Abstract
Accumulating evidence indicates that mitotic checkpoint serine/threonine kinase B (BUB1B) plays a critical role in multiple types of cancer. However, the biological function and molecular regulatory mechanism of BUB1B in glioblastoma (GBM) remain unclear. In the present study, we identified that BUB1B expression was enriched in GBM tumors and was functionally required for tumor proliferation both in vitro and in vivo. Clinically, BUB1B expression was associated with poor prognosis in GBM patients and BUB1B-dependent radioresistance in GBM was decreased by targeting BUB1B via shRNAs. Mechanistically, forkhead box protein M1 (FOXM1) transcriptionally regulated BUB1B expression by binding to and then activating the BUB1B promoter. Therapeutically, we found that FOXM1 inhibitor attenuated tumorigenesis and radioresistance of GBM both in vitro and in vivo. Altogether, BUB1B promotes tumor proliferation and induces radioresistance in GBM, indicating that BUB1B could be a potential therapeutic target for GBM.
Keywords: glioblastoma, BUB1B, FOXM1, radioresistance
Introduction
Glioblastoma (GBM) is the most common intra-parenchymal and lethal brain cancer while GBM patients are left behind without any curable therapy to date (1,2). The current standard therapy for GBM involves maximal surgical resection followed by radiotherapy and tomozolomide (TMZ) chemotherapy. However, this treatment strategy fails to eliminate a subset of tumor cells that escape from therapeutic insults and finally results in tumor recurrence, leading to reduced survival in these patients (3,4). Among these treatments, radiation (IR) plays a major role in the treatment of GBM patients. Factually, the efficacy of this therapeutic modality is often limited by the occurrence of radioresistance (5). However, the molecular mechanisms responsible for the radioresistance of human GBM are not yet clear.
Mitotic checkpoint serine/threonine kinase B (BUB1B) is the mammalian homolog of yeast Mad3, but differs significantly since BUB1B has a kinase domain that is absent in Mad3 (6). A reecent study showed that complete deletion of BUB1B in the mouse germline results in early embryonic death (7). Additionally, BUB1B− (haplo-insufficient) mice display increased megakaryopoiesis and increased chromosome instability, as well as susceptibility to cancer (8,9). Moreover, reduction in the BUB1B level or inhibition of BUB1B kinase activity in human cancer cells results in massive chromosome loss and apoptotic cell death (10). Another study recently identified BUB1B as an essential element for the growth and survival of rhabdomyosarcoma cells using a bar-coded, tetracycline-inducible shRNA library screening and found that suppression of forkhead box protein M1 (FOXM1) either by shRNAs or FOXM1 inhibitor siomycin A resulted in reduction of BUB1B expression and decreased cell growth (11). FOXM1 is a transcription factor and is well known to play an essential role in the regulation of a wide spectrum of biological processes including cell proliferation, cell cycle progression, cell differentiation, DNA damage repair, tissue homeostasis, angiogenesis and apoptosis (12,13). Recent evidence shows that FOXM1 expression is elevated in human GBM and is essential for maintaining neural, progenitor and GBM stem cells as a master regulator of metastasis (14). In addition, other studies indicate that radiation induces stimulation of FOXM1 expression which is dependent on STAT3 phosphorylation (15). Moreover, the FOXM1-RFC5 axis has been proven to mediate TMZ resistance, and thiostrepton may serve as a potential therapeutic agent against TMZ resistance in glioma cells (16). Based on these findings, it has been proven that BUB1B plays an essential role in tumor proliferation and is correlated with poor patient prognosis in multiple types of cancer including breast, gastric, colorectal and prostate cancer (17–20). However, the functional role and mechanism of FOXM1/BUB1B signaling in therapeutic resistance remain unclear.
In the present study, we identified that BUB1B expression was enriched in GBM tumors and was functionally required for tumor proliferation both in vitro and in vivo. Clinically, BUB1B expression was associated with poor prognosis in GBM patients and BUB1B-dependent radioresistance in GBM was decreased by targeting BUB1B via shRNAs. Mechanistically, FOXM1 transcriptionally regulated BUB1B expression by binding to and then activating the BUB1B promoter. Therapeutically, we found that FOXM1 inhibitor attenuated tumorigenesis and radioresistance of GBM both in vitro and in vivo. Collectively, BUB1B promotes tumor proliferation and induces radioresistance in GBM, implying that BUB1B is a potential therapeutic target for GBM.
Materials and methods
Ethics
Protocols for the usage of experimental animals (nude mice), human samples and cell lines in the present study were approved by the Scientific Ethics Committee of Shaanxi Provincial People's Hospital (Xi'an, China).
Reagents and antibodies
The following reagents and primary antibodies were used in the present study: DMEM-F12, fetal bovine serum (FBS), Accutase solution (Sigma-Aldrich, Billerica, MA, USA), alamarBlue solution (all from Thermo Fisher Scientific, Waltham, MA, USA), RIPA buffer, phosphatase inhibitor cocktail, protease inhibitor cocktail (all from Sigma-Aldrich), Bradford protein assay (Bio-Rad Laboratories, Hercules, CA, USA), albumin from bovine serum (BSA; Sigma-Aldrich), PageRuler Plus prestained protein (Thermo Fisher Scientific), iScript Reverse Transcription Supermix for qRT-PCR (Bio-Rad Laboratories), BUB1B overexpression lentivirus (pLenti-GIII-CMV-GFP-2A-Puro; BC018739), shBUB1B lentivirus (piLenti-siRNA-GFP, iV002138), shFOXM1 lentivirus (piLenti-siRNA-GFP, iV008091) [all from Applied Biological Materials (abm) Inc., Richmond, BC, Canada], siomycin A (ALX-380-243-MC05; Enzo Life Sciences, Farmingdale, NY, USA), Alexa Fluor® 488 Annexin V/Dead Cell Apoptosis kit (Thermo Fisher Scientific). Anti-BUB1B (rabbit; #4116; Cell Signaling Technology, Inc., Danvers, MA, USA), anti-FOXM1 (rabbit; ab184637; Abcam, Cambridge, MA, USA), and β-actin (A5316; mouse; Sigma-Aldrich) were used.
In vitro cell cultures
GBM cells U87, U251 and U138 and HEK-293T cells were provided by Shaanxi Provincial People's Hospital. Cell lines were cultured in DMEM/F12 medium containing 10% FBS supplement (vol %). The culture medium was replaced every 5–10 days. Normal human astrocytes were used as a control sample in the present study. Radiation for in vitro GBM cells was performed using Thermo Fisher t ICx radiation equipment according to the manufacturer's protocol.
In vitro cell proliferation assay
GBM cells were transduced with the BUB1B or FOXM1 shRNA lentivirus for 3 days, and the cells were dissociated into a single-cell suspension with Accutase. After the live cell number was measured by the Trypan blue method with the Countess Automated Cell Counter (Thermo Fisher Scientific), 200 µl of the cell suspension containing 2,000 cells was added into 96-well plates. The cell number was evaluated by alamarBlue according to the manufacturer's protocol every 2 days after seeding.
Luciferase assays of the BUB1B promoter
We amplified the −350/−216 bp region of the human BUB1B promoter from RD genomic DNA by PCR using the primers containing the restriction sites XhoI and HindIII, respectively. These nucleotide sequences of primers were as follows: sense primer, 5′-CTCGAGTAAGCCTGCTGCACTTCCAC-3′ and antisense primer, 5′-AAGCTTCTCCTCCGTGCTCTCGCGTCT-3′ for the −350/−216 bp fragment. These fragments were ligated into a TOPO® TA vector (Thermo Fisher Scientific), and then cloned into a pGL4.18 firefly luciferase reporter vector (Promega, Madison, WI, USA) after digestion with XhoI and HindIII. PCR-based site-directed mutagenesis was performed to generate a single point mutation in FOXM1 binding sites of the BUB1B promoter region using QuickChange II XL Site-Directed Mutagenesis kit (Agilent Technologies, La Jolla, CA, USA). Sequences were verified by DNA sequencing. HEK293T or U87 cells were transfected using Lipofectamine™ 2000 with 1 µg of empty vector, or BUB1B promoter luciferase reporter and cultured for 2 weeks. Luciferase assays were performed using Bright-Glo reagent (Promega) on the Victor3 counter (PerkinElmer, Walthan, MA, USA).
RNA isolation and quantitative real-time PCR
RNA was isolated using RNeasy Mini kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. RNA concentration was determined using NanoDrop 2000c (Thermo Fisher Scientific). cDNA was synthesized using iScript Reverse Transcription Supermix for qRT-PCR according to the manufacturer's protocol. The reverse-transcribed cDNA was analyzed by quantitative RT-PCR (qRT-PCR), GAPDH or 18S was used as an internal control. Each qRT-PCR included a 10 µl reaction mixture/well that included 2.5 µl cDNA, 0.5 µl forward primer (0.5 µM), 0.5 µl reverse primer (0.5 µM), 1.5 µl of DNase/RNase-free distilled water, and 5 µl SYBR-Green reagent (Qiagen). The following cycles were performed during DNA amplification: 94°C for 2 min, 40 cycles of 94°C (30 sec), 60°C (30 sec), and 72°C (40 sec). The primer sequences were as follows: FOXM1 forward, TCTCCTCTTTCCCTGGTCCT and FOXM1 reverse, ATAGCAAGCGAGTCCGCATT; BUB1B forward, CCAGGCTTTCTGGTGCTTAG and BUB1B reverse, GTGCTTCCCAGTTTCACTCC; GAPDH forward, CGGAG TCAACGGATTTGGTCGTAT and GAPDH reverse, AGCCT TCTCCATGGTGGTGAAGAC; 18S forward, GGCCCTGT AATTGGAATGAGTC and 18S reverse, CCAAGATCCAACT ACGAGCTT.
Western blotting
The cell lysates were prepared in RIPA buffer containing 1% protease and 1% phosphatase inhibitor cocktail on ice. Protein concentrations were determined using the Bradford method. Equal amounts of protein lysates (10 µg/lane) were fractionated on NuPAGE Novex 4–12% Bis-Tris Protein gel and transferred to a polyvinylidene difluoride (PVDF) membrane (both from Thermo Fisher Scientific). Subsequently, the membranes were blocked with 5% skimmed milk for 1 h and then treated with the relevant antibody at 4°C overnight. Protein expression was visualized with ECL Western Blotting System (GE Healthcare Life Sciences, Pittsburgh, PA, USA). β-actin served as a loading control.
Flow cytometric analysis
Cells were harvested after the incubation period and washed in cold phosphate-buffered saline (PBS) for 3 times. The washed cells were re-centrifuged (from step 2), the supernatant was discarded and 5×105 cells were suspended in 100 µl 1X Annexin-binding buffer. Alexa Fluor® 488 Annexin V (5 µl) was added and 1 µl 100 µg/ml propidium iodide (PI) working solution was prepared according to the protocol. The cells were incubated at room temperature for 15 min, and then 400 µl 1X Annexin-binding buffer was added, mixed gently and the samples were kept on ice. The stained cell population was analyzed by measuring the fluorescence emission at 530 and 575 nm with 488 nm excitation.
In vivo intracranial xenograft tumor models
Six-week-old nude mice (female) were purchased from Xi'an Jiaotong University and were used for GBM intracranial implantation. All animal experiments were carried out at Shaanxi Provincial People's Hospital. The GBM suspension (1×105 cells in 5 µl of PBS) transduced with the non-target or shBUB1B lentivirus was injected into the brains of nude mice after anesthesia. At least 5 mice were used for each group. Drug treatment was carried out through tail vein injection, and started from 5 days after tumor cells were implanted. Mice were monitored once a day for symptoms related to tumor growth including an arched back, unsteady gait, paralysis of legs and body weight loss. Mice were euthanized by an overdose of anesthesia of ketamine and xylazine after a total body weight loss of 40% or severe symptoms were observed.
Statistical analysis
All data are presented as mean ± SD. The number of replicates for each experiment is stated in the figure legends. Statistical differences between 2 groups were evaluated by two-tailed t-test. Comparison among multiple groups was performed by one-way analysis of variance (one-way ANOVA) followed by Dunnett's post test. The statistical significance of the Kaplan-Meier survival plot was determined by log-rank analysis. Flow cytometric results were analyzed with χ2 analysis. Statistical analysis was performed by Prism 6 (GraphPad Prism 5.0; GraphPad Software, Inc., La Jolla, CA, USA), unless mentioned otherwise in the figure legends. P<0.05 was considered to indicate a statistically significant result.
Results
BUB1B expression is highly enriched in GBM cells
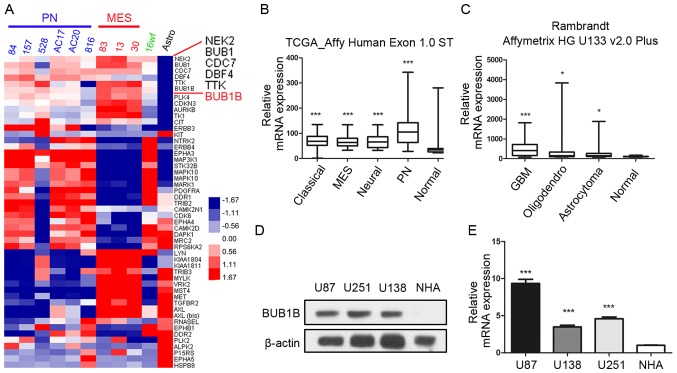
In the present study, we first sought to characterize the role of BUB1B in GBM tumors. To this end, we analyzed the microarray data from a microarray database published in 2013 (21) and found that BUB1B was one of the most upregulated kinase-encoding genes in GBM samples compared to normal human astrocyte (NHA) cells (Fig. 1A). Additionally, we found that BUB1B expression was significantly enriched in all the 4 subgroups of GBM according to The Cancer Genome Atlas (TCGA) database [classical, mesenchymal (MEN), neural and proneural (PN)] compared with non-tumor tissue (Fig. 1B). Furthermore, the data from the Rembrandt database also demonstrated that BUB1B mRNA expression in GBM was significantly higher than that in non-tumor and other types of glioma (Fig. 1C). To confirm this, western blotting using 3 GBM cell lines (U87, U251 and U138) and NHAs was performed. The results exhibited higher expression of BUB1B protein in the GBM cells (Fig. 1D). Similar to the protein expression, qRT-PCR data demonstrated the same result (Fig. 1E). Taken together, these data indicated that BUB1B is preferentially expressed in GBM.
Figure 1.
BUB1B expression is highly enriched in GBM cells. (A) Genome-wide transcriptome microarray analysis from Mao's database (21) showed that BUB1B was one of the most upregulated kinase encoding genes in GBM samples compared to normal tissue. (B) Analysis of the TCGA database indicated that BUB1B was highly expressed in all the 4 subgroups of GBM compared with the non-tumor tissue (***P<0.001, with one-way ANOVA followed by Dunnett's post test). (C) Analysis of the Rembrandt database demonstrated that BUB1B expression in GBM was significantly higher than that in non-tumor and other types of glioma (*P<0.05, ***P<0.001, with one-way ANOVA followed by Dunnett's post test). (D) Western blotting indicated that BUB1B protein expression was enriched in 3 GBM cell lines compared with that noted in normal astrocytes. β-actin served as a control. (E) qRT-PCR showed that BUB1B mRNA expression was elevated in 3 GBM cell lines (U138, U251 and U87) compared with that noted in normal astrocytes (NHA) (n=3, ***P<0.001, with one-way ANOVA followed by Dunnett's post test). MEN, mesenchymal; PN, proneural.
BUB1B is functionally required for GBM proliferation both in vitro and in vivo
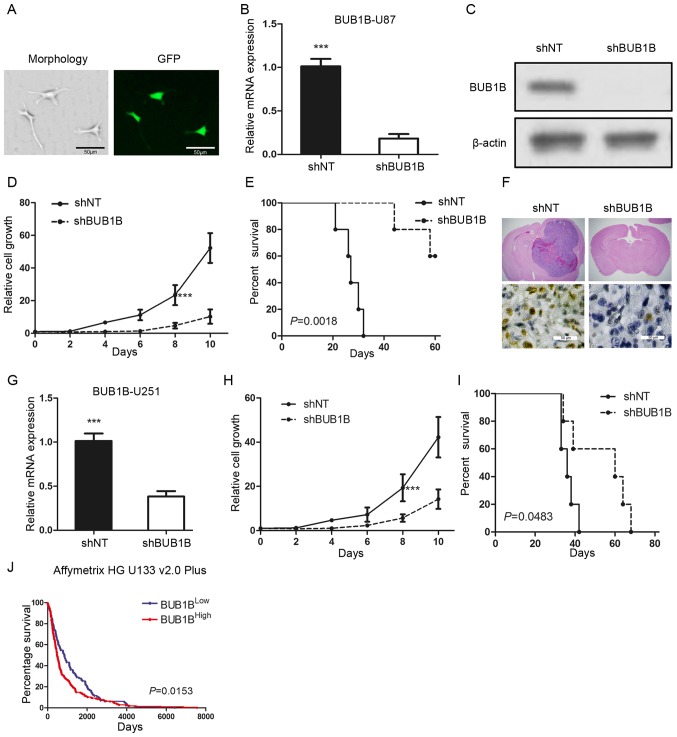
To examine the biological role of BUB1B in GBM, we selected one in vitro GBM cell line (U87) which had a relatively higher BUB1B expression and transduced the cells with either a lentiviral shRNA clone for BUB1B (shBUB1B) or a non-targeting shRNA (shNT). The efficiency for lentiviral infection was confirmed by GFP fluorescence (Fig. 2A). qRT-PCR and western blot analysis indicated that BUB1B expression was significantly reduced in the shBUB1B U87 cells (Fig. 2B and C). Furthermore, in vitro cell growth kinetics of the shBUB1B-transfected U87 cells was proportionally diminished to the reduction levels of BUB1B (Fig. 2D). Next, we investigated the effect of BUB1B knockdown on in vivo tumor formation. To this end, we used U87 cell-derived mouse intracranial tumor models. A longer survival was observed in the mouse xenografted shBUB1B group, highlighting potent antitumorigenic effects of BUB1B knockdown (Fig. 2E). We then harvested U87-implanted mouse brains and H&E or IHC staining for BUB1B was performed. The results indicated that the mice xenografted with shNT-transfected U87 cells rapidly formed lethal hypervascular GBM-like tumors while BUB1B expression was decreased in the shBUB1B-transfected U87 cell xenografted mouse brains (Fig. 2F). Similar results were obtained when using U251 GBM cells (Fig. 2G-I). Moreover, the survival analysis from the Rembrandt database indicated that overall survival was significantly longer in the BUB1BLow expression group than that noted in the BUB1BHigh expression group (Fig. 2J). Collectively, these findings indicate that BUB1B is required for GBM proliferation both in vitro and in vivo.
Figure 2.
BUB1B is functionally required for GBM proliferation both in vitro and in vivo. (A) Fluorescence image shows that the pGFP-shBUB1B lentivirus was successfully transfected into U87 cells. (B) qRT-PCR of U87 cells transduced with shNT or shBUB1B (n=3, ***P<0.001, with t-test). (C) Western blotting of U87 cells transduced with shNT or shBUB1B. β-actin served as a control. (D) In vitro cell growth assay showed that shBUB1B reduced cell proliferation of U87 cells (n=6, ***P<0.001, with one-way ANOVA). (E) Kaplan-Meier analysis of nude mice harboring intracranial tumors derived from U87 cells transduced with shNT or shBUB1B (n=5, P=0.0018, with log-rank test). (F) Representative images of H&E (upper panel) or BUB1B (lower panel) stained mouse brain section after the intracranial transplantation of U87 cells transduced with shNT or shBUB1B. (G) qRT-PCR of U251 cells transduced with shNT or shBUB1B (n=3, ***P<0.001, with t-test). (H) In vitro cell growth assay showed that hBUB1B reduced cell proliferation of U251 cells (n=6, ***P<0.001, with one-way ANOVA). (I) Kaplan-Meier analysis of nude mice harboring intracranial tumors derived from U251 cells transduced with shNT or shBUB1B (n=5, P=0.0483, with log-rank test). (J) Analysis of the Rembrandt data revealed an inverse correlation between BUB1B expression and post-surgical survival of GBM patients (P=0.0153, with log-rank test).
FOXM1-binding region is critical for the activation of the BUB1B promoter
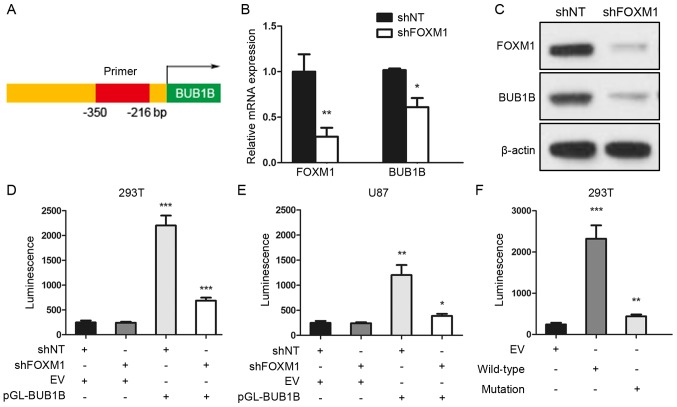
To further assess the regulatory mechanism of BUB1B in GBM, we performed a luciferase reporter assay with constructs driven by a human BUB1B promoter (Fig. 3A). qRT-PCR and western blot analyses indicated that BUB1B expression was significantly reduced after FOXM1 inhibition via shRNA (Fig. 3B and C). Thus, we performed luciferase assay to measure the BUB1B promoter activity with reduced expression of FOXM1 in HEK293T or U87 cells. The region (−350/−216 bp) of the human BUB1B promoter was constructed and cloned into a luciferase reporter vector based on a previous study (11). Then, empty vector (EV) or reporter for BUB1B promoter (pGL-BUB1B) was transduced into HEK293T or U87 cells followed by shNT or shFOXM1 transfection. As expected, shRNA-mediated knockdown of FOXM1 resulted in a marked decrease in transcriptional activity of the FOXM1-binding region in both HEK293T and U87 cells (Fig. 3D and E). Moreover, we transiently transfected either EV or BUB1B promoter wild-type (WT) or BUB1B promoter mutation type (mutation) into HEK293T cells, and luciferase assay showed a marked decrease in transcriptional activity of the mutated BUB1B promoter in the HEK293T cells (Fig. 3F). Altogether, these data suggest that the FOXM1-binding region was critical for BUB1B promoter activation in GBM cells.
Figure 3.
FOXM1-binding region is critical for the activation of the BUB1B promoter. (A) Schematic drawing of the promoter region of human BUB1B. (B) qRT-PCR for FOXM1 and BUB1B expression in U87 cells transduced with shNT or shFOXM1 (n=3, *P<0.05, **P<0.01, with t-test). (C) Western blotting of U87 cells transduced with shNT or shFOXM1. β-actin served as a control. (D and E) Luciferase activity assay showed that shRNA-mediated knockdown of FOXM1 resulted in a marked decrease in transcription activity of the FOXM1-binding region in both (D) HEK293T and (E) U87 cells (n=3, *P<0.05, **P<0.01, ***P<0.001, with one-way ANOVA followed by Dunnett's post test). (F) Luciferase activity assay showed a decrease in transcription activity of mutated BUB1B promoter in the HEK293T cells (n=3, **P<0.01, ***P<0.001, with one-way ANOVA followed by Dunnett's post test).
BUB1B-mediated radioresistance is essential for GBM recurrence
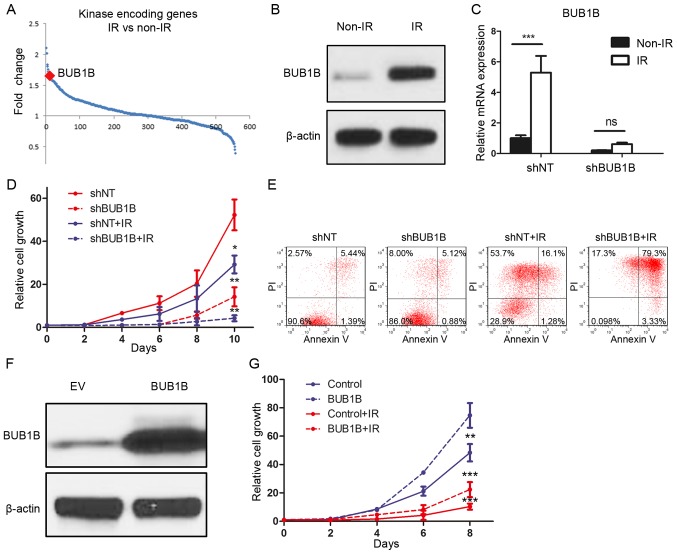
To identify the functional role of BUB1B in the radioresistance of GBM, DNA microarray analysis focusing on 668 kinase-encoding genes in GBM cell lines was performed. The results demonstrated that BUB1B was one of the top genes which was upregulated after radiation of 12 Gy (Fig. 4A). To confirm this, we treated U87 cells with radiotherapy of 12 Gy or without and purified protein at 72 h after treatment. Western blot analysis indicated that the BUB1B expression level was significantly elevated after radiation (Fig. 4B). To investigate whether BUB1B is necessary for GBM cells to maintain radioresistance, we then combined BUB1B knockdown along with radiotherapy. qRT-PCR results showed that the post-radiation elevation of BUB1B was reduced by shBUB1B (Fig. 4C). In vitro cell proliferation was markedly decreased by shBUB1B when combined with radiation (Fig. 4D). Moreover, we performed flow cytometry for apoptosis with Annexin V (AV) antibody and PI using shBUB1B-transduced U87 cells with (12 Gy) or without radiation. The results indicated that the percentages of cells undergoing early (AV+/PI−) and late (AV+/PI+) apoptosis were both markedly increased after radiation when combined with BUB1B knockdown in comparison to radiation alone (Fig. 4E). Finally, BUB1B or EV was overexpressed in U138 cells (Fig. 4F). In vitro cell proliferation assay showed that both cell growth and radioresistance of U138 cells were enhanced by exogenous BUB1B (Fig. 4G). Taken together, BUB1B-dependent radioresistance was essential for GBM tumorigenesis and recurrence after therapeutic treatment.
Figure 4.
BUB1B mediates the radioresistance of GBM. (A) Genome-wide transcriptome microarray analysis showed that BUB1B was one of the most upregulated kinases in GBM cells post-radiation (12 Gy) compared to naïve GBM cells. (B) Western blotting for BUB1B protein expression in U87 cells post-radiation (12 Gy) compared to naïve U87 cells. β-actin served as a control. (C) qRT-PCR showed that the post-radiation elevation of BUB1B was reduced by shBUB1B (n=3; ns, P>0.05; ***P<0.001, with t-test). (D) In vitro cell growth assay for U87 cells pre-transduced with shNT or shBUB1B, and then treated with or without radiation (12 Gy) (n=6; *P<0.05, **P<0.01, with one-way ANOVA). (E) Flow cytometric analysis for apoptosis using U87 cells pre-transduced with shNT or shBUB1B and then treated with or without radiation (12 Gy) (P<0.05, with χ2 analysis). (F) Western blotting for BUB1B protein expression in U138 cells transduced with empty vector or exogenous BUB1B overexpression vector. β-actin served as a control. (G) In vitro cell growth assay for U138 cells pre-transduced with empty vector or exogenous BUB1B overexpression vector and then treated with or without radiation (12 Gy) (n=6; **P<0.01, ***P<0.001, with one-way ANOVA).
FOXM1 inhibitor reduces tumorigenicity and radioresistance of GBM
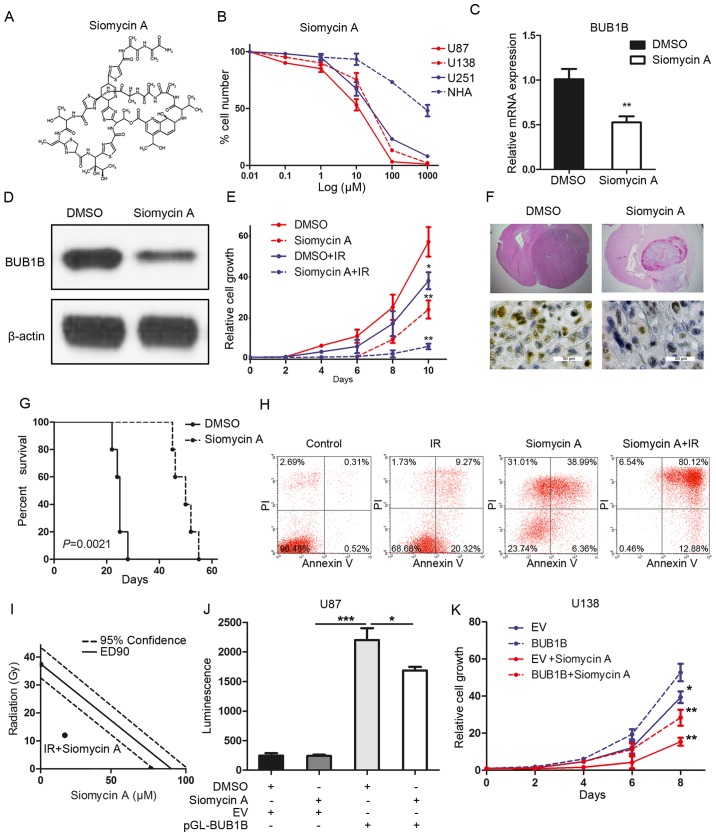
Based on the inhibitory effects of BUB1B silencing on GBM cell proliferation and tumorigenicity, we sought to identify a potential target for the clinical treatment of GBM. To this end, FOXM1 inhibitor siomycin A was used to clarify the effects of BUB1B inhibition on GBM (Fig. 5A). To characterize the efficacy of this inhibitor, we investigated the IC50 value of siomycin A in different GBM cell lines. As expected, in vitro sensitivity of these GBM cells was correlated with the BUB1B expression level, and normal astrocytes showed more resistance to the FOXM1 inhibitor (Fig. 5B). qRT-PCR and western blotting indicated that siomycin A treatment significantly reduced the expression of BUB1B in the U87 cells (Fig. 5C and D). Furthermore, in vitro cell proliferation was significantly decreased by siomycin A when combined with radiation compared with radiotherapy alone (Fig. 5E). More importantly, systemic treatment of the U87-derived mouse model with siomycin A significantly attenuated tumor growth, thereby extending the survival of tumor-bearing mice compared to the vehicle-treated counterparts (Fig. 5F and G). Flow cytometric analysis indicated that the percentage of cells undergoing apoptosis was markedly increased after radiation when combined with siomycin A treatment in comparison to that noted following radiation alone (Fig. 5H). To address whether siomycin A combined with radiation enhances cell apoptosis due to synergism or addictive effects, isobologram analysis was performed. The results indicated that siomycin A (20 µM) increased the radiosensitivity of the U87 cells via a synergistic effect (Fig. 5I). Additionally, luciferase assay showed that the BUB1B promoter activity was decreased by siomycin A treatment in the U87 cells (Fig. 5J). Finally, in vitro cell proliferation assay showed that both siomycin A-mediated reduction in the cell growth of U138 cells could be partially, but not completely, rescued by exogenous BUB1B (Fig. 5K). In conclusion, siomycin A reduced the tumorigenicity and radioresistance of GBM via inhibition of the FOXM1/BUB1B signaling pathway.
Figure 5.
Siomycin A reduces tumorigenicity and radioresistance of GBM. (A) Chemical structure of siomycin A. (B) In vitro cell viability assay for siomycin A with 3 GBM cell lines (U138, U251 and U87) compared with normal astrocytes (NHA) (n=6, P<0.05, with one-way ANOVA). (C) qRT-PCR for BUB1B expression in U87 cells treated with or without siomycin A in U87 cells (n=3, **P<0.01, with t-test). (D) Western blotting for BUB1B expression in U87 cells treated with or without siomycin A. (E) In vitro cell growth assay showed that siomycin A decreased cell proliferation of U87 cells when combined with radiation (n=6, *P<0.05, **P<0.01, with one-way ANOVA). (F) Representative images of H&E (upper panel) or BUB1B (lower panel) stained mouse brain section after the intracranial transplantation of U87 cells and then followed by continuous 7-day siomycin A treatment or placebo by tail vein injection. (G) Kaplan-Meier analysis was performed for the comparison of survival in U87 cell-implanted mice treated with or without siomycin A (n=5, P=0.0021, with log-rank test). (H) Flow cytometric analysis of apoptosis using U87 cells pre-treated with siomycin A and then treated with or without radiation (12 Gy) (P<0.05, with χ2 analysis). (I) Isobologram analysis results for siomycin A (20 µM) combined with radiation in U87 cells. (J) Luciferase activity assay showed that siomycin A treatment resulted in a decrease in transcription activity of FOXM1-binding region in U87 cells (n=3, *P<0.05, ***P<0.001, with one-way ANOVA followed by Dunnett's post test). (K) In vitro cell growth assay for U138 cells pre-transduced with empty vector or exogenous BUB1B overexpression vector and then treated with or without siomycinA (n=6, *P<0.05, **P<0.01, with one-way ANOVA).
Discussion
Accumulating data indicate that BUB1B is a critical protein involved in the growth and survival of multiple types of cancer including breast, gastric, colorectal and prostate cancer (17–20). Our findings revealed that knockdown of BUB1B led to the loss of GBM in vitro cell growth and in vivo tumorigenicity. Furthermore, we identified the forkhead box protein M1 (FOXM1)/BUB1B pathway as a pivotal regulator of radioresistance in GBM besides its main function in cell mitosis. In addition, deletion of this axis induced cell apoptosis when combined with radiotherapy. These findings showed us that targeting of BUB1B could become an efficient supporting therapy for GBM.
FOXM1 is a typical proliferation-associated transcription factor. However, the transcriptional downstream targets of FOXM1 in tumor recurrence and therapy resistance remain to be determined (22). FOXM1 was reported as an upregulator of enhancer of zeste homolog 2 (EZH2) and is functionally required for glioma stem cells to maintain stemness and therapy resistance via forming a complex with MELK (23). Wan et al (11) demonstrated that BUB1B is a direct transcriptional target of FOXM1 and FOXM1 expression drives a BUB1B promoter construct containing a consensus FOXM1 binding site (−350/−216 bp region). Similarly, in the present study, using a luciferase reporter assay we found that FOXM1 regulates the activity of the BUB1B promoter in GBM and this cell signaling pathway is essential for tumor proliferation and tumorigenicity in GBM cells.
Recent evidence indicates that the FOXM1-Sox2 signaling axis promotes clonogenic growth and radioresistance of GBM, suggesting that FOXM1 targeting combined with radiation may be a potentially effective therapeutic approach for GBM (24). Moreover, FOXM1 also contributes to the maintanence of stemness and radioresistance in GBM by upregulation of the DNA damage repair signaling or MELK-mediated oncogenic regulation of EZH2 (23,25). Nonetheless, whether there are other mechanisms involved in FOXM1-dependent tumorigenesis and therapy resistance in GBM warrants further investigation. Herein, we found that blocking the FOXM1/BUB1B pathway using a selective FOXM1 inhibitor siomycin A reduced tumor growth and enhanced the radiosensitivity of GBM cells, indicating that FOXM1 promotes radioresistance of GBM cells, at least partially, dependent on its transcriptional regulation of BUB1B. However, whether or not BUB1B promotes tumor growth and radioresistance in GBM through its kinase activity warrents further investigation. Further study is needed to design and synthesize a specific small-molecule inhibitor directly targeting BUB1B to show more efficiency for suppressing tumor growth and radioresistance.
Endoreduplication is the replication of the genome during the cell cycle without the subsequent completion of mitosis and/or cytokinesis (26). A wide variety of agents including microtubule inhibitors, topoisomerase II inhibitors and DNA damaging agents have been reported as able to induce endoreduplication (27). Endoreduplication can be associated with cell differentiation but also frequently occurs in malignant cells and may play a role in maintaining cell fate (28,29). Numerous cell cycle-related proteins including p21, aurora B, cyclin-dependent kinase 1 (CDK1), cyclin B1 and B2, have been identified to be involved in endoreduplication (11,30). A recent study evaluated the effects of FOXM1 knockdown on breast cancer cells and showed both endoreduplication and or mitotic catastrophe (31). Another study showed that BUB1B knockdown induces endoreduplication and mitotic catastrophe in rhabdomyosarcoma (11). However, regardless of these findings, it is still unclear whether the function of BUB1B in GBM depends on endoreduplication, or not. Thus, further study should be performed to answer these questions.
Altogether, in the present study, we identified that BUB1B expression was enriched in GBM tumors and was functionally required for tumor proliferation both in vitro and in vivo. Clinically, BUB1B expression was associated with poor prognosis in GBM patients and BUB1B-dependent radioresistance in GBM could be decreased by targeting BUB1B via shRNAs. Mechanistically, FOXM1 transcriptionally regulated BUB1B expression by binding to and then activating the BUB1B promoter. Therapeutically, we found that FOXM1 inhibitor attenuated tumorigenesis and radioresistance of GBM both in vitro and in vivo. Altogether, BUB1B promotes tumor proliferation and induces radioresistance in GBM, indicating that BUB1B is a potential therapeutic target for GBM.
Acknowledgements
The present study was supported by the Natural Science Foundation of Shaanxi Province, China (2012K17-01-03).
References
- 1.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 2.Meyer MA. Malignant gliomas in adults. N Engl J Med. 2008;359:1850. doi: 10.1056/NEJMc086380. [DOI] [PubMed] [Google Scholar]
- 3.Kim SH, Ezhilarasan R, Phillips E, Gallego-Perez D, Sparks A, Taylor D, Ladner K, Furuta T, Sabit H, Chhipa R, et al. Serine/threonine kinase MLK4 determines mesenchymal identity in glioma stem cells in an NF-κB-dependent manner. Cancer Cell. 2016;29:201–213. doi: 10.1016/j.ccell.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng P, Wang J, Waghmare I, Sartini S, Coviello V, Zhang Z, Kim SH, Mohyeldin A, Pavlyukov MS, Minata M, et al. FOXD1-ALDH1A3 signaling is a determinant for the self-renewal and tumorigenicity of mesenchymal glioma stem cells. Cancer Res. 2016;76:7219–7230. doi: 10.1158/0008-5472.CAN-15-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noda SE, El-Jawahri A, Patel D, Lautenschlaeger T, Siedow M, Chakravarti A. Molecular advances of brain tumors in radiation oncology. Semin Radiat Oncol. 2009;19:171–178. doi: 10.1016/j.semradonc.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Taylor SS, Ha E, McKeon F. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J Cell Biol. 1998;142:1–11. doi: 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, Kumar R, Jenkins RB, de Groen PC, Roche P, et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet. 2004;36:744–749. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- 8.Dai W, Wang Q, Liu T, Swamy M, Fang Y, Xie S, Mahmood R, Yang YM, Xu M, Rao CV. Slippage of mitotic arrest and enhanced tumor development in mice with BubR1 haploinsufficiency. Cancer Res. 2004;64:440–445. doi: 10.1158/0008-5472.CAN-03-3119. [DOI] [PubMed] [Google Scholar]
- 9.Wang Q, Liu T, Fang Y, Xie S, Huang X, Mahmood R, Ramaswamy G, Sakamoto KM, Darzynkiewicz Z, Xu M, et al. BUBR1 deficiency results in abnormal megakaryopoiesis. Blood. 2004;103:1278–1285. doi: 10.1182/blood-2003-06-2158. [DOI] [PubMed] [Google Scholar]
- 10.Kops GJ, Foltz DR, Cleveland DW. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc Natl Acad Sci USA. 2004;101:8699–8704. doi: 10.1073/pnas.0401142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wan X, Yeung C, Kim SY, Dolan JG, Ngo VN, Burkett S, Khan J, Staudt LM, Helman LJ. Identification of FoxM1/Bub1b signaling pathway as a required component for growth and survival of rhabdomyosarcoma. Cancer Res. 2012;72:5889–5899. doi: 10.1158/0008-5472.CAN-12-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raychaudhuri P, Park HJ. FoxM1: A master regulator of tumor metastasis. Cancer Res. 2011;71:4329–4333. doi: 10.1158/0008-5472.CAN-11-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monteiro LJ, Khongkow P, Kongsema M, Morris JR, Man C, Weekes D, Koo CY, Gomes AR, Pinto PH, Varghese V, et al. The Forkhead Box M1 protein regulates BRIP1 expression and DNA damage repair in epirubicin treatment. Oncogene. 2013;32:4634–4645. doi: 10.1038/onc.2012.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu M, Dai B, Kang SH, Ban K, Huang FJ, Lang FF, Aldape KD, Xie TX, Pelloski CE, Xie K, et al. FoxM1B is overexpressed in human glioblastomas and critically regulates the tumorigenicity of glioma cells. Cancer Res. 2006;66:3593–3602. doi: 10.1158/0008-5472.CAN-05-2912. [DOI] [PubMed] [Google Scholar]
- 15.Maachani UB, Shankavaram U, Kramp T, Tofilon PJ, Camphausen K, Tandle AT. FOXM1 and STAT3 interaction confers radioresistance in glioblastoma cells. Oncotarget. 2016;7:77365–77377. doi: 10.18632/oncotarget.12670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng WX, Han X, Zhang CL, Ge L, Du FY, Jin J, Gong AH. FoxM1-mediated RFC5 expression promotes temozolomide resistance. Cell Biol Toxicol. 2017 Feb 9; doi: 10.1007/s10565-017-9381-1. (Epub ahead of print). doi: 10.1007/s10565-017-9381-1. [DOI] [PubMed] [Google Scholar]
- 17.Hudler P, Britovsek NK, Grazio SF, Komel R. Association between polymorphisms in segregation genes BUB1B and TTK and gastric cancer risk. Radiol Oncol. 2016;50:297–307. doi: 10.1515/raon-2015-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mansouri N, Movafagh A, Sayad A, Heidary Pour A, Taheri M, Soleimani S, Mirzaei HR, Shargh Alizadeh S, Azargashb E, Bazmi H, et al. Targeting of BUB1b gene expression in sentinel lymph node biopsies of invasive breast cancer in iranian female patients. Asian Pac J Cancer Prev. 2016;17(S3):317–321. doi: 10.7314/APJCP.2016.17.S3.317. [DOI] [PubMed] [Google Scholar]
- 19.Hahn MM, Vreede L, Bemelmans SA, van der Looij E, van Kessel AG, Schackert HK, Ligtenberg MJ, Hoogerbrugge N, Kuiper RP, de Voer RM. Prevalence of germline mutations in the spindle assembly checkpoint gene BUB1B in individuals with early-onset colorectal cancer. Genes Chromosomes Cancer. 2016;55:855–863. doi: 10.1002/gcc.22385. [DOI] [PubMed] [Google Scholar]
- 20.Fu X, Chen G, Cai ZD, Wang C, Liu ZZ, Lin ZY, Wu YD, Liang YX, Han ZD, Liu JC, et al. Overexpression of BUB1B contributes to progression of prostate cancer and predicts poor outcome in patients with prostate cancer. Onco Targets Ther. 2016;9:2211–2220. doi: 10.2147/OTT.S101994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mao P, Joshi K, Li J, Kim SH, Li P, Santana-Santos L, Luthra S, Chandran UR, Benos PV, Smith L, et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci USA. 2013;110:8644–8649. doi: 10.1073/pnas.1221478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Z, Zhang G, Kong C. FOXM1 participates in PLK1-regulated cell cycle progression in renal cell cancer cells. Oncol Lett. 2016;11:2685–2691. doi: 10.3892/ol.2016.4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim SH, Joshi K, Ezhilarasan R, Myers TR, Siu J, Gu C, Nakano-Okuno M, Taylor D, Minata M, Sulman EP, et al. EZH2 protects glioma stem cells from radiation-induced cell death in a MELK/FOXM1-dependent manner. Stem Cell Reports. 2015;4:226–238. doi: 10.1016/j.stemcr.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee Y, Kim KH, Kim DG, Cho HJ, Kim Y, Rheey J, Shin K, Seo YJ, Choi YS, Lee JI, et al. FoxM1 promotes stemness and radioresistance of glioblastoma by regulating the master stem cell regulator Sox2. PLoS One. 2015;10:e0137703. doi: 10.1371/journal.pone.0137703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ganguly R, Mohyeldin A, Thiel J, Kornblum HI, Beullens M, Nakano I. MELK-a conserved kinase: Functions, signaling, cancer, and controversy. Clin Transl Med. 2015;4:11. doi: 10.1186/s40169-014-0045-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edgar BA, Orr-Weaver TL. Endoreplication cell cycles: More for less. Cell. 2001;105:297–306. doi: 10.1016/S0092-8674(01)00334-8. [DOI] [PubMed] [Google Scholar]
- 27.Cortés F, Mateos S, Pastor N, Domínguez I. Toward a comprehensive model for induced endoreduplication. Life Sci. 2004;76:121–135. doi: 10.1016/j.lfs.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 28.Lee HO, Davidson JM, Duronio RJ. Endoreplication: Polyploidy with purpose. Genes Dev. 2009;23:2461–2477. doi: 10.1101/gad.1829209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaźmierczak A. Endoreplication in Anemia phyllitidis coincides with the development of gametophytes and male sex. Physiol Plant. 2010;138:321–328. doi: 10.1111/j.1399-3054.2009.01323.x. [DOI] [PubMed] [Google Scholar]
- 30.Kim JA, Lee J, Margolis RL, Fotedar R. SP600125 suppresses Cdk1 and induces endoreplication directly from G2 phase, independent of JNK inhibition. Oncogene. 2010;29:1702–1716. doi: 10.1038/onc.2009.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wonsey DR, Follettie MT. Loss of the forkhead transcription factor FoxM1 causes centrosome amplification and mitotic catastrophe. Cancer Res. 2005;65:5181–5189. doi: 10.1158/0008-5472.CAN-04-4059. [DOI] [PubMed] [Google Scholar]