Abstract
Drug resistance is the major factor contributing to the failure of chemotherapy in non-small cell lung cancer (NSCLC) patients. Emerging evidence suggests that autophagy plays a vital role in the chemoresistance of many types of tumors. However, the exact mechanism underlying the chemoresistance of NSCLC is still elusive, and it is unclear whether lncRNA-XIST is involved in autophagy and chemoresistance of NSCLC. In the present study, we demonstrated that lncRNA-XIST was overexpressed in NSCLC tumor samples, and knockdown of lncRNA-XIST significantly decreased autophagy by regulation of ATG7 as determined by qPCR and by western blotting. Furthermore, we found that miR-17 was upregulated following knockdown of lncRNA-XIST, and miR-17 mimics decreased the protein levels of ATG7 by directly targeting the 3-untranslated region of ATG7 mRNA as determined by RT-qPCR and by western blotting. Furthermore, we found that the expression level of lncRNA-XIST was markedly increased in cisplatin-resistant A549 cells as determined by q-PCR. Knockdown of lncRNA-XIST restored the chemosensitivity of cisplatin-resistant A549 cells to cisplatin, which was reversed by miR-17 inhibitor and overexpression of ATG7 as determined by CCK8 assays. This study provides evidence that lncRNA-XIST may be a potential marker of poor response to cisplatin chemotherapy in NSCLC patients and the pathway ‘lncRNA-XIST/miR-17/autophagy’ may be a promising target for patients with chemoresistant NSCLC.
Keywords: NSCLC, chemoresistance, lncRNA-XIST, autophagy, miR-17
Introduction
Non-small cell lung cancer (NSCLC) is the leading cause of cancer-related mortality in the world, accounting for 85% of all lung cancer cases. According to previous reports, most patients with NSCLC present with advanced cancer at diagnosis and patients with advanced NSCLC have an average 5-year survival rate of approximately 15% (1). Although efforts and improvements have been made in the diagnosis and therapy for NSCLC, the prognosis of patients is still not optimistic. Drug resistance is the leading factor contributing to the failure of chemotherapy treatment (2–5). The high mortality of patients with NSCLC and the poor prognosis associated with NSCLC has propelled us to improve our understanding of the mechanisms underlying the chemoresistance of NSCLC. More important, establishing effective therapeutic targets for chemotherapy is urgent.
Recently, many studies have shown that the majority of human genome transcripts are transcribed into non-coding RNAs including small ncRNAs and long non-coding RNAs (6,7). Long non-coding RNAs (lncRNAs) are a class of non-coding RNA transcripts with a length of more than 200 nucleotides (8). To date, growing evidence has shown that lncRNAs play important roles in cancer development and progression (9). Mounting evidence has shown that lncRNAs are involved in diverse pathological processes such as autophagy, proliferation and apoptosis (10–13). The role and function of lncRNAs in chemoresistance have also been widely investigated. In breast cancer, Xue et al (14) reported that lncRNA-HOTAIR was overexpression in tamoxifen-resistant cancer samples and contributed to tamoxifen-resistance by promoting ER signaling. In addition, in HER2-positive breast cancers, lncRNA-ATB was suggested to lead to trastuzumab resistance by directly binding miRNA200c and inducing EMT (15). Zhang et al (16) also demonstrated that long non-coding RNA PVT-1 was upregulated in cisplatin-resistant gastric cancer tissues and cisplatin-resistant gastric cells. In in vitro experiments, knockdown of PVT-1 weakened the resistance to cisplatin of cisplatin-resistant cell lines. Several studies have also reported that many types of lncRNAs participate in the drug resistance of NSCLC. Liu et al (17) previously documented that lncRNA-HOTAIR conferred cisplatin resistance to A549 cells by downregulating the expression of p21WAF1/CIP1. Liu et al (18) showed that lncRNA-MEG3 was decreased in cisplatin-resistant lung cancer cells and overexpression of MEG3 restored chemosensitivity to cisplatin of cisplatin-resistant cells both in vitro and in vivo. Recently, the levels of lncRNA-XIST were found to be increased in tumor tissues and serum from NSCLC patients (19), indicating that it may play an important role in NSCLC. However, the role and function of lncRNA-XIST in drug resistance remain unclear.
Autophagy is a conserved metabolic process, which is rapidly induced to ensure the survival of cancer cells facing a harsh tumor microenvironment such as hypoxia, starvation or exposure to chemotherapy drugs (20–23). Many studies have demonstrated that autophagy is involved in the development and progression of cancer such as promotion of cancer cell growth, cancer cell resistance to chemotherapeutic drugs and cancer cell metastasis (24). In hepatocellular carcinoma (HCC) cells, Xiong et al (25) and colleagues demonstrated that lncRNA-HULC could induce autophagy to attenuate the chemosensitivity of HCC cells. However, the role of lncRNA-XIST in autophagy is not fully understood, and whether lncRNA-XIST affects the chemosensitivity of NSCLC through autophagy remains to be fully clarified.
In the present study, we showed that lncRNA-XIST was upregulated in NSCLC tissues compared with the adjacent normal tissues and in DDP-resistance A549 cell lines. In addition, we found that knockdown of lncRNA-XIST decreased basal autophagy and autophagic flux in NSCLC cells. Furthermore, we demonstrated that knockdown of lncRNA-XIST impaired the chemoresistance of NSCLC dependent on autophagy, suggesting that the lncRNA-XIST/autophagy pathway may be a potential target for NSCLC chemotherapy.
Materials and methods
Non-small cell lung cancer and adjacent normal tissues
Fifty paired NSCLC tissues and adjacent normal specimens were obtained from Tongji Hospital, Huazhong University of Science and Technology from October 2014 to November 2016. Informed content for the present study was obtained from those patients participating in this investigation prior to the study. This project was also approved by the Ethics Committee of Tongji Hospital. All tissue samples were stored in liquid nitrogen.
Total RNA extraction and real-time polymerase chain reaction (qPCR)
Total RNA was obtained from tissue samples or cancer cell lines using TRIzol reagent (Invitrogen, Shanghai, China). Next, cDNA was synthesized from 5 to 10 µg of total RNA using the Super Master Mix synthesis kit (Takara Bio, Shiga, Japan). Then, we examined the mRNA expression of lncRNA-XIST and GAPDH in tissue samples or cancer cell lines using TaqMan Universal Master Mix II (Applied Biosystems, Foster City, CA, USA). GAPDH was considered as the endogenous control. Data were analyzed using the 2−∆∆CT method. The qPCR conditions used were as follows: 10 min at 95°C, 40 cycles of 15 sec at 95°C and 1 min at 60°C. The sequences of qPCR used in the present study were as follows: GAPDH forward primer, 5-GGAGCGAGATCCCTCCAA AAT-3 and reverse primer, 5-GGCTGTTGTCATACTTC TCATGG-3; lncRNA-XIST forward primer, 5-ACGCTG CATGTGTCCTTAG-3 and reverse primer, 5-GAGCCT CTTATAAGCTGTTTG-3.
Western blotting
We collected cancer cells according to the requirement of the corresponding experiment. Then, we immediately lysed the cell sediment in RIPA buffer with 1% PMSF. Briefly, the protein samples were separated on SDS-PAGE gel and transferred onto PVDF membranes. The membranes were incubated with the corresponding primary antibody overnight at 4°C. Next, the membranes were incubated with the secondary antibody and visualized using ECL substrates.
Confocal microscopy
For quantification of autophagy, A549 cells were transiently co-transfected with GFP-LC3B and shRNA-lncRNA-XIST or control-shRNA. Briefly, after transfection for 48 h, the cells were washed with phosphate-buffered saline (PBS) at 4°C twice. Then, the cells were carefully fixed with 4% paraformaldehyde and then incubated with 4′,6-diamidino-2-phenylindole (DAPI) for 5 min at room temperature after washing the cells twice with PBS. Subsequently, the stained cells were observed using Zeiss LSM 510 laser confocal microscopy. The autophagy dots were counted in 20 cells randomly. Finally, the average autophagy dots in each cell were analyzed.
Cell lines and cell culture
Human cell lines A549 and H1299 were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA. All cell lines were cultured in Dulbeccos modified Eagles medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Invitrogen) at 37°C in a humidified incubator. The cisplatin-resistant DDP A549 cell line was obtained from the Bank of Cancer Cell Lines of the Chinese Academy of Medical Science (Beijing, China) and 2 µg/ml cisplatin was added to the medium to maintain its drug resistance phenotype.
Reagents and antibodies
Primary antibodies against LC3B and ATG7 were purchased from Cell Signaling Technology (Danvers, MA, USA) and SQSTM and GAPDH antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Corresponding secondary antibodies were purchased from Abcam (Cambridge, MA, USA). Chloroquine (CQ) was purchased from Sigma-Aldrich (St. Louis, MO, USA). The mimics and inhibitors of miRNA, the negative controls of mimics and inhibitors (NC) were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). Lipofectamine 2000 reagent was obtained from Invitrogen.
Construction of plasmids and transfection
The total DNA fragment coding ATG7 was cloned into the pCDNA3.1(+) vector. The DNA fragment containing the predicted miR-17 target site (ATG7-WT) of 3′-UTR ATG7 mRNA and the mutation DNA fragment of the predicted miR-17 target site (ATG7-Mut) separately was inserted into a pmirG1O Dual-Luciferase miRNA Target Expression Vector. The different plasmids were co-transfected into cells as indicated using Lipofectamine 2000 reagent. In addition, luciferase assays were performed according to the manufacturers protocol included in the Dual-Luciferase reporter assay system (Promega, Madison, WI, USA). Specific shRNA oligonucleotides targeting lncRNA-XIST (Sh-X) and negative control (Ctrl) were obtained from Shanghai Genepharma Co., Ltd. (Shanghai, China) and this sequence was cloned into the pLKO.1-Puro vector according to the manufacturer's instructions. The pmirG1O Dual-Luciferase miRNA Target Expression Vector and the pLKO.1-Puro vector were respectively obtained from Promega and Addgene.
Cell viability assay
Cells were plated into 96-well plates at 3,000 cells/well. After transfection of the plasmids and treatment with cisplatin according to the experiment, 10 µl of Cell Counting Kit-8 (CCK-8/WST-8; Sigma-Aldrich) was added to the plates and incubation was carried out for 3 h. Subsequently, the absorbance of each plate was examined at 450 nm.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 5. Data were shown as mean ± SD. Differences between the groups were estimated using the Students t-test, the χ2 test or the Pearson correlation coefficient. P<0.05 was considered to indicate a statistically significant result.
Results
lncRNA-XIST is highly expressed and associated with autophagy in NSCLC
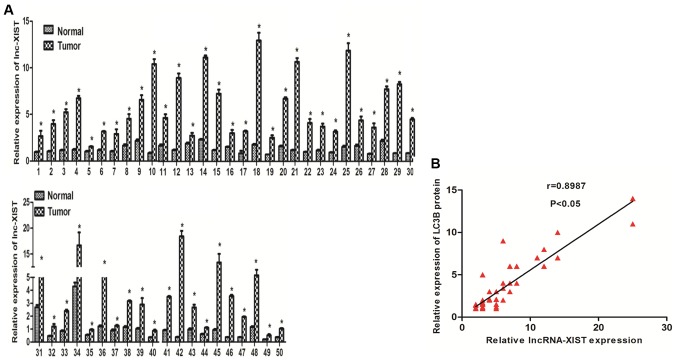
Previous studies have shown that lncRNA-XIST is upregulated in many types of cancers (26,27). To evaluate the role of lncRNA-XIST in NSCLC, we examined the mRNA levels of lncRNA-XIST in paired NSCLC fresh tumor and normal tissues. As shown in Fig. 1A, we found that lncRNA-XIST was highly expressed in the tumor samples compared with the adjacent normal tissues. In addition, we further investigated the association between lncRNA-XIST and clinical features of the NSCLC patients. Our results indicated that lncRNA-XIST is positively associated with TNM stage (Table I). Those results strongly implied that lncRNA-XIST may play an important role in the development and progression of NSCLC. Recent evidence suggests that autophagy is involved in tumor development and progression. To further estimate whether there is a relationship between lncRNA-XIST and autophagy, we found that there was a positive correlation between lncRNA-XIST and LC3B (an autophagy marker) in 50 NSCLC samples using correlation analysis (Fig. 1B), and the protein expression level of LC3B was also found to be associated with TNM stage of the NSCLC cases (Table I) indicating that both lncRNA-XIST and autophagy is involved in the same segment of tumor progression and there is the possibility that the formation and function of autophagy must be tightly regulated to promote NSCLC progression. Furthermore, in accordance with previous research, the LC3B protein level was overexpressed in the NSCLC tumor samples compared with the adjacent normal tissues (data not shown). The above data prompted us to hypothesize that lncRNA-XIST regulates autophagy to promote the progression of NSCLC.
Figure 1.
lncRNA-XIST is highly expressed and associated with autophagy marker LC3B in NSCLC. (A) Analysis of lncRNA-XIST mRNA expression in 50 paired NSCLC tumor and adjacent normal tissues. (B) Spearmans rank correlation coefficient was used to examine the correlation between lncRNA-XIST and LC3B in 50 NSCLC tumor samples. *P<0.05.
Table I.
Correlation between clinicopathological characteristics and expression of lncRNA-XIST and LC3B in the NSCLC patients (n=50).
| Relative lncRNA-XIST expression | Relative LC3B expression | |||||
|---|---|---|---|---|---|---|
| Characteristics | High | Low | P-value | High | Low | P-value |
| Age (years) | 0.563 | 0.782 | ||||
| <45 | 15 (55.6) | 11 (47.8) | 14 (51.9) | 13 (56.5) | ||
| ≥45 | 12 (44.4) | 12 (52.2) | 13 (48.1) | 10 (43.5) | ||
| Sex | 0.773 | 0.783 | ||||
| Female | 12 (42.9) | 8 (36.4) | 12 (54.5) | 14 (50) | ||
| Male | 16 (57.1) | 14 (63.6) | 10 (45.5) | 14 (50) | ||
| Tumor size (cm) | 0.158 | 0.571 | ||||
| ≥5 | 16 (69.6) | 13 (48.1) | 13 (56.5) | 12 (44.4) | ||
| <5 | 7 (30.4) | 14 (51.9) | 10 (39.1) | 15 (55.6) | ||
| LNM | 0.156 | 0.272 | ||||
| Positive | 16 (61.5) | 9 (37.5) | 14 (60.9) | 12 (44.4) | ||
| Negative | 10 (38.5) | 15 (62.5) | 9 (39.1) | 15 (55.6) | ||
| TNM stage | 0.045 | 0.044 | ||||
| I–II | 7 (28) | 15 (60) | 8 (38.1) | 20 (69) | ||
| III–IV | 18 (72) | 10 (40) | 13 (61.9) | 9 (31) | ||
Data are expressed as n (%). LNM, lymph node metastasis.
Knockdown of lncRNA-XIST inhibits basal autophagy and autophagic flux in NSCLC cells
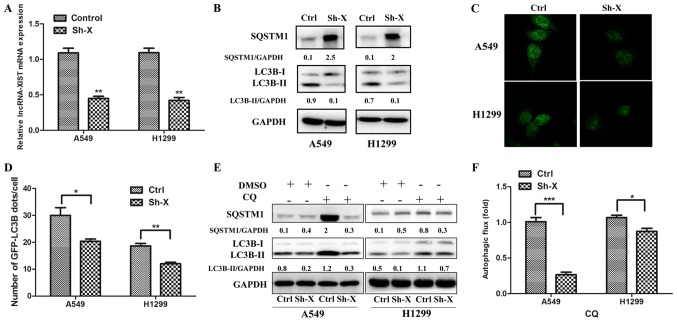
To assess the relationship between lncRNA-XIST and autophagy, we separately co-transfected the A549 and H1299 cells with the shRNA-lncRNA-XIST (Sh-X) plasmid and GFP-LC3B plasmid, the control-shRNA plasmid and GFP-LC3B plasmid. As shown in Fig. 2A, the mRNA levels of lncRNA-XIST were downregulated in the A549 and H1299 cells following transfection of Sh-X as determined by qPCR. Furthermore, using western blot analysis, we verified that knockdown of lncRNA-XIST markedly reduced the LC3B-II protein accumulation and increased SQSTM1 protein expression in the A549 and H1299 cells (Fig. 2B), which was accumulated when autophagic flux was inhibited. In addition, using immunofluorescence technique, we observed that the number of GFP-LC3B autophagic dots/cell was lower in the Sh-X groups than that in the control groups (Fig. 2C and D). To investigate the role of lncRNA-XIST in autophagic flux, we added CQ to the cells for assessing the level of autophgic flux affected by lncRNA-XIST. As shown in Fig. 2E and F, the LC3B-II and SQSTM1 protein accumulation were lower in Sh-X groups than in the control groups following treatment with CQ.
Figure 2.
Knockdown of lncRNA-XIST inhibits basal autophagy and autophagic flux in NSCLC cells. (A) Efficiency of shRNA sequences against lncRNA-XIST was examined by quantitative real-time PCR (qPCR) both in A549 and H1299 cells. Data are shown as the mean ± SD of three independent experiments. (B) A549 and H1299 cells were transfected with shRNA-lncRNA-XIST (Sh-X) or the control plasmid (Ctrl). After 72 h, the expression of SQSTM1, LC3BI, LC3BII and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were detected by western blotting in the cells. (C and D) A549 and H1299 cells were co-transfected with shRNA-lncRNA-XIST (Sh-X) and GFP-LC3B plasmid or the control plasmid (Ctrl) for 72 h, and the number of GFP-LC3B dots were examined by (C) confocal microscopy (D) and quantified for each cell line. Data are showed as mean ± SD of three independent experiments. (E) A549 and H1299 cells were transfected with Sh-X or Ctrl for 72 h and treated with or without 25 µM CQ for 4 h. The expression of LC3B-II and SQSTM1 was examined by western blotting. (F) The autophagic flux was shown by the quantitation of the rate of LC3BII/GAPDH in cells cultured with 25 µM CQ. Data are shown as mean ± SD of three independent experiments. *P<0.05; **P<0.01; ***P<0.001.
lncRNA-XIST regulates autophagy through ATG7
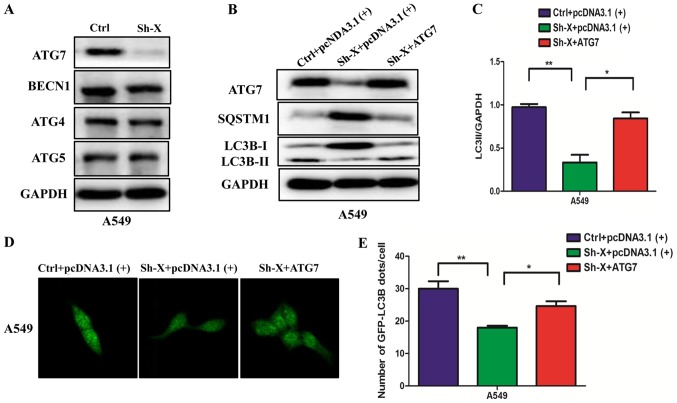
Next, to understand the molecular mechanism of the influence of lncRNA-XIST on autophagy, we examined the changes in the protein levels of different autophagy proteins in A549 cells transfected with Sh-X or the control siRNA. Unexpectedly, among the different autophagy proteins, the ATG7 protein was markedly decreased in the Sh-X group compared with the control group (Fig. 3A). Furthermore, we performed ‘rescue experiment’ to understand whether lncRNA-XIST regulates autophagy through ATG7. We co-transfected the A549 cells with the ATG7 plasmid and shRNA-lncRNA-XIST (Sh-X) plasmid into NSCLC cells for expressing similar ATG7 protein levels compared with the control. Using western blot analysis and immunofluorescence assays, we found that ATG7 rescued the levels of autophagy proteins and the number of GFP-LC3B dots was suppressed by the knockdown lncRNA-XIST (Fig. 3B-E).
Figure 3.
lncRNA-XIST regulates autophagy through ATG7. (A) A549 cells were transfected with Sh-X or the control plasmid (Ctrl) for 72 h, and the expression of ATG7, ATG5, ATG4 and BECN1 was examined by western blotting. GAPDH was used as the normalization control. (B) A549 cells were co-transfected with Sh-X and ATG7 plasmid or control plasmid for 72 h, and the LC3B-II accumulation and SQSTM1 degradation were detected by western blotting. (C) The rate of LC3BII/GAPDH (autophagic flux) is shown. (D and E) A549 cells were co-transfected with Sh-X, GFP-LC3B plasmid and ATG7 plasmid for 72 h and the number of GFP-LC3B dots were examined by (D) confocal microscopy and (E) quantified for each cell. Data are shown as mean ± SD of three independent experiments. *P<0.05; **P<0.01.
lncRNA-XIST inhibits the expression of miR-17 to modulate ATG7
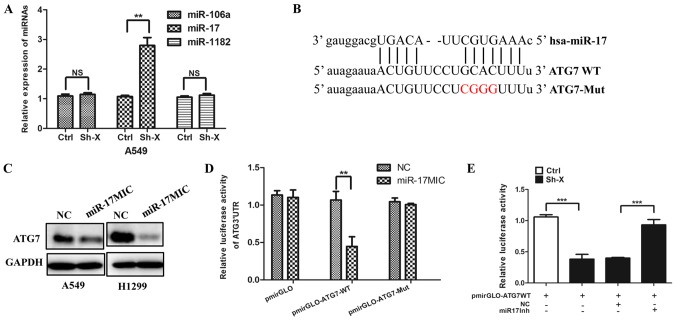
Recently, mounting evidence has demonstrated that the interaction between lncRNAs and miRNAs may be a mechanism of lncRNAs involved in cancer progression (28,29). Thus, using several online bioinformatic prediction websites, we chose the candidate miRNAs: miR-106a, miR-17 and miR-1182, which may be involved in autophagy. To confirm whether lncRNA-XIST regulates the expression of the above miRNAs, we transfected the Sh-X plasmid and the control plasmid into cells, respectively. As shown in Fig. 4A, knockdown of lncRNA-XIST significantly increased the expression of miR-17. Furthermore, we analyzed the potential binding site in the 3′-UTR region of ATG7 by miR-17 using the TargetScan website. The target site of ATG7 is shown in Fig. 4B. In accordance with the above hypothesis, we found that miR-17 mimics downregulated the expression of ATG7 (Fig. 4C) and Dual-Luciferase reporter assays indicated that miRNA-17 can directly target ATG7 (Fig. 4D). More importantly, we further found that knockdown of lncRNA-XIST decreased the luciferase activity of ATG7 3′-UTR and the decreased luciferase activity of ATG7 3′-UTR was reversed by miR-17 inhibitor (Fig. 4E).
Figure 4.
lncRNA-XIST inhibits the expression of miR-17 to modulate ATG7. (A) A549 cells were transfected with Sh-X or the control plasmid (Ctrl) for 72 h, and the expression of miR106, miR-17 and miR-1182 were examined by quantitative real-time PCR. Data are showed as mean ± SD of three independent experiments. (B) Schematic representation of the predicted miR-17 target site and the mutation miR-17 binding site in the 3′-UTR of human ATG7 mRNA. (C) A549 and H1299 cells were transfected with miR-17 mimics (miR-17MIC) and the negative control (NC) for 48 h. The expression of ATG7 was examined by western blotting. GAPDH was used as the normalization control. (D) Luciferase reporter activity for the 3′-UTR of ATG7 and the mutation binding site cloned in pmriGLO and transfected in A549 cells with NC and miR-17MIC, respectively. Luciferase activity was examined after 48 h. (E) The control or knockdown lncRNA-XIST A549 cells were co-transfected the miR-17 inhibitor (miR-17Inh) and NC with pmirGLO, pmirGLO-ATG7WT for 24 h. Then the dual-luciferase reporter assay was performed. NS, not significant. **P<0.01, ***P<0.001.
Knockdown of lncRNA-XIST attenuates the chemoresistance of NSCLC cells via suppression of autophagy
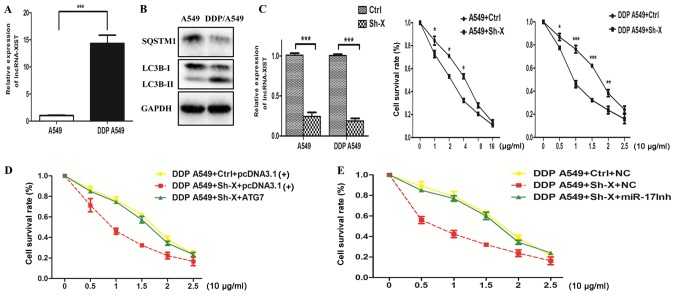
The above data showed that lncRNA-XIST may be involved in NSCLC progression and regulates autophagy activity. Since previous studies have demonstrated that autophagy plays an important role in chemoresistance, this phenomenon prompted us to hypothesize that the association between lncRNA-XIST and autophagy can influence the chemoresistance of NSCLC cells. Thus, we firstly examined the mRNA expression of lncRNA-XIST in both cisplatin (DDP)-resistant DDP A549 cells and parental A549 cells. As shown in Fig. 5A and B, we found that the mRNA expression of lncRNA-XIST was higher in the DDP A549 cells than the parental A549 cells and the autophagy activity was also enhanced in the DDP A549 cells compared to that in the parental A549 cells. Cell viability assays indicated that knockdown of lncRNA-XIST could strongly restore the response to cisplatin of the DDP A549 cells and parental A549 cells (Fig. 5C). More importantly, knockdown of lncRNA-XIST increased DDP-mediated inhibition of cell survival, which was reversed by an miR-17 inhibitor and overexpression of ATG7, respectively (Fig. 5D and E). Taken together, the above results demonstrated that knockdown of lncRNA-XIST attenuated the chemoresistance of NSCLC cells via suppression of autophagy.
Figure 5.
Knockdown of lncRNA-XIST attenuates the chemoresistance of NSCLC cells via suppression of autophagy. (A) Analysis of lncRNA-XIST mRNA expression in parental A549 and DDP-resistance A549 cells (DDP A549). (B) Analysis of the autophagy activity in parental A549 cells and DDP A549 cells. (C) Both DDP A549 and parental A549 cells were transfected with Ctrl and Sh-X plasmid, respectively. After 24 h, DDP A549 cells were treated with cisplatin (0, 5, 10, 15, 20 and 25 µg/ml) and parental A549 cells were with cisplatin (0, 1, 2, 4, 8 and 16 µg/ml) for 48 h, and then cell viability was examined by CCK-8 assays. Data are shown as mean ± SD of three independent experiments. (D and E) DDP A549 cells were co-transfected with Ctrl and Sh-X plasmid or ATG7 plasmid and miR-17 inhibitor (miR-17Inh), respectively, and then, cell viability was examined by CCK-8 assays. Data are shown as the mean ± SD of three independent experiments. *P<0.05; **P<0.01; ***P<0.001.
Discussion
In the present study, we found that overexpression of lncRNA-XIST was positively associated with NSCLC TNM stage. In addition, clinical data showed that the expression of lncRNA-XIST was closely related with the protein levels of LC3B in clinical tumor samples. We revealed a novel molecular mechanism by which lncRNA-XIST regulates autophagy and the potential role in the chemoresistance of NSCLC. lncRNA-XIST may be considered as a vital therapeutic target in NSCLC.
Previous studies have shown that lncRNAs are involved in the progression of many types of cancer and affect disease prognosis (30–32). Recently, lncRNAs have been considered as a novel mechanism underlying the resistance of many types of cancer. Aberrant expression of lncRNAs has been identified in many human cancers or in chemoresistant cancer cells. Targeting lncRNAs may be a promising strategy for patients with chemoresistance (33–37). Consistent with these previous studies, we found that lncRNA-XIST was upregulated in NSCLC tissues compared with that in the adjacent normal samples. Furthermore, a positive correlation between the expression of lncRNA-XIST and NSCLC TNM stage was observed in clinical data, indicating that lncRNA-XIST may be involved in NSCLC progression. However, the exact role and function of lncRNA-XIST in NSCLC progression and chemoresistance remains unclear.
Autophagy, a conserved metabolic process for cell homeostasis, has been verified as a protective mechanism for chemoresistance of many cancer cells. In response to chemotherapy, autophagy is activated as a protective way for maintaining cancer cell survival (38,39). For example, Xiong et al and colleagues demonstrated that overexpression of lncRNA-HULC attenuated the chemosensitivity of HCC cells by upregulation of autophagy (25). In the present study, we found that knockdown of lncRNA-XIST strongly decreasedautophagy activity and the levels of ATG7, an important autophagic protein. Rescue experiments further demonstrated that the regulation by lncRNA-XIST of autophagy depends on ATG7. To ascertain the exact molecular mechanism underlying the regulation of autophagy by lncRNA-XIST, we used online biologic websites, and found that miR-106a, miR-17 and miR-1182 may be the best target candidates of lncRNA-XIST. Dual-Luciferase reporter assays showed that miR-17 directly targets ATG7, suggesting that lncRNA-XIST may regulate autophagy via the miR-17/ATG7 signaling pathway. It is known that autophagy plays a vital role in chemoresistance and inhibition autophagy can enhance the chemosensitivity of cancer cells. These results prompted us to hypothesize whether lncRNA-XIST effects the chemoresistance of cancer cells by regulation of autophagy. We found that the expression level of lncRNA-XIST was markedly increased in DDP-resistant A549 cells compared with that in the corresponding parental A549 cells. Furthermore, cell viability assays showed that knockdown of lncRNA-XIST restored the chemosensitivity of DDP-resistant A549 cells to cisplatin. In contrast, overexpression of ATG7 could reverse the influence of the knockdown of lncRNA-XIST on the chemosensitivity of DDP-resistant A549 cells to cisplatin, indicating that lncRNA-XIST modulates the chemoresistance of cancer cells by regulation of autophagy. Of course, further research should be focused on other molecular mechanisms involved in chemoresistance regulated by lncRNA-XIST in NSCLC.
In summary, our present data showed that dysregulation of lncRNA-XIST plays a vital role in NSCLC progression and cisplatin resistance of NSCLC cells, indicating that its overexpression may confer increased cisplatin chemoresistance via autophagy induced by the miR-17/ATG7 signaling pathway. Taken together, this study suggests that lncRNA-XIST may be act as a potential marker of a poor response to cisplatin of NSCLC and a novel therapeutic target for NSCLC chemotherapy.
Acknowledgements
The present study was supported by grants from the National Natural Science Foundation (no. 81301505).
Glossary
Abbreviations
- NSCLC
non-small cell lung cancer
- lncRNA
long non-coding RNA
- CQ
chloroquine
- CCK-8
Cell Counting Kit-8
- 3′-UTR
3′-untranslated region
- ATG7
autophagy associated gene 7
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
References
- 1.Hirsch FR, Suda K, Wiens J, Bunn PA., Jr New and emerging targeted treatments in advanced non-small-cell lung cancer. Lancet. 2016;388:1012–1024. doi: 10.1016/S0140-6736(16)31473-8. [DOI] [PubMed] [Google Scholar]
- 2.Maas KW, El Sharouni SY, Smit EF, Schramel FM. Sequencing chemotherapy, radiotherapy and surgery in combined modality treatment of stage III nonsmall cell lung cancer. Curr Opin Pulm Med. 2007;13:297–304. doi: 10.1097/MCP.0b013e32819f834a. [DOI] [PubMed] [Google Scholar]
- 3.Richard PJ, Rengan R. Oligometastatic non-small-cell lung cancer: Current treatment strategies. Lung Cancer (Auckl) 2016;7:129–140. doi: 10.2147/LCTT.S101639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosell R, Karachaliou N. Lung cancer in 2014: Optimizing lung cancer treatment approaches. Nat Rev Clin Oncol. 2015;12:75–76. doi: 10.1038/nrclinonc.2014.225. [DOI] [PubMed] [Google Scholar]
- 5.Woods NT, Monteiro AN, Thompson ZJ, Amankwah EK, Naas N, Haura EB, Beg AA, Schabath MB. Interleukin polymorphisms associated with overall survival, disease-free survival, and recurrence in non-small cell lung cancer patients. Mol Carcinog. 2015;54(Suppl 1):E172–E184. doi: 10.1002/mc.22275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Serghiou S, Kyriakopoulou A, Ioannidis JP. Long noncoding RNAs as novel predictors of survival in human cancer: A systematic review and meta-analysis. Mol Cancer. 2016;15:50. doi: 10.1186/s12943-016-0535-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weng M, Wu D, Yang C, Peng H, Wang G, Wang T, Li X. Noncoding RNAs in the development, diagnosis, and prognosis of colorectal cancer. Transl Res. 2016;181:108–120. doi: 10.1016/j.trsl.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borensztein M, Syx L, Ancelin K, Diabangouaya P, Picard C, Liu T, Liang JB, Vassilev I, Galupa R, Servant N. Xist-dependent imprinted X inactivation and the early developmental consequences of its failure. Nat Struct Mol Biol. 2017;24:226–233. doi: 10.1038/nsmb.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo JY, White E. Autophagy, metabolism, and cancer. Cold Spring Harb Symp Quant Biol. 2017;81:73–78. doi: 10.1101/sqb.2016.81.030981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li D, Liu X, Zhou J, Hu J, Zhang D, Liu J, Qiao Y, Zhan Q. LncRNA HULC modulates the phosphorylation of YB-1 through serving as a scaffold of ERK and YB-1 to enhance hepatocarcinogenesis. Hepatology. 2016;65:1612–1627. doi: 10.1002/hep.29010. [DOI] [PubMed] [Google Scholar]
- 12.Li L, Chen H, Gao Y, Wang YW, Zhang GQ, Pan SH, Ji L, Kong R, Wang G, Jia YH, et al. Long noncoding RNA MALAT1 promotes aggressive pancreatic cancer proliferation and metastasis via the stimulation of autophagy. Mol Cancer Ther. 2016;15:2232–2243. doi: 10.1158/1535-7163.MCT-16-0008. [DOI] [PubMed] [Google Scholar]
- 13.Liu Z, Wei X, Zhang A, Li C, Bai J, Dong J. Long non-coding RNA HNF1A-AS1 functioned as an oncogene and autophagy promoter in hepatocellular carcinoma through sponging hsa-miR-30b-5p. Biochem Biophys Res Commun. 2016;473:1268–1275. doi: 10.1016/j.bbrc.2016.04.054. [DOI] [PubMed] [Google Scholar]
- 14.Xue X, Yang YA, Zhang A, Fong KW, Kim J, Song B, Li S, Zhao JC, Yu J. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene. 2016;35:2746–2755. doi: 10.1038/onc.2015.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi SJ, Wang LJ, Yu B, Li YH, Jin Y, Bai XZ. LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget. 2015;6:11652–11663. doi: 10.18632/oncotarget.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang XW, Bu P, Liu L, Zhang XZ, Li J. Overexpression of long non-coding RNA PVT1 in gastric cancer cells promotes the development of multidrug resistance. Biochem Biophys Res Commun. 2015;462:227–232. doi: 10.1016/j.bbrc.2015.04.121. [DOI] [PubMed] [Google Scholar]
- 17.Liu Z, Sun M, Lu K, Liu J, Zhang M, Wu W, De W, Wang Z, Wang R. The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregualtion of p21WAF1/CIP1 expression. PLoS One. 2013;8:e77293. doi: 10.1371/journal.pone.0077293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J, Wan L, Lu K, Sun M, Pan X, Zhang P, Lu B, Liu G, Wang Z. The long noncoding RNA MEG3 contributes to cisplatin resistance of human lung adenocarcinoma. PLoS One. 2015;10:e0114586. doi: 10.1371/journal.pone.0114586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tantai J, Hu D, Yang Y, Geng J. Combined identification of long non-coding RNA XIST and HIF1A-AS1 in serum as an effective screening for non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8:7887–7895. [PMC free article] [PubMed] [Google Scholar]
- 20.Hu X, Xuan Y. Bypassing cancer drug resistance by activating multiple death pathways - a proposal from the study of circumventing cancer drug resistance by induction of necroptosis. Cancer Lett. 2008;259:127–137. doi: 10.1016/j.canlet.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 21.Levine B, Klionsky DJ. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/S1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 22.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferraresi A, Phadngam S, Morani F, Galetto A, Alabiso O, Chiorino G, Isidoro C. Resveratrol inhibits IL-6-induced ovarian cancer cell migration through epigenetic up-regulation of autophagy. Mol Carcinog. 2017;56:1164–1181. doi: 10.1002/mc.22582. [DOI] [PubMed] [Google Scholar]
- 25.Xiong H, Ni Z, He J, Jiang S, Li X, He J, Gong W, Zheng L, Chen S, Li B, et al. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene. 2017;36:3528–3540. doi: 10.1038/onc.2016.521. [DOI] [PubMed] [Google Scholar]
- 26.Ma L, Zhou Y, Luo X, Gao H, Deng X, Jiang Y. Long non-coding RNA XIST promotes cell growth and invasion through regulating miR-497/MACC1 axis in gastric cancer. Oncotarget. 2017;8:4125–4135. doi: 10.18632/oncotarget.13670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao Y, Ma J, Xue Y, Wang P, Li Z, Liu J, Chen L, Xi Z, Teng H, Wang Z, et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 2015;359:75–86. doi: 10.1016/j.canlet.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 28.Batista PJ, Chang HY. Long noncoding RNAs: Cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ulitsky I, Bartel DP. lincRNAs: Genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du Y, Kong G, You X, Zhang S, Zhang T, Gao Y, Ye L, Zhang X. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem. 2012;287:26302–26311. doi: 10.1074/jbc.M112.342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hafner SJ, Talvard TG, Lund AH. Long noncoding RNAs in normal and pathological pluripotency. Semin Cell Dev Biol. 2016 doi: 10.1016/j.semcdb.2016.07.011. [DOI] [PubMed] [Google Scholar]
- 32.Li SP, Xu HX, Yu Y, He JD, Wang Z, Xu YJ, Wang CY, Zhang HM, Zhang RX, Zhang JJ, et al. LncRNA HULC enhances epithelial-mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget. 2016;7:42431–42446. doi: 10.18632/oncotarget.9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bian Z, Jin L, Zhang J, Yin Y, Quan C, Hu Y, Feng Y, Liu H, Fei B, Mao Y, et al. LncRNA-UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204-5p. Sci Rep. 2016;6:23892. doi: 10.1038/srep23892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee H, Kim C, Ku JL, Kim W, Yoon SK, Kuh HJ, Lee JH, Nam SW, Lee EK. A long non-coding RNA snaR contributes to 5-fluorouracil resistance in human colon cancer cells. Mol Cells. 2014;37:540–546. doi: 10.14348/molcells.2014.0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Özeş AR, Miller DF, Özeş ON, Fang F, Liu Y, Matei D, Huang T, Nephew KP. NF-κB-HOTAIR axis links DNA damage response, chemoresistance and cellular senescence in ovarian cancer. Oncogene. 2016;35:5350–5361. doi: 10.1038/onc.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi K, Yan IK, Wood J, Haga H, Patel T. Involvement of extracellular vesicle long noncoding RNA (linc-VLDLR) in tumor cell responses to chemotherapy. Mol Cancer Res. 2014;12:1377–1387. doi: 10.1158/1541-7786.MCR-13-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Zhang D, Wu K, Zhao Q, Nie Y, Fan D. Long noncoding RNA MRUL promotes ABCB1 expression in multidrug-resistant gastric cancer cell sublines. Mol Cell Biol. 2014;34:3182–3193. doi: 10.1128/MCB.01580-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jing Z, Han W, Sui X, Xie J, Pan H. Interaction of autophagy with microRNAs and their potential therapeutic implications in human cancers. Cancer Lett. 2015;356:332–338. doi: 10.1016/j.canlet.2014.09.039. [DOI] [PubMed] [Google Scholar]
- 39.Liu JJ, Lin M, Yu JY, Liu B, Bao JK. Targeting apoptotic and autophagic pathways for cancer therapeutics. Cancer Lett. 2011;300:105–114. doi: 10.1016/j.canlet.2010.10.001. [DOI] [PubMed] [Google Scholar]