39). The expression level of β-catenin is associated with the malignant degree of prostate cancer, lung cancer, and colorectal cancer (40). Claudin is a vital protein in cell membrane tight junction, whose abnormal expression leads to structural damage and functional impairment of epithelial cells (41). The expression of claudin is closely related to the occurrence and development of a wide variety of tumors (42–44). Claudin and β-catenin were highly expressed in PC-07-0015, PC-07-0038 and PC-07-0045. The motility of the 3 cells may be related to the expression of β-catenin and claudin. Similar to most malignant tumors, the basis of tumor growth and metastasis is the formation of tumor blood vessels, which is also designated as ‘angiogenesis’ (45,46). In the endothelial tube formation assay, following treatment with the culture media of PC-07-000, PC-07-0038 and PC-07-0049, the HUVECs were capable of forming integrated tubes. On the contrary, the culture media of PC-07-0037 and PC-07-0045 were unable to induce endothelial tube formation.
Chemotherapeutic drugs may have serious side effects, such as gastrointestinal reactions, liver and kidney damage, and bone marrow suppression, even though they are able to kill tumors. Thus, if we can evaluate the sensitivity of these drugs before clinical trial, we can administer them to the drug-sensitive population for effective treatment, while avoiding or minimizing the side effects of the drugs. Gemcitabine is the first line drug for advanced pancreatic cancer, which was certified by the U.S. Food and Drug Administration (FDA) in 1997. However, the clinical response rate of gemcitabine is still lower than 10%. Over 90% of the patients are resistant to gemcitabine. Long-term administration of gemcitabine has been shown to lead to acquired drug resistance and side-effect in sensitive patients. In traditional chemotherapeutic regimens, identification of the drug sensitivity of patients before taking medication was unknown. Patients diagnosed with pancreatic cancer were all recommended to receive gemcitabine treatment. Many patients did not have a satisfactory outcome, yet suffered the side effects of gemcitabine instead. Thus, the sensitivity of patients to gemcitabine should be evaluated before clinical application. Only the drug sensitive population should be administered gemcitabine. In our experiment, we found that the sensitivity of the cells to gemcitabine was inconsistent. When PC-07-0049 was treated with 10 µM gemcitabine for 72 h, cell proliferation was significantly inhibited. Moreover, cell debris and cell necrosis increased. However, when PC-07-0037 was treated with 10 µM gemcitabine for 72 h, no obvious cell proliferation inhibition was observed, and cell morphology appeared normal. As for the in vivo tests, when the PDX models subcutaneously inoculated with the 2 cells were treated with the same concentration of gemcitabine, the volume of the tumor xenografts in the drug-resistant PC-07-0037 PDX model administered gemcitabine did not differ from that of the untreated control. In contrast, the tumor volume of the drug-sensitive PC-07-0049 PDX model (3,318.63±549.47 mm3) was obviously reduced as compared to that of the untreated control (1,579.87±274.8 mm3). Therefore, we concluded that we can selectively and specifically apply therapeutic agents to patients, according to the results of the sensitivity test performed in the PDX models. This will strategically not only increase therapeutic efficacy of chemotherapy but also reduce its side effects.
It has become an urgent issue that drug resistance greatly reduces therapeutic effects. Recent studies have identified various proteins that may lead to gemcitabine resistance by means of proteomics, RNA-seq and whole-genome siRNA library to measure the expression spectrum of gemcitabine resistance between resistant and sensitive strains. We tested and verified the mRNA expression levels of these proteins in the primary cells and found that, compared to the gemcitabine-ensitive cells, the gemcitabine-resistant cells had a high level of MCF2L expression. MCF2L, also named DBS/DBLs Big Sister, belongs to the DBL family. MC2L is one of the guanine nucleotide exchange factor and potentially links pathways that signal through RAC1, RHOA and CDC42 (47). It can catalyze guanine nucleotide exchange on RHOA and CDC42 and interacts specifically with the GTP-bound form of RAC1, suggesting that it functions as an effector of RAC1. There are two types of endogenous DBS subtypes: the molecular weight of the first type is 130 kDa (DBS-130), which is located in the Golgi apparatus; the other one (DBS-80) is located in the endoplasmic reticulum. A previous study has demonstrated that the DBS-130 inhibitors can reduce the motility of MDA-MB-231 cells in Transwell and scratch assays (48,49). DBS becomes activated and highly tumorigenic via truncation of the N-terminus, and the activation of DBS can promote cell proliferation during the development of hemocytes (50). However, the role of MCF2L/DBS in the drug resistance of tumor cells has not yet been studied. Our study found that the gemcitabine-resistant cells had a high level of MCF2L expression, as compared to the gemcitabine-sensitive cells. Consequently, we conclude that MCF2L may play an important role in gemcitabine resistance, which still needs to be further confirmed.
Limitations of the study. Pancreatic cancer is deadly and often does not cause any signs or symptoms at the early stage. The patients are usually diagnosed at the late stage when they feel something wrong with their bodies (e.g. indigestion, heartburn, unexplained weight loss, and abdominal pain). Currently, there is no standard diagnostic tool or established early detection method for pancreatic cancer. The delay in diagnosis leads to loss of therapeutic golden chance. Gemcitabine is a recommended chemotherapeutic agent used in the treatment of pancreatic cancer. Unfortunately, this treatment is very challenging due to the occurrence of chemoresistance. In this regard, many patients do not show a satisfactory outcome, but suffer the side effects of gemcitabine instead. The purpose of this study was to establish a PDX model for pre-clinical assessment of chemoresistance in pancreatic cancer rather than for development of a clinical diagnostic strategy. We expect that future improvement of the technique for establishing the PDX model can make this pre-clinical platform more useful for exploring the mechanisms of chemoresistance. Regarding the pitfalls of this study, we recognize that inadequate acquisition of the patient follow-up information weakened the value and clinical significance of this study; understanding of the correlation between patient responsiveness and therapeutic efficacy may help unravel the underlying mechanisms of gemcitabine resistance. Therefore, our future goal is to pursue further studies on the downstream genes associated with gemcitabine resistance.
In conclusion, the in vivo PDX models and in vitro primary cell models derived from the clinical samples provide a powerful support for the susceptibility testing of gemcitabine and drug development. MCF2L can be considered to play an important role in gemcitabine resistance.
References
- 1.Weissmueller S, Manchado E, Saborowski M, Morris JP, IV, Wagenblast E, Davis CA, Moon SH, Pfister NT, Tschaharganeh DF, Kitzing T, et al. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor β signaling. Cell. 2014;157:382–394. doi: 10.1016/j.cell.2014.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li D, Xie K, Wolff R, Abbruzzese JL. Pancreatic cancer. Lancet. 2004;363:1049–1057. doi: 10.1016/S0140-6736(04)15841-8. [DOI] [PubMed] [Google Scholar]
- 3.Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, Kamiyama M, Hruban RH, Eshleman JR, Nowak MA, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Minami K, Shinsato Y, Yamamoto M, Takahashi H, Zhang S, Nishizawa Y, Tabata S, Ikeda R, Kawahara K, Tsujikawa K, et al. Ribonucleotide reductase is an effective target to overcome gemcitabine resistance in gemcitabine-resistant pancreatic cancer cells with dual resistant factors. J Pharmacol Sci. 2015;127:319–325. doi: 10.1016/j.jphs.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 5.Xu CP, Xue XJ, Liang N, Xu DG, Liu FJ, Yu XS, Zhang JD. Effect of chemoradiotherapy and neoadjuvant chemoradiotherapy in resectable pancreatic cancer: A systematic review and meta-analysis. J Cancer Res Clin Oncol. 2014;140:549–559. doi: 10.1007/s00432-013-1572-4. [DOI] [PubMed] [Google Scholar]
- 6.Wennerström AB, Lothe IM, Sandhu V, Kure EH, Myklebost O, Munthe E. Generation and characterisation of novel pancreatic adenocarcinoma xenograft models and corresponding primary cell lines. PLoS One. 2014;9:e103873. doi: 10.1371/journal.pone.0103873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kato M, Shimada Y, Tanaka H, Hosotani R, Ohshio G, Ishizaki K, Imamura M. Characterization of six cell lines established from human pancreatic adenocarcinomas. Cancer. 1999;85:832–840. doi: 10.1002/(SICI)1097-0142(19990215)85:4<832::AID-CNCR10>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 8.Smith HS. In vitro properties of epithelial cell lines established from human carcinomas and nonmalignant tissue. J Natl Cancer Inst. 1979;62:225–230. [PubMed] [Google Scholar]
- 9.Chen WH, Horoszewicz JS, Leong SS, Shimano T, Penetrante R, Sanders WH, Berjian R, Douglass HO, Martin EW, Chu TM. Human pancreatic adenocarcinoma: In vitro and in vivo morphology of a new tumor line established from ascites. In Vitro. 1982;18:24–34. doi: 10.1007/BF02796382. [DOI] [PubMed] [Google Scholar]
- 10.Koay EJ, Truty MJ, Cristini V, Thomas RM, Chen R, Chatterjee D, Kang Y, Bhosale PR, Tamm EP, Crane CH, et al. Transport properties of pancreatic cancer describe gemcitabine delivery and response. J Clin Invest. 2014;124:1525–1536. doi: 10.1172/JCI73455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, Madhu B, Goldgraben MA, Caldwell ME, Allard D, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moir JA, Mann J, White SA. The role of pancreatic stellate cells in pancreatic cancer. Surg Oncol. 2015;24:232–238. doi: 10.1016/j.suronc.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 13.Zagorac S. Identification and functional characterization of epigenetic determinants of pancreatic CSCs. http://hdl.handle.net/10486/669541 PhD dissertation, la Universidad Autónoma de Madrid, 11–16–2015. 2015 [Google Scholar]
- 14.Ku JL, Yoon KA, Kim WH, Jang Y, Suh KS, Kim SW, Park YH, Park JG. Establishment and characterization of four human pancreatic carcinoma cell lines. Genetic alterations in the TGFBR2 gene but not in the MADH4 gene. Cell Tissue Res. 2002;308:205–214. doi: 10.1007/s00441-001-0510-y. [DOI] [PubMed] [Google Scholar]
- 15.Rückert F, Aust D, Böhme I, Werner K, Brandt A, Diamandis EP, Krautz C, Hering S, Saeger HD, Grützmann R, et al. Five primary human pancreatic adenocarcinoma cell lines established by the outgrowth method. J Surg Res. 2012;172:29–39. doi: 10.1016/j.jss.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 16.Briske-Anderson MJ, Finley JW, Newman SM. The influence of culture time and passage number on the morphological and physiological development of Caco-2 cells. Proc Soc Exp Biol Med. 1997;214:248–257. doi: 10.3181/00379727-214-44093. [DOI] [PubMed] [Google Scholar]
- 17.Chang-Liu CM, Woloschak GE. Effect of passage number on cellular response to DNA-damaging agents: Cell survival and gene expression. Cancer Lett. 1997;113:77–86. doi: 10.1016/S0304-3835(97)04599-0. [DOI] [PubMed] [Google Scholar]
- 18.Esquenet M, Swinnen JV, Heyns W, Verhoeven G. LNCaP prostatic adenocarcinoma cells derived from low and high passage numbers display divergent responses not only to androgens but also to retinoids. J Steroid Biochem Mol Biol. 1997;62:391–399. doi: 10.1016/S0960-0760(97)00054-X. [DOI] [PubMed] [Google Scholar]
- 19.Mohamed A, Blanchard MP, Albertelli M, Barbieri F, Brue T, Niccoli P, Delpero JR, Monges G, Garcia S, Ferone D, et al. Pasireotide and octreotide antiproliferative effects and sst2 trafficking in human pancreatic neuroendocrine tumor cultures. Endocr Relat Cancer. 2014;21:691–704. doi: 10.1530/ERC-14-0086. [DOI] [PubMed] [Google Scholar]
- 20.Murakami S, Ajiki T, Hori Y, Okazaki T, Fukumoto T, Ku Y. Establishment of a novel cell line from intraductal papillary neoplasm of the bile duct. Anticancer Res. 2014;34:2203–2209. [PubMed] [Google Scholar]
- 21.Kong D, Nishino N, Shibusawa M, Kusano M. Establishment and characterization of human pancreatic adenocarcinoma cell line in tissue culture and the nude mouse. Tissue Cell. 2007;39:217–223. doi: 10.1016/j.tice.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 22.Lawson T, Ouellette M, Kolar C, Hollingsworth M. Culture and immortalization of pancreatic ductal epithelial cells. Methods Mol Med. 2005;103:113–122. doi: 10.1385/1-59259-780-7:113. [DOI] [PubMed] [Google Scholar]
- 23.Park JY, Hong SM, Klimstra DS, Goggins MG, Maitra A, Hruban RH. Pdx1 expression in pancreatic precursor lesions and neoplasms. Appl Immunohistochem Mol Morphol. 2011;19:444–449. doi: 10.1097/PAI.0b013e318206d958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fendrich V, Lauth M. The role of pancreatic and duodenal homeobox 1 as a therapeutic target in pancreatic cancer. Expert Opin Ther Targets. 2014;18:1277–1283. doi: 10.1517/14728222.2014.945427. [DOI] [PubMed] [Google Scholar]
- 25.Moll R, Franke WW, Schiller DL, Geiger B, Krepler R. The catalog of human cytokeratins: Patterns of expression in normal epithelia, tumors and cultured cells. Cell. 1982;31:11–24. doi: 10.1016/0092-8674(82)90400-7. [DOI] [PubMed] [Google Scholar]
- 26.Merjava S, Brejchova K, Vernon A, Daniels JT, Jirsova K. Cytokeratin 8 is expressed in human corneoconjunctival epithelium, particularly in limbal epithelial cells. Invest Ophthalmol Vis Sci. 2011;52:787–794. doi: 10.1167/iovs.10-5489. [DOI] [PubMed] [Google Scholar]
- 27.Liu F, Chen Z, Wang J, Shao X, Cui Z, Yang C, Zhu Z, Xiong D. Overexpression of cell surface cytokeratin 8 in multidrug-resistant MCF-7/MX cells enhances cell adhesion to the extracellular matrix. Neoplasia. 2008;10:1275–1284. doi: 10.1593/neo.08810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munz M, Baeuerle PA, Gires O. The emerging role of EpCAM in cancer and stem cell signaling. Cancer Res. 2009;69:5627–5629. doi: 10.1158/0008-5472.CAN-09-0654. [DOI] [PubMed] [Google Scholar]
- 29.Went PT, Lugli A, Meier S, Bundi M, Mirlacher M, Sauter G, Dirnhofer S. Frequent EpCam protein expression in human carcinomas. Hum Pathol. 2004;35:122–128. doi: 10.1016/j.humpath.2003.08.026. [DOI] [PubMed] [Google Scholar]
- 30.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deer EL, González-Hernández J, Coursen JD, Shea JE, Ngatia J, Scaife CL, Firpo MA, Mulvihill SJ. Phenotype and genotype of pancreatic cancer cell lines. Pancreas. 2010;39:425–435. doi: 10.1097/MPA.0b013e3181c15963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53:549–554. doi: 10.1016/0092-8674(88)90571-5. [DOI] [PubMed] [Google Scholar]
- 33.Yin X, Su J, Zhou X, Guo J, Wei W, Wang Z. K-ras-driven engineered mouse models for pancreatic cancer. Discov Med. 2015;19:15–21. [PubMed] [Google Scholar]
- 34.Moore PS, Sipos B, Orlandini S, Sorio C, Real FX, Lemoine NR, Gress T, Bassi C, Klöppel G, Kalthoff H, et al. Genetic profile of 22 pancreatic carcinoma cell lines. Analysis of K-ras, p53, p16 and DPC4/Smad4. Virchows Arch. 2001;439:798–802. doi: 10.1007/s004280100474. [DOI] [PubMed] [Google Scholar]
- 35.Dong M, Nio Y, Yamasawa K, Toga T, Yue L, Harada T. p53 alteration is not an independent prognostic indicator, but affects the efficacy of adjuvant chemotherapy in human pancreatic cancer. J Surg Oncol. 2003;82:111–120. doi: 10.1002/jso.10186. [DOI] [PubMed] [Google Scholar]
- 36.Dave JM, Bayless KJ. Vimentin as an integral regulator of cell adhesion and endothelial sprouting. Microcirculation. 2014;21:333–344. doi: 10.1111/micc.12111. [DOI] [PubMed] [Google Scholar]
- 37.Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci. 2011;68:3033–3046. doi: 10.1007/s00018-011-0735-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hong SM, Li A, Olino K, Wolfgang CL, Herman JM, Schulick RD, Iacobuzio-Donahue C, Hruban RH, Goggins M. Loss of E-cadherin expression and outcome among patients with resectable pancreatic adenocarcinomas. Mod Pathol. 2011;24:1237–1247. doi: 10.1038/modpathol.2011.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang J, Dokurno P, Tonks NK, Barford D. Crystal structure of the M-fragment of α-catenin: Implications for modulation of cell adhesion. EMBO J. 2001;20:3645–3656. doi: 10.1093/emboj/20.14.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Al-Rawi R. β-Catenin as a biomarker in diagnosis of tumors with special emphasis on colorectal carcinoma. Biom J. 2017;3:1. [Google Scholar]
- 41.Overgaard CE, Mitchell LA, Koval M. Roles for claudins in alveolar epithelial barrier function. Ann NY Acad Sci. 2012;1257:167–174. doi: 10.1111/j.1749-6632.2012.06545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiurillo MA. Role of the Wnt/β-catenin pathway in gastric cancer: An in-depth literature review. World J Exp Med. 2015;5:84–102. doi: 10.5493/wjem.v5.i2.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Itallie CM, Anderson JM. Claudin interactions in and out of the tight junction. Tissue Barriers. 2013;1:e25247. doi: 10.4161/tisb.25247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jamieson C, Sharma M, Henderson BR. Targeting the β-catenin nuclear transport pathway in cancer. Semin Cancer Biol. 2014;27:20–29. doi: 10.1016/j.semcancer.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 45.Folkman J. Fighting cancer by attacking its blood supply. Sci Am. 1996;275:150–154. doi: 10.1038/scientificamerican0996-150. [DOI] [PubMed] [Google Scholar]
- 46.Xu Z, Pothula SP, Wilson JS, Apte MV. Pancreatic cancer and its stroma: A conspiracy theory. World J Gastroenterol. 2014;20:11216–11229. doi: 10.3748/wjg.v20.i32.11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Snyder JT, Rossman KL, Baumeister MA, Pruitt WM, Siderovski DP, Der CJ, Lemmon MA, Sondek J. Quantitative analysis of the effect of phosphoinositide interactions on the function of Dbl family proteins. J Biol Chem. 2001;276:45868–45875. doi: 10.1074/jbc.M106731200. [DOI] [PubMed] [Google Scholar]
- 48.Fitzpatrick ER, Hu T, Ciccarelli BT, Whitehead IP. Regulation of vesicle transport and cell motility by Golgi-localized Dbs. Small GTPases. 2014;5:1–12. doi: 10.4161/sgtp.28570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Z, Adams HC, III, Whitehead IP. The rho-specific guanine nucleotide exchange factor Dbs regulates breast cancer cell migration. J Biol Chem. 2009;284:15771–15780. doi: 10.1074/jbc.M901853200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klinger MB, Guilbault B, Kay RJ. The RhoA- and CDC42-specific exchange factor Dbs promotes expansion of immature thymocytes and deletion of double-positive and single-positive thymocytes. Eur J Immunol. 2004;34:806–816. doi: 10.1002/eji.200324400. [DOI] [PubMed] [Google Scholar]