Abstract
Although the introduction of tyrosine kinase inhibitors greatly improved the survival of patients with chronic myeloid leukemia (CML), drug resistance remains a problem. Thus, mechanism-based novel therapeutic targets warrant exploration. Recently, epidermal growth factor receptor kinase substrate 8 (EPS8), which has been identified as an oncogene and plays an important role in a broad spectrum of solid tumours, was reported to be related to poor prognosis or chemoresistance in acute leukemia patients. However, its role in CML remains unclear. In the present study, using q-RT-PCR, we demonstrated that CML patients expressed a higher level of EPS8 mRNA in bone marrow mononuclear cells than healthy controls. Then, to determine the effect of EPS8 on the biological functions of CML cells, EPS8 expression was knocked down in the human CML cell line K562. Reduced proliferation, increased apoptosis, impaired adhesion and migration were observed in K562 cells after EPS8 silencing. Notably, attenuation of EPS8 increased chemosensitivity both in imatinib-sensitive K562 cells and in the imatinib-resistant murine BCR-ABL+ 32D-p210BCR/ABL-T315I cells. Mechanistically, knockdown of EPS8 downregulated p-BCR/ABL and its downstream AKT/mTOR signalling pathway. Finally, knockdown of EPS8 attenuated K562 cell proliferation in BALB/c nude mice. These data indicated that EPS8 regulated the proliferation, apoptosis and chemosensitivity in BCR-ABL positive cells via the BCR-ABL/PI3K/AKT/mTOR pathway. Targeting EPS8 alone or combined with a tyrosine kinase inhibitor may be a promising alternative therapeutic strategy.
Keywords: epidermal growth factor receptor kinase substrate 8, chronic myeloid leukemia, apoptosis, drug resistant, mTOR
Introduction
Chronic myeloid leukemia (CML) is a BCR-ABL oncogene-driven malignant disease, characterized by markedly elevated immature myeloid cells in bone marrow and peripheral blood. CML cells progress slowly in the chronic phase (CP) for about one to three years, and then proliferate more rapidly stepping into the accelerated phase (AP) with an increase in blast numbers. The accelerated phase lasts for only several months and eventually converts to acute leukemia in the blast crisis phase (BC) with more aggressive characteristics than de novo acute leukemia. Most CML patients respond well to the tyrosine kinase inhibitor (TKI) imatinib in the chronic phase, however, ~20–30% patients develop resistance to imatinib (1–3). Some of the patients are de novo resistant to imatinib, others exhibit a good response in the beginning, however this response is lost with the progression of this disease. Almost half of the imatinib-resistant patients develop point mutations in the BCR/ABL gene during the course of TKI treatment. Other drug resistance mechanisms include BCR-ABL amplification, additional acquired gene mutation and drug efflux (4,5). Second and third generation tyrosine kinase inhibitors such as dasatinib, ponatinib, are able to overcome imatinib resistance in some patients. However, some mechanisms, for example, BCR/ABL point mutation T315I-mediated resistance cannot be overcome by current available clinical drugs thus highlighting the need for further research on the mechanism of leukemogenesis of CML cells in order to explore novel mechanism-based strategies with high efficacy and low toxicity.
Epidermal growth factor receptor kinase substrate 8 (EPS8) is a cytoplasmic protein that acts as a substrate of receptor and non-receptor tyrosine kinases such as EGFR, FGFR, VEGFR and Src kinase. EPS8 functionally serves as an adaptor protein associating with diverse partner proteins to form complexes that regulate multiple signalling pathways. Physiologically, EPS8 forms a complex with Abi-1 and SOS-1 to regulate the Rac signalling pathway promoting cytoskeletal remodelling. EPS8 also plays a role in membrane flow, pseudopodium formation, morphogenesis of microvilli, stereocilia function and length, cellular adhesion and motility (6). Furthermore, EPS8 has been identified as an oncogene, as it enables cellular transformation in vitro and tumour formation in vivo upon overexpression (7). EPS8 has been documented to be highly expressed in a broad spectrum of solid tumours, such as squamous carcinoma, cervical cancer, colon carcinoma, and breast cancer (8–12). However, only a few studies have addressed the role of EPS8 in haematological malignancies. Microarray analysis by Kang et al revealed that a high level of EPS8 predicted a poor prognosis of infant acute lymphoblastic leukemia (ALL) patients with MLL rearrangements (13). In addition, we previously determined that increased expression of EPS8 mRNA in bone marrow was related to a poor response to chemotherapy and a poor prognosis in acute myeloid leukemia (AML) and ALL patients (14,15). However, it remains unclear whether EPS8 is implicated in CML and how EPS8 regulates the biological functions of CML cells.
In the present study we performed q-RT-PCR to demonstrate that CML patients expressed higher EPS8 mRNA than healthy controls in bone marrow mononuclear cells. Then, we knocked down the expression of EPS8 in the CML cell line K562 and observed that attenuated EPS8 reduced proliferation, increased apoptosis, arrested the cell cycle at the G1 phase and reduced adhesion and migration. Notably, silencing EPS8 increased chemosensitivity both in the imatinib sensitive cell line K562 and the resistant cell line 32D-p210BCR/ABL-T315I. Mechanistically, knockdown of EPS8 downregulated p-BCR/ABL and its downstream AKT/mTOR signalling pathway. Notably, knockdown of EPS8 attenuated K562 cell proliferation in BALB/c nude mice. Collectively, these data revealed that EPS8 regulated the cell biology of CML. Targeting EPS8 alone or combined with TKI may be promising therapeutic strategies for refractory and relapsed CML patients.
Materials and methods
Cell lines and human samples
Bone marrow mononuclear cells were collected from patients with CML at the Department of Hematology of Zhujiang Hospital, Southern Medical University from 2013 to 2015. Some of the RNA samples were purchased from KingMed Diagnostics (Guangzhou, China). In total, 113 cases of CML (male n=60, female n=53) including 50 cases of chronic phase (CP), 21 cases of accelerated phase (AP) and 21 cases of blast crisis phase (BC) as well as 21 CML patients in complete remission (CR) and 21 normal control were enrolled. In these cases 82 CML patients had clinical data of their quantitative BCR-ABL-p210 level presented as the percentage of p210 to Abl as assessed by qRT-PCR and blast percentage in bone marrow. All the patients had signed informed consents. The study was approved by the Ethics Committee of Zhujiang Hospital, Southern Medical University.
The human K562, KBM5, MEG01 cell lines were purchased from Jennio Biotech Co. (Guangzhou, China) and maintained in the laboratory of the Department of Hematology, Zhujiang Hospital, Southern Medical University. The murine 32D-p210-T315I and 32D-p210-WT myeloid precursor cell lines were kindly provided as gifts by Professor Lin Qiu from the Chinese Academy of Medical Sciences. The cells were cultured in RPMI-1640 containing 10% fetal calf serum (FCS) at 37°C and 5% v/v CO2.
Establishment of the stably transfected cell lines
A shRNA targeting EPS8 (CCCTATTGAATAAGGAC) or a scrambled shRNA was inserted into the pLVX lentiviral vector carrying the puromycin resistance gene to construct the transfer vector pLVX-shRNA-EPS8-puro. Then, the pLVX-shRNA-EPS8-puro vector and the control pLVX-GFP-puro vector were transfected into 293T cells, along with the packaging vector to envelop the lentivirus. The K562 cells were transfected with the lentivirus carrying the pLVX-shRNA-EPS8-puro vector or control vector, followed by puromycin selection for ~3 weeks to establish the stably transfected K562-shRNA-EPS8 and K562-GFP cell lines. Similarly, two efficient shRNA sequences targeting EPS8 and a scrambled shRNA were synthesized to establish the stably transfected 32D-p210-T315I-shRNA1-EPS8, T315I-shRNA2-EPS8 and T315I-scramble cell lines, respectively.
Quantitative real-time PCR
Total RNA was prepared using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA). RNase-free DNase I (Promega, Madison, WI, USA) was used to remove the genomic DNA. Reverse transcription was performed with the M-MLV Reverse transcriptase cDNA synthesis kit (Promega). Quantitative real-time PCR was performed with SYBR Green qPCR SuperMix (Invitrogen) on an ABI PRISM 7500 Sequence Detection System according to the standard SYBR-Green PCR protocol. 18S rRNA was used as the internal control. Triplicate reactions were performed as follows: 40 cycles of a two-step PCR (95°C for 15 sec, 60°C for 32 sec) after initial denaturation (95°C for 2 min). The data are presented as the fold change in expression, as determined with the 2−ΔΔCt method (17). The primer sequences were as follows: EPS8 forward, 5′-GATGGAGGAAGTGCAAGATG and reverse, 5′-GACTGTAACCACGTCTTCACA; 18S rRNA forward, 5′-CCTGGATACCGCAGCTAGGA and reverse, 5′-GCGGCGCAATACGAATGCCCC.
Cell proliferation assay
Cells (1×104 cells/well) were plated in 96-well plates in quadruplicate and cultured in 10% FCS-containing medium with or without the indicated concentrations of drugs. At 0, 20, 44 and 68 h, 10 µl of CCK-8 solution was added to the cells, which were then incubated for an additional 4 h. Then, the OD450 values were obtained using a microplate reader.
Apoptosis assay
Cells (5×105) were collected, and 1.24 µl of Annexin V-APC was added to the cells. The cells were incubated at room temperature for 15 min, centrifuged at 1,000 × g for 5 min, and the supernatant was discarded. The cells were then resuspended in 0.5 ml of cold 1X binding buffer. Ten microliters of 7-ADD or propidium iodide (PI) was added on ice, and the cells were analyzed by flow cytometry. The percentage of apoptotic cells (Annexin V+) was quantified.
Cell cycle analysis
Cells (1×106) were collected and washed twice with cold PBS. Then, cold 70% ethanol was added to the cells, which were incubated overnight at 4°C. The following day, the cells were washed once with PBS, and 500 µl of PBS containing 50 µl/ml PI, 100 µg/ml RNase A, and 0.2% Triton X-100 were added, and the cells were incubated for 30 min at 4°C in the dark. DNA content was detected by flow cytometry.
Adhesion and migration assay
The 96-well plates were coated with fibronectin (30 mg/l) and dried overnight. PBS (20 µl) containing 3% BSA was added to the 96-well plate at 37°C for 2 h, and then removed. Two hundred microliters of cells (0.5×106 cells/ml) suspended in RPMI-1640 and 10% FBS were added to the 96-well plates and incubated for 1.5 h; the plate was then washed gently. The cells that adhered to the bottom of the 96-well plate were observed under a microscope, and the number of adherent cells was assessed using the CCK-8 assay.
The cell migration assay was performed as previously described. Briefly, the cells were suspended in serum-free RPMI-1640 containing 0.1% BSA at a concentration of 1×106 cells/ml. A suspension of 0.1 ml of cells was added to the upper chamber, and the cells were allowed to migrate to the lower chamber, which contained RPMI-1640 and 10% FBS, for 6–8 h at 37°C in a 5% CO2 incubator. The cells that had migrated to the lower chamber were observed under a microscope. The number of cells in the lower chamber was assessed using the CCK-8 assay.
Chemotherapy drug sensitivity assay
The cells (1×104 cells/100 µl/well) were plated onto 96-well culture plates in quadruplicate in the presence of different concentrations of either daunorubicin or imatinib for the indicated time-points. Cell viability was assayed by adding CCK-8 solution. The percentage of cell viability was evaluated by assessing the absorbance at 450 nm and normalized to the corresponding untreated control.
Western blotting
Total protein lysates were separated on sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a polyvinylidene fluoride (PVDF) membranes. The blots were subjected to a standard immunodetection procedure using specific antibodies. The signals were enhanced with a chemiluminescence substrate and detected using an image analyser. The primary antibodies used were as follows: ERK (rabbit, polyclonal, 1:1,000; cat. no. ab196883), p-ERK (rabbit, polyclonal, 1:1,000; cat. no. ab214362) (both from Abcam, Cambridge, UK); STAT5 (rabbit, monoclonal, 1:1,000; cat. no. 94205), p-STAT5 (rabbit, polyclonal, 1:1,000; cat. no. 9351), Bcl-2 (rabbit, polyclonal, 1:1,000; cat. no. 2872) (all from CST, Danvers, MA, USA); Mcl-1 (rabbit, polyclonal, 1:1,000; cat. no. 16225-1-AP; Proteintech, Rosemont, IL, USA); PTEN (rabbit, monoclonal, 1:1,000; cat. no. ab32199), EPS8 (rabbit, polyclonal, 1:1,000; cat. no. ab96144) (both from Abcam); phospho-BCR/ABL (rabbit, polyclonal, 1:500; cat no. 3901), BCR/ABL (rabbit, polyclonal, 1:1,000; cat. no. 3902), β-actin (mouse, monoclonal, 1:1,000; cat. no. 3700), mTOR (rabbit, polyclonal, 1:1,000; cat. no. 2972), phospho-mTOR-s2448 (rabbit, polyclonal, 1:1,000; cat. no. 2971), Akt (rabbit, polyclonal, 1:1,000; cat. no. 9272), phospho-Akt-t308 (rabbit, polyclonal, 1:1,000; cat. no. 5110), phospho-Akt-s473 (rabbit, monoclonal, 1:1,000; cat. no. 4060), GSK3β (rabbit, monoclonal, 1:1,000, cat. no. 5676), phospho-GSK3β-s9 (rabbit, monoclonal, 1:1,000; cat. no. 9322) (all from CST); 4EBP1 (mouse, polyclonal, 1:1,000; cat. no. 60246-1-Ig; Proteintech); phospho-4EBP1-T37/46 (rabbit, polyclonal, 1:1,000; cat. no. 9459; CST); and GAPDH (mouse, monoclonal, 1:1,000; cat. no. KC-5G4; KangChen Bio-tech).
Xenograft model
BALB/c nu/nu mice were purchased from Southern Medical University. Nude mice, 6–8 weeks, were implanted subcutaneously with 1×107 K562-scramble or K562-shRNA-EPS8 cells, irrespectively. Tumor volume was assessed every 3 days. Mice were sacrificed on day 24 after inoculation and the subcutaneous tumors were weighed. All the procedures were performed according to an approved Southern Medical University, Animal Care and Use Committee protocol.
Statistical analyses
The data are reported as the mean ± standard deviations (SDs). One-way ANOVA followed by the Student-Newman-Keuls' pairwise multiple comparison test were used to compare the EPS8 mRNA expression among CML patients at different phases with healthy donors, as well as differences obtained in the proliferation, apoptosis, cell cycle, adhesion and migration assays. Spearman's rank correlation coefficient was used to analyze the correlation between EPS8 mRNA expression and clinical characteristics. A two-tailed independent Student's t-test was used to compare the means in the chemotherapy drug sensitivity assay. Differences were considered significant when P<0.05.
Results
CML patients express a high level of EPS8 mRNA
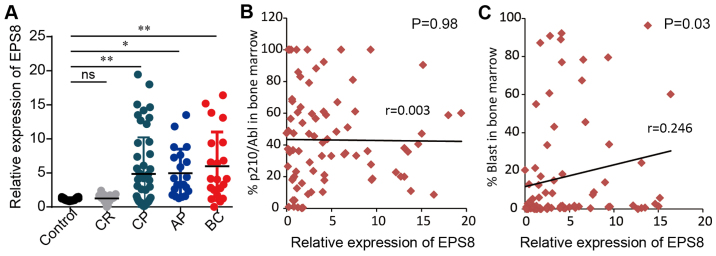
A published microarray data of 91 cases of CML patients reported by Radich et al (16) revealed an increase in EPS8 mRNA expression of CML patients compared to healthy controls, although the diffence was not statistically significant. Furthermore, there was a significant increase in EPS8 expression in the blast crisis phase of CML patients compared to healthy controls. To validate the result of these microarray data, we assessed EPS8 mRNA expression in bone marrow mononuclear cells from 113 cases of CML in CP (50 cases), AP (21 cases), BC (21 cases), CR (21 cases), and 21 healthy controls using quantitative real-time PCR. CML patients in CP, AP and BC exhibited significantly higher expression of EPS8 than those in complete remission and healthy donors (P<0.05). Moreover, there was a tendency for patients in BC to have increased EPS8 expression compared to patients in CP and AP (P>0.05) (Fig. 1A). We further performed correlation analysis between EPS8 expression and clinical features. There was no significant difference in EPS8 expression between the sex (P=0.74), age (P=0.85) and BCR-ABL level (P=0.98) of CML patients, however there was a correlation to blast percentage in bone marrow (P=0.03) (Fig. 1B and C; Table I). Collectively, these data revealed that EPS8 may play a role in CML leukemsogenesis.
Figure 1.
EPS8 mRNA expression is elevated in CML patients. (A) Bone marrow mononuclear cells were collected from 113 cases of CML patients and 21 normal controls. qRT-PCR was performed to assess the expression of EPS8 mRNA. The ratio of EPS8 and 18srRNA was expressed using the 2−ΔΔCt method. The relative expression of EPS8 mRNA was compared among CP (n=50), AP (n=21), BC (n=21) of CML and healthy controls (n=21). (B and C) Correlation analysis was performed between EPS8 mRNA expression with (B) the BCR-ABL transcript or (C) the blast percentage in bone marrow. *P<0.05 vs. the healthy control; **P<0.01 vs. the healthy control. CML, chronic myeloid leukemia; CP, chronic phase; AP, accelerated phase; BC, blast crisis phase; ns, non-significant.
Table I.
Correlation analysis between EPS8 expression and the clinical features of CML patients.
| Clinical features | Eps8 expression (mean rank) | P-value |
|---|---|---|
| Male (n=60) | 49.83 | 0.74 |
| Female (n=53) | 48.99 | |
| Correlation coefficient (Spearmans r) | ||
| Age (years) | 0.019 | 0.85 |
| p210 | 0.003 | 0.98 |
| Blast percentage | 0.246 | 0.03 |
EPS8, epidermal growth factor receptor kinase substrate 8; CML, chronic myeloid leukemia.
Knockdown of EPS8 inhibits CML cell proliferation, induces apoptosis and arrests the cell cycle in the G1 phase
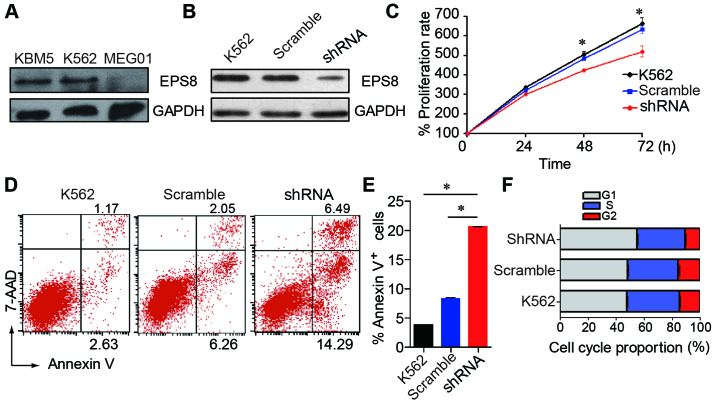
To evaluate the effect of EPS8 on the biological function of CML cells, we knocked down the expression of EPS8 in CML cell line K562. EPS8 expression was detected in three human CML cell lines. The K562 and KBM5 cells expressed a higher level of EPS8, while the MEG01 cells exhibited a lower expression (Fig. 2A). Then, the K562 cells were transfected with a lentivirus carrying an EPS8-targeted shRNA vector to knockdown EPS8. As a result, EPS8 expression was attenuated in the K562-shRNA-EPS8 cells that were stably transduced with the lentivirus (Fig. 2B).
Figure 2.
Effect of EPS8 on proliferation, apoptosis and the cell cycle of K562 cells. (A) The expression of EPS8 was revealed in human CML cell lines using western blotting. (B) The expression levels of the EPS8 protein in the K562-shRNA-EPS8 and the control cells were analysed by western blotting. (C) Cells were plated in 96-well plates and the OD values were assessed at indicated time-points using the CCK-8 assay. (D and E) The K562 cells and their derivatives were stained with Annexin V-APC and 7-AAD, and apoptosis was assessed by flow cytometry. (F) The K562 cells and their derivatives were stained with PI, and then the cell cycle was analysed by flow cytometry. Plots are representative of 3 independent experiments. *P<0.05 vs. the control. CML, chronic myeloid leukemia.
To investigate the role of EPS8 in CML cells, we assessed the proliferation, apoptosis and cell cycle after EPS8 silencing. We first examined the proliferation of K562-shRNA-EPS8 cells and determined that knockdown of EPS8 reduced the proliferation of the K562 cells (Fig. 2C). Then, we assessed cell apoptosis and the results revealed that the K562-shRNA-EPS8 cells exhibited enhanced apoptosis compared with their parental cells (Fig. 2D and E). Then, the cell cycle was evaluated and it was determined that more K562-shRNA-EPS8 cells were present in the G1 phase, and fewer cells were present in the S phase compared with the corresponding control cells (Fig. 2F). These data revealed that EPS8 regulated the proliferation, apoptosis and cell cycle in CML cells.
Attenuated EPS8 expression leads to impaired adhesion and migration
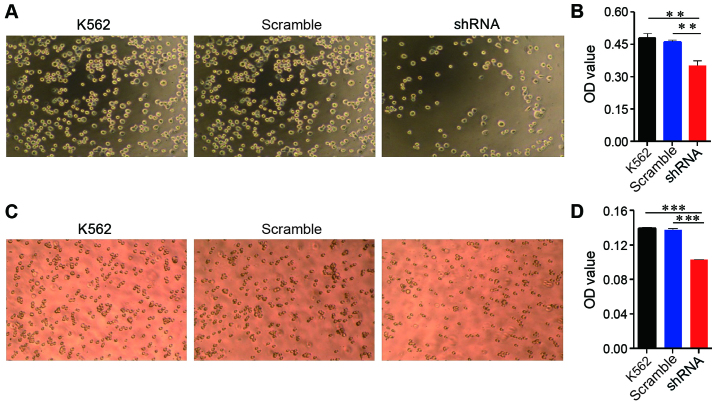
EPS8 plays an important role in cytoskeletal remodelling and cellular motility; therefore, we evaluated the effect of EPS8 on adhesion and migration of K562 cells. The K562 cells and their derived cells were plated on fibronectin-coated 96-well plates for 1.5 h and then washed. Less K562-shRNA-EPS8 cells remained on the fibronectin-coated plates compared with the control cells (Fig. 3A and B). Cellular migration was assessed using the Transwell assay, in which the cells were placed into the upper chamber, which contained serum-free medium, and the lower chamber contained medium with 10% FBS. Fewer K562-shRNA-EPS8 cells were observed in the lower chamber compared with the K562-scramble cells demonstrating that the attenuation of EPS8 expression impaired the migration of the K562 cells (Fig. 3C and D).
Figure 3.
EPS8 increases the adhesion and migration of leukemia cells. (A and B) K562 cells and their derivatives (1×105 cells/well) were suspended in RPMI-1640 with 10% FBS and then added to a fibronectin-coated 96-well plate, incubated for 1.5 h and washed gently. The cells that adhered to the bottom of the plate were observed under a microscope. (A) The representative plots of the indicated strains under the microscope. (B) The number of adhered cells was assessed using the CCK-8 assay. (C and D) Transwell chambers were used to assess migration. Cells ~1×105 were placed on the upper layer of a cell permeable membrane in serum-free RPMI-1640 containing 0.1% bovine serum albumin and allowed to migrate to the lower chamber, which contained RPMI-1640 and 10% FBS, for 6–8 h at 37°C. The cells that had migrated to the lower chamber were observed under a microscope. (C) The representative plots of the indicated strains under the microscope. (D) The number of migrating cells was calculated using the CCK-8 assay. Plots are representative of 3 independent experiments. **P<0.01 vs. the control; ***P<0.001 vs. the control.
EPS8 is implicated in the chemosensitivity of CML cells
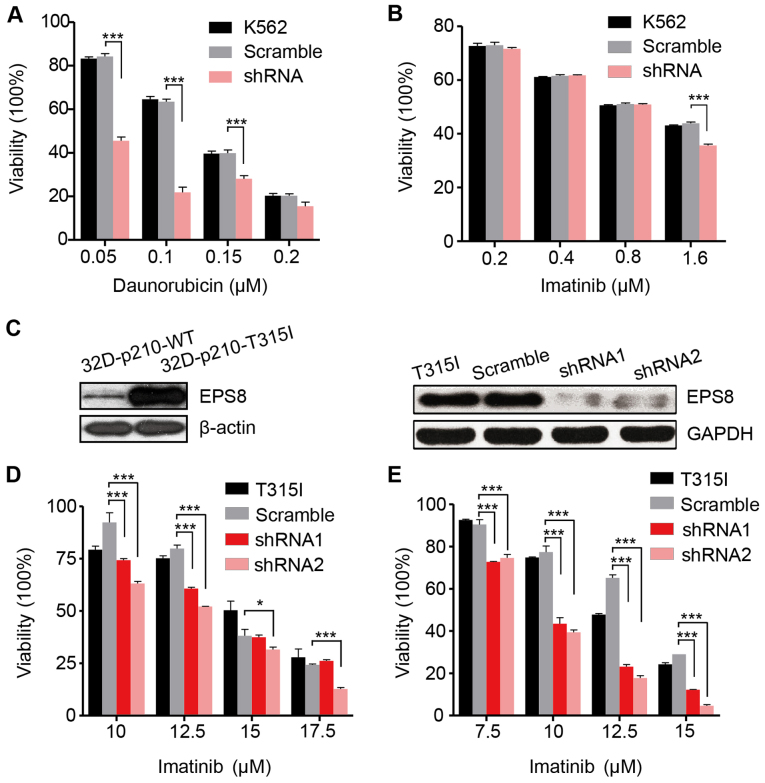
We then investigated the effect of EPS8 on the chemosensitivity of the K562 cells. Daunorubicin is a widespread used common chemotherapy drug to treat myeloid leukemia. In the present we determined that the viability of the K562-shRNA-EPS8 cells was significantly reduced in the presence of indicated concentrations of daunorubicin indicating that EPS8 knockdown increases their chemosensitivity (Fig. 4A). However, as an imatinib-sensitive cell line, the K562-shRNA-EPS8 cells did not exhibit further increased sensitivity to imatinib except in the high drug concentration (Fig. 4B).
Figure 4.
Knockdown of EPS8 increases sensitivity to chemotherapy. K562 cells and their derivatives (1×104 cells/100 µl/well) were added to 96-well culture plates and treated with the indicated concentrations of (A) daunorubicin and (B) imatinib for 48 h. Viability was assessed with the CCK-8 assay. (C) EPS8 expression in 32D-p210-WT and 32D-p210-T315I cells were assessed by western blotting. LV-EPS8-shRNA1 and LV-EPS8-shRNA2 vectors were transfected into 32D-p210-T315I cells, respectively. EPS8 expression was revealed in 32D-p210-T315I-shRNA1-EPS8, T315I-shRNA2-EPS8, and the control cell lines. (D and E) 32D-p210-T315I cells and their derived cells (1×104 cells/100 µl/well) were added to 96-well culture plates and treated with the indicated concentrations of imatinib for (D) 24 h and (E) 48 h, respectively. Viability was assessed with the CCK-8 assay. Plots are representative of 3 independent experiments. ***P<0.001 vs. the scramble control.
To investigate whether knockdown of EPS8 can overcome imatinib resistance, we employed a murine imatinib-resistant cell line 32D-p210T325I-BCR/ABL (32D-p210-T315I), which was generated by transfecting myeloid precursor cells with the vector carrying prototype imatinib-resistant BCR/ABL point mutation T315I in CML patients. Compared to the imatinib sensitive cell line 32D-P210BCR/ABL (32D-p210-WT) which expressed native BCR/ABL protein p210, 32D-p210-T315I cells expressed a higher level of EPS8 protein (Fig. 4C). To silence EPS8 expression, LV-EPS8-shRNA1 and LV-EPS8-shRNA2 vectors were transfected into 32D-p210-T315I cells, respectively. EPS8 expression was considerably decreased compared with the controls (Fig. 4C). The results revealed that knockdown of EPS8 reduced the cell viability in a dose- and time-dependent manner. Following 24 and 48 h of incubation, cell viability in the EPS8-shRNA1 or EPS8-shRNA2 groups was significantly lower than the scramble group. Therefore, downregulation of EPS8 increased the sensitivity of 32D-p210-T315I cells to imatinib (Fig. 4D and E).
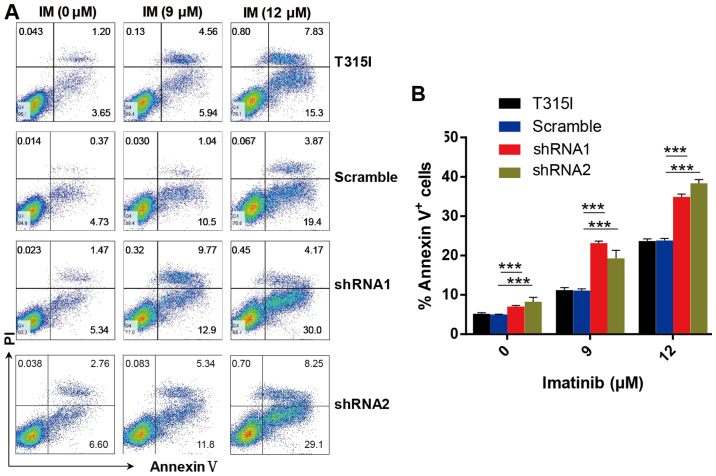
In accordance with these results, we determined that knockdown of EPS8 notably increased apoptosis in imatinib-treated 32D-p210-T315I cells in a dose-dependent manner. Apoptosis was markedly increased in EPS8-shRNA cells compared with the control groups after imatinib treatment for 24 h (Fig. 5A and B). These results indicated that EPS8 protected imatinib-resisitant CML cells against drug-induced apoptosis.
Figure 5.
EPS8 knockdown increases imatinib induced apoptosis of 32D-p210-T315I cells. (A) The 32D-p210-T315I cells and their derived cells were stained with Annexin V-APC and 7-AAD, and apoptosis was assessed by flow cytometry. (B) Statistical analysis of the percentage of apoptotic cells in each group. Plots are representative of 3 independent experiments. ***P<0.001 vs. the scramble control.
EPS8 regulates apoptosis and proliferation signalling pathways in CML cells
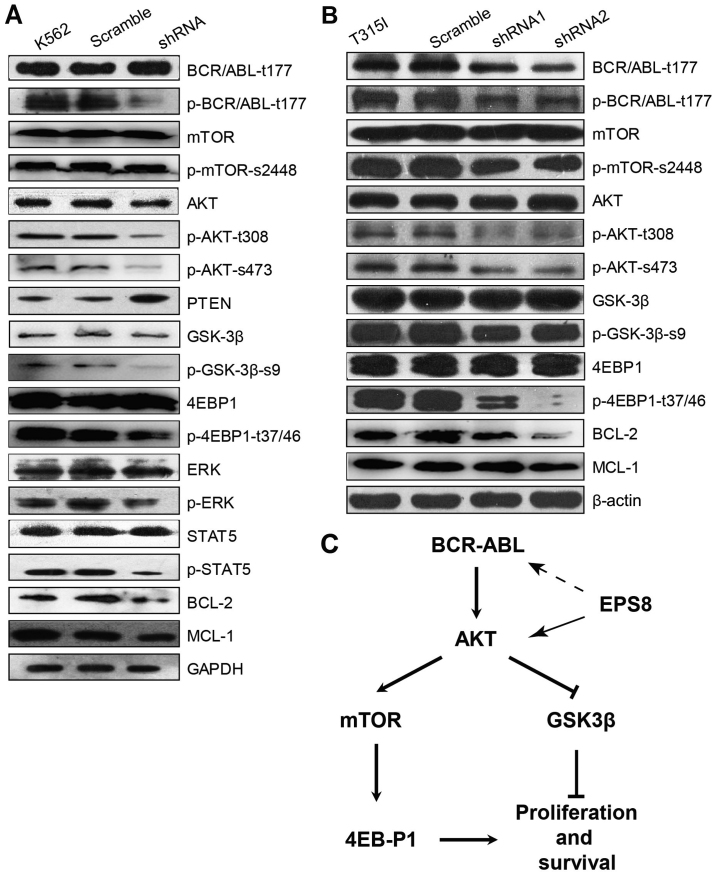
To determine the molecular mechanism through which EPS8 regulates CML cells, key molecules in main signalling pathways involved in leukemogenesis were assessed. Notably, phosphorylated (p)-BCR-ABL was significantly decreased in EPS8-knockdown K562 cells compared with the control group, suggesting that EPS8 regulated BCR-ABL. Furthermore, our results illustrated that p-AKT and phosphorylation of its downstream proteins mTOR, eukaryotic translation initiation factor 4E binding protein 1 (EIF4EBP1) and glycogen synthase kinase 3β (GSK3β), as well as, p-STAT5 and p-ERK levels were reduced in K562-shRNA-EPS8 cells compared with the control cells. Since Mcl-1 and Bcl-2 function as anti-apoptotic markers in various cancers, we explored whether their expression was affected by EPS8. Indeed, we found that the expression of Mcl-1 and Bcl-2 was suppressed by EPS8 silencing. These data revealed that EPS8 inhibition-induced apoptosis was related with the reduction of anti-apoptotic factors and inhibition of proliferation and anti-apoptotic pathway activity (Fig. 6A).
Figure 6.
Silencing of EPS8 downregulates the protein levels of BCR-ABL and its downstream Akt/mTOR signaling pathway. (A) K562, K562-scramble and K562-shRNA-EPS8 cells were lysed. Indicated proteins were blotted. GAPDH served as a loading control. (B) 32D-p210T315I-shRNA1-EPS8, T315I-shRNA2-EPS8, T315I-scramble and T315I cells were lysed. Indicated proteins were blotted. β-actin was used as a loading control. (C) A schematic diagram depicting the possible EPS8 regulation pathway in BCR-ABL positive cells. The solid arrow represents regulation, the dash arrow represents indirect regulation.
Given that EPS8 regulated the anti-apoptotic signalling and increased imatinib-induced apoptosis in imatinib-resistant 32D-p210-T315I cells, we next investigated the effect of EPS8 on BCR-ABL expression and its downstream PI3K/Akt signalling pathway in 32D-p210-T315I cells and their derived cells. In accordance with K562-shRNA-EPS8 cells, 32D-p210-T315I-shRNA-EPS8 cells exhibited decreased total BCR-ABL and p-BCR-ABL proteins. Furthermore, EPS8 silencing led to a decrease of p-Akt (T308, S476), p-4EBP1 (T37/46), p-mTOR (S2448) and p-GSK3-β (Fig. 6B). Collectively, these data revealed that EPS8 knockdown interfered with the BCR-ABL/PI3K/AKT/mTOR pathway (Fig. 6C).
Knockdown of EPS8 attenuated proliferation of K562 cells in vivo
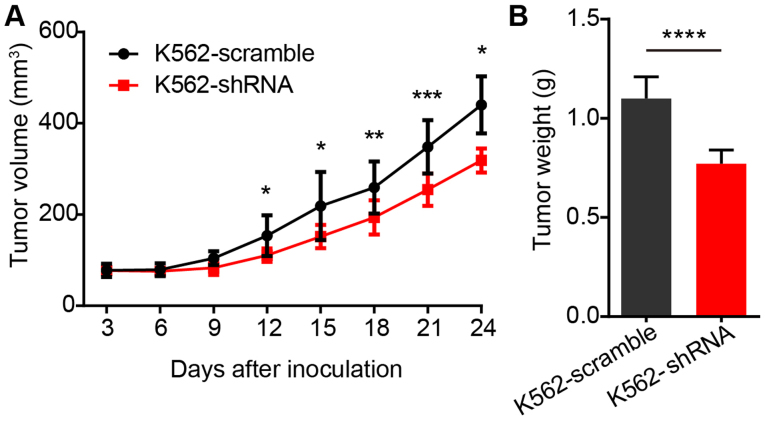
Finally, we determined the efficacy of the inhibition of EPS8 on the proliferation of BCR-ABL positive cells in vivo. Since no EPS8 inhibitor was available, we utilized the shRNA to knockdown EPS8 in K562 cells in a xenograft model. BALB/c nude mice were implanted subcutaneously with 1×107 K562-scramble or K562-shRNA-EPS8 cells, and the tumor volume was observed every 3 days. The results revealed that the mice bearing K562-shRNA-EPS8 cells had a significantly smaller tumor volume compared to those bearing the K562-scramble cells (Fig. 7A). Consistently, tumor weight in mice harboring K562-shRNA-EPS8 cells at 24 days after inoculation was significantly lower than that in the control mice (Fig. 7B).
Figure 7.
Knockdown of EPS8 attenuated the proliferation of K562 cells in vivo. BALB/c nude mice (n=8 for each group) were implanted subcutaneously with 1×107 K562-scramble or K562-shRNA-EPS8 cells. (A) The tumor volume was assessed every 3 days. (B) Mice were sacrificed on day 24 after inoculation and the subcutaneous tumors were weighed. Plots are representative of 2 independent experiments. *P<0.05 vs. the scramble control, **P<0.01, ***P<0.001; ****P<0.0001.
Discussion
EPS8 is a cytoplasmic non-receptor kinase and is well recognized as an oncogene in a variety of solid tumours (6). However, its role in haematological malignant diseases is less documented. A gene expression profiling study of 97 cases of infant ALL identified EPS8, along with six other genes as prognostic factors. The high-EPS8 cohort had an event-free survival of 20%, while the low-EPS8 cohort had a rate of 65% (13). Our previous study revealed similar results. We found that EPS8 expression in AML patients was elevated compared with the control group. Moreover, EPS8 expression was negatively related to the response of AML patients to chemotherapy (15). Furthermore, we addressed the expression of the EPS8 mRNA in 107 cases of ALL patients, and there was a correlation between the expression of EPS8 and MDR1, as well as EPS8 and WT1. A high EPS8/MDR1/WT1 expression profile was associated with a higher risk of relapse. In addition, patients with high expression of EPS8/MDR1/WT1 had a shorter event-free survival (14). Notably, consistent with the reported CML microarray data, we revealed in our present study that our cohort CML patients had a higher expression of EPS8 compared to the controls, indicating that EPS8 plays a role in CML disease. In addition, our data demonstrated that EPS8 mRNA expression was not correlated with BCR-ABL, suggesting that EPS8 may be a BCR-ABL independent factor for CML patients. However, our study did not contain follow-up clinical information, therefore future investigation to address the relationship between EPS8 expression and drug resistance or survival in CML patients is warranted.
To determine the role in CML cells, we knocked down the expression of EPS8 in CML cell line K562 and determined that silencing of EPS8 led to reduced proliferation, increased apoptosis and cell cycle arrest in the G1 phase which was consistent with other studies on EPS8. Xu et al reported that the stable overexpression of EPS8 accelerated proliferation and reduced the serum withdrawal-induced apoptosis of pituitary tumour cells (19). Wang et al found that EPS8 expression was increased in oral squamous carcinoma compared with normal keratinocytes. The NH4 cell line exhibited mildly elevated EPS8 expression and reduced invasion, while the NH12 cell line, which expressed a high level of EPS8, was extremely aggressive. EPS8 overexpression in NH4 cells led to proliferation that was similar to that of NH12 cells (20). Similar results were observed in other cancers. The results in the present study indicated that EPS8 was involved in CML as well.
One of the characteristics of leukemia cells is invasion to other organs. Cytoskeletal proteins have an important role in AML cell migration (21). EPS8 is an important regulator of the cytoskeleton and is reported to be related to the migration and invasion of many solid tumours by regulating MMP9, E-cadherin and N-cadherin (7). EPS8 was demonstrated to upregulate the expression of CXCL5 and CXCL12 via the transcription factor FOXM1 to enhance migration (22). Yap et al demonstrated that EPS8 promoted oral squamous cancer cell migration and invasion by activating Rac in an integrin-dependent manner (23). EPS8, Abi-1 and SOS-1 form a tricomplex and induce Rac-specific guanine nucleotide exchange factor (GEF) activity, which has been revealed to play a critical role in ovarian cancer metastasis (9). In the present study the ability of adhesion and migration was attenuated in K562-shRNA-EPS8 cells compared with the control, which was consistent with previous research indicating that EPS8 may contribute to migration from bone marrow and infiltration to other organs.
Gorsic et al revealed that EPS8 knockdown increased the chemosensitivity of lymphoblastic cell lines, small cell lung cancer cell lines and bladder cancer cell lines to cisplatin (24). Chen et al reported that EPS8 reduced the chemosensitivity of cervical cancer cells to cisplatin and paclitaxel (25). Notably, our results demonstrated that EPS8 regulated the sensitivity of CML cells, including imatinib-sensitive and imatinib-resistant cells, to chemotherapy and tyrosine kinase inhibitors.
To explore the molecular mechanism involved in the EPS8-mediated effect on the response of CML cells to imatinib, we detected BCR-ABL and its downstream proteins in imatinib-sensitive cell line K562 and imatinib-resistant cell line 32D-p210-T315I and their derivatives. EPS8 is a critical signaling molecule and integrates multiple pathways. EPS8 has been reported to regulate PI3K/AKT/mTOR (19,20) and MEK/ERK (8) signalling which control cell proliferation, survival, differentiation and apoptosis. Consistent with the evidence that EPS8 knockdown inactived Akt and downstream 4EBP1 phosphorylation in colon cancer cells (26), our study provided an extension of this notion, revealing for the first time that the phosphorylation of Akt (S308, S476), GSK3-β (S9), and 4EBP1 (T37/46) were decreased in CML cells after EPS8 silencing. Notably, we determined that the active form of the oncoprotein BCR-ABL was decreased after EPS8 knockdown, highlighting the critical role of EPS8 in BCR-ABL positive cells. The exact underlying mechanism revealing how EPS8 regulates BCR-ABL remains unclear. Although EPS8 is an adaptor protein, it may be less likely that EPS8 directly combines with BCR-ABL to form a complex since recently Reckel et al performed a quantitative comparative proteomic study in p190 or p210 transfected BaF3 cells and EPS8 was not identified as an interactor of BCR-ABL (27). Despite the fact that BCR-ABL is an oncoprotein which is capable of autophosphorylation, proliferation and survival signalling can also regulate its phosphorylation. It is more possible that knockdown of EPS8 leads to attenuation of proliferation and survival signalling, which subsequently decreases the phosphorylation of BCR-ABL. A future study is warranted to test this hypothesis.
In conclusion, our data revealed that EPS8 regulated multiple biological functions such as proliferation, apoptosis, the cell cycle, drug sensitivity of CML cells possibly by mediating the regulation of the BCR-ABL/AKT/mTOR signalling pathway. Strategies that target EPS8 in CML patients may help to overcome resistance to tyrosine kinase inhibitors. The EPS8 inhibitor alone or combined with a tyrosine kinase inhibitor may be promising customized strategies to treat refractory and resistant CML patients.
Acknowledgements
The present study was supported by the Major Program for Health Medical Collaborative Innovation of Guangzhou (grant no. 201704020216), the Startup Project for Clinical Trials of Southern Medical University (grant no. LC2016ZD027), the Special Project for the Development of Science and Technology of Guangdong Province (grant no. 2016A020213005), the Competitive Allocation Project of the Comprehensive Strategic Cooperation of the Chinese Academy of Sciences of Guangdong Province (grant no. 2013B091500072), the National Science Foundation of China (no. 81372249 and 81300431) and the Youth Scientific Fund of Southern Medical University (no. PY2014N072).
References
- 1.Huang R, Kang Q, Liu H, Li Y. New insights into the molecular resistance mechanisms of chronic myeloid leukemia. Curr Cancer Drug Targets. 2016;16:323–345. doi: 10.2174/1568009615666150921141004. [DOI] [PubMed] [Google Scholar]
- 2.Druker BJ, Guilhot F, O'Brien SG, Gathmann I, Kantarjian H, Gattermann N, Deininger MW, Silver RT, Goldman JM, Stone RM, et al. IRIS Investigators Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 3.Hochhaus A, O'Brien SG, Guilhot F, Druker BJ, Branford S, Foroni L, Goldman JM, Müller MC, Radich JP, Rudoltz M, et al. IRIS Investigators Six-year follow-up of patients receiving imatinib for the first-line treatment of chronic myeloid leukemia. Leukemia. 2009;23:1054–1061. doi: 10.1038/leu.2009.38. [DOI] [PubMed] [Google Scholar]
- 4.Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, Sawyers CL. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 5.Thomas J, Wang L, Clark RE, Pirmohamed M. Active transport of imatinib into and out of cells: Implications for drug resistance. Blood. 2004;104:3739–3745. doi: 10.1182/blood-2003-12-4276. [DOI] [PubMed] [Google Scholar]
- 6.Li YH, Xue TY, He YZ, Du JW. Novel oncoprotein EPS8: A new target for anticancer therapy. Future Oncol. 2013;9:1587–1594. doi: 10.2217/fon.13.104. [DOI] [PubMed] [Google Scholar]
- 7.Maa MC, Hsieh CY, Leu TH. Overexpression of p97Eps8 leads to cellular transformation: Implication of pleckstrin homology domain in p97Eps8-mediated ERK activation. Oncogene. 2001;20:106–112. doi: 10.1038/sj.onc.1204069. [DOI] [PubMed] [Google Scholar]
- 8.Chen C, Liang Z, Huang W, Li X, Zhou F, Hu X, Han M, Ding X, Xiang S. Eps8 regulates cellular proliferation and migration of breast cancer. Int J Oncol. 2015;46:205–214. doi: 10.3892/ijo.2014.2710. [DOI] [PubMed] [Google Scholar]
- 9.Chen H, Wu X, Pan ZK, Huang S. Integrity of SOS1/EPS8/ABI1 tri-complex determines ovarian cancer metastasis. Cancer Res. 2010;70:9979–9990. doi: 10.1158/0008-5472.CAN-10-2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding X, Zhou F, Wang F, Yang Z, Zhou C, Zhou J, Zhang B, Yang J, Wang G, Wei Z, et al. Eps8 promotes cellular growth of human malignant gliomas. Oncol Rep. 2013;29:697–703. doi: 10.3892/or.2012.2160. [DOI] [PubMed] [Google Scholar]
- 11.Schoenherr C, Serrels B, Proby C, Cunningham DL, Findlay JE, Baillie GS, Heath JK, Frame MC. Eps8 controls Src- and FAK-dependent phenotypes in squamous carcinoma cells. J Cell Sci. 2014;127:5303–5316. doi: 10.1242/jcs.157560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun J, Jin LY, Wang HY, Zhou JQ, Li YJ, He YZ, Li YH. Correlation of Eps8 with proliferation, metastasis and prognosis of malignant tumors. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2013;21:493–497. doi: 10.7534/j.issn.1009-2137.2013.02.050. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 13.Kang H, Wilson CS, Harvey RC, Chen IM, Murphy MH, Atlas SR, Bedrick EJ, Devidas M, Carroll AJ, Robinson BW, et al. Gene expression profiles predictive of outcome and age in infant acute lymphoblastic leukemia: A Children's Oncology Group study. Blood. 2012;119:1872–1881. doi: 10.1182/blood-2011-10-382861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He YZ, Liang Z, Wu MR, Wen Q, Deng L, Song CY, Wu BY, Tu SF, Huang R, Li YH. Overexpression of EPS8 is associated with poor prognosis in patients with acute lymphoblastic leukemia. Leuk Res. 2015;39:575–581. doi: 10.1016/j.leukres.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, Cai SH, Xiong WY, He YJ, Deng L, Li YH. Real-time quantitative polymerase chain reaction assay for detecting the eps8 gene in acute myeloid leukemia. Clin Lab. 2013;59:1261–1269. doi: 10.7754/Clin.Lab.2013.121107. [DOI] [PubMed] [Google Scholar]
- 16.Radich JP, Dai H, Mao M, Oehler V, Schelter J, Druker B, Sawyers C, Shah N, Stock W, Willman CL, et al. Gene expression changes associated with progression and response in chronic myeloid leukemia; Proc Natl Acad Sci USA; 2006; pp. 2794–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 19.Xu M, Shorts-Cary L, Knox AJ, Kleinsmidt-DeMasters B, Lillehei K, Wierman ME. Epidermal growth factor receptor pathway substrate 8 is overexpressed in human pituitary tumors: Role in proliferation and survival. Endocrinology. 2009;150:2064–2071. doi: 10.1210/en.2008-1265. [DOI] [PubMed] [Google Scholar]
- 20.Wang H, Patel V, Miyazaki H, Gutkind JS, Yeudall WA. Role for EPS8 in squamous carcinogenesis. Carcinogenesis. 2009;30:165–174. doi: 10.1093/carcin/bgn252. [DOI] [PubMed] [Google Scholar]
- 21.Capala ME, Vellenga E, Schuringa JJ. ELMO1 is upregulated in AML CD34+ stem/progenitor cells, mediates chemotaxis and predicts poor prognosis in normal karyotype AML. PLoS One. 2014;9:e111568. doi: 10.1371/journal.pone.0111568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, Teh MT, Ji Y, Patel V, Firouzabadian S, Patel AA, Gutkind JS, Yeudall WA. EPS8 upregulates FOXM1 expression, enhancing cell growth and motility. Carcinogenesis. 2010;31:1132–1141. doi: 10.1093/carcin/bgq058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yap LF, Jenei V, Robinson CM, Moutasim K, Benn TM, Threadgold SP, Lopes V, Wei W, Thomas GJ, Paterson IC. Upregulation of Eps8 in oral squamous cell carcinoma promotes cell migration and invasion through integrin-dependent Rac1 activation. Oncogene. 2009;28:2524–2534. doi: 10.1038/onc.2009.105. [DOI] [PubMed] [Google Scholar]
- 24.Gorsic LK, Stark AL, Wheeler HE, Wong SS, Im HK, Dolan ME. EPS8 inhibition increases cisplatin sensitivity in lung cancer cells. PLoS One. 2013;8:e82220. doi: 10.1371/journal.pone.0082220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen YJ, Shen MR, Chen YJ, Maa MC, Leu TH. Eps8 decreases chemosensitivity and affects survival of cervical cancer patients. Mol Cancer Ther. 2008;7:1376–1385. doi: 10.1158/1535-7163.MCT-07-2388. [DOI] [PubMed] [Google Scholar]
- 26.Maa MC, Lee JC, Chen YJ, Chen YJ, Lee YC, Wang ST, Huang CC, Chow NH, Leu TH. Eps8 facilitates cellular growth and motility of colon cancer cells by increasing the expression and activity of focal adhesion kinase. J Biol Chem. 2007;282:19399–19409. doi: 10.1074/jbc.M610280200. [DOI] [PubMed] [Google Scholar]
- 27.Reckel S, Hamelin R, Georgeon S, Armand F, Jolliet Q, Chiappe D, Moniatte M, Hantschel O. Differential signaling networks of Bcr-Abl p210 and p190 kinases in leukemia cells defined by functional proteomics. Leukemia. 2017;31:1502–1512. doi: 10.1038/leu.2017.36. [DOI] [PMC free article] [PubMed] [Google Scholar]