Abstract
Cytotoxic T-lymphocyte antigen-4 (CTLA-4) is a critical negative regulator of immune responses. CTLA-4 is rapidly upregulated following T-cell activation, and then binds to B7 molecules with a higher affinity than CD28. CTLA-4 may abolish the initiation of the responses of T cells by raising the threshold of signals required for full activation of T cells, and it also may terminate ongoing T-cell responses. This regulatory role has led to the development of monoclonal antibodies (mAbs) designed to block CTLA-4 activity for enhancing immune responses against cancer. mAbs have several disadvantages including high production cost and unstable behavior. Nanobodies (Nbs) are single-domain antigen-binding fragments derived from the camelid heavy-chain antibodies, which are highly attractive in cancer immunotherapy due to their small size, high specificity, and stability. We selected CTLA-4-specific Nbs from a high quality dromedary camel immune library by phage display technology. Four positive colonies were sequenced and classified based on the amino acids sequences in the CDR3 region. These Nbs recognized unique epitopes on CTLA-4 and displayed high binding rates when used on PHA-stimulated human T cells. Treatment of B16 melanoma-bearing C57BL/6 mice with anti-CTLA-4 nanobody 16 (Nb16) delayed melanoma growth and prolonged the survival time of mice. These data indicate that anti-CTLA-4 Nbs selected from a high quality phage display library may be effective for the treatment of patients with tumors.
Keywords: CTLA-4, nanobody, immunotherapy, melanoma, phage-displayed library
Introduction
Regulatory T-cell receptors serve as immunotherapeutic targets for enhancing activation of antitumor immune responses or reversing immunosuppressive mechanisms of tumor resistance to immune surveillance and destruction. Cytotoxic T-lymphocyte antigen-4 (CTLA-4), an essential inhibitory regulator, is responsible for the early stages of T-cell expansion that opposes the action of CD28-mediated costimulation (1). Following T-cell activation, CTLA-4 is rapidly upregulated, then binds to B7 molecules with a higher affinity than CD28 (2,3). CTLA-4 may abolish the initiation of the responses of T cells by raising the threshold of signals required for full activation of T cells, and it also may terminate the ongoing T-cell responses. Based on the significant regulatory effect of CTLA-4 on immune responses, antibodies against either mouse or human CTLA-4 have been developed for boosting immunological responses against cancer (4). Anti-CTLA-4 antibodies have been confirmed to confer a blockage effect on CTLA-4/B7 interactions in vivo, and they can enhance T-cell responses to peptides, superantigens, and parasites (5). It has been shown that anti-CTLA-4 antibodies are able to induce the rejection of newly implanted murine tumors (6,7). In addition, promising results have been observed in clinical trials of anti-human CTLA-4 monoclonal antibodies (mAbs) for the treatment of late-stage metastatic melanoma (8,9).
Based on their high affinity and specificity, mAbs have become ideal therapeutic strategies for research, diagnosis and clinical applications (10,11). The traditional immunoglobulin G (IgG) molecules consist of two identical heavy chains and light chains, forming the antigen binding site together. However, the complex structure, costly production and unstable behavior of IgG greatly limit their practical applications (12–14). Recently, single domain antibodies (sdAbs; also called nanobodies; Nbs) have emerged as small (~15 kDa) antigen-binding fragments which are derived from camelid heavy-chain antibodies (15). They present several advantages including good solubility, thermal stability, and high expression yield (16–18). Furthermore, Nbs have a natural tendency for binding epitopes that are inaccessible to conventional antibodies (19). Nbs have been evaluated in vitro and in vivo and have been proven to be a valuable tool for optical molecular imaging of HER2-positive breast cancer (20). They are capable of selectively targeting HGF-producing tumors. Furthermore, treatment of U87 MG-bearing mice with these Nbs resulted in inhibition of tumor growth and ultimately caused cures (21). Consequently, these unique advantages make Nbs an attractive and valuable approach for the diagnosis and treatment of tumors.
In the present study, we successfully constructed an immune phage display library against CTLA-4 with the size of 1.85×108 colonies and generated characteristic anti-CTLA-4 Nbs. We further demonstrated their valuable properties of high binding rates and anti-melanoma activity.
Materials and methods
Reagents and materials
The human CTLA-4 protein was purchased from Abcam (Cambridge, UK). Freund's incomplete adjuvant was purchased from Sigma-Aldrich (St. Louis, MO, USA).
Density gradient centrifugation with Ficoll-Paque™ Plus (GE Healthcare, Beijing, China). Fast Track 2.0 kit and ThermoScript RT-PCR kit were provided by Invitrogen (Carsbad, CA, USA). OligodT primers were obtained from Thermo Fisher Scientific, Inc. (Waltham, MA, USA). Restriction enzymes PstI and NotI were provided by New England BioLabs (NEB) (Ipswich, MA, USA). Anti-mouse IgG-alkaline phosphatase, NI-NTA Superflow sepharose column and bisphosphate and phytohemagglutinin (PHA) were acquired from Sigma-Aldrich. The VCSM13 helper phages, TG1 and WK6 cells were kindly provided by Professor Serge Muyldermans (Laboratory of Cellular and Molecular Immunology, Vrije Universiteit Brussel, Brussels, Belgium). Anti-HA tag antibody and mouse anti-human CTLA-4 mAb were purchased from Abcam (clone, 16B12) and BD Pharmingen (San Diego, CA, USA; clone, BNI3), respectively.
Cells and animals
B16/BL6 cells were obtained from the National Center for International Research of Biological Targeting Diagnosis and Therapy of Guangxi Medical University. B16/BL6 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) (both from Gibco, Grand Island, NY, USA) and 1% double antibiotics (penicillin/streptomycin) at 37°C in a 5% CO2 incubator.
C57BL/6 mice were obtained from Vital River Company (Beijing, China) and were raised in specific pathogen-free (SPF) conditions. All animal experiments were carried out according to the guidelines of the Federation of European Laboratory Animal Science Associations. All protocols were approved by the Animal Ethics Committee of Guangxi Medical University.
Dromedary camel immunization
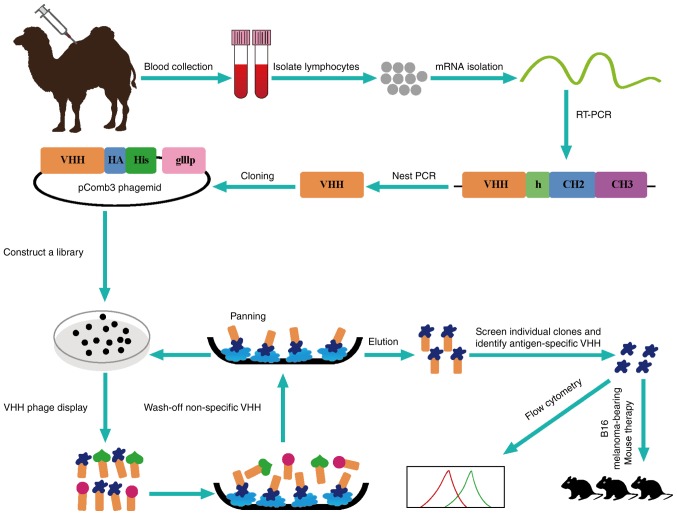
A healthy dromedary camel was immunized subcutaneously 7 times at 7-day intervals with human CTLA-4 protein (1 mg, 1 ml) mixed with an equal volume of Freund's incomplete adjuvant for stimulating antigen-specific B cells expressing Nbs (22). The peripheral blood lymphocytes were extracted from 100 ml blood sample to construct the library after the last injection.
Library construction
Lymphocytes were isolated from peripheral blood by density gradient centrifugation. The total RNA was extracted from ~107 lymphocytes by using Fast Track 2.0 kit and mRNA (40 µg) was used to synthesize cDNA strands using a ThermoScript RT-PCR kit with oligodT primers. To avoid contamination of VH genes, the variable regions of heavy-chain immunoglobulins (VHH) were amplified by 2-steps nested PCR. The first step PCR was performed with a template of the first-strand cDNA using the primers CALL001 and CALL002 (23). This protocol consisted of an initial denaturation step at 94°C for 7 min, followed by 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min for 30 cycles, and a final extension step at 72°C for 10 min. The first PCR products consist of ~700 bp fragments and were used as the template for the second step PCR. Then, the VHH encoding gene fragments were amplified by second PCR using degenerated primers including PstI and NotI restriction sites. The amplified products were ligated into phagemid pComb3 after digesting by restriction enzymes PstI and NotI and then electro-transformed into competent E. coli TG1 cells (24). The transformants were plated onto 2X YT medium which contained 2% glucose and 100 µg/ml ampicillin. The transformants were subsequently cultured at 37°C overnight. After gradient dilution, the size of the library was measured by the number of colonies. Twenty-four colonies were chosen to detect the insertion rate of the library by PCR.
Selection of Nbs by phage display
The Nbs against CTLA-4 were selected by phage display. VHH library was amplified and infected with VCSM13 helper phages with 3 consecutive rounds of bio-panning (24). The CTLA-4 protein (20 µg) in buffer (100 mM NaHCO3, pH 8.2) was used as an antigen to coat microtiter plates of 96-wells at 4°C overnight. After blocking with 0.1% casein in phosphate-buffered saline (PBS) for 2 h, the wells were incubated with displayed phages by PBS for 1 h at room temperature. The specific phages were then eluted with 100 mM triethylamine for 10 min and neutralized with 1.0 M Tris-HCl (pH 7.4) immediately. The exponentially growing culture of TG1 cells (OD600=0.4–0.6) was infected with the eluted phages. Then, they were incubated in constant temperature incubator at 37°C for 30 min. The helper phages VCSM13 were added to rescue the phages. The process represented one round of bio-panning and these rescued phage particles were used in the next round of panning. After 3 rounds, the CTLA-4-specific phages were enriched gradually.
To obtain positive colonies, 96 individual colonies were selected randomly for PE-ELISA and cultured in 1 ml Terrific broth (TB) containing 100 µg/ml ampicillin for 3 h. Then, isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to induce the expression of Nbs overnight at 28°C. The supernatant of cells was collected after an osmotic shock and added into the plate wells which were coated with human CTLA-4 protein in advance for 1 h, followed by incubation with mouse anti-HA tag antibody for another 1 h, and subsequently incubated with anti-mouse IgG-alkaline phosphatase. The chromogenic solution containing bisphosphate (pNPP) was added after washing with PBS with 0.05% Tween-20 (PBST). The absorbance was read using an ELISA reader at 405 nm. Finally, the positive colonies were sequenced and classified according to the amino acid sequence in the CDR3 region.
Expression and purification
For expressing Nbs, the recombinant phagemids were transformed into E. coli WK6 electrocompetent cells from the TG1 strain. These cells were cultured at 37°C in TB medium containing 0.1% glucose, 2 M MgCl2 and ampicillin (100 µg/ml). The cultures were induced with 1 mM IPTG and incubated overnight at 28°C when the optical density (OD) reached 0.6–1. The periplasmic proteins were extracted by osmotic shock and then purified by immobilized metal affinity chromatography (IMAC) by NI-NTA superflow sepharose columns in a gradient of increasing imidazole concentration (pH 7.0) (25). The purity of eluted proteins was checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Flow cytometric experiments
Human peripheral blood mononuclear cells (PBMCs) were isolated with Ficoll-Hypaque density-gradient centrifugation of whole blood from healthy donors included in the present study (informed consent has been provided). The PBMCs were suspended in RPMI-1640 medium supplemented with 10% FBS for 1 h. The nonadherent cells were removed, and then T cells were isolated by nylon-wool separation.
All flow cytometric experiments were performed at 4°C. PHA (1×106) (10 µg/ml) stimulated human T cells were saturated with PBS/2% BSA solution during 30 min with shaking to avoid nonspecific binding. Nbs (1 µg) were added to cells in PBS/2% BSA and incubated for 30 min. After 3 washes in PBS/2% BSA, cells were incubated for 30 min with 1 µg of PE anti-HA tag antibody (26,27). After 3 last washes in PBS, binding was detected by flow cytometry. Mouse anti-human CTLA-4 mAb was used as a positive control. An irrelevant Nb (anti-CD105) served as the negative control. Data were analyzed using FlowJo software 10.0.7 FlowJo LLC, Ashland, OR, USA).
B16 melanoma tumor challenge and treatments
C57BL/6 mice were subcutaneously injected with 1×105 B16/BL6 melanoma cells in the right flank. On days 7, 10, 13 and 16, the mice were treated intraperitoneally with Nb16 (100 µg in 100 µl). Control groups received a corresponding dose of anti-CTLA-4 mAb, irrelevant Nb and PBS intraperitoneally.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 6.02 (GraphPad Software, Inc., San Diego, CA, USA). Data were analyzed by two-way analysis of variance (ANOVA) or log-rank (Mantel-Cox) test. P<0.05 was considered statistically significant.
Results
Construction of the VHH library
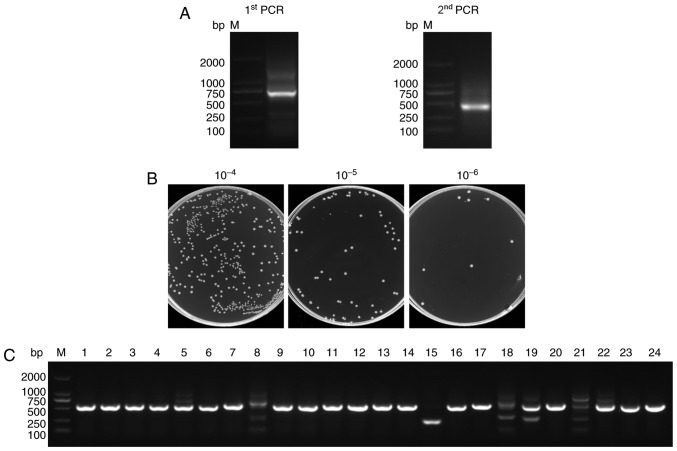
After a healthy camel was immunized with the human CTLA-4 protein 7 times, peripheral blood lymphocytes were isolated and the VHH genes were amplified from the lymphocyte cDNA (Fig. 1). The first step PCR products contained the 700 bp fragments of the VH-CH2 exons and served as templates for the second step PCR that generated 400 bp fragments of the VHH exons (Fig. 2A).
Figure 1.
Schematic of the strategy for constructing the immunized library.
Figure 2.
Construction of the VHH library. (A) The VHH genes were obtained by two-step PCR. (B) The library size was measured by counting the number of colonies after gradient dilution. (C) Twenty-four colonies were randomly selected to estimate the correct insertion rate by PCR amplification.
To construct the library, PstI and NotI sites were introduced at the 5′ and 3′ ends of the VHH fragments, respectively. In total, 10 µg of VHH fragments and 20 µg of linearized pComb3 vector were used for the ligation. Then, the recombinant plasmids were transformed into TG1 cells by 30 electroporation transformations. The size of the library was calculated by counting the number of colonies after gradient dilution. Its size was found to reach 1.85×108 colonies (Fig. 2B) which enabled the acquisition of Nbs with high specificity and sequence diversity. Twenty-four individual colonies were selected randomly for PCR analysis and the PCR results showed a library insertion rate of 95% (Fig. 2C). All of these suggested that a high-quality immunized phage display library was successfully constructed for the subsequent selection of the CTLA-4-specific Nbs.
Library screening and selection of CTLA-4-specific Nbs
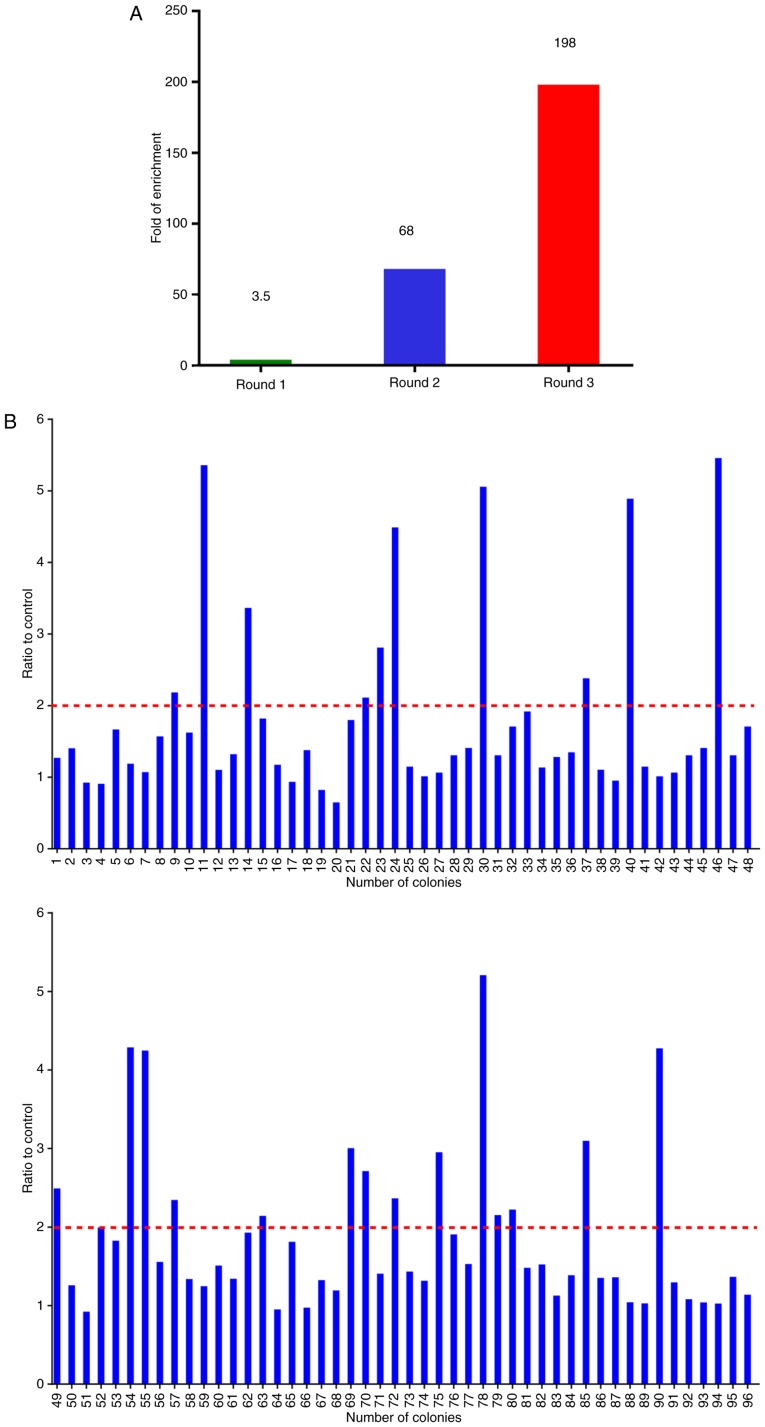
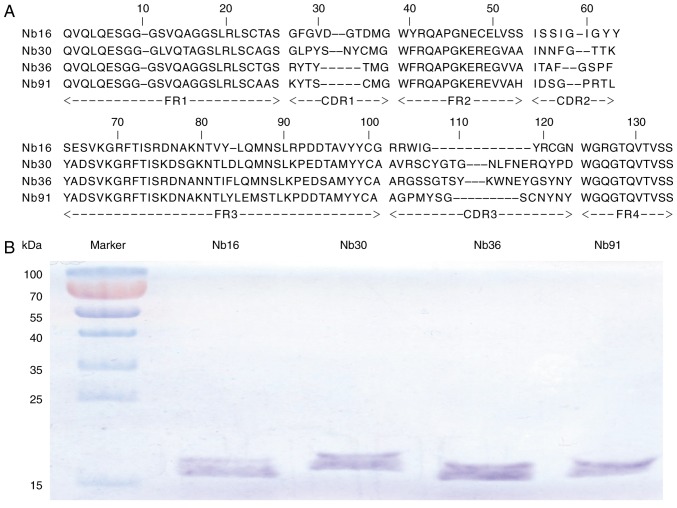
The CTLA-4-specific Nbs were identified by bio-panning from the phage display library using ~5×1011 phages. Consecutive rounds of bio-panning were performed for enriching the phages expressing CTLA-4-specific VHHs. After 3 rounds of panning, specific VHHs were enriched 198-fold compared with the negative control (Fig. 3A). Subsequently, 96 colonies were randomly chosen for PE-ELISA. Nbs existed in the supernatant of the cultured cells were disrupted by osmotic shock (25). Twenty-four colonies were selected as positive colonies whose binding ratios were >2 (Fig. 3B). After PE-ELISA, the sequences of positive colonies were analyzed and then divided into 4 families (Nb16, Nb30, Nb36 and Nb91) according to the variety of amino acid sequences in CDR3 (Fig. 4A) (28).
Figure 3.
Selection of nanobodies (Nbs) against CTLA-4 by phage display library. (A) Enrichment (198-fold) of CTLA-4-specific VHHs after 3 rounds of panning. (B) Identification of 24 positive clones following PE-ELISA of 96 clones. An optical density (OD) ratio >2 was considered as positive.
Figure 4.
Phage display library derived against CTLA-4 nanobodies (Nbs). (A) Four types of different amino acid sequences of anti-CTLA-4 VHHs were identified. (B) SDS-PAGE analysis of purified Nbs.
Expression and purification of the Nbs
VHH fragments in the phage display vector pComb3 were transformed into E. coli WK6 strains. These cells cannot suppress the amber stop codon between VHH and gene III on pComb3. Upon IPTG induction, soluble Nbs were expressed in the periplasmic region of WK6 cells. The induced Nbs were further purified by NI-NTA superflow sepharose columns. SDS-PAGE analysis showed that Nbs had single bands with high purity (Fig. 4B). The molecular weights of 4 Nbs were 15.30, 16.41, 16.12 and 15.48 kDa, respectively.
Flow cytometric analysis
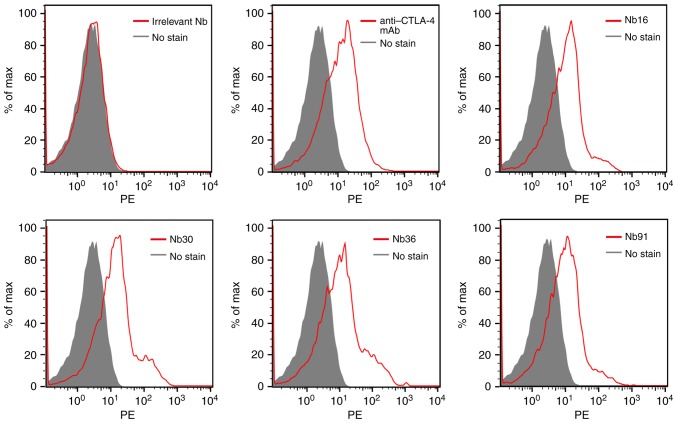
To analyze the ability of the selected nanobody to bind PHA-stimulated human T cells, flow cytometry was performed. Our results demonstrated that all of the 4 selected Nbs were able to recognize CTLA-4-expressing T cells, while the irrelevant Nb could not bind to activated T cells (Fig. 5).
Figure 5.
Flow cytometric analysis of nanobodies (Nbs) on PHA-stimulated T cells. Anti-CTLA-4 mAb and irrelevant Nb were used as positive and negative controls, respectively. mAb, monoclonal antibody.
Anti-CTLA-4 Nb16 therapy is effective against B16 melanoma tumors
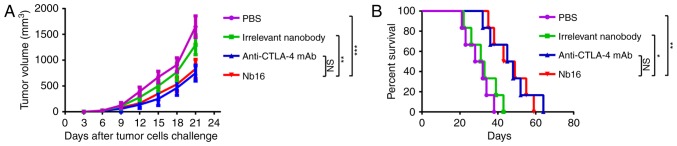
To test the contribution of Nb16, C57BL/6 mice were treated on days 7, 10, 13 and 16 with a B16/BL6 challenge (1×105 cells). Treatment of mice with Nb16 clearly delayed melanoma tumor growth (Fig. 6A) and prolonged the survival time of melanoma-bearing mice (Fig. 6B).
Figure 6.
Anti-melanoma activity of anti-CTLA-4 Nb16. (A) Melanoma tumor growth curves of C57BL/6 mice. (B) Survival percentage of melanoma-bearing mice. Each curve represents three independent experiments of 6 mice/group. *P<0.05, **P<0.01, ***P<0.001; NS indicates no significant difference.
Discussion
In the present study, CTLA-4-specific nanobodies (Nbs) were selected from the repertoire of an immunized dromedary camel with phage display technology. The VHH fragments encoded 4 types of CTLA-4-specific Nbs in phage plasmid pComb3 were directly transformed into WK6 from TG1 strains. SDS-PAGE analysis showed a band ~15 kDa after purification of soluble Nbs by NI-NTA SuperFlow sepharose columns. We expressed and isolated Nbs of high quality. We used anti-CTLA-4 Nbs in vitro by flow cytometry and subsequently demonstrated that they had a good combination with PHA stimulated and CTLA-4-positive human T cells. These Nbs showed a strong binding ability to CTLA-4. After treating B16/BL6 melanomas, we found that Nb16 delayed melanoma tumor growth and prolonged the survival time of melanoma-bearing mice.
CTLA-4 is a key surface receptor on T-lymphocytes that modulates immune responses. Antibody blockade of CTLA-4 has been shown to enhance antitumor immune responses in murine tumor models (29). Nbs present other advantages including their chemical and thermal stability, specificity, and high yield (30). Furthermore, their substantial sequence identity (>80%) with human VH sequences and the small size of 15 kDa make Nbs less likely to elicit immune responses in humans. Nbs have been reported to have low immunogenicity; humanization of Nbs has also been previously described (31). In clinical trials, anti-CTLA-4 antibodies have shown efficacy in treating prostate cancer, malignant melanoma and lung cancer (small-cell and non-small-cell) (32,33).
In summary, immunotherapy has emerged as a prominent modality in the treatment of tumors (34). For the first time, the present study shows that anti-CTLA-4 Nb16 has an intrinsic value for delaying melanoma tumor. This suggests the possibility that anti-CTLA-4 Nbs may also be advantageous for the treatment of other types of cancer or even in the clinic (35). Next, in subsequent research by us, it will be constructed into a polymer, to further improve its efficacy.
Acknowledgements
The present study was supported, in part, by grants from the Programs for Changjiang Scholars and Innovative Research Team in the University (no. IRT_15R13), the National Natural Scientific Foundation of China (nos. 81372452, 81430055, 81560494 and 81773254), the International Cooperation Project of the Ministry of Science and Technology of China (no. 2015DFA31320), the Project for Innovative Research Team in Guangxi Natural Science Foundation (2015GXNSFFA139001), the Project of Science and Technology of Guangxi (nos. 14125008-2-12 and 1599005-2-10), and the Project for International Nanobody Research Center of Guangxi (no. GuiKe-AD17195001).
References
- 1.Peggs KS, Quezada SA, Chambers CA, Korman AJ, Allison JP. Blockade of CTLA-4 on both effector and regulatory T cell compartments contributes to the antitumor activity of anti-CTLA-4 antibodies. J Exp Med. 2009;206:1717–1725. doi: 10.1084/jem.20082492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zamarin D, Holmgaard RB, Subudhi SK, Park JS, Mansour M, Palese P, Merghoub T, Wolchok JD, Allison JP. Localized oncolytic virotherapy overcomes systemic tumor resistance to immune checkpoint blockade immunotherapy. Sci Transl Med. 2014;6:226ra32. doi: 10.1126/scitranslmed.3008095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butte MJ, Keir ME, Phamduy TB, Sharpe AH, Freeman GJ. Programmed death-1 ligand 1 interacts specifically with the B7-1 costimulatory molecule to inhibit T cell responses. Immunity. 2007;27:111–122. doi: 10.1016/j.immuni.2007.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Motoshima T, Komohara Y, Horlad H, Takeuchi A, Maeda Y, Tanoue K, Kawano Y, Harada M, Takeya M, Eto M. Sorafenib enhances the antitumor effects of anti-CTLA-4 antibody in a murine cancer model by inhibiting myeloid-derived suppressor cells. Oncol Rep. 2015;33:2947–2953. doi: 10.3892/or.2015.3893. [DOI] [PubMed] [Google Scholar]
- 5.Hurwitz AA, Foster BA, Kwon ED, Truong T, Choi EM, Greenberg NM, Brug MB, Allison JP. Combination immunotherapy of primary prostate cancer in a transgenic mouse model using CTLA-4 blockade. Cancer Res. 2000;60:2444–2448. [PubMed] [Google Scholar]
- 6.Van Elsas A, Hurwitz AA, Allison JP. Combination immunotherapy of B16 melanoma using anti-cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) and granulocyte/macrophage colony-stimulating factor (GM-CSF)-producing vaccines induces rejection of subcutaneous and metastatic tumors accompanied by autoimmune depigmentation. J Exp Med. 1999;190:355–366. doi: 10.1084/jem.190.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leach DR, Krummel MF, Allison JP. Enhancement of antitumor immunity by CTLA-4 blockade. Science. 1996;271:1734–1736. doi: 10.1126/science.271.5256.1734. [DOI] [PubMed] [Google Scholar]
- 8.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ribas A, Camacho LH, Lopez-Berestein G, Pavlov D, Bulanhagui CA, Millham R, Comin-Anduix B, Reuben JM, Seja E, Parker CA, et al. Antitumor activity in melanoma and anti-self responses in a phase I trial with the anti-cytotoxic T lymphocyte-associated antigen 4 monoclonal antibody CP-675,206. J Clin Oncol. 2005;23:8968–8977. doi: 10.1200/jco.2005.23.16_suppl.7524. [DOI] [PubMed] [Google Scholar]
- 10.Fernández LÁ, Muyldermans S. Recent developments in engineering and delivery of protein and antibody therapeutics. Curr Opin Biotech. 2011;22:839–842. doi: 10.1016/j.copbio.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 11.Saerens D, Frederix F, Reekmans G, Conrath K, Jans K, Brys L, Huang L, Bosmans E, Maes G, Borghs G, Muyldermans S. Engineering camel single-domain antibodies and immobilization chemistry for human prostate-specific antigen sensing. Anal Chem. 2005;77:7547–7555. doi: 10.1021/ac051092j. [DOI] [PubMed] [Google Scholar]
- 12.Hassanzadeh-Ghassabeh G, Devoogdt N, De Pauw P, Vincke C, Muyldermans S. Nanobodies and their potential applications. Nanomedicine. 2013;8:1013–1026. doi: 10.2217/nnm.13.86. [DOI] [PubMed] [Google Scholar]
- 13.Market E, Papavasiliou FN. V(D)J recombination and the evolution of the adaptive immune system. PLoS Biol. 2003;1:E16. doi: 10.1371/journal.pbio.0000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holt LJ, Herring C, Jespers LS, Woolven BP, Tomlinson IM. Domain antibodies: proteins for therapy. Trends Biotechnol. 2003;21:484–490. doi: 10.1016/j.tibtech.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 15.Muyldermans S. Nanobodies: Natural single-domain antibodies. Annu Rev Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 16.Gueorguieva D, Li S, Walsh N, Mukerji A, Tanha J, Pandey S. Identification of single-domain, Bax-specific intra-bodies that confer resistance to mammalian cells against oxidative-stress-induced apoptosis. FASEB J. 2006;20:2636–2638. doi: 10.1096/fj.06-6306fje. [DOI] [PubMed] [Google Scholar]
- 17.Pérez JM, Renisio JG, Prompers JJ, van Platerink CJ, Cambillau C, Darbon H, Frenken LG. Thermal unfolding of a llama antibody fragment: A two-state reversible process. Biochemistry. 2001;40:74–83. doi: 10.1021/bi0009082. [DOI] [PubMed] [Google Scholar]
- 18.Muyldermans S. Single domain camel antibodies: Current status. J Biotechnol. 2001;74:277–302. doi: 10.1016/s1389-0352(01)00021-6. [DOI] [PubMed] [Google Scholar]
- 19.De Genst E, Silence K, Decanniere K, Conrath K, Loris R, Kinne J, Muyldermans S, Wyns L. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies; Proc Natl Acad Sci USA; 2006; pp. 4586–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kijanka M, Warnders FJ, El Khattabi M, Lub-de Hooge M, van Dam GM, Ntziachristos V, de Vries L, Oliveira S, van Bergen En Henegouwen PM. Rapid optical imaging of human breast tumour xenografts using anti-HER2 VHHs site-directly conjugated to IRDye 800CW for image-guided surgery. Eur J Nucl Med Mol Imaging. 2013;40:1718–1729. doi: 10.1007/s00259-013-2471-2. [DOI] [PubMed] [Google Scholar]
- 21.Vosjan MJ, Vercammen J, Kolkman JA, Stigter-van Walsum M, Revets H, van Dongen GA. Nanobodies targeting the hepatocyte growth factor: Potential new drugs for molecular cancer therapy. Mol Cancer Ther. 2012;11:1017–1025. doi: 10.1158/1535-7163.MCT-11-0891. [DOI] [PubMed] [Google Scholar]
- 22.De Meyer T, Eeckhout D, De Rycke R, De Buck S, Muyldermans S, Depicker A. Generation of VHH antibodies against the Arabidopsis thaliana seed storage proteins. Plant Mol Biol. 2014;84:83–93. doi: 10.1007/s11103-013-0118-0. [DOI] [PubMed] [Google Scholar]
- 23.Conrath KE, Lauwereys M, Galleni M, Matagne A, Frère JM, Kinne J, Wyns L, Muyldermans S. Beta-lactamase inhibitors derived from single-domain antibody fragments elicited in the camelidae. Antimicrob Agents Chemother. 2001;45:2807–2812. doi: 10.1128/AAC.45.10.2807-2812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vincke C, Gutiérrez C, Wernery U, Devoogdt N, Hassanzadeh-Ghassabeh G, Muyldermans S. Generation of single domain antibody fragments derived from camelids and generation of manifold constructs. Methods Mol Biol. 2012;907:145–176. doi: 10.1007/978-1-61779-974-7_8. [DOI] [PubMed] [Google Scholar]
- 25.Zhu M, Gong X, Hu Y, Ou W, Wan Y. Streptavidin-biotin-based directional double Nanobody sandwich ELISA for clinical rapid and sensitive detection of influenza H5N1. J Transl Med. 2014;12:352. doi: 10.1186/s12967-014-0352-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nevoltris D, Lombard B, Dupuis E, Mathis G, Chames P, Baty D. Conformational nanobodies reveal tethered epidermal growth factor receptor involved in EGFR/ErbB2 predimers. ACS Nano. 2015;9:1388–1399. doi: 10.1021/nn505752u. [DOI] [PubMed] [Google Scholar]
- 27.Hernot S, Unnikrishnan S, Du Z, Shevchenko T, Cosyns B, Broisat A, Toczek J, Caveliers V, Muyldermans S, Lahoutte T, et al. Nanobody-coupled microbubbles as novel molecular tracer. J Control Release. 2012;158:346–353. doi: 10.1016/j.jconrel.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muyldermans S, Baral T, Retamozzo VC, De Baetselier P, De Genst E, Kinne J, Leonhardt H, Magez S, Nguyen V, Revets H, et al. Camelid immunoglobulins and nanobody technology. Vet Immunol Immunopathol. 2009;128:178–183. doi: 10.1016/j.vetimm.2008.10.299. [DOI] [PubMed] [Google Scholar]
- 29.Curran MA, Montalvo W, Yagita H, Allison JP. PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors; Proc Natl Acad Sci USA; 2010; pp. 4275–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dumoulin M, Conrath K, Van Meirhaeghe A, Meersman F, Heremans K, Frenken LG, Muyldermans S, Wyns L, Matagne A. Single-domain antibody fragments with high conformational stability. Protein Sci. 2002;11:500–515. doi: 10.1110/ps.34602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vu KB, Ghahroudi MA, Wyns L, Muyldermans S. Comparison of llama VH sequences from conventional and heavy chain antibodies. Mol Immunol. 1997;34:1121–1131. doi: 10.1016/S0161-5890(97)00146-6. [DOI] [PubMed] [Google Scholar]
- 32.Vaneycken I, Govaert J, Vincke C, Caveliers V, Lahoutte T, De Baetselier P, Raes G, Bossuyt A, Muyldermans S, Devoogdt N. In vitro analysis and in vivo tumor targeting of a humanized, grafted nanobody in mice using pinhole SPECT/micro-CT. J Nucl Med. 2010;51:1099–1106. doi: 10.2967/jnumed.109.069823. [DOI] [PubMed] [Google Scholar]
- 33.Vincke C, Loris R, Saerens D, Martinez-Rodriguez S, Muyldermans S, Conrath K. General strategy to humanize a camelid single-domain antibody and identification of a universal humanized nanobody scaffold. J Biol Chem. 2009;284:3273–3284. doi: 10.1074/jbc.M806889200. [DOI] [PubMed] [Google Scholar]
- 34.Gao Y, Gao W, Chen X, Cha N, Wang X, Jia X, Wang B, Ren M, Ren J. Enhancing the treatment effect on melanoma by heat shock protein 70-peptide complexes purified from human melanoma cell lines. Oncol Rep. 2016;36:1243–1250. doi: 10.3892/or.2016.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vosjan MJ, Vercammen J, Kolkman JA, Stigter-van Walsum M, Revets H, van Dongen GA. Nanobodies targeting the hepatocyte growth factor: Potential new drugs for molecular cancer therapy. Mol Cancer Ther. 2012;11:1017–1025. doi: 10.1158/1535-7163.MCT-11-0891. [DOI] [PubMed] [Google Scholar]