Abstract
Non-small cell lung cancer (NSCLC) is the most common type of lung cancer and a tumor with a broad spectrum of targeted therapies already available or in clinical trials. Thus, molecular characterization of the tumor using next generation sequencing (NGS) technology, has become a key tool for facilitating treatment decisions and the clinical management of NSCLC patients. The performance of a custom 23 gene multiplex amplification hot spot panel, based on Ion AmpliSeq™ technology, was evaluated for the analysis of tumor DNA extracted from formalin-fixed and paraffin-embedded (FFPE) tissues. Furthermore, the Ion AmpliSeq™ RNA Fusion Lung Cancer Research Panel was used for fusion RNA transcript analysis. The mutation spectrum of the tumors was determined in a cohort of 502 patients with NSCLC using the aforementioned targeted gene panels. The panel used for tumor DNA analysis in this study exhibited high rates (100%) of sensitivity, specificity and reproducibility at a mutation allelic frequency of 3%. At least one DNA mutation was detected in 374 patients (74.5%) and an RNA fusion was identified in 16 patients, (3.2%). In total, alterations in a cancer-driver gene were identified (including point mutations, gene rearrangements and MET amplifications) in 77.6% of the tumors tested. Among the NSCLC patients, 23% presented a mutation in a gene associated with approved or emerging targeted therapy. More specifically, 13.5% (68/502) presented a mutation in a gene with approved targeted therapy (EGFR, ALK, ROS1) and 9.4% (47/502) had an alteration in a gene related to emerging targeted therapies (ERBB2, BRAF, MET and RET). Furthermore, 51.6% of the patients had a mutation in a gene that could be related to an off label therapy or indicative for access to a clinical trial. Thus, the targeted NGS panel used in this study is a reliable approach for tumor molecular profiling and can be applied in personalized treatment decision making for NSCLC patients.
Keywords: next generation sequencing, non-small cell lung cancer, predictive markers, targeted therapy
Introduction
Lung cancer is the primary cause of cancer-associated mortality worldwide in men and the second leading cause of cancer-related death in women (1). The main type of lung cancer is non-small cell lung cancer (NSCLC) which accounts for ~85% of all lung cancer cases (2).
Treatment options for lung cancer include surgery, radiation therapy and chemotherapy. Although chemotherapy remains an important form of treatment, novel drug development has focused on molecular-targeted therapies. The effectiveness of such targeted therapies rely on the presence or absence of alterations in gene(s) that encode for the protein targeted by the agent or for proteins involved in the molecular pathway targeted (3).
This targeted therapeutical approach necessitates the molecular analysis of the tumor in order to select patients with increased probability of response to the treatment given. Until recently, the routine screening of tumors for mutations was confined by limited tissue availability, cost, time and labor demanding methods such as Sanger sequencing (4). More recently, the introduction of next generation sequencing (NGS) methods has resolved this issue by simultaneously sequencing thousands of short DNA sequences in a massively parallel manner, thus offering a cost effective approach for detecting multiple genetic alterations for many samples simultaneously. Moreover, the sensitivity obtained by NGS is superior to that of conventional sequencing technology, making possible the detection of mutations that are present in very low percentages in a background of normal DNA, which is extremely important for the detection of somatic mutations (5,6).
Large scale application of this technology in tumor molecular characterization led to the generation of databases that catalog the NGS cancer genome. Such databases include the COSMIC database, International Genome Consortium (ICGC) and the Cancer Genome Atlas (TCGA) project (7–10). Additionally, interpretation of somatic variants is feasible due to several available knowledge resources and online interpretation tools with My Cancer Genome being the first of such public somatic variant interpretation resources (11). Alternatively, commercially available software and bioinformatics tools such as Ion Torrent™ Oncomine™ Knowledgebase Reporter (https://www.thermofisher.com/) and QCI™ Interpret for Somatic Cancer (www.qiagenbioinformatics.com) are now available for clinical testing laboratories. These tools facilitate the interpretation and reporting of genomic variants identified by NGS, enabling the easy identification of actionable variants and the association with treatment options and open clinical trials.
Currently, the most widely used platforms are those offered by Illumina, Inc. (San Diego, CA, USA) and by Thermo Fisher Scientific (Waltham, MA, USA). The Illumina platform involves bridge amplification, to clonally amplify the fragments that are then sequenced using sequencing-by-synthesis (SBS) chemistry. The Thermo Fisher Scientific platform uses the Ion semiconductor sequencing, detecting the protons released as nucleotides are incorporated during synthesis. Both ΝGS platforms have all the features required to carry out simultaneous analysis of a large number of genes with actionable alterations in tumor tissue and thus a more precise molecular characterization of the tumor. Such targeted NGS panels for somatic mutation detection, include actionable cancer genes aiming to increase the percentage of patients with detectable actionable somatic gene alterations that can be used to guide treatment decision (12–14).
The integration of these NGS multi-gene panels in the health care setting is of great clinical importance and increases the necessity of appropriate guidelines in order to assure their optimal performance and accuracy (15,16).
In the present study, a custom 23 gene Ion AmpliSeq Panel for DNA analysis and the Ion AmpliSeq™ RNA Fusion Lung Cancer Research Panel for fusion RNA transcript analysis were applied in a cohort of patients with NSCLC. The Ion AmpliSeq Panel for DNA analysis was selected since it includes all the major mutations occurring in lung cancer. It includes the same hot spot regions of the 22 gene Colon and Lung V2 panel with the addition on certain amplicons to detect exon 14 skipping mutations in the MET gene and amplicons for exons 2 and 3 of the HRAS gene (13,14). Since, these targeted NGS panels contain all important genes related to targeted therapies in NSCLC, they were used in the the mutation spectrum identification of tumors in newly diagnosed NSCLC patients.
Materials and methods
Tumor samples
A total of 512 tumors collected at various institutions between January 2014 and December 2015, from patients with newly diagnosed non-small cell lung cancer (NSCLC), were included in this study. All available clinical factors were evaluated. Tumor classification was performed using the 2015 World Health Organization (WHO) Classification of Lung Tumors (17). Informed consent was obtained from all patients before testing. The study was approved by the Ethics Committee of the Pulmonary Department, Oncology Unit, ‘G. Papanikolaou’ General Hospital, Aristotle University of Thessaloniki, Thessaloniki, Greece.
Tissue selection and DNA extraction
Hematoxylin and eosin-stained sections of formalin-fixed and paraffin-embedded (FFPE) tumor biopsies from all samples were reviewed to ensure tumor cell content of >75% when possible, and the tumor area was marked by a pathologist. Genomic DNA was extracted from unstained 10-µm thick FFPE sections using the QIAmp FFPE tissue kit (Qiagen, Antwerp, Belgium). After extraction, the concentration of all samples was measured with the use of a spectrophotometer (NanoDrop2000, Thermo Fisher Scientific).
Ion AmpliSeq next generation sequencing
The NGS for DNA analysis was conducted using a custom 23 gene Ion AmpliSeq panel which was based on the Ion AmpliSeq Colon and Lung Cancer Research Panel v2 with an additional amplicon in the MET gene (to include the exon 14 skipping mutations) and two amplicons of exons 2 and 3 of the HRAS gene. Fusion RNA transcript analysis was performed using the Ion AmpliSeq RNA Fusion Lung Cancer Research Panel (Thermo Fisher Scientific). Details of the target regions of the 23-gene panel are available upon request.
The genes analyzed include AKT1 (NM_05163), ALK (NM_004304), BRAF (NM_004333), CTNNB1 (NM_001904), DDR2 (NM_001014796), EGFR (NM_005228), ERBB2 (NM_004448), ERBB4 (NM_005235), FBXW7 (NM_033632), FGFR1 (NM_023110), FGFR2 (NM_022970), FGFR3 (NM_000142), KRAS (NM_033360), MAP2K1 (NM_002755), MET (NM_001127500), NOTCH1 (NM_017617), NRAS (NM_002524), PIK3CA (NM_006218), PTEN (NM_000314), SMAD4 (NM_005359), STK11 (NM_000455), TP53 (NM_000546) and HRAS (001130442).
The Ion AmpliSeq RNA Fusion Lung Cancer Research Panel targets over 70 fusion transcripts associated with lung cancer research. It enables the analysis of the major ALK, RET, ROS1, and NTRK1 fusion transcripts, in addition to targets designed to detect 5′ and 3′ ALK gene expression. The panel also includes 5 positive control genes.
Library preparation
DNA or RNA concentrations were measured using the Qubit™ 2.0 Fluorometer in combination with the Qubit dsDNA HS assay kit (Thermo Fischer Scientific). cDNA was generated with SuperScript® VILO™ cDNA Synthesis kit (Thermo Fischer Scientific) from 10 ng of total RNA. For DNA library construction, 10 ng of DNA from each of the 502 FFPE samples was utilized.
An amplicon library was thus generated from total DNA/cDNA using the Ion AmpliSeq Library kit 2.0 (Thermo Fischer Scientific) according to the manufacturer's instructions. Briefly, amplicon amplification was performed using Ion AmpliSeq HiFi Master Mix (Thermo Fischer Scientific). The amplicons were then digested with FUPA reagent and barcoded with the IonCode™ Barcode Adapters 1–384 kit (Thermo Fischer Scientific). Subsequently, the amplified products were purified from the other reaction components using Agencourt AMPure XP PCR purification system (Beckman Coulter, Inc., Brea, CA, USA).
RNA libraries were quantified using the Qubit 2.0 Fluorometer and the Qubit dsDNA HS assay kit. Each library (20 pM) was multiplexed. For libraries that originated from genomic DNA, the Ion Library Equalizer™ kit method was used for normalizing library concentration at ~100 pM without the need for special instrumentation for quantification. Finally, equal volumes of each normalized DNA library and 20 pM of each RNA library were combined and clonally amplified on Ion Sphere™ particles (ISP) by emulsion PCR performed on the Ion One Touch™ 2 instrument with the Ion PI™ Hi-Q™ OT2 200 kit (Thermo Fisher Scientific) according to the manufacturer's instructions.
Quality control was performed using the Ion Sphere Quality Control kit (Thermo Fisher Scientific) to ensure that 10–30% of template positive ISP were generated in the emulsion PCR. Finally, the template-positive Ion PI Ion Sphere™ Particles were enriched using the Ion OneTouch™ ES instrument, loaded on an Ion PI Chip v3 and sequenced on an Ion Proton™ Sequencer with the Ion PI Hi-Q Sequencing 200 kit according to the manufacturer's instructions.
NGS data analysis was performed with Ion Reporter™ 5.0 software directly from within Torrent Suite™ 5.0.4 software (Thermo Fisher Scientific) along with the commercial analysis software Sequence Pilot version 4.3.0 (JSI Medical Systems GmbH, Ettenheim, Germany). The coverage analysis was performed using the coverage analysis plug-in v5.0.4.0. The statistics generated from this plug in were used to evaluate the quality of each library in the sequencing run. NGS amplification for each library was considered successful when a minimum average of 500 reads or greater was achieved across all target regions and the number of mapped reads was >150,000. Copy number variation (CNV) analysis was performed using the Ion Reporter Software directly from within the Torrent Suite Software (Thermo Fisher Scientific). CNVs are reported based on their copy number relative to the control sample used. The software reports all possible CNVs assigning a score, with scores >10 indicating high-confidence CNVs. This value is used as threshold for identifying a copy number amplification.
High resolution melting curve and Sanger sequencing analysis
EGFR exon 18, 19, 20, 21, KRAS and NRAS exon 2,3,4 and BRAF exon 11 and 15 mutation analysis was carried out by high resolution melting curve (HRM) analysis followed by sequencing analysis as previously described (18,19). For the Sanger sequencing reaction, PCR amplification products were purified using the NucleoFast® 96 PCR Clean-up kit (Macherey-Nagel, Düren, Germany), according to the manufacturer's protocol. The purified product (7 µl) was used for the sequencing reaction using the BigDye® Terminator v1.1 Cycle Sequencing kit (Applied Biosystems, Inc., Foster City, CA, USA). Sequencing reaction products were purified prior to electrophoresis using the Montage™ SEQ96 Sequencing Reaction kit (EMD Millipore Corporation, Billerica, MA, USA). Sequencing analysis was performed on an Applied Biosystems 3130 Genetic Analyzer (Applied Biosystems, Inc.).
TruSeq Custom Amplicon Targeted NGS assay
TruSeq Custom Amplicon Library Preparation (Illumina, Inc.) allows targeted sequencing of the genomic regions spanning upwards of 600 kb with up to 1,536 amplicons in a single multiplex reaction.
A pool of custom upstream and down primers was designed. The oligos were specific for amplification of specific regions involved in somatic mutations in different cancers. In total, 17 targets were amplified, using 42 amplicons, according to the manufacturer's protocol. The amplicons were located in BRAF, NRAS, BRAF, HRAS, cKIT, PDGFRA and EGFR genes. The library preparation was performed as previously described (18). Sequencing was carried out on the MiSeq sequencer (Illumina, Inc.). NGS data analysis was performed using Illumina's genomics computing environment BaseSpace.
Real-time PCR for ERBB2 and MET amplification
Copy number variation analysis of MET and ERBB2 gene amplification compared to a reference gene was carried out using TaqMan® Copy Number Assays (Hs01432482_cn and Hs00817646_cn) and TaqMan Copy Number Reference Assay (using TERT as endogenous reference gene). The real-time PCR quantification was performed on the Cobas 4800 (Roche Molecular Biochemicals). DNAs obtained from FFPE tissues were used. The qPCR reaction mixture contained 4 µl genomic DNA template (diluted to a concentration of 5 ng/µl), 10 µl of 2X TaqMan Genotyping Master Mix, 1 µl of the TaqMan copy number target assay (MET or ERBB2), 1 µl of the TaqMan Copy Number Reference Assay (TERT), and 4 µl of nuclease-free water. Each sample was run in a minimum of three replicates. Calculation of the relative amounts of the target gene compared to the reference gene was performed by Cobas 4800 Relative Quantification software (Roche Molecular Biochemicals). The final results are expressed as a ratio of the target gene to the reference gene copies in the sample, normalized with the ratio of target gene:reference gene copies in the Calibrator DNA (which was set to one). A ratio of <2.0 was assumed to be negative for gene over-amplification, a ratio of ≥2.0 was assumed to be positive for gene over-amplification.
Method sensitivity
The mutation detection limit was determined using genomic DNA reference standards with defined allelic frequencies (Horizon Diagnostics, Cambridge, UK).
Calculation of the NGS mutation detection limit was carried out using two EGFR multiplex reference standards that cover mutations at codons 719 (p.G719S), 746–750 (A746-E750del), 790 (p.T790M), 858 (p.L858R) and 861 (p.L861Q) spanning exons 19, 20 and 21. These standards were manufactured using five engineered EGFR mutant cell lines and mixed to generate a 5% and 1% mutant EGFR allelic frequency, respectively. A 3% reference standard was created by adding equimolecular quantities of the 5% and 1% mutant EGFR allelic frequency standards. Additionally, a third Quantitative Multiplex FFPE Reference Standard (Horizon Diagnostics) was used. This standard covers mutations at codons 719 (p.G719S), 746–750 (A746-E750del), 790 (p.T790M), 858 (p.L858R), with a mutant EGFR allelic frequency of 24.5, 2, 1 and 3%, respectively. A custom DNA Reference Standard (Horizon Diagnostics), containing KRAS (p.G12D, p.G13D, p.Q61H), NRAS (p.Q61R) and BRAF (p.V600E) mutations at a 5% mutant allelic frequency was also used. All reference standards were run in triplicate in three different runs.
Furthermore, a reference standard positive for EML4-ALK (Variant 1), CCDC6-RET and SLC34A2-ROS1 (Horizon) was used for the evaluation of the Ion AmpliSeq RNA Fusion Lung Cancer Research Panel.
Results
Test performance and comparison of methods
The performance of our customized AmpliSeq 23 gene panel was first evaluated using high resolution melting curve (HRM) analysis and sequencing in 100 consecutive samples in exons 18, 19, 20, 21 of the EGFR gene, exons 2, 3, 4 of the KRAS and NRAS genes and exons 11 and 15 of the BRAF gene. The concordance between HRM and NGS was 100% for all mutations detected in a percentage >5%. These 100 samples were also genotyped using a TruSeq Custom Amplicon assay (Illumina, Inc.) that targets hotspot regions in 7 genes (KRAS, NRAS, BRAF, cKIT, PDGFRA, EGFR, HRAS). A total of 21/100 (21%) samples tested were not efficiently amplified using the TruSeq Custom Amplicon assay, compared to only 1/100 (1%) of the samples tested using the AmpliSeq panel. This was attributed to the better DNA quality and quantity required by the TruSeq Custom Amplicon assay. In the 79 samples that were efficiently amplified by both methods, a full concordance was observed in the genes that were common to both panels (KRAS, NRAS, HRAS, BRAF, and EGFR). Two samples, presented detectable mutations by both NGS platforms (KRAS p.Q61H and EGFR p.L858R, respectively), but were classified as normal using the HRM method. Since the mutation rate in both samples was <4%, it was assumed that the discordance was due to the lower sensitivity of the HRM method.
All three methods applied can reliably be used for somatic mutation detection. However, the NGS-based approaches had increased sensitivity compared to the HRM method. Between the two NGS-based techniques used, the AmpliSeq technology had the advantage of being compatible with lower DNA concentrations, thus it was the method of choice for tumor molecular profiling in our cohort.
The sensitivity of the AmpliSeq method was also evaluated using 5 Horizon reference standards. The samples were analyzed in triplicate in 3 different experiments. In every experiment the same DNA was used for the construction of 3 different libraries. All EGFR, KRAS, NRAS and BRAF mutations present at mutant allelic frequency >3% in the reference standards were always detected by the AmpliSeq panel used in all three experiments performed in triplicate. Concerning the 1% EGFR reference standard, the performance was variable between the 3 replicates as well as between the three experiments. In all three experiments, p.G719S was not detected. The A746-E750del mutation was detectable in 2/3 replicates in the first experiment, in 2/3 replicates in the second and in 1/3 replicates in the third experiment. Similarly, p.L861Q was detected in 1/3 replicates in the first, in 1/3 replicates in the second and in 2/3 replicates in the third experiment. p.T790M was detected in 2/3 replicates in the first experiment, in 1/3 replicates in the second experiment and in 2/3 replicates in the third experiment. All EGFR mutations present in the a third Quantitative Multiplex FFPE Reference Standard (Horizon Diagnostics) were detected in all three NGS runs with the exception of the 790 mutation (1% mutant allelic frequency) which was inconsistently detected (in 50% of cases). Based on these results, we defined the mutant allelic frequency detection limit at 3%, since at this allelic frequency all variants were consistently detected.
The performance evaluation of the AmpliSeq RNA Fusion Lung Cancer Research Panel was carried out using 8 samples previously identified as positive for ALK translocation (by FISH) and one sample positive for ROS translocation. Additionally a Reference Standard positive for EML4-ALK (Variant 1), CCDC6-RET and SLC34A2-ROS1 was analyzed in triplicate in three different experiments. In all cases the correct gene translocation was detected.
Mutation distribution and patient characteristics
A total of 512 NSCLC patients were included in this study. In 10 patients, no amplification was obtained due to insufficient DNA quantity/quality, thus they were excluded from the study. In total, 502 NSCLC patients were eligible for NGS analysis (including the 99 patients of the evaluation cohort). Among these, 374 (74.5%) were male and 128 (25.5%) were female. The mean age of diagnosis was 66 years. The majority of tumors with known histology were classified as adenocarcinomas (85%).
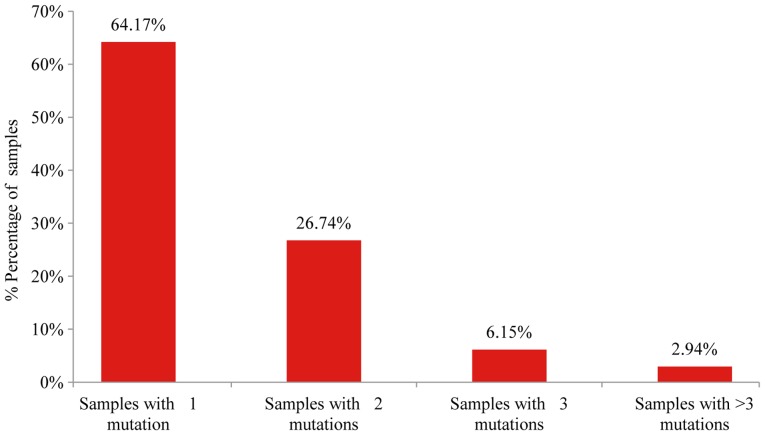
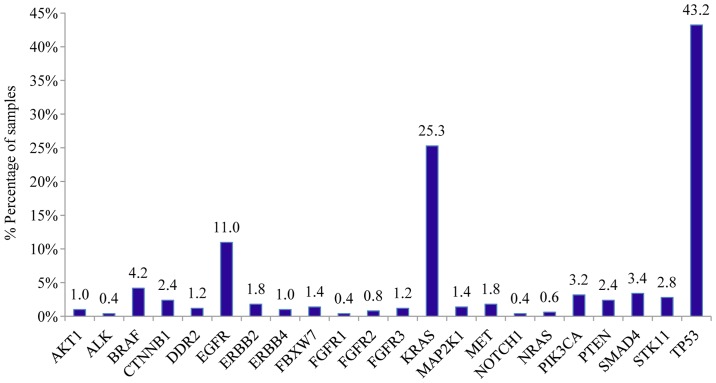
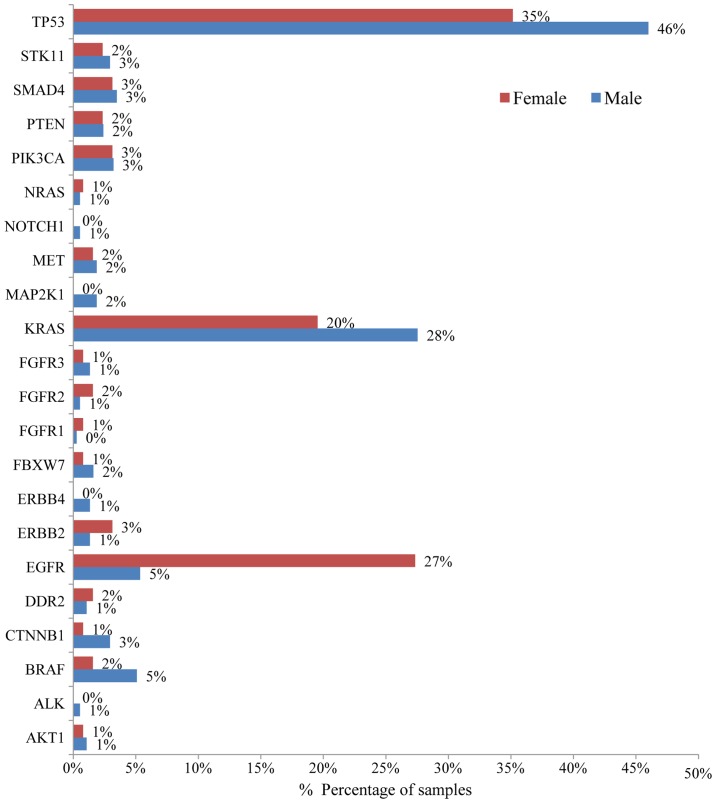
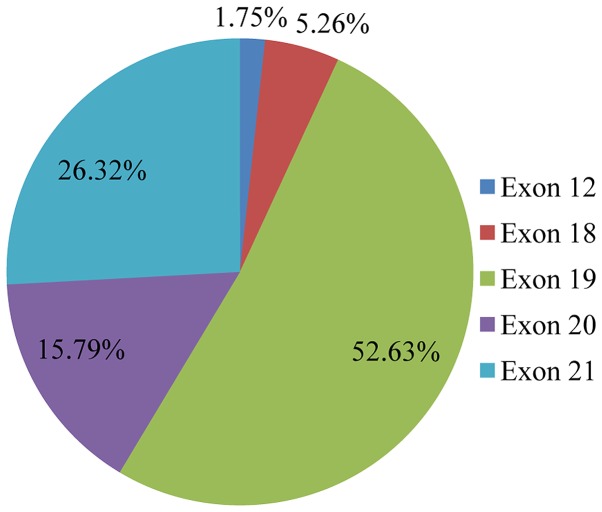
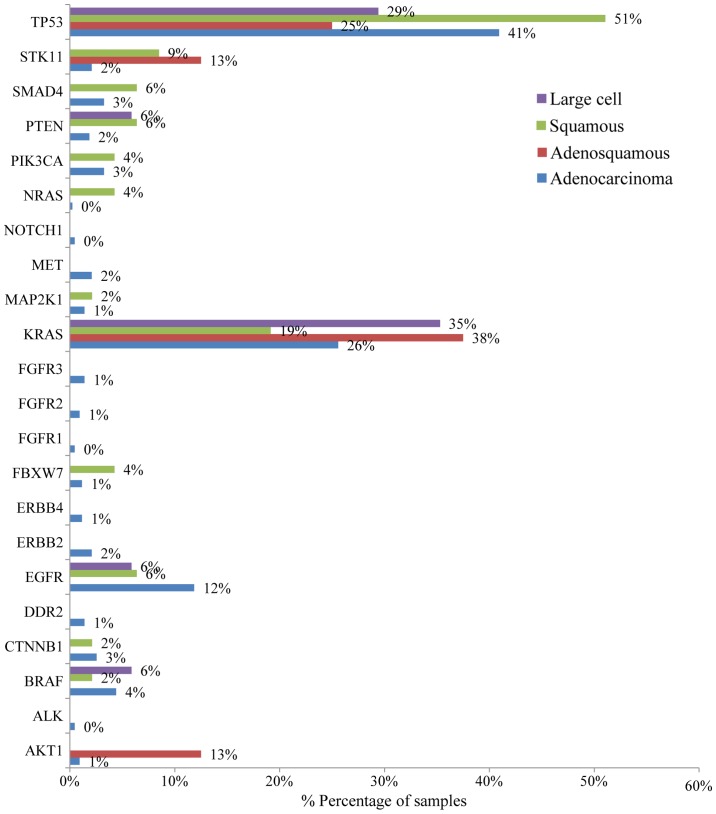
Mutation analysis of the 23 gene NGS panel revealed the presence of at least one mutation in 74.5% of the tumors tested (374/502). Among these 64.2% (240/374) presented only one mutation, 26.74% (100/374) two, 6.1% (23/374) three and 2.9% (11/374) more than three mutations (Fig. 1). In accordance with previous studies, TP53 mutations were the most common alterations (detected in 43% of the patients), followed by KRAS and EGFR gene mutations detected in 25% and 11% of the cases, respectively (Fig. 2) (20). The most notable difference in mutation frequency between male and female patients was observed as expected for the EGFR gene. In male patients the mutation frequency was much lower (5%) than in female patients (27%), indicating a gender-related EGFR mutation frequency (P<0.001) (Fig. 3). EGFR mutation distribution was 1 in exon 12 (1.75%), 3 in exon 18 (5.26%), 30 in exon 19 (52.63%), 9 in exon 20 (15.79%) and 15 in exon 21 (26.32%) (Fig. 4). In 1 sample, 2 concomitant exon 18 mutations were identified: G719S (drug-sensitive)+E709A compound mutation. In vitro the double-mutant receptor has been shown less sensitive to EGFR TKIs (21). All except one mutations detected in exon 19 were deletions. In one sample an insensitizing primary mutation was identified in exon 19 (p.D761Y) (22). p.L858R was the predominant mutation in exon 21 accounting for 90% of the mutations detected in that exon. Histopathology of the tumor appeared to be related to the EGFR mutation rates as well (Fig. 5). The greater mutation percentage was observed in adenocarcinomas tumors. On the contrary, adenosquamous, squamous and large cell NSCLC were associated with reduced mutation rates. This was the case for both males and female.
Figure 1.
Percentage of tumors with one, two, three or more detectable mutations among the 374 NSCLC tumors with detected mutations.
Figure 2.
Bar chart showing the distribution of mutated genes in 502 NSCLC patients.
Figure 3.
Bar chart showing mutation frequency among male (blue) and female (red) NSCLC patients.
Figure 4.
Pie chart showing the epidermal growth factor receptor EGFR mutation spectrum in NSCLC patients. Percentages were calculated out of the total mutated tumors.
Figure 5.
Bar chart showing mutation frequency according to the NSCLC patient histology.
KRAS gene somatic mutations were also present in a high percentage of NSCLC patients and are associated with poorer prognosis and resistance to EGFR-TKIs (19,23). In our cohort, a KRAS mutation was observed in 25.3% (127/502) of all tumors analyzed. Among the KRAS mutant patients, the majority of mutations was observed in exon 2 of the KRAS gene (111/127, 87.4%), while a KRAS exon 3 mutation was observed in 7.81% (10/127) and an exon 4 mutation was observed in 4.69% (6/127) of the cases.
Of note, 79.4% of the KRAS exon 2 mutations detected were transversion mutations (substitution of a purine for a pyrimidine or conversely, G→T or G→C), which are known to be smoking-associated (24), while the percentage of transition mutations (substitution of a purine for a purine, e.g., G→A or a pyrimidine for a pyrimidine, C→T) was limited to 20.6% of the patients.
BRAF is an important new therapeutic target in NSCLC (25–27). In our study, a mutation was observed in 4.18% (21/502) of the cases. Among the BRAF mutant cases, the majority was observed in exon 11 of BRAF (12/21, 57.14%), while in 42.86% (9/21) of the BRAF mutant patients an exon 15 mutation was observed. However, only in 4 cases the V600E mutation which is associated with targeted therapy was detected. Thus, the V600E mutation percentage in NSCLC patients was only 0.8% (4/502).
Other genes with a significant mutation frequency in the NSCLC cohort tested included ERBB2, MET, CTNBB1 and PTEN (with mutation rates for each one of ~2%), PIK3CA, SMAD4, STK11 (each one with mutation frequency ~2%). At a frequency ~1%, mutations were also observed for AKT1, DDR2, ERBB4, FBWX7, FGFR1, FGFR3, MAP2K1 and NRAS.
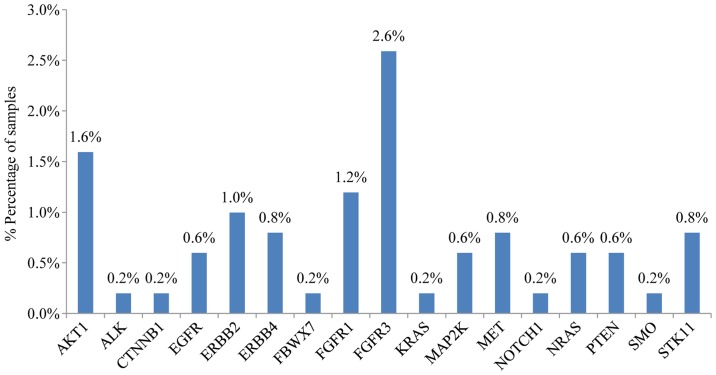
In addition to the presence of somatic mutations, this 23 gene panel was also accessed for the detection of copy number variations (CNV) in cancer-driver genes. Regarding NSCLC, copy number amplifications (CNA) have been observed for MET, ERBB2, FGFR1 and FGFR3 that represent viable treatment targets for approved or investigational therapies (28–31) (Fig. 6). The presence of MET and ERBB2 amplification was also confirmed by real-time PCR in all samples.
Figure 6.
Bar chart showing gene CNA (copy number amplification) in 502 NSCLC patients.
The analysis of the major ALK, RET, ROS1, and NTRK1 fusion transcripts using the Ion AmpliSeq RNA Fusion Lung Cancer Research Panel, revealed the presence of an ALK fusion in 2.2% (11/502) of the samples while a ROS or RET fusion was present in 0.4% and 0.6% of the cases, respectively (2/502 and 3/502) (Fig. 7). A complete list of gene mutation fusions and copy number amplifications are available upon request.
Figure 7.
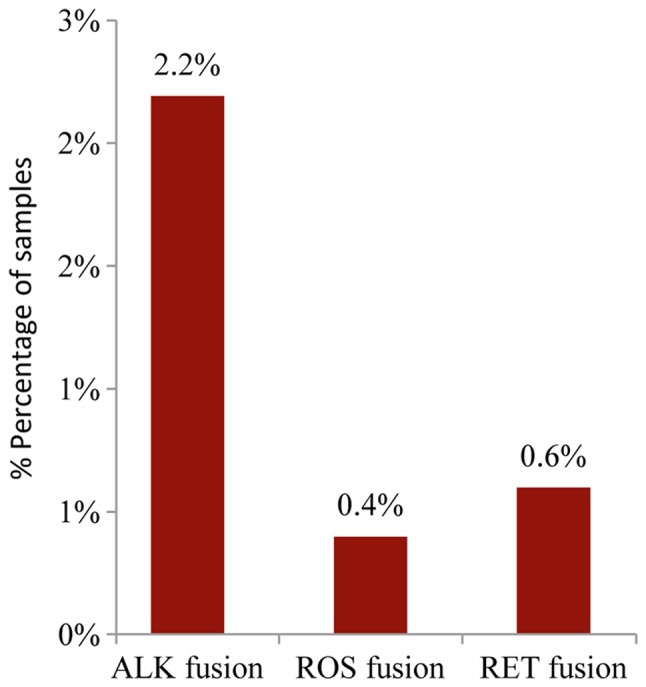
Bar chart showing gene fusion frequencies in 502 NSCLC tumors.
Discussion
During the past few years, cancer treatment has been redirected toward the use of targeted therapeutics, with their effectiveness depending on tumor-specific alterations. Consequently, the management of neoplastic diseases has changed substantially, since the treatment decision is no longer unspecific but depends on the molecular profile of the tumor for patient selection (32).
A main prerequisite for the accurate molecular characterization of the tumor is the selection of the optimal analysis method. Single gene analysis methods are time consuming and require a large amount of tumor DNA, which in the majority of the cases is not available. Furthermore, the number of genetic alterations associated with targeted therapies is constantly increasing. Thus, global analysis of the genetic profile of tumors is crucial and cannot be achieved by sequential analysis of a few genes (33).
Targeted NGS, involving gene panels, is a quick and cost effective multiple gene analysis method that allows a more comprehensive tumor mutation profiling. The selection of the optimal NGS methodology should be based on its performance using a limited and bad quality DNA extracted from FFPE tissues. Additionally, a careful selection of the genes included in the panels should be conducted in order to include all important treatment-related targets. Thus, targeted NGS panels for somatic mutation detection include actionable cancer genes and allow the determination of the tumor molecular profile of the patient (34,35).
In the present study, a targeted amplicon-based NGS assay was used for simultaneous analysis of tumor-specific alterations in 23 genes commonly mutated in lung and colorectal cancers. This panel used the Ion AmpliSeq technology offered by Thermo Fisher Scientific and was selected since it has several advantages that make it an attractive option of diagnostic utility. AmpliSeq-based panels are currently available or can be custom made. They are cost effective, have good performance, are compatible with low DNA concentration and have very low failure rates even with DNAs of relative low quality such as those extracted from FFPE samples. Consequently, they can be an attractive option and of clinical value for somatic mutation analysis (13,14,35).
The gene panel used in this study contained all important genes with actionable mutations related to NSCLC. Therefore, it was deemed optimal for clinical use in this tumor type. Additionally, it exhibited high rates (100%) of sensitivity and specificity at a low mutation frequency of 3%. This is extremely important for somatic mutation detection since, due to tumor heterogeneity, the mutation rate in tumor tissue could be extremely low. Furthermore, it allows the detection of different types of alteration in one workflow (mutations, copy number changes, fusions, expression).
The aim of such an approach is the determination of the molecular profile (gene mutations) of the tumor, the determination of the drug (if it exists) that targets the mutated gene(s) or the pathway that the gene(s) is involved in and the determination of gene interaction (in case of multiple mutations). Moreover, patients can be assigned to clinical trials associated with their molecular profile and consequently have more potential treatment options in addition to FDA-approved targeted therapies. This could be particularly important for patients who have failed conventional therapy. In fact, there is an increasing number of clinical trials in which patient access depends on the molecular profile of the tumor determined by NGS technology (36–38). There are two main approaches. The so-called ‘basket’ trials enroll patients with multiple tumor types and assign different therapies based on their tumor genetic alterations. The National Cancer Institute (NCI)-Molecular Analysis for Therapy Choice (NCI MATCH) Trial (39), and the Targeted Agent and Profiling Utilization Registry (TAPUR) (40), which is sponsored by the American Society of Clinical Oncology (ASCO) are two main examples of such trials in previously treated advanced solid tumors. Alternatively, the ‘umbrella’ trials enroll patients with a single tumor type, and assign various treatments according to their tumor molecular profile. Examples of such an approach constitute the Lung Master Protocol (Lung MAP) trial in recurrent stage IV squamous cell lung cancer (41), the National Lung Matrix Trial (42) and the Biomarker-Integrated Targeted Therapy Study in Previously Treated Patients with Advanced NSCLC (BATTLE-2) (43).
NSCLC is a tumor type with the most available targeted therapies. Genes with approved targeted agents include EGFR, ALK, and ROS1 (3,44). Additionally, specific genetic alterations in NSCLC are now recognized as optimal targets for agents approved in other tumor types. These agents target genes that are frequently altered in NSCLC such as KRAS, ERBB2, MET and RET, that are recommended as emerging new biomarkers by the National Comprehensive Cancer Network (NCCN) guidelines. Furthermore, potential targets, such as BRAF, PIK3CA, FGFR1, DDR2, PTEN are currently being tested in clinical trials (45,46). Therefore, the number of predictive biomarkers for novel targeted drugs entering into clinical practice is rapidly increasing. The selected gene panel includes the most commonly mutated genes in lung and colorectal cancer. Mutational status of some of the genes tested may be associated with the activity of certain approved drugs. This is obvious when accessing data from MyCancer Genome knowledge database (www.mycancergenome.org/), that provides reliable information concerning important cancer-related genes and their correlation with treatment options. As indicated in Table I, multiple approved therapies targeting important cancer-related genes or their pathways are available. However, each agent has received approval for a particular tumor type(s) and not all agents targeting specific genes have sufficient clinical evidence in a patient tumor type. In principal, the molecular profile of a tumor could be indicative of the treatment that should be followed if an approved targeted therapy can be related to the patients genetic profile or if clinical trials exist (37,38). Thus, particular caution should be made when using the genetic information retrieved from these panels.
Table I.
Approved agents targeting important cancer-related genes or their pathways.
| Agent | Target(s) | FDA-approved indication(s) |
|---|---|---|
| Cetuximab (Erbitux) | EGFR (HER1/ERBB1) | Colorectal cancer (KRAS wild-type), squamous cell cancer of the head and neck |
| Panitumumab (Vectibix) | EGFR (HER1/ERBB1) | Colorectal cancer (KRAS wild-type) |
| Regorafenib (Stivarga) | KIT, PDGFRβ, RAF, RET, VEGFR1/2/3 | Colorectal cancer, gastrointestinal stromal tumors |
| Crizotinib (XALKori) | ALK, MET, ROS1 | Non-small cell lung cancer (with ALK fusion or ROS1 gene alteration) |
| Alectinib (Alecensa) | ALK | Non-small cell lung cancer (with ALK fusion) |
| Ceritinib (Zykadia) | ALK | Non-small cell lung cancer (with ALK fusion) |
| Gefitinib (Iressa) | EGFR (HER1/ERBB1) | Non-small cell lung cancer [with EGFR exon 19 deletions or exon 21 substitution (L858R) mutations] |
| Afatinib (Gilotrif) | EGFR (HER1/ERBB1), HER2 (ERBB2/neu) | Non-small cell lung cancer [with EGFR exon 19 deletions or exon 21 substitution (L858R) mutations] |
| Erlotinib (Tarceva) | EGFR (HER1/ERBB1) | Non-small cell lung cancer [with EGFR exon 19 deletions or exon 21 substitution (L858R) mutations], pancreatic cancer |
| Osimertinib (Tagrisso) | EGFR | Non-small cell lung cancer (with EGFR T790M mutation) |
| Necitumumab (Portrazza) | EGFR (HER1/ERBB1) | Squamous non-small cell lung cancer |
| Ponatinib (Iclusig) | ABL, FGFR1-3, FLT3, VEGFR2 | Chronic myelogenous leukemia, acute lymphoblastic leukemia (Philadelphia chromosome-positive) |
| DaBRAFenib (Tafinlar) | BRAF | Melanoma (with BRAF V600 mutation) |
| Vemurafenib (Zelboraf) | BRAF | Melanoma (with BRAF V600 mutation) |
| Vandetanib (Caprelsa) | EGFR (HER1/ERBB1), RET, VEGFR2 | Medullary thyroid cancer |
| Cabozantinib [Cabometyx (tablet), Cometriq (capsule)] | FLT3, KIT, MET, RET, VEGFR2 | Medullary thyroid cancer, renal cell carcinoma |
| Ado-trastuzumab emtansine (Kadcyla) | HER2 (ERBB2/neu) | Breast cancer (HER2+) |
| Pertuzumab (Perjeta) | HER2 (ERBB2/neu) | Breast cancer (HER2+) |
| Trastuzumab (Herceptin) | HER2 (ERBB2/neu) | Breast cancer (HER2+), gastric cancer (HER2+) |
| Lapatinib (Tykerb) | HER2 (ERBB2/neu), EGFR (HER1/ERBB1) | Breast cancer (HER2+) |
| Cobimetinib (Cotellic) | MEK | Melanoma (with BRAF V600E or V600K mutation) |
| Trametinib (Mekinist) | MEK | Melanoma (with BRAF V600 mutation) |
| Everolimus (Afinitor) | mTOR | Pancreatic, gastrointestinal, or lung origin neuroendocrine tumor, renal cell carcinoma, breast cancer (HR+, HER2−), non-resectable subependymal giant cell astrocytoma associated with tuberous sclerosis |
| Temsirolimus (Torisel) | mTOR | Renal cell carcinoma |
| Sorafenib (Nexavar) | VEGFR, PDGFR, KIT, RAF | Hepatocellular carcinoma, thyroid carcinoma, renal cell carcinoma |
The information listed in this table was retrieved from MyCancer Genome Site (www.mycancergenome.org/).
Molecular tumor profiling using our multigene panel was feasible for the majority of the patients tested with a limited amplification failure rate of approximately 2%. At least one DNA mutation was detected in 374 patients (74.5%). In 16 patients, (3.2%) an RNA fusion was identified. In accordance to previous studies, the most commonly mutated gene in our cohort was the TP53 gene (43%), followed by KRAS and EGFR gene mutations detected in 25 and 11% of the cases (20). As expected, great differences in EGFR mutation frequency were observed between male (5%) and female patients (27%) (18). The mutual exclusivity of EGFR, KRAS, BRAF and HER2 genes, as well as ALK, ROS1 and RET fusions has been already described and it was also confirmed in our cohort (47).
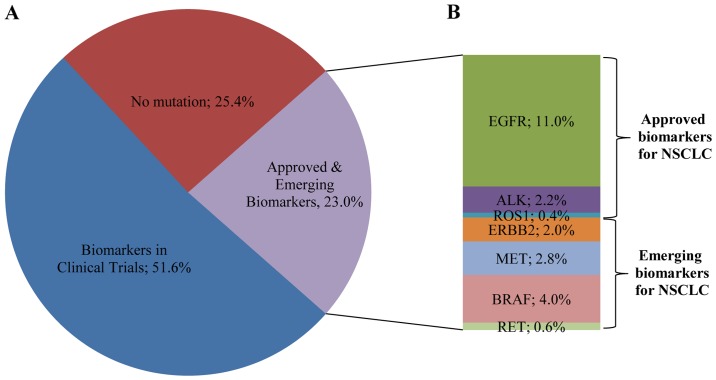
In the present study, 23% of the patients presented a mutation in a gene associated with approved or emerging treatments. More specifically, 13.5% (68/502) of the patients presented a mutation in a gene with approved targeted therapy (EGFR, ALK, ROS1) and 9.4% (47/502) of the patients had an alteration in a gene related to emerging targeted therapies according to the NCCN guidelines (Fig. 8). These alterations include ERBB2, BRAF and MET mutations, MET amplification and RET rearrangements. Thus, the information provided by this multigene analyses, facilitates a physician's decision concerning the selection of the appropriate treatment options. The remaining 51.6% (259/502) of the patients had a mutation in a gene that could be related to an off label therapy or indicate access to a clinical trial (Fig. 8). The identification of such alterations could be valuable in cases with advanced tumors that have progressed on standard therapies since it offers these patients the opportunity to be enrolled in ongoing clinical trials.
Figure 8.
(A) Percentage of patients with mutations associated with approved, emerging and clinical trial biomarkers. (B) Distribution of somatic gene alterations among NSCLC patients eligible for approved or emerging therapies for NSCLC patients.
In conclusion, the multigene NGS panel applied was able to identify alterations in a cancer-driver gene (including point mutations, gene rearrangements and MET amplifications) in 77.6% of the tumors tested. The method used has a vast applicability in tumor molecular characterization of NSCLC, allowing the simultaneous detection of actionable DNA alterations in 23 genes in addition to 4 gene fusions in these patients. Furthermore, due to its cost and time effectiveness, it can be efficiently applied for molecular characterization of tumors in order to increase the percentage of patients with gene alterations related to approved, emerging or ongoing clinical trials for targeted therapy.
References
- 1.Torre LA, Siegel RL, Jemal A. Lung Cancer Statistics. Adv Exp Med Biol. 2016;893:1–19. doi: 10.1007/978-3-319-24223-1_1. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D, Bray F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries in 2012. Eur J Cancer. 2013;49:1374–1403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 3.Tsao AS, Scagliotti GV, Bunn PA, Jr, Carbone DP, Warren GW, Bai C, de Koning HJ, Yousaf-Khan AU, McWilliams A, Tsao MS, et al. Scientific advances in lung cancer 2015. J Thorac Oncol. 2016;11:613–638. doi: 10.1016/j.jtho.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 4.Gagan J, Van Allen EM. Next-generation sequencing to guide cancer therapy. Genome Med. 2015;7:80. doi: 10.1186/s13073-015-0203-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diaz Z, Aguilar-Mahecha A, Paquet ER, Basik M, Orain M, Camlioglu E, Constantin A, Benlimame N, Bachvarov D, Jannot G, et al. Next-generation biobanking of metastases to enable multidimensional molecular profiling in personalized medicine. Mod Pathol. 2013;26:1413–1424. doi: 10.1038/modpathol.2013.81. [DOI] [PubMed] [Google Scholar]
- 6.Meldrum C, Doyle MA, Tothill RW. Next-generation sequencing for cancer diagnostics: A practical perspective. Clin Biochem Rev. 2011;32:177–195. [PMC free article] [PubMed] [Google Scholar]
- 7.Pavlopoulou A, Spandidos DA, Michalopoulos I. Human cancer databases (Review) Oncol Rep. 2015;33:3–18. doi: 10.3892/or.2014.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, et al. COSMIC: Exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chin L, Hahn WC, Getz G, Meyerson M. Making sense of cancer genomic data. Genes Dev. 2011;25:534–555. doi: 10.1101/gad.2017311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu TJ, Schriml LM, Chen QR, Colbert M, Crichton DJ, Finney R, Hu Y, Kibbe WA, Kincaid H, Meerzaman D, et al. Generating a focused view of disease ontology cancer terms for pan-cancer data integration and analysis. Database (Oxford) 2015;2015:bav032. doi: 10.1093/database/bav032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dumur CI. Available resources and challenges for the clinical annotation of somatic variations. Cancer Cytopathol. 2014;122:730–736. doi: 10.1002/cncy.21471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L, Li Y, Li S, Hu N, He Y, Pong R, Lin D, Lu L, Law M. Comparison of next-generation sequencing systems. J Biomed Biotechnol. 2012;2012:251364. doi: 10.1155/2012/251364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tops BB, Normanno N, Kurth H, Amato E, Mafficini A, Rieber N, Le Corre D, Rachiglio AM, Reiman A, Sheils O, et al. Development of a semi-conductor sequencing-based panel for genotyping of colon and lung cancer by the Onconetwork consortium. BMC Cancer. 2015;15:26. doi: 10.1186/s12885-015-1015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'Haene N, Le Mercier M, De Nève N, Blanchard O, Delaunoy M, El Housni H, Dessars B, Heimann P, Remmelink M, Demetter P, et al. Clinical validation of targeted next generation sequencing for colon and lung cancers. PLoS One. 2015;10:e0138245. doi: 10.1371/journal.pone.0138245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jennings LJ, Arcila ME, Corless C, Kamel-Reid S, Lubin IM, Pfeifer J, Temple-Smolkin RL, Voelkerding KV, Nikiforova MN. Guidelines for validation of next-generation sequencing-based oncology panels: A joint consensus recommendation of the Association for Molecular Pathology and College of American Pathologists. J Mol Diagn. 2017;19:341–365. doi: 10.1016/j.jmoldx.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shahsiah R, DeKoning J, Samie S, Latifzadeh SZ, Kashi ZM. Validation of a next generation sequencing panel for detection of hotspot cancer mutations in a clinical laboratory. Pathol Res Pract. 2017;213:98–105. doi: 10.1016/j.prp.2016.11.016. [DOI] [PubMed] [Google Scholar]
- 17.Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, Chirieac LR, Dacic S, Duhig E, Flieder DB, et al. WHO Panel: The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol. 2015;10:1243–1260. doi: 10.1097/JTO.0000000000000630. [DOI] [PubMed] [Google Scholar]
- 18.Papadopoulou E, Tsoulos N, Tsirigoti A, Apessos A, Agiannitopoulos K, Metaxa-Mariatou V, Zarogoulidis K, Zarogoulidis P, Kasarakis D, Kakolyris S, et al. Determination of EGFR and KRAS mutational status in Greek non-small-cell lung cancer patients. Oncol Lett. 2015;10:2176–2184. doi: 10.3892/ol.2015.3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Negru S, Papadopoulou E, Apessos A, Stanculeanu DL, Ciuleanu E, Volovat C, Croitoru A, Kakolyris S, Aravantinos G, Ziras N, et al. KRAS, NRAS and BRAF mutations in Greek and Romanian patients with colorectal cancer: A cohort study. BMJ Open. 2014;4:e004652. doi: 10.1136/bmjopen-2013-004652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scoccianti C, Vesin A, Martel G, Olivier M, Brambilla E, Timsit JF, Tavecchio L, Brambilla C, Field JK, Hainaut P. European Early Lung Cancer Consortium: Prognostic value of TP53, KRAS and EGFR mutations in nonsmall cell lung cancer: The EUELC cohort. Eur Respir J. 2012;40:177–184. doi: 10.1183/09031936.00097311. [DOI] [PubMed] [Google Scholar]
- 21.Tam IY, Leung EL, Tin VP, Chua DT, Sihoe AD, Cheng LC, Chung LP, Wong MP. Double EGFR mutants containing rare EGFR mutant types show reduced in vitro response to gefitinib compared with common activating missense mutations. Mol Cancer Ther. 2009;8:2142–2151. doi: 10.1158/1535-7163.MCT-08-1219. [DOI] [PubMed] [Google Scholar]
- 22.Costa DB. Kinase inhibitor-responsive genotypes in EGFR mutated lung adenocarcinomas: Moving past common point mutations or indels into uncommon kinase domain duplications and rearrangements. Transl Lung Cancer Res. 2016;5:331–337. doi: 10.21037/tlcr.2016.06.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kempf E, Rousseau B, Besse B, Paz-Ares L. KRAS oncogene in lung cancer: Focus on molecularly driven clinical trials. Eur Respir Rev. 2016;25:71–76. doi: 10.1183/16000617.0071-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahrendt SA, Decker PA, Alawi EA, Zhu Yr YR, Sanchez-Cespedes M, Yang SC, Haasler GB, Kajdacsy-Balla A, Demeure MJ, Sidransky D. Cigarette smoking is strongly associated with mutation of the K-ras gene in patients with primary adenocarcinoma of the lung. Cancer. 2001;92:1525–1530. doi: 10.1002/1097-0142(20010915)92:6<1525::AID-CNCR1478>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 25.Cardarella S, Ogino A, Nishino M, Butaney M, Shen J, Lydon C, Yeap BY, Sholl LM, Johnson BE, Jänne PA. Clinical, pathologic, and biologic features associated with BRAF mutations in non-small cell lung cancer. Clin Cancer Res. 2013;19:4532–4540. doi: 10.1158/1078-0432.CCR-13-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Villaruz LC, Socinski MA, Abberbock S, Berry LD, Johnson BE, Kwiatkowski DJ, Iafrate AJ, Varella-Garcia M, Franklin WA, Camidge DR, et al. Clinicopathologic features and outcomes of patients with lung adenocarcinomas harboring BRAF mutations in the Lung Cancer Mutation Consortium. Cancer. 2015;121:448–456. doi: 10.1002/cncr.29042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen-Ngoc T, Bouchaab H, Adjei AA, Peters S. BRAF alterations as therapeutic targets in non-small-cell lung cancer. J Thorac Oncol. 2015;10:1396–1403. doi: 10.1097/JTO.0000000000000644. [DOI] [PubMed] [Google Scholar]
- 28.Dutt A, Ramos AH, Hammerman PS, Mermel C, Cho J, Sharifnia T, Chande A, Tanaka KE, Stransky N, Greulich H, et al. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS One. 2011;6:e20351. doi: 10.1371/journal.pone.0020351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jorge SE, Schulman S, Freed JA, VanderLaan PA, Rangachari D, Kobayashi SS, Huberman MS, Costa DB. Responses to the multitargeted MET/ALK/ROS1 inhibitor crizotinib and co-occurring mutations in lung adenocarcinomas with MET amplification or MET exon 14 skipping mutation. Lung Cancer. 2015;90:369–374. doi: 10.1016/j.lungcan.2015.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mar N, Vredenburgh JJ, Wasser JS. Targeting HER2 in the treatment of non-small cell lung cancer. Lung Cancer. 2015;87:220–225. doi: 10.1016/j.lungcan.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 31.Collisson EA, Campbell JD, Brooks AN, Berger AH, Lee W, Chmielecki J, Beer DG, Cope L, Creighton CJ, Danilova L, et al. Cancer Genome Atlas Research Network: Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirsch FR, Suda K, Wiens J, Bunn PA., Jr New and emerging targeted treatments in advanced non-small-cell lung cancer. Lancet. 2016;388:1012–1024. doi: 10.1016/S0140-6736(16)31473-8. [DOI] [PubMed] [Google Scholar]
- 33.Kamps R, Brandão RD, Bosch BJ, Paulussen AD, Xanthoulea S, Blok MJ, Romano A. Next-generation sequencing in oncology: Genetic diagnosis, risk prediction and cancer classification. Int J Mol Sci. 2017;18:E308. doi: 10.3390/ijms18020308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takeda M, Sakai K, Terashima M, Kaneda H, Hayashi H, Tanaka K, Okamoto K, Takahama T, Yoshida T, Iwasa T, et al. Clinical application of amplicon-based next-generation sequencing to therapeutic decision making in lung cancer. Ann Oncol. 2015;26:2477–2482. doi: 10.1093/annonc/mdv475. [DOI] [PubMed] [Google Scholar]
- 35.de Leng WW, Gadellaa-van Hooijdonk CG, Barendregt-Smouter FA, Koudijs MJ, Nijman I, Hinrichs JW, Cuppen E, van Lieshout S, Loberg RD, de Jonge M, et al. Targeted next generation sequencing as a reliable diagnostic assay for the detection of somatic mutations in tumours using minimal DNA amounts from formalin fixed paraffin embedded material. PLoS One. 2016;11:e0149405. doi: 10.1371/journal.pone.0149405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siu LL, Conley BA, Boerner S, LoRusso PM. Next-generation sequencing to guide clinical trials. Clin Cancer Res. 2015;21:4536–4544. doi: 10.1158/1078-0432.CCR-14-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rashdan S, Gerber DE. Going into BATTLE: Umbrella and basket clinical trials to accelerate the study of biomarker-based therapies. Ann Transl Med. 2016;4:529. doi: 10.21037/atm.2016.12.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sundar R, Chénard-Poirier M, Collins DC, Yap TA. Imprecision in the era of precision medicine in non-small cell lung cancer. Front Med (Lausanne) 2017;4:39. doi: 10.3389/fmed.2017.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colwell J. NCI-MATCH trial draws strong interest. Cancer Discov. 2016;6:334. doi: 10.1158/2159-8290.CD-NB2016-018. [DOI] [PubMed] [Google Scholar]
- 40.Markman M. Maurie Markman on the Groundbreaking TAPUR Trial. Oncology (Williston Park) 2017;31:158–168. [PubMed] [Google Scholar]
- 41.Herbst RS, Gandara DR, Hirsch FR, Redman MW, LeBlanc M, Mack PC, Schwartz LH, Vokes E, Ramalingam SS, Bradley JD, et al. Lung master protocol (Lung-MAP)-A biomarker-driven protocol for accelerating development of therapies for squamous cell lung cancer: SWOG S1400. Clin Cancer Res. 2015;21:1514–1524. doi: 10.1158/1078-0432.CCR-13-3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Middleton G, Crack LR, Popat S, Swanton C, Hollingsworth SJ, Buller R, Walker I, Carr TH, Wherton D, Billingham LJ. The National Lung Matrix Trial: Translating the biology of stratification in advanced non-small-cell lung cancer. Ann Oncol. 2015;26:2464–2469. doi: 10.1093/annonc/mdv394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim C, Giaccone G. Lessons learned from BATTLE-2 in the war on cancer: The use of Bayesian method in clinical trial design. Ann Transl Med. 2016;4:466. doi: 10.21037/atm.2016.11.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chan BA, Hughes BG. Targeted therapy for non-small cell lung cancer: Current standards and the promise of the future. Transl Lung Cancer Res. 2015;4:36–54. doi: 10.3978/j.issn.2218-6751.2014.05.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rothschild SI. Targeted therapies in non-small cell lung cancer-beyond EGFR and ALK. Cancers (Basel) 2015;7:930–949. doi: 10.3390/cancers7020816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zer A, Leighl N. Promising targets and current clinical trials in metastatic non-squamous NSCLC. Front Oncol. 2014;4:329. doi: 10.3389/fonc.2014.00329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kohno T, Nakaoku T, Tsuta K, Tsuchihara K, Matsumoto S, Yoh K, Goto K. Beyond ALK-RET, ROS1 and other oncogene fusions in lung cancer. Transl Lung Cancer Res. 2015;4:156–164. doi: 10.3978/j.issn.2218-6751.2014.11.11. [DOI] [PMC free article] [PubMed] [Google Scholar]