Abstract
Increasing evidence indicates that BAF53a is crucial for embryonic development and maintenance of stemness, and may be associated with epithelial-mesenchymal transition (EMT), which suggests its involvement in cancer progression. However, the role of BAF53a in glioma remains unknown. In the present study, BAF53a was found to be highly expressed in glioma tissues and was associated with poor overall survival (OS) and progression-free survival (PFS) in glioma patients. A multivariate Cox regression analysis revealed that BAF53a might be an independent prognostic factor for OS and PFS in glioma patients. Further functional analysis indicated that BAF53a overexpression could promote proliferation and increase the motility and invasion of U87 glioma cells, whereas BAF53a knockdown had the opposite effect. In addition, BAF53a expression was associated with the levels of E-cadherin and vimentin expression in glioma tissues. This was further confirmed in U87 cells expressing different levels of BAF53a; BAF53a overexpression was concomitant with decreased E-cadherin and increased vimentin expression, whereas BAF53a knockdown showed the opposite pattern of expression. Taken together, these results suggest that BAF53a may be a novel prognostic factor for glioma patients, and that BAF53 may facilitate glioma progression by promoting proliferation, invasion, and associate with EMT. Therefore, BAF53a could be a potential promising biomarker and a target for the treatment of glioma.
Keywords: glioma, BAF53a, epithelial-mesenchymal transition, tumor progression, prognosis
Introduction
Glioma is among the most common and lethal malignant brain tumors in humans (1). Despite advancements in surgical, radiotherapeutic and chemotherapeutic treatments, the survival of glioma patients remains very poor, and the majority of patients die within 2 years of diagnosis due to cancer progression (2). According to the World Health Organization (WHO) grading criteria, glioma is classified into four grades (I–IV), and survival rate has been reported to worsen with increasing grade; the 5-year survival rate is 30–70% in patients with grade I/II, compared with 3% in patients with grade IV disease (3). However, the detailed mechanisms underlying these differences in survival rates between different tumor grades remain obscure. Therefore, it is necessary to better understand the molecular mechanisms of glioma progression and thus develop novel therapeutic strategies.
Recently, studies have suggested that the epithelial-mesenchymal transition (EMT) plays an important role in cancer progression (4,5). EMT is the biological process in which cells are converted from an epithelial phenotype into a mesenchymal-like phenotype, which is involved in development and tissue regeneration. The hallmarks of EMT are decreased expression of epithelial markers (E-cadherin, β-catenin, etc.) and increased expression of mesenchymal markers (vimentin, N-cadherin, etc.), as well as changes in cell morphology (6). Numerous studies have confirmed that EMT is involved in the invasion and metastasis of cancers, including glioma. Patients with glioma that exhibit EMT-related changes typically have decreased survival times, and glioma cells that undergo EMT have stronger migratory and invasive potentials (7–9). Hence, further exploration of the association between EMT and glioma is critical for clarifying the mechanism of glioma progression.
BAF53a (also known as ACTL6A or ARP4) is a subunit of the Brg/Brm-associated factor (BAF) complex, which is crucial for embryonic development and maintenance of stemness in stem or progenitor cells (10–12). Therefore, BAF53a may also be associated with EMT. Recently, studies have reported that the aberrant expression of BAF53a is correlated with the progression of rhabdomyosarcoma, osteosarcoma, hepatocellular carcinoma and head and neck squamous cell carcinoma (HNSCC) (13–16). Based on its biological function in stem and progenitor cells, BAF53a is considered to promote cancer progression via EMT. However, the role of BAF53a in the invasion and metastasis of glioma remains unclear to date.
In the present study, we report for the first time that a high level of BAF53a expression in glioma specimens is associated with poor prognosis, and that the overexpression of BAF53a can promote the proliferation and invasion of glioma cells. Furthermore, BAF53a expression was found to be significantly associated with the expression of EMT markers, which indicates that BAF53a may promote the metastasis of glioma through EMT. Taken together, these findings preliminarily indicate that BAF53a may be a novel prognostic marker and promising therapeutic target for glioma.
Materials and methods
Patient specimens and cell cultures
The 121 glioma tissue samples were randomly obtained from the Department of tissue, Xiangya Hospital between January 2009 and December 2012. All the gliomas were pathology confirmed by 2 independent pathologists according to the 2007 WHO classification. All patients eligible for this study acceptted surgery and were regularly followed-up, and collected detailed clinicopathological data and survival data. Magnetic resonance imaging (MRI) or contrast-enhanced MRI was performed every 6 months. OS was defined as the time from the surgery to the death or the last follow-up visit. PFS was defined as the time from the surgery to the first evidence of recurrence, progression, or death. All patients signed a written informed consent. The study was approved by the ethics committee of Xiangya Hospital in accordance with the Declaration of Helsinki.
The human primary glioma cell line U87 was obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China), and cultured in DMEM (Gibco, NY, USA) with 10% fetal bovine serum (Gibco) at 37°C in a humidified atmosphere of 5% CO2.
RNA isolation and real-time PCR
Total RNA of glioma tissues and cells were isolated with TRIzol reagent (Invitrogen, CA, USA) following the manufacturer's instructions. The concentration and purity of all RNA samples were determined by NanoDrop Spectrophotometer (NanoDrop Technologies, TX, USA). Then, the cDNA was synthesized by reverse transcription of total RNA using cDNA synthesis kit (Roche Life Sciences, Switzerland), and real-time PCR reaction was conducted using the SYBR Green PCR kit (Roche Life Sciences) and performed on ABI PRISM 7100 Sequence Detection system (Applied Biosystems, CA, USA) following the manufacturer's instructions. GAPDH served as control. The target mRNA expression was calculated using the 2−∆∆Ct method. Each experiment was repeated 3 times. The primers were as follows: BAF53a (forward, 5′-CCAGGTCTCTATGGCAGTGTAA-3′; reverse, 5′-CGTAAGGTGACAAAAGGAAGGTA-3′); GAPDH (forward, 5′-GTCTCCTCTGACTTCAACAGCG-3′; reverse, 5′-ACCACCCTGTTGCTGTAGCCAA-3′); E-cadherin (forward, 5′-GCCTCCTGAAAAGAGAGTGGAAG-3′; reverse, 5′-TGGCAGTGTCTCTCCAAATCCG-3′); vimentin (forward, 5′-AGGCAAAGCAGGAGTCCACTGA-3′; reverse, 5′-ATCTGGCGTTCCAGGGACTCAT-3′).
Protein extraction and western blotting
Fresh tissues or cells were dissolved by RIPA lysis buffer (Invitrogen) and PMSF (Zhongshan Goldenbridge Biotechnology, Shanghai, China). Then isolated by centrifugation and quantified by Bradford Protein assay kit (Beyotime Institute of Biotechnology, Shanghai, China). Next the protein was separated by 10% SDS-PAGE and transferred to the PVDF membrane (Sigma, MO, USA). The membranes were blocked with 5% calf serum and incubated with appropriate primary antibody, followed by incubation with secondary antibody. Lastly, the signal was detected using enhanced chemiluminescence regents (Thermo Scientific, MA, USA) and photographed by camera and image processing system (Bio-Rad, CA, USA).
Immunohistochemistry
The glioma tissues were embedded in paraffin, and sectioned into slides with a thickness of 4 µm. Then, sections were dewaxed in xylene and rehydrated in graded ethanol. Antigen retrieval was accomplished using citrate buffer at pH 6.0. Then 3% H2O2 was used to block endogenous peroxidase activity, and goat serum incubation was used to block the non-specific antigen binding, followed by incubation with appropriate concentration of primary antibody at 4°C overnight. Then, the sections were incubated with the corresponding secondary antibody for 30 min at room temperature after rinsing three times in PBS. The antigen-antibody interactions was detected by DAB and counterstained by hematoxylin, followed by dehydrated in graded ethanols and mounted. The results were evaluated by two independent pathologists blinded to the study. The staining intensity of BAF53a was scored according to the percentage of positive stained tumor cells, as negative (−) <5% of tumor cells stained positive, (+) 0–25%, (++) 26–75%, >76% of tumor cells stained positive.
Lentiviral vectors production and cellular transduction
Full-length human BAF53a overexpression clone lentivirus and short hairpin RNAs (shRNA) lentivirus and their control vectors were constructed by a biotechnology company (GeneChem, Shanghai, China). Cells were cultured in 6-well plates before transfection until 80–90% confluency within 24 h. Then the lentivirus were transfected into cells according to the manufacturer's instructions. Puromycin (2 µg/ml) was used to select stable clones if necessary. After 48–72-h transfection, the cells were harvested for subsequent assays. Transfection efficiency was assessed by fluorescence microscope, real-time PCR and western blotting.
Reagents
Mitomycin-C and puromycin were purchased from ApexBio, Technology LLC A4452, A3740; ApexBio, TX, USA). The Matrigel matrix was purchased from Corning Life Sciences (354248; Corning, MA, USA). The primary antibodies against BAF53a (sc-137062), E-cadherin (sc-71007), vimentin (sc-80975), and β-actin (sc-130300) were purchased from Santa Cruz Biotechnology (CA, USA).
Proliferation assays
The proliferation potential was measured by MTT assay and colony formation assay. For MTT assays, the cells were grown into 96-well plates with a density of 5×103 cells/well. Each day 6 wells of each group were added 100 µl MTT (0.5 mg/ml; Sigma) reagent per well and incubated at 37°C for 4 h. Subsequently, the absorbance values were measured at 570 nm. For colony formation assays, 2×103 cells/well were seeded into 6-well plates and cultured for 14 days. Then the cell colonies were dyed with crystal violet and counted. The results were recorded as mean ± SD of three repeats.
Migration and invasion assays
The migration potential was evaluated by wound-healing and Transwell assay. The cells were cultured into 6-well plates at a density of 2×105 cells/well, when grew to 90% confluence, cells were incubated with mitomycin-C (10 µg/ml) for 1 h to suppress proliferation, and starved in serum-free medium for 24 h. When cells were confluent monolayer artificial wound was scraped by a 10-µl pipette tip. The migration gap of the wound was assessed after 24 or 48 h. The migration potential was evaluated by Transwell system. The cells treated with mitomycin-C (10 µg/ml) for 1 h at 37°C in serum-free medium were placed into them with a density of 1×105 cells/insert. The DMEM with 10% FBS was added into the lower chamber. After incubation for 24–48 h, the cells in the upper membrane of insert were removed and the cells adhering to the lower membrane of the inserts were stained with crystal violet and counted by microscope after rinsing. Then the invasion potential was evaluated by Transwell-Matrigel system. The culture upper inserts were coated with Matrigel, the subsequence processes were carried out as Transwell assay. All the experiments were performed in triplicates.
Statistical analysis
Statistical analysis was conducted with SPSS version 18.0 (SPSS, Inc., Chicago, IL, USA). Student's two-tailed t-test was used to determine the statistical significance of measurement data. Survival curves were performed by the Kaplan-Meier method and compared by the log-rank test. Univariate and multivariate Cox regression model was used for survival analysis. The continuous data are shown as the mean ± SEM. A P-value of <0.05 was considered statistically significant.
Results
BAF53a is highly expressed in glioma specimens
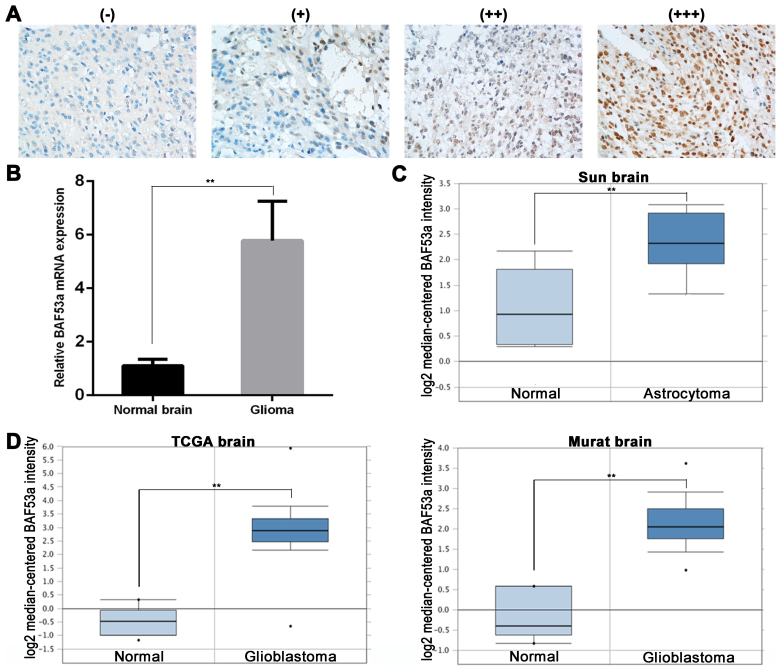
To examine the role of BAF53a in glioma, we initially detected the protein expression of BAF53a in tumor tissues by immunohistochemistry (IHC). BAF53a was predominantly expressed in the nucleus. We defined high BAF53a expression as strongly (+++) or moderately (++) positive staining, and low BAF53a expression as weakly positive (+) or negative staining (Fig. 1A). BAF53a was positively expressed in the majority of glioma tissues (102/121), of which 79 exhibited high expression (Table I). Additionally, BAF53a mRNA expression was examined in 10 randomly selected cases of glioma and 10 normal brain tissues obtained from resection following trauma. These results indicated that BAF53a mRNA expression levels in glioma tissues were significantly higher than those in normal brain tissues (P<0.01; Fig. 1B).
Figure 1.
BAF53a expression is upregulated in glioma. (A) The representative images of BAF53a expression is detected by IHC in glioma tissues (scored as - to +++). Magnification, ×400. (B) The real-time PCR results show that the expression level of BAF53a mRNA in glioma tissues was higher than that in normal brain tissues. The expression profile of BAF53a in Oncomine database: BAF53a gene copy number in astrocytoma is higher than normal brain tissues in Sun brain data (C), and BAF53a gene copy number in glioblastomas is significantly higher than normal brain tissues in TCGA brain data and Murat brain data (D). *P<0.05; **P<0.01.
Table I.
BAF53a expression and clinicopathologic features of 121 glioma cases.
| BAF53a expression | ||||
|---|---|---|---|---|
| Variables | Total n (121) | Low (42) | High (79) | P-value |
| Sex | ||||
| Female | 53 | 17 | 36 | |
| Male | 68 | 25 | 43 | 0.591 |
| Age (years) | ||||
| ≤50 | 72 | 29 | 43 | |
| >50 | 49 | 13 | 36 | 0.119 |
| Tumor size (cm) | ||||
| ≤4 | 66 | 25 | 41 | |
| >4 | 55 | 17 | 38 | 0.423 |
| Necrosis | ||||
| Absence | 71 | 26 | 45 | |
| Presence | 50 | 16 | 34 | 0.599 |
| KPS score | ||||
| ≤90 | 82 | 31 | 51 | |
| >90 | 39 | 11 | 28 | 0.299 |
| WHO grade | ||||
| I and II | 53 | 24 | 29 | |
| III and IV | 68 | 18 | 50 | 0.031 |
KPS, Karnofsky performance status; WHO, World Health Organization.
Subsequently, the expression profile of BAF53a was explored using the Oncomine™ database (https://www.oncomine.org). The results showed that the gene copy number of BAF53a in astrocytoma was higher than that in normal brain tissue (Sun brain: fold change, 2.460; P<0.01; Fig. 1C). Furthermore, the BAF53a gene copy number in glioblastoma was significantly higher than that in normal brain tissue (TCGA brain: fold change, 11.006; P<0.01; Murat brain: fold change, 5.298; P=0.002; Fig. 1D). These results enable us to speculate that BAF53 may be involved in the progression of glioma.
BAF53a expression is associated with poor prognosis in patients with glioma
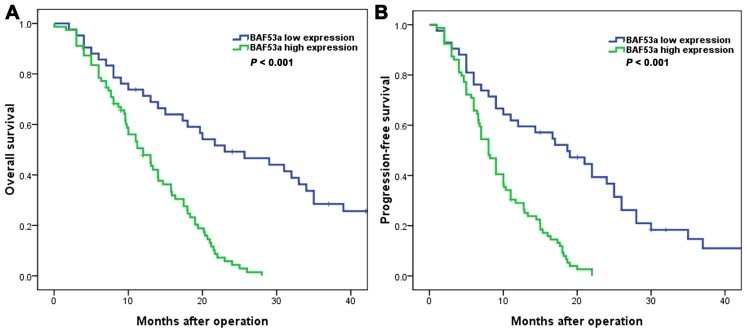
We next explored the detailed associations between BAF53a protein expression (determined by IHC) and the clinicopathological features and prognosis of glioma patients. The results indicated that high BAF53a expression was significantly correlated with WHO grade (P=0.031; Table I). Survival analysis showed that the BAF53a low expression group had favorable overall survival (OS) [median OS time, 23.0 (95% CI, 11.6–34.4) vs. 12.0 (95% CI, 8.9–15.1) months; P<0.001; Fig. 2A] and progression-free survival (PFS) [median PFS time, 18.7 (95% CI, 10.6–26.8) vs. 8.0 (95% CI, 6.7–9.3) months, P<0.001; Fig. 2B] compared with the BAF53a high expression group. Furthermore, univariate and multivariate Cox regression analyses indicated that BAF53a expression was an independent prognostic factor for the OS and PFS of glioma patients (HR=1.752, P=0.013 and HR=1.874, P 0.008, respectively; Tables II and III). These results indicate that BAF53a may be useful as a prognostic marker for patients with glioma.
Figure 2.
The survival curve of glioma patients analyzed according to the expression of BAF53a. (A) Results showed that glioma patients in the high BAF53a expression group had poorer overall survival than those in the low BAF53a expression group. (B) Glioma patients in the high BAF53a expression group had poorer progression-free survival than those in the low BAF53a expression group.
Table II.
The univariate and multiple Cox regression analyses of overall survival (OS) in glioma patients.
| Univariable analysis | Multivariable analysis | |||
|---|---|---|---|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Sex (male vs. female) | 1.322 (0.492–2.485) | 0.251 | ||
| Age (≤50 vs. >50 years) | 1.183 (0.642–1.934) | 0.426 | ||
| Tumor size (≤4 vs. >4 cm) | 0.613 (0.382–1.267) | 0.137 | ||
| Necrosis (absence vs. presence) | 0.714 (0.433–1.162) | 0.325 | ||
| KPS score (≤90 vs. >90) | 0.682 (0.310–1.254) | 0.185 | ||
| WHO grade (III and IV vs. I and II) | 3.126 (1.643–5.233) | <0.001 | 2.304 (1.192–3.542) | 0.002 |
| BAF53a expression (high vs. low) | 1.937 (1.326–3.174) | 0.006 | 1.752 (1.226–2.638) | 0.013 |
HR, hazard risk ratio; CI, confidence interval. Bold, statistically significant.
Table III.
The univariate and multiple Cox regression analyses of progression-free survival (PFS) in glioma patients.
| Univariable analysis | Multivariable analysis | |||
|---|---|---|---|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Sex (male vs. female) | 1.167 (0.643–1.823) | 0.572 | ||
| Age (≤50 vs. >50 years) | 1.094 (0.735–1.541) | 0.714 | ||
| Tumor size (≤4 vs. >4 cm) | 0.582 (0.221–1.142) | 0.095 | 0.623 (0.334–1.095) | 0.142 |
| Necrosis (absence vs. presence) | 0.802 (0.455–1.372) | 0.483 | ||
| KPS score (≤90 vs. >90) | 0.713 (0.386–1.197) | 0.224 | ||
| WHO grade (III and IV vs. I and II) | 4.075 (1.825–7.437) | <0.001 | 3.194 (1.426–5.273) | <0.001 |
| BAF53a expression (high vs. low) | 2.056 (1.274–3.312) | 0.003 | 1.874 (1.411–2.942) | 0.008 |
BAF53a promotes the proliferation of glioma cells in vitro
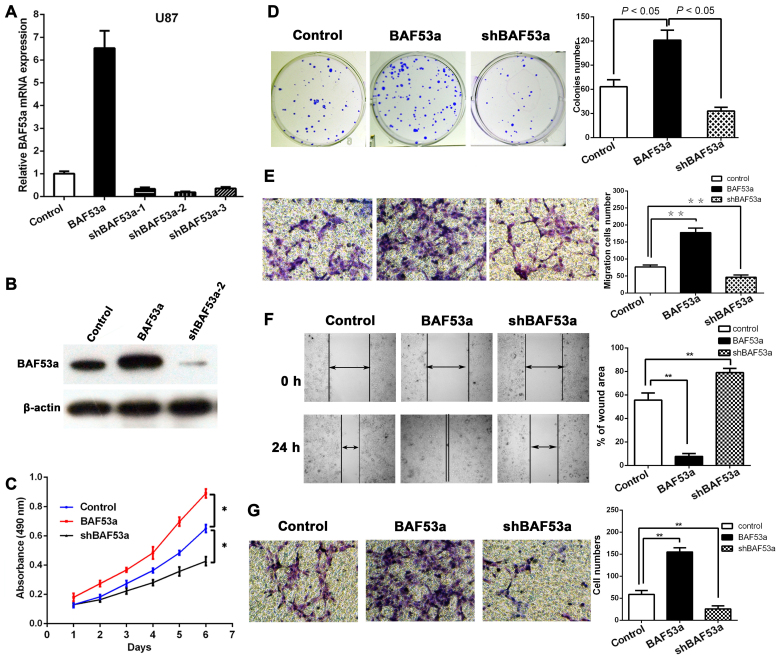
In order to study the functional role of BAF53a in glioma, stable BAF53a-overexpressing U87BAF53a and BAF53a-knockdown U87shBAF53a cells and control cells transfected with empty vector were established. The transfection efficiency in each cell type was confirmed by qPCR and western blotting (Fig. 3A and B). Subsequently, the cells were subjected to MTT and colony formation assays. The MTT assay revealed that BAF53a overexpression could markedly increase U87 cell growth, while BAF53a knockdown had the opposite effect as compared with control cells (P<0.05, respectively; Fig. 3C). The colony formation assay also showed that the U87BAF53a cells were significantly larger and greater in number than the control cells, while U87shBAF53a cells exhibited the opposite characteristics (P<0.05, respectively; Fig. 3D). These results indicated that BAF53a could promote the proliferation of glioma cells.
Figure 3.
BAF53a promotes proliferation, migration and invasion of glioma cells. The overexpression and knockdown interfered efficiency of BAF53a detected by real-time PCR (A) and western blotting (B), results showed shBAF53a-2 had the best knockdown effect and the overexpression effect was also satisfactory. (C) The MTT assays showed BAF53a overexpression promoted glioma cell proliferation, while BAF53a knockdown inhibited glioma cell proliferation. (D) The colony formation assays also showed BAF53a expression promoted glioma cell proliferation. (E) The Transwell assay showed BAF53a expression facilitated migration of glioma cells. (F) The wound healing assay showed BAF53a expression provided the faster wound healing capacities of glioma cells. (G) The Transwell-Matrigel chamber assay showed BAF53a expression facilitated invasion of glioma cells.
BAF53a promotes the migration and invasion of glioma cells in vitro
Wound healing and Transwell assays were used to assess the migratory ability of U87 cells expressing varying levels of BAF53a. The results showed that BAF53a-overexpressing U87BAF53a cells exhibited increased migration and faster wound healing capacities, whereas U87shBAF53a cells exhibited decreased migration and slower wound healing capacities when compared with control cells (Fig. 3E and F). A Transwell-Matrigel chamber assay accordingly showed that U87BAF53a cells had a greater capacity for invasion across the Matrigel membrane compared with the control cells, whereas U87shBAF53a cells had reduced invasive ability (Fig. 3G). These results indicated that BAF53a could promote the invasion capacity of glioma cells and potentially facilitate the metastasis of glioma.
BAF53a expression is associated with EMT in glioma
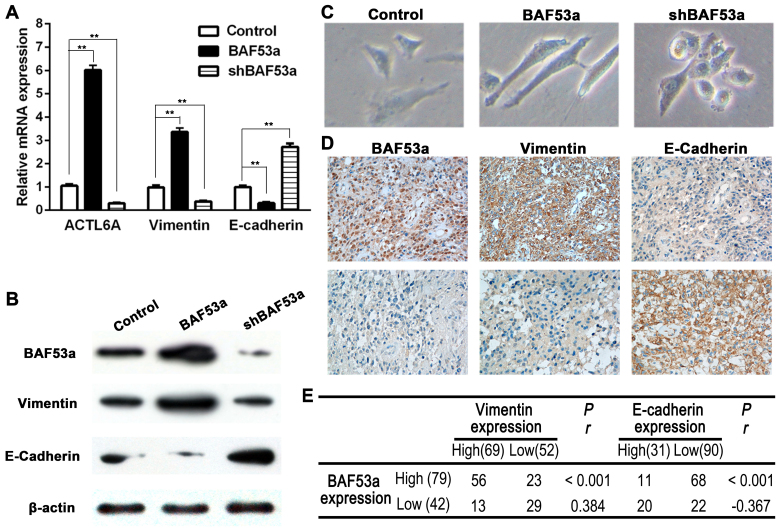
As previous studies have reported a possible link between BAF53a and EMT in other physiological processes and cancer progression, we aimed to explore this association in glioma. The expression levels of two EMT markers, E-cadherin and vimentin, were detected in cells expressing different levels of BAF53a. The qPCR and western blotting results showed decreased expression of the epithelial marker E-cadherin and increased expression of the mesenchymal marker vimentin in U87BAF53a cells. By contrast, the opposite pattern of expression was demonstrated in U87shBAF53a cells (Fig. 4A and B). Alterations in cellular morphology assessed by phase contrast microscopy also revealed that U87BAF53a cells had more pseudopodia and were elongated, indicating a mesenchymal phenotype, whereas U87shBAF53a cells had an oval or cobblestone-like appearance (Fig. 4C). E-cadherin and vimentin expression were also detected in glioma tissues by IHC. The results revealed that E-cadherin expression was frequently absent in glioma, whereas vimentin was consistently expressed (Fig. 4D). The Spearman rank correlation coefficient indicated that BAF53a expression was positively correlated with the expression of vimentin (r=0.384, P<0.001) and negatively correlated with the expression of E-cadherin (r=−0.367, P<0.001) (Fig. 4E). Taken together, these results suggest that BAF53a may promote glioma progression via EMT.
Figure 4.
BAF53a associates with EMT in glioma. The expression correlation of BAF53a, vimentin and E-cadherin in glioma cells detected by real-time PCR (A) and western blotting (B), results showed that vimentin expression was upregulated along with BAF53a overexpression, but E-cadherin expression was decreased; when BAF53a was knocked down, they had inverse expression. (C) The phase contrast images of BAF53a-interfered U87 cells, results showed cells became mesenchymal-like in BAF53a overexpression, and cells changed to oval or cobblestone-like appearance in BAF53a knockdown. (D) The representative images of expression correlation of BAF53a, vimentin and E-cadherin in glioma tissues detected by IHC. (E) The Spearman's rank correlation test indicated that BAF53a and vimentin expression were positively correlated, while BAF53a and E-cadherin expression were negatively correlated.
Discussion
The findings of the present study revealed that BAF53a is a potential valuable prognostic marker for glioma patients, and that it may promote glioma cell proliferation, migration and invasion and be associated with EMT.
Notably, the present study demonstrated the functional role of BAF53a in promoting glioma progression. The subunits of the BAF complex are known to serve important roles in many physiological and pathological processes in various cell types and organs. At present, the association of BAF53a with cancer development and progression is attracting increasing attention. In 2002, a study demonstrated that BAF53a was critical for the oncogenic activity of c-Myc (17). Additionally, studies of rhabdomyosarcoma, hepatocellular carcinoma and squamous cell carcinoma have also provided evidence that BAF53a is essential for cancer invasion and metastasis, and is associated with poor prognosis (13,15,16). Studies have also demonstrated that the subunits of the BAF complex, including ARID2, ARID5B and BAF47, play important roles in a variety of cancer types (18–20). These results are consistent with those of the present study of glioma, which showed that BAF53a may be a cancer-promoting gene.
According to previous studies, BAF53a knockdown can significantly impair neural stem/progenitor cell proliferation capacity and their differentiation into neurons (12,21). Another study showed that BAF53a was overexpressed in fibroblasts and blocked the neuronal conversion of fibroblasts (22). A study by Krasteva et al also found that the deletion of BAF53a resulted in embryonic lethality and proliferation defects of hematopoietic stem cells (11). Furthermore, a recent study by Bao et al showed that ectopic expression of BAF53a promoted the proliferation and progenitor state of epidermal cells (23). These studies suggest that BAF53a can control differentiation and proliferation in embryonic and adult stem cells, as well as during embryogenesis. It is well established that cancer progression shares many similar events with embryogenesis, such as cell growth, differentiation and migration. Therefore, genes that are crucial for embryonic or stem cell development are often associated with cancer progression. Accordingly, the present study has verified the hypothesis that BAF53a can promote glioma progression and lead to poor prognosis.
When cancer cells undergo EMT, they acquire increased motility and invasive capacity, as well as resistance to chemoradiotherapy; EMT thereby promotes metastasis and results in poor prognosis. In glioma, various genes and non-coding RNAs, such as AC1MMRY2, MLK4 and miR-200a/b, are reportedly able to induce EMT, particularly in glioblastoma (24,25). Notably, the present data indicated that alteration of BAF53a expression could induce morphological changes in glioma cells, as well as influencing the expression levels of epithelial and mesenchymal markers. This suggested that BAF53a may induce EMT in glioma and function as an EMT-related transcriptional factor. These results illustrate a novel biological function of BAF53a in glioma, which is consistent with previous studies of osteosarcoma and HCC (14,16). Additionally, a recent study in HNSCC found that BAF53a could activate YAP1, which can induce EMT in various cancer types, and these findings confirm the role of BAF53a in EMT (13,26).
EMT inhibitors, such as BGB324 and A83-01, are designed to target receptors in the cytomembrane; however, targeting EMT-related genes in the cell nucleus may be a more precise approach. Therefore, the present results provide the basis to potentially develop a novel targeted therapy.
The mechanism by which BAF53a induces EMT remains unclear. Previous studies found that BAF53a could interact with p63, SALL4, FGF4 and KLF4 to maintain the stemness and undifferentiated state of stem/progenitor cells, which is important for the acquisition of mesenchymal characteristics (10,13,23). A study of HCC showed that BAF53a could regulate the transcription of SOX2 to further activate the Notch pathway to induce EMT (16). However, the detailed molecular mechanism regarding the contribution of BAF53a to EMT in glioma still requires further research.
The present study had certain limitations. First, the BAF53a expression profile and its prognostic significance were only retrospectively analyzed in glioma tissues and in a single institution; this need to be expanded to different cancer types and validated in an external cohort in the future. Second, the association between BAF53 and EMT requires further in vitro and in vivo experiments to support the hypothesis. Third, further research into the precise molecular mechanisms of BAF53 in promoting glioma progression and EMT is necessary.
In conclusion, the present study suggests that high BAF53a expression predicts poor prognosis of glioma patients, and that BAF53a can promote invasion and EMT of glioma cells. Therefore, BAF53a may be a useful prognostic marker and potential promising target for glioma therapeutics.
References
- 1.Wen PYKS, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 2.Chen R, Smith-Cohn M, Cohen AL, Colman H. Glioma subclassifications and their clinical significance. Neurotherapeutics. 2017;14:284–297. doi: 10.1007/s13311-017-0519-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang X, Yang H, Gong B, Jiang C, Yang L. Combined gene expression and protein interaction analysis of dynamic modularity in glioma prognosis. J Neurooncol. 2012;107:281–288. doi: 10.1007/s11060-011-0757-4. [DOI] [PubMed] [Google Scholar]
- 4.Alderton GK. Metastasis: Epithelial to mesenchymal and back again. Nat Rev Cancer. 2013;13:3. doi: 10.1038/nrc3428. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Kahlert UD, Maciaczyk D, Doostkam S, Orr BA, Simons B, Bogiel T, Reithmeier T, Prinz M, Schubert J, Niedermann G, et al. Activation of canonical WNT/β-catenin signaling enhances in vitro motility of glioblastoma cells by activation of ZEB1 and other activators of epithelial-to-mesenchymal transition. Cancer Lett. 2012;325:42–53. doi: 10.1016/j.canlet.2012.05.024. [DOI] [PubMed] [Google Scholar]
- 8.Zhang L, Zhang W, Li Y, Alvarez A, Li Z, Wang Y, Song L, Lv D, Nakano I, Hu B, et al. SHP-2-upregulated ZEB1 is important for PDGFRα-driven glioma epithelial-mesenchymal transition and invasion in mice and humans. Oncogene. 2016;35:5641–5652. doi: 10.1038/onc.2016.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kahlert UD, Nikkhah G, Maciaczyk J. Epithelial-to-mesenchymal(-like) transition as a relevant molecular event in malignant gliomas. Cancer Lett. 2013;331:131–138. doi: 10.1016/j.canlet.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 10.Lu W, Fang L, Ouyang B, Zhang X, Zhan S, Feng X, Bai Y, Han X, Kim H, He Q, et al. Actl6a protects embryonic stem cells from differentiating into primitive endoderm. Stem Cells. 2015;33:1782–1793. doi: 10.1002/stem.2000. [DOI] [PubMed] [Google Scholar]
- 11.Krasteva V, Buscarlet M, Diaz-Tellez A, Bernard MA, Crabtree GR, Lessard JA. The BAF53a subunit of SWI/SNF-like BAF complexes is essential for hemopoietic stem cell function. Blood. 2012;120:4720–4732. doi: 10.1182/blood-2012-04-427047. [DOI] [PubMed] [Google Scholar]
- 12.Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saladi SV, Ross K, Karaayvaz M, Tata PR, Mou H, Rajagopal J, Ramaswamy S, Ellisen LW. ACTL6A is co-amplified with p63 in squamous cell carcinoma to drive YAP activation, regenerative proliferation, and poor prognosis. Cancer Cell. 2017;31:35–49. doi: 10.1016/j.ccell.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun W, Wang W, Lei J, Li H, Wu Y. Actin-like protein 6A is a novel prognostic indicator promoting invasion and metastasis in osteosarcoma. Oncol Rep. 2017;37:2405–2417. doi: 10.3892/or.2017.5473. [DOI] [PubMed] [Google Scholar]
- 15.Taulli R, Foglizzo V, Morena D, Coda DM, Ala U, Bersani F, Maestro N, Ponzetto C. Failure to downregulate the BAF53a subunit of the SWI/SNF chromatin remodeling complex contributes to the differentiation block in rhabdomyosarcoma. Oncogene. 2014;33:2354–2362. doi: 10.1038/onc.2013.188. [DOI] [PubMed] [Google Scholar]
- 16.Xiao S, Chang RM, Yang MY, Lei X, Liu X, Gao WB, Xiao JL, Yang LY. Actin-like 6A predicts poor prognosis of hepatocellular carcinoma and promotes metastasis and epithelial-mesenchymal transition. Hepatology. 2016;63:1256–1271. doi: 10.1002/hep.28417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park J, Wood MA, Cole MD. BAF53 forms distinct nuclear complexes and functions as a critical c-Myc-interacting nuclear cofactor for oncogenic transformation. Mol Cell Biol. 2002;22:1307–1316. doi: 10.1128/MCB.22.5.1307-1316.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li M, Zhao H, Zhang X, Wood LD, Anders RA, Choti MA, Pawlik TM, Daniel HD, Kannangai R, Offerhaus GJ, et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat Genet. 2011;43:828–829. doi: 10.1038/ng.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R, Benz CC, et al. Cancer Genome Atlas Research Network: Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadoch C, Crabtree GR. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell. 2013;153:71–85. doi: 10.1016/j.cell.2013.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bao X, Tang J, Lopez-Pajares V, Tao S, Qu K, Crabtree GR, Khavari PA. ACTL6a enforces the epidermal progenitor state by suppressing SWI/SNF-dependent induction of KLF4. Cell Stem Cell. 2013;12:193–203. doi: 10.1016/j.stem.2012.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim SH, Ezhilarasan R, Phillips E, Gallego-Perez D, Sparks A, Taylor D, Ladner K, Furuta T, Sabit H, Chhipa R, et al. Serine/threonine kinase MLK4 determines mesenchymal identity in glioma stem cells in an NF-κB-dependent manner. Cancer Cell. 2016;29:201–213. doi: 10.1016/j.ccell.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi Z, Zhang J, Qian X, Han L, Zhang K, Chen L, Liu J, Ren Y, Yang M, Zhang A, et al. AC1MMYR2, an inhibitor of dicer-mediated biogenesis of Oncomir miR-21, reverses epithelial-mesenchymal transition and suppresses tumor growth and progression. Cancer Res. 2013;73:5519–5531. doi: 10.1158/0008-5472.CAN-13-0280. [DOI] [PubMed] [Google Scholar]
- 26.Shao DD, Xue W, Krall EB, Bhutkar A, Piccioni F, Wang X, Schinzel AC, Sood S, Rosenbluh J, Kim JW, et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell. 2014;158:171–184. doi: 10.1016/j.cell.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]