Abstract
Background:
Early-life exposure to lead is associated with deficits in neurodevelopment and with hematopoietic system toxicity. DNA methylation may be one of the underlying mechanisms for the adverse effects of prenatal lead on the offspring, but epigenome-wide methylation data for low levels of prenatal lead exposure are lacking.
Objectives:
We investigated the association between prenatal maternal lead exposure and epigenome-wide DNA methylation in umbilical cord blood nucleated cells in Project Viva, a prospective U.S.-based prebirth cohort with relatively low levels of lead exposure.
Methods:
Among 268 mother–infant pairs, we measured lead concentrations in red blood cells (RBC) from prenatal maternal blood samples, and using HumanMethylation450 Bead Chips, we measured genome-wide methylation levels at 482,397 CpG loci in umbilical cord blood and retained 394,460 loci after quality control. After adjustment for batch effects, cell types, and covariates, we used robust linear regression models to examine associations of prenatal lead exposure with DNA methylation in cord blood at epigenome-wide significance level [false discovery rate ].
Results:
The mean [standard deviation (SD)] maternal RBC lead level was 1.22 (0.63) . CpG cg10773601 showed an epigenome-wide significant negative association with prenatal lead exposure ( per doubling increase in lead exposure; ) and was annotated to C-Type Lectin Domain Family 11, Member A (CLEC11A), which functions as a growth factor for primitive hematopoietic progenitor cells. In sex-specific analyses, we identified more CpGs with among female infants () than among male infants (). One CpG (cg24637308), which showed a strong negative association with prenatal lead exposure among female infants ( per doubling increase in lead exposure; ), was annotated to Dynein Heavy Chain Domain 1 gene (DNHD1) which is highly expressed in human brain. Interestingly, there were strong correlations between blood and brain methylation for CpG (cg24637308) based on another independent set of samples with a high proportion of female participants.
Conclusion:
Prenatal low-level lead exposure was associated with newborn DNA methylation, particularly in female infants. https://doi.org/10.1289/EHP1246
Introduction
Lead is an environmental pollutant whose widespread use has caused extensive environmental contamination and health effects throughout the world (WHO 2015). Lead affects multiple body systems, and there is no known level of lead exposure that is considered safe (WHO 2015). Lead can pass freely across the placenta (Chuang et al. 2001), and as a result, there is substantial concern about the potential adverse effects of prenatal lead exposure on the developing fetus. Fetal and early-life exposure to lead, even at low levels, has been associated with a range of adverse health outcomes, including preterm delivery and small-for-gestational-age birth (Jelliffe-Pawlowski et al. 2006; Perkins et al. 2014), poor growth in childhood (Afeiche et al. 2011), impaired cognitive and behavioral functions (Boucher et al. 2014; Liu et al. 2014; Virgolini et al. 2008), and hematopoietic system toxicity (Jacob et al. 2000; Serrani et al. 1997). Interestingly, sex differences in lead exposure–related health effects have been reported in previous studies (Afeiche et al. 2011; Bunn et al. 2001; Perkins et al. 2014; Schneider et al. 2013; Virgolini et al. 2008), suggesting sexually dimorphic responses to lead exposure between males and females.
Previous animal, in vitro, and epidemiological studies have suggested that DNA methylation may be one of the mechanisms by which lead exposure exerts its biological effects. For example, lead exposure induced dose- and sex-specific responses in DNA methylation at several specific loci that were responsible for phenotypes including weight and coat color in mice (Faulk et al. 2013). An in vitro study showed that lead exposure reduces the global DNA methylation level by noncompetitive inhibition and alteration of DNA methyltransferase (DNMT) expression (Sanchez et al. 2017), whereas additional animal studies also found that lead exposure affects the expression of maintenance enzyme DNMT1 and the de novo methyltransferase DNMT3a in the brain (Schneider et al. 2012; Schneider et al. 2013). Specifically, an in vitro study demonstrated that lead exposure induced changes in the methylation status of genes involved in neurogenetic signaling pathways in human embryonic stem cells and altered their neuronal differentiation (Senut et al. 2014). These studies provide evidence that lead exposure may affect both global and brain genomic methylation. In addition, a recent epidemiological study found that the epigenome-wide DNA methylation profile in umbilical cord blood was associated with high levels of prenatal lead exposure in 127 mother–infant pairs in lead-endemic regions outside the United States (Engström et al. 2015).
Although existing evidence supports the role of lead exposure in modifying DNA methylation, little is known about potential effects of relatively low levels of lead exposure in utero—common to much of the U.S. population—on DNA methylation. It is also unknown whether variation in DNA methylation associated with prenatal lead exposure may last over time, which may help elucidate the long-term health effects of lead exposure. We aimed to investigate these questions by performing an epigenome-wide association study (EWAS) of low-level maternal lead exposure [ in red blood cells (RBC)] and DNA methylation profiles in umbilical cord blood samples from 268 mother–infant pairs enrolled in Project Viva, a U.S.-based prebirth cohort, overall and separately among boys and girls. We also attempted to replicate the observed prenatal lead exposure–cord blood DNA methylation associations using data collected during midchildhood for 240 children (7–10 y of age).
Methods
Study Subjects
Study subjects were mother–infant pairs in Project Viva, a prospective prebirth cohort with 2,128 women recruited between 1999 and 2002 from Atrius Harvard Vanguard Medical Associates in eastern Massachusetts. Eligibility criteria included English speaking, singleton pregnancy, and of gestation. All women provided written informed consent, and the research was approved by the Institutional Review Board at Harvard Pilgrim Health Care.
We collected blood samples from 1,614 women at an average of 27.9 wk of gestation and analyzed blood samples for 952 women for lead, selecting those with children who participated in follow-up visits in midchildhood. We measured genome-wide DNA methylation using the Infinium HumanMethylation450 (HM450K) BeadChip (Illumina, Inc.) for 507 umbilical cord blood samples collected at delivery and 473 child blood samples collected during midchildhood. A total of 485 cord blood samples and 460 midchildhood blood samples passed quality-control (QC) procedures (see Table S1). Among these, 269 samples had data on both maternal RBC lead and DNA methylation in cord blood, and 241 samples had data on both maternal RBC lead and DNA methylation in midchildhood blood samples. We excluded one sample with undetectable maternal RBC lead, leaving 268 cord blood samples and 240 midchildhood blood samples for the data analysis. Participants with cord blood samples and midchildhood blood samples (, with 131 overlapped between delivery and midchildhood) were generally similar in baseline characteristics compared with those who were not included (, data not shown). However, women included in this analysis were more likely to be white (76% vs. 65%) and to have a college degree (70% vs. 64%) than those who were excluded.
Sample Collection
All blood samples were collected in vacutainer tubes containing ethylenediaminetetraacetic acid (EDTA) and were put on ice immediately. The tubes were centrifuged at for 10 min at 4°C to separate plasma, nucleated cells (including leukocytes and nucleated RBC in cord blood and leukocytes in maternal blood), and RBC within 24 h after collection. We extracted genomic DNA from the nucleated cells with commercially available PureGene Kits (Fisher, Catalog Nos. A407-4, A416-4; Qiagen, Catalog Nos.158908, 158912, 158924). Sample aliquots were then stored at until analysis.
Measurement of RBC Lead
Lead concentrations in RBC from prenatal blood samples were measured at the Trace Metals Laboratory at Harvard T.H. Chan School of Public Health in Boston, Massachusetts. RBC samples were weighed and digested for 24 h in of concentrated nitric acid and of 30% hydrogen peroxide per of RBC. Samples were then diluted to a volume of with deionized water and were measured using a dynamic reaction cell–inductively coupled plasma mass spectrometer (Elan DRC II; PerkinElmer) for lead concentrations. Quality-control measures included analysis of initial and continuous calibration verification standards, 1-ppb lead standard, procedural blanks, QC standard [National Institute of Standards and Technology Standard Reference Material (NIST SRM) 1643d, Trace Elements in Water (NIST SRM955b, Lead in Blood)]. Results were calculated as the mean of five replicate measurements. The limit of detection for this procedure was in RBC.
Epigenome-Wide DNA Methylation Profiling and Quality Controls
Extracted DNA underwent bisulfite modification using the Zymo EZ DNA Methylation kit (Zymo Research). We randomized samples of bisulfite-modified DNA ( for each sample) across different plates and BeadChips to ensure balance by sex and to reduce the influence of batch effects. Epigenome-wide DNA methylation analysis using the HM450K microarray interrogates 482,397 methylation loci at single-nucleotide resolution. The microarray measurements were processed at Illumina, Inc. We preprocessed data using the minfi package in R software (R Project for Statistical Computing). We excluded samples if they a) were replicates; b) were of low quality with individual call rate ; c) had genotype mismatch; or d) had sex mismatch (see Table S1). We excluded methylation probes if a) they were on either the X or the Y chromosome; b) they were nonCpG probes; c) they had nonsignificant detection p-values () for of the samples; d) they had been identified as cross-hybridizing with other genomic locations (Chen et al. 2013); or e) single nucleotide polymorphism (SNP)-associated probes at either the single base extension or within the target region for SNPs that have a minor allele frequency (MAF) . A total of 394,460 autosomal probes passed the QC. We performed background correction and dye-bias equalization using the normal-exponential out-of-band (noob) correction method (Triche et al. 2013). We performed a probe type normalization using the beta-mixture quantile (BMIQ) method in the minfi package in R (Teschendorff et al. 2013).
Covariates
We collected maternal demographics including age, race/ethnicity, educational level, parity history, and pregnancy health information including smoking status and prepregnancy weight and height, as well as infant sex using self-administered questionnaires and interviews during pregnancy. We collected information on delivery date from medical records. Prepregnancy body mass index (BMI; kilograms per square meter) was calculated from self-reported prepregnancy weight and height. Women reported their last menstrual period (LMP) at enrollment, and we calculated gestational age by subtracting the LMP date from the delivery date or by ultrasound when available and when it differed from the LMP by more than 10 d (Gillman et al. 2004). To estimate cell-type distribution in blood samples, we used a reference panel of nucleated cells (leukocytes and nucleated RBC) isolated from cord blood (Bakulski et al. 2016) for cord blood samples and an adult leukocyte reference panel for blood samples collected in midchildhood as implemented in minfi (Aryee et al. 2014; Reinius et al. 2012). Leukocytes included T cells, T cells, natural killer (NK) cells, monocytes, and B cells.
Statistical Analysis
We accounted for batch effects from plates and for other potential sources of technical variability in the noob-adjusted and BMIQ-normalized cord blood methylation data using the ComBat adjustment (Johnson et al. 2007). We visually inspected the effectiveness of the removal of batch effects using principal component analysis before and after technical batch adjustment.
ComBat-adjusted cord blood methylation -values were to M-values to be more appropriate for differential analysis of DNA methylation (Du et al. 2010). Robust linear regression models were constructed using the MASS package in R, with M-values included as dependent variables and maternal RBC lead levels as the main predictor, with adjustment for other potential confounders. Covariates included child sex, gestational age (weeks; as a continuous variable), maternal prepregnancy BMI (kilograms per square meter; as a continuous variable), age at enrollment (years; as a continuous variable), parity history (nulliparous, nonnulliparous), educational level (college graduate, noncollege graduate), smoking status during pregnancy (never, former, current smoker), race (white, nonwhite), and blood cell type distribution in cord blood.
To account for multiple testing, we only report top loci with a false discovery rate (FDR)-corrected , as has commonly been implemented in previous EWAS analyses (Engström et al. 2015; Senut et al. 2014). To facilitate interpretation of the results, we report effect estimates in the main text using the -value scale for top loci with an based on the robust linear regression. We further stratified the EWAS by child sex, using the same criterion of to determine the top loci. We tested the interaction between child sex and lead by adding a multiplicative term in the model along with their main effect terms for all significant loci meeting the criterion .
We attempted to replicate the identified associations for top loci in cord blood by further examining associations of prenatal lead exposure with DNA methylation at the same top CpGs in midchildhood blood samples. For these analyses, we again performed robust linear regression analyses with adjustment for the same covariates [but using Houseman blood cell type algorithms based on an adult blood cell reference panel (Aryee et al. 2014; Reinius et al. 2012)] as well as for child age at blood sample collection. We drew visualized regional DNA comethylation patterns for identified potential target genes using the coMET package in R. To address the potential residual confounding due to smoking, we first performed sensitivity analyses excluding infants with sustained maternal smoking during pregnancy () according to a definition reported elsewhere (Joubert et al. 2016). We further created a methylation score predicting smoking status by calculating the linear combination of methylation values of 28 CpGs as described elsewhere (Reese et al. 2017), and we adjusted for the score as a quartile-covariate instead of adjusting for the smoking status in the EWAS analyses. Finally, we used the bumphunter package from Bioconductor to perform a bumphunting analysis, as previously described (Jaffe et al. 2012), to detect any differentially methylated regions (DMRs) relative to prenatal maternal lead exposure. Briefly, we fit models with adjustment for the same covariates as in the primary EWAS analyses and ran bumphunter on the -values using RBC-lead as the exposure of interest. All statistical analyses were performed using R 3.2.3 (R Project for Statistical Computing).
Results
Among the 268 mothers included in the primary analysis, 138 (51.5%) had female infants (Table 1). The mean age of women at enrollment was 31.9 y; 51.1% were nulliparous, 79% were white, and 69.0% were college graduates. The mean maternal RBC blood lead level was [standard deviation (SD), 0.63; range, 0.29–4.97] and was generally similar between mothers of female () and male () infants (Wilcoxon rank test , see Table S2). This level approximates to a whole-blood level of based on the estimate that RBC concentrations are the products of the whole-blood level divided by hematocrit ( in women) (Chuang et al. 2001). Characteristics of the 240 mother–child pairs with child blood samples in midchildhood were largely similar. The mean age of children in midchildhood was 7.9 y (SD, 0.8; range, 7–10).
Table 1.
Characteristics of Project Viva mother–infant pairs with umbilical cord blood and midchildhood blood methylation measurement.
| Characteristic | Birth () | Midchildhood () |
|---|---|---|
| Maternal | ||
| Prepregnancy BMI, : Mean (SD) | 24.2 (5.0) | 24.5 (4.9) |
| Age at enrollment, years: Mean (SD) | 31.9 (4.7) | 32.1 (5.3) |
| Nulliparous: (%) | 131 (48.9) | 113 (47.1) |
| College graduate: (%) | 185 (69.0) | 169 (70.4) |
| Smoking status: (%) | ||
| Never | 178 (66.4) | 168 (70.0) |
| Former | 55 (20.5) | 47 (19.6) |
| During pregnancy | 35 (13.1) | 25 (10.4) |
| Maternal white race/ethnicity: (%) | 212 (79.1) | 173 (72.1) |
| 2nd-trimester Pb, : Mean (SD) | 1.22 (0.63) | 1.20 (0.53) |
| Child | ||
| Female: (%) | 138 (51.5) | 121 (50.4) |
| Gestational age at birth, wk: Mean (SD) | 39.7 (1.6) | 39.5 (1.7) |
| Age at blood collection, years: Mean (SD) | – | 7.9 (0.8) |
Note: BMI, body mass index; Pb, lead; SD, standard deviation.
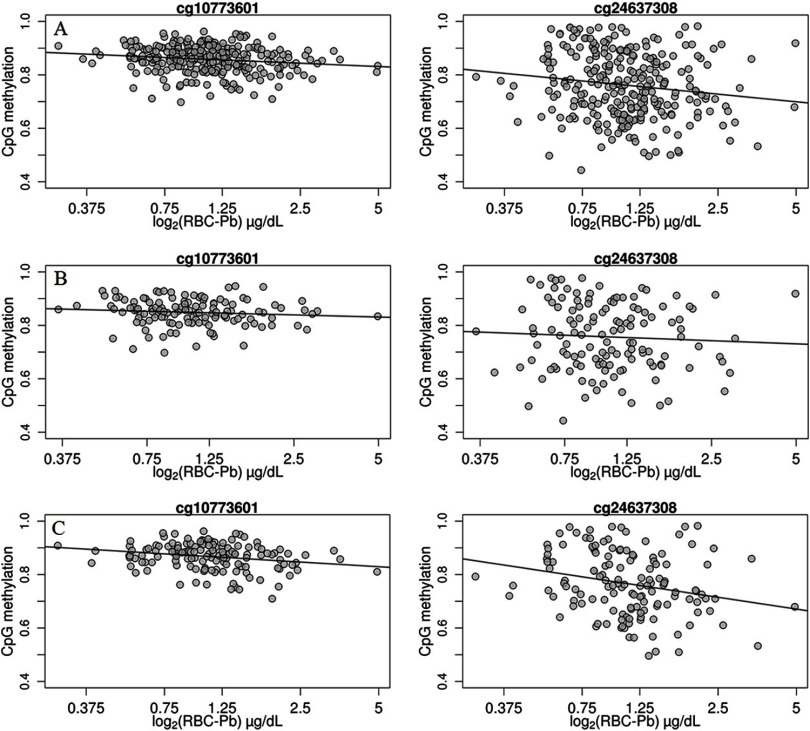
Our analysis showed that the p-value distribution of all CpGs was generally similar to the theoretical distribution among all newborns and female infants (in quantile–quantile plots, values were 0.9308 and 1.0192, respectively) and was slightly different from the theoretical distribution among male infants (; see Figure S1). We found that methylation levels at four CpGs demonstrated significant negative associations () with maternal RBC lead among all newborns (Table 2; see also Figure S2A). Among these CpGs, one (cg10773601) was annotated to bps from the transcription start site of C-Type Lectin Domain Family 11, Member A (CLEC11A). For each doubling increase in maternal RBC lead levels, there was a 1.4% decrease in the methylation level of cg10773601 in cord blood (). This association appeared to be linear over the lead exposure range (Figure 1A). This effect estimate attenuated to in midchildhood blood samples (; see Table S3). Effect estimates for the other three loci in cord blood and midchildhood blood samples were generally minimal and were not annotated to any genes.
Table 2.
CpGs with DNA methylation levels in umbilical cord blood associated with prenatal maternal red blood cell lead concentrations in analyses for all newborns, male infants, and female infants (selected based on for EWAS).
| CpG | Chr | Position | Relation to island | Gene | Relation to gene | Average methylation (%)a | All newborns | Male infants | Female infants | p-Value for interaction | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Effect estimate (SE)b | p-Value | Effect estimate (SE)b | p-Value | Effect estimate (SE)b | p-Value | ||||||||
| Top loci identified for all newborns | |||||||||||||
| cg02272457 | chr6 | 42427790 | Open Sea | – | – | 99.1 | (0.02) | (0.03) | (0.02) | 0.88 | |||
| cg20324491 | chr7 | 149128397 | N_Shore | – | – | 94.2 | (0.1) | (0.2) | (0.1) | 0.55 | |||
| cg22112000 | chr7 | 5647182 | Island | – | – | 98.5 | (0.02) | (0.03) | (0.02) | 0.29 | |||
| cg10773601 | chr19 | 51226046 | N_Shore | CLEC11A | TSS1500 | 85.8 | (0.3) | (0.4) | (0.4) | 0.52 | |||
| Top loci identified for male infants | |||||||||||||
| cg22512536 | chr5 | 154134507 | Island | LARP1 | Body | 1.9 | 0.1 (0.03) | 0.1 (0.03) | (0.04) | 0.002 | |||
| cg08964024 | chr12 | 14923326 | Island | – | – | 1.6 | 0.1 (0.02) | 0.1 (0.02) | 0.01 (0.02) | 0.002 | |||
| Top loci identified for female infants | |||||||||||||
| cg04295372 | chr1 | 101184332 | Open Sea | VCAM1 | TSS1500 | 41.9 | 0.9 (0.3) | (0.6) | 1.6 (0.3) | 0.03 | |||
| cg09356083 | chr1 | 2430057 | N_Shelf | PLCH2 | Body | 85.1 | (0.1) | (0.2) | (0.1) | 0.12 | |||
| cg20816789 | chr1 | 27462916 | Open Sea | SLC9A1 | Body | 87.0 | (0.1) | (0.3) | (0.2) | 0.11 | |||
| cg22446399 | chr2 | 27326580 | Open Sea | CGREF1 | Body | 95.9 | (0.1) | (0.2) | (0.1) | 0.06 | |||
| cg27403609 | chr2 | 11101403 | Island | – | – | 91.8 | (0.3) | 0.1 (0.4) | (0.3) | 0.004 | |||
| cg07248017 | chr4 | 8230689 | Open Sea | SH3TC1 | Body | 71.1 | (0.2) | (0.3) | (0.2) | 0.009 | |||
| cg06239355 | chr5 | 32714010 | Island | NPR3 | Body | 1.7 | 0.1 (0.02) | 0.03 (0.04) | 0.1 (0.02) | 0.04 | |||
| cg15601915 | chr5 | 1065775 | S_Shore | SLC12A7 | Body | 85.7 | (0.1) | (0.2) | (0.2) | 0.001 | |||
| cg17599748 | chr6 | 31589597 | S_Shore | SNORA38 | TSS1500 | 68.1 | (0.3) | 0.4 (0.4) | (0.3) | 0.002 | |||
| cg08673909 | chr7 | 1329592 | Island | – | – | 2.1 | (0.04) | (0.1) | (0.05) | 0.07 | |||
| cg10090217 | chr7 | 45151583 | S_Shore | TBRG4 | TSS1500 | 2.2 | (0.02) | (0.03) | (0.02) | 0.03 | |||
| cg16565528 | chr8 | 91010920 | N_Shelf | – | – | 83.1 | 0.8 (0.3) | (0.4) | 1.8 (0.4) | 0.05 | |||
| cg03152353 | chr9 | 139417194 | Island | NOTCH1 | Body | 56.6 | (0.2) | (0.3) | (0.2) | 0.02 | |||
| cg13817920 | chr9 | 96587843 | N_Shore | – | – | 89.0 | (0.1) | (0.2) | (0.1) | 0.10 | |||
| cg13791644 | chr10 | 119794366 | Open Sea | RAB11FIP2 | Body | 93.2 | 0.5 (0.2) | (0.3) | 1.0 (0.2) | 0.01 | |||
| cg18454045 | chr10 | 88391658 | N_Shore | – | – | 69.2 | (0.2) | 0.1 (0.5) | (0.3) | 0.004 | |||
| cg11127561 | chr11 | 125462151 | N_Shore | STT3A | TSS1500 | 1.6 | 0.1 (0.02) | 0.03 (0.02) | 0.1 (0.02) | 0.005 | |||
| cg17971003 | chr11 | 107582804 | OpenSea | SLN | TSS200 | 93.3 | 0.7 (0.2) | 0.4 (0.2) | 1.0 (0.2) | 0.18 | |||
| cg24637308 | chr11 | 6592297 | Island | DNHD1 | Body | 76.2 | (0.9) | (1.4) | (0.9) | 0.12 | |||
| cg05959994 | chr13 | 114253916 | Open Sea | TFDP1 | Body | 98.6 | (0.04) | 0.1 (0.05) | (0.04) | ||||
| cg11203293 | chr13 | 25777762 | Open Sea | – | – | 60.5 | (0.5) | 0.4 (0.7) | (0.4) | 0.001 | |||
| cg04545835 | chr14 | 102675570 | Open Sea | WDR20 | Body | 97.1 | (0.1) | 0.002 (0.2) | (0.1) | 0.004 | |||
| cg08131309 | chr15 | 40399131 | N_Shore | BMF | TSS1500 | 23.2 | (0.3) | (0.4) | (0.3) | 0.002 | |||
| cg04730825 | chr16 | 16116191 | Open Sea | ABCC1 | Body | 58.7 | (0.3) | 0.1 (0.4) | (0.4) | 0.02 | |||
| cg26686608 | chr16 | 88705716 | N_Shore | IL17C | Body | 41.5 | 0.5 (0.2) | (0.4) | 1.2 (0.3) | 0.0007 | |||
| cg00461015 | chr17 | 42295635 | N_Shore | UBTF | 5'UTR | 1.9 | (0.03) | (0.04) | (0.03) | 0.07 | |||
| cg04571282 | chr17 | 44108753 | Open Sea | KIAA1267 | 3'UTR | 98.3 | (0.03) | (0.1) | (0.04) | 0.13 | |||
| cg11252953 | chr17 | 80358829 | N_Shelf | C17orf101 | Body | 7.6 | 0.4 (0.2) | 0.2 (0.3) | 0.8 (0.2) | 0.13 | |||
| cg15922057 | chr17 | 25784714 | S_Shore | – | – | 31.8 | (0.3) | 0.2 (0.4) | (0.3) | 0.001 | |||
| cg06753949 | chr19 | 15334309 | Island | – | – | 1.9 | (0.03) | (0.05) | (0.04) | 0.27 | |||
| cg07780528 | chr19 | 35630334 | N_Shelf | FXYD1 | 5'UTR | 23.3 | (0.1) | (0.2) | (0.2) | 0.15 | |||
| cg11610754 | chr19 | 18722595 | Island | TMEM59L | TSS1500 | 1.7 | 0.03 (0.01) | (0.02) | 0.1 (0.01) | 0.03 | |||
| cg18598117 | chr19 | 941126 | Island | ARID3A | Body | 84.6 | (0.3) | 0.1 (0.8) | (0.4) | 0.02 | |||
| cg22217660 | chr19 | 50765301 | S_Shelf | MYH14 | Body | 92.2 | (0.1) | 0.1 (0.3) | (0.2) | 0.04 | |||
| cg03373781 | chr20 | 30582113 | N_Shore | XKR7 | Body | 74.8 | (0.2) | (0.3) | (0.2) | 0.02 | |||
| cg07341934 | chr20 | 57463711 | Island | GNAS | 3'UTR | 60.1 | (0.3) | (0.5) | (0.3) | 0.002 | |||
| cg21307155 | chr20 | 3216740 | N_Shore | SLC4A11 | Body | 87.5 | (0.1) | 0.02 (0.2) | (0.1) | 0.008 | |||
| cg17174023 | chr22 | 50987453 | Island | KLHDC7B | 1st Exon | 9.0 | (0.2) | 0.2 (0.4) | (0.3) | 0.05 | |||
Note: Body, gene body; Chr, Chromosome; FDR, false discovery rate; SE, standard error; TSS200, within 200 bps from transcription start site; TSS1500, within 1500 bps from transcription start site; UTR, untranslated region.
Average methylation in umbilical cord blood among all newborns.
The percent change in DNA methylation level for every doubling increase in prenatal maternal red blood cell lead concentrations. Adjusted for child sex, maternal age, maternal prepregnancy body mass index, maternal race, maternal educational level, maternal smoking status, parity, gestational age, and differential nucleated cell proportions (monocytes, T cells, T cells, natural killer cells, B cells, and nucleated red blood cells).
Figure 1.
Scatterplots for prenatal red blood cell lead levels and cg10773601 (left panel) and cg24637308 (right panel) methylation levels of umbilical cord blood DNA among all newborns (A), male infants (B), and female infants (C).
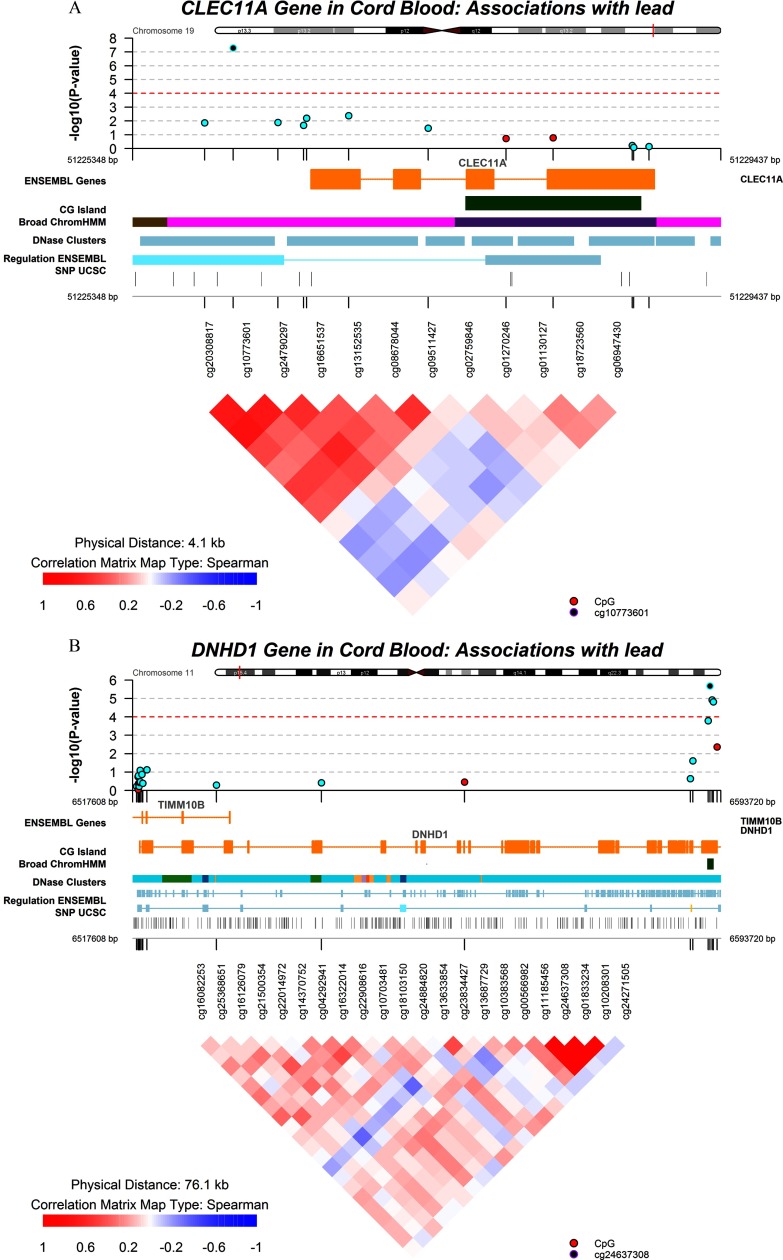
We identified somewhat different top loci in sex-specific EWAS analyses, and most of the loci had for interaction tests between infant sex and lead exposure (Table 2). Among male infants, methylation levels at only two CpGs in cord blood showed significant but small positive associations with maternal RBC lead at (Table 2; see also Figure S2B). Neither of these two CpGs showed an association with prenatal lead in blood DNA collected in midchildhood (see Table S3). Among female infants, maternal RBC lead levels were negatively associated with methylation levels of 29 CpGs and were positively associated with DNA methylation at 9 CpGs in cord blood at (Table 2; see also Figure S2C). One CpG (cg24637308) showed a strong negative association with prenatal lead exposure (the effect estimate was for each doubling of maternal RBC lead, ). This association also appeared to be linear over the lead exposure range (Figure 1C) and was consistent in midchildhood, with each doubling of maternal RBC lead associated with 4.1% lower methylation in midchildhood females (). The CpG cg24637308 is located in the body of the Dynein Heavy Chain Domain 1 gene (DNHD1) and is annotated to a CpG island). Effect estimates for the other loci among female infants were either minor or were substantially attenuated in midchildhood. Visualization of regional DNA comethylation patterns for top loci showed that most CpGs around the DNHD1 gene had similar changes in the same direction as that of the top CpG cg24637308 in association with lead exposure, although they did not pass the FDR adjustment (Figure 2). Lists of annotated CpGs with uncorrected in EWAS analyses and scatterplots for prenatal RBC lead levels and methylation levels at top loci other than cg10773601 and cg24637308 as shown in Table 2 for all newborns, male infants, and female infants have been included in the Supplemental Material for reference (see Tables S4–S6; see also Figure S3). Sensitivity analyses excluding infants with sustained maternal smoking yielded generally similar results (see Tables S7–S9). Results from analyses with adjustment for the methylation score predicting smoking status were also generally consistent with the original results, and major top CpGs, including cg10773601 and cg24637308, still met (see Tables S10–S12).
Figure 2.
Visualization of regional DNA comethylation patterns for genes CLEC11A (A) and DNHD1 (B) in umbilical cord blood among all newborns. Black dots indicate the identified key loci (cg10773601 and cg24637308), blue dots indicate CpGs in the same direction, and red dots indicate CpGs in the opposite direction associated with lead exposure.
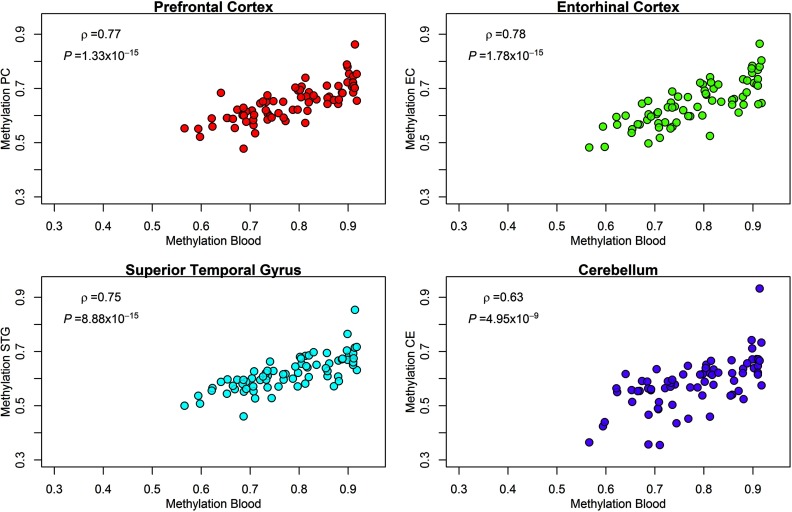
Because DNA methylation is tissue-specific, we were interested in examining the consistency of methylation patterns between our surrogate tissue (blood) and the potential target tissue (e.g., brain). We therefore investigated blood–brain DNA methylation correlations for top loci based on an independent sample dominated by adult females (the female:male ratio was approximately 2:1) (Hannon et al. 2015) and found that blood DNA methylation at the cg24637308 locus (DNHD1) was highly correlated with methylation at the same locus in the prefrontal cortex (), the entorhinal cortex (), the superior temporal gyrus () and the cerebellum () (Figure 3).
Figure 3.
Scatterplots and correlations for cg24637308 (DNHD1 gene) methylation levels of blood DNA and four brain regions: prefrontal cortex (PC, ), entorhinal cortex (EC, ), superior temporal gyrus (STG, ), and cerebellum (CE, ), based on an independent data set dominated by females (the female:male ratio was approximately 2:1) from (Hannon et al. 2015).
We also investigated the top loci identified by a previous EWAS analysis on prenatal lead exposure and cord blood DNA methylation (Engström et al. 2015) based on our data set and found that one of the loci (cg05374025) was significantly associated with prenatal lead exposure in our samples (; see Table S13).
Bumphunting analysis identified only one small DMR with two CpGs in chromosome 13 (chr13: 50194554–50194643) associated with RBC lead for all newborns [family-wise error rate ) and two DMRs in chromosome 6 (chr6: 29648271–29649084) and chromosome 5 (chr5: 135415819–135416613) for male infants (; see Table S14). However, these regions did not contain any of the top loci reported in Table 2.
Discussion
In this study of 268 U.S. mother–infant pairs, we found that prenatal RBC lead levels were associated with differential DNA methylation levels at several CpGs in umbilical cord blood. We also observed notable sex differences: RBC lead levels were associated with DNA methylation in more CpGs in female infants than in male infants. Most of the identified CpGs showed lower methylation levels in association with higher prenatal lead exposure. Estimated whole blood lead levels in our sample () were slightly below the U.S. national average for women () (Jones et al. 2010). Our results suggest that even at very low levels, prenatal lead exposure may be significantly associated with altered DNA methylation profiles in newborns.
Lead is a multiple-source pollutant well known for its adverse effects, and the main target organs of lead are the hematopoietic, nervous, and renal systems (Papanikolaou et al. 2005). Interestingly, we found that in utero lead exposure was associated with altered methylation levels at CpGs located within or near important regulatory genes of hematopoietic and nervous functions (i.e., CLEC11A and DNHD1). Lead is well known to interfere with hemoglobin synthesis and to affect erythrocyte morphology and survival (Sun et al. 2012). The gene CLEC11A encodes a secreted sulfated glycoprotein (CLEC11A) that functions as a growth factor for primitive hematopoietic progenitor cells (Mio et al. 1998). CLEC11A stimulates the proliferation and differentiation of hematopoietic precursor cells from various lineages, including RBCs, lymphocytes, granulocytes, and macrophages (Hiraoka et al. 2001).
DNHD1 (DYNC1H1) is a member of the dynein heavy chain family and is highly expressed in the brain. Cytoplasmic dynein complex comprises subunits assembled on the DNHD1 dimer and is particularly important for neurons because it carries essential signals and organelles from distal sites to the cell body (Schiavo et al. 2013). Our results on DNHD1 suggest that methylation changes in this gene may be potential targets of in utero lead exposure. Interestingly, there were strong correlations between blood–brain methylation for cg24637308 (DNHD1) based on another independent set of samples with a high proportion of female participants (Hannon et al. 2015). Furthermore, the significant inverse association among female infants seemed consistent through midchildhood, highlighting the possibility of long-term programming effects of lead exposure. In support of our findings, previous studies have documented a strong link between lead exposure and impaired cognitive and behavioral functions (Boucher et al. 2014; Liu et al. 2014; Virgolini et al. 2008).
Our study found substantial sex differences in lead exposure–associated methylation changes. It is plausible that the slightly higher RBC levels in mothers of female infants than in male infants may be partially responsible for the more significant CpGs found in female infants in the study, although the difference in exposure levels did not reach statistical significance. Similarly, our previous study on prenatal maternal lead and birth outcomes and another epidemiological study on prenatal lead and childhood body weight also did not find appreciable differences in lead exposure levels between the sexes despite the presence of different effect estimates associated with lead exposure in males and females (Afeiche et al. 2011; Perkins et al. 2014). In light of the fact that several toxicological studies also observed appreciable effect differences between the sexes at the same doses of lead exposure (Bunn et al. 2001; Virgolini et al. 2008), it is possible that the variation in the lead-associated effect spectrum over the sexes may be associated with sex-specific biological mechanisms rather than with the lead exposure doses. In addition, it has been shown that sex does influence genomic methylation status (Liu et al. 2010), yet little is known about sex differences in the regulation of the epigenome. Interestingly, a previous animal study found that the effects of lead exposure on DNMTs and DNA-binding proteins in the hippocampus appear to be different in males and females and are further differentiated by the period of development during which exposure occurs (Schneider et al. 2013). The authors hypothesized that an interaction between genes and sex hormones during development, modified by lead exposure, may make specific brain regions such as the hippocampus differentially susceptible to methylation-associated modulation of transcriptional regulation. It is possible that the potential effects of lead exposure on epigenetic mechanisms, as observed in previous studies and in the present study, may contribute to a differential susceptibility between males and females to the adverse health outcomes reported in previous epidemiological and toxicological studies (Afeiche et al. 2011; Bunn et al. 2001; Perkins et al. 2014; Virgolini et al. 2008).
A previous study also investigated the epigenome-wide association of prenatal lead exposure with cord blood DNA methylation profiles in a birth cohort in rural Bangladesh () with high environmental lead levels, and the authors identified a set of loci with , among which some were located around the gene glycoprotein VI (GP6) (Engström et al. 2015). However, only one of these loci (cg05374025) reached statistical significance in our samples. It is notable that prenatal lead exposure levels were much higher in the Bangladesh cohort (mean maternal RBC lead level, ) than in our U.S. samples (mean maternal RBC lead level, ). In addition, several important participant characteristics differed substantially between the two study populations, including ethnic background and BMI (the median maternal BMI was in the Bangladesh cohort and in our cohort). Therefore, the potential adverse effect spectra and the action patterns of prenatal lead exposure may vary substantially between these two cohorts.
Our study has several strengths. We were able to prospectively investigate the methylation changes in association with prenatal lead exposure among male infants and female infants separately. We collected both cord blood samples and midchildhood blood samples, allowing us to examine the consistency of in utero lead exposure–DNA methylation associations over time. In addition, we were able to control for a number of covariates based on the detailed cohort follow-up information.
Our study also has several limitations. First, the distribution of lead exposure was relatively narrow, and the exposure levels were quite low. However, this range gave us the opportunity to investigate the potential effects of very low-level exposure, which represents common exposure levels in the United States and in other Western countries. Our results may not be generalizable to populations with different exposure ranges, or to lower-income and racial/ethnic minority populations. Second, our study measured maternal lead in RBC instead of in whole blood. Nevertheless, this is likely to be an accurate biomarker because 99% of whole-blood lead is found in RBC; thus, RBC lead is very highly correlated with whole-blood concentrations (Leggett 1993). Third, although lead is known to affect multiple organs, DNA methylation was measured using unfractionated blood collected at delivery and midchildhood—the only available biological sample. Whether the associations reported herein may reflect associations between in utero lead exposure and methylation at target organs is uncertain. However, peripheral blood is the major carrier of lead absorbed into the human body, and peripheral blood cells contact and react directly with the internal forms of lead (Chiesa et al. 2006). Furthermore, there are strong correlations between blood and brain methylation for the CpG cg24637308 on the body of the DNHD1 gene, suggesting that peripheral blood may serve well as a surrogate tissue for the target organ brain for certain key regulatory loci. Fourth, we were unable to investigate the exact biological mechanisms behind the observed methylation changes associated with prenatal lead exposure. Although lead exposure has been found to reduce the global DNA methylation level by noncompetitive inhibition and alteration of DNMT (Sanchez et al. 2017), this would not explain why not all affected genes are in the negative direction. Further studies are needed to investigate this issue in detail. Fifth, we were unable to validate the methylation changes associated with lead exposure using another method, such as gene expression assays or pyrosequencing assays, owing to the unavailability of extra biological samples. Nevertheless, the visualization of regional DNA comethylation patterns for potential target genes reveals generally consistent change patterns for CpGs in these genes, strengthening the validity of the reported results. Finally, we aimed to examine the potential effects of in utero lead exposure and did not include lead exposures after birth and before midchildhood; whether the associations between prenatal lead exposure and methylation in midchildhood may be confounded by other exposures is unknown. However, early pregnancy is a critical time window for epigenetic programming in the fetus, and it is during this time period that adverse in utero exposures are of most interest in relation to DNA methylation programming.
Conclusion
Based on an EWAS of relatively low levels of prenatal lead exposure and DNA methylation in a prospective prebirth cohort, we identified several CpGs of biological relevance for effects of in utero exposure to lead. There were sex-specific differences in the top CpGs associated with prenatal lead exposure, suggesting a potential susceptibility difference in relation to lead exposure between males and females. Further studies are needed to replicate our findings and to investigate their relevance to specific adverse health outcomes. Pending further investigation, our results suggest that prenatal lead exposure, even at very low levels, may modify the DNA methylation profiles in offspring, providing a mechanistic explanation for previously reported adverse postpartum health outcomes of prenatal lead exposure.
Supplemental Material
Supplemental Material
Acknowledgments
We thank the Project Viva participants and staff. We also thank E. Hannon and J. Mill for providing assistance in the analysis of blood–brain DNA methylation correlations.
This work was supported by Nationals Institutes of Health (NIH) grants R37 HD 034568, R01 ES016314, RC1 HD063590, K24 HD069408, R01 NR013945, R01 HL111108 and by the Harvard T.H. Chan School of Public Health/National Institute of Environmental Health Sciences (HSPH-NIEHS) Center for Environmental Health New Investigator Fund (P30ES000002). S.W. is supported by startup funding from the “Young Thousand Talents” Program of China and Peking University Health Science Center (no. BMU20160549).
References
- Afeiche M, Peterson KE, Sánchez BN, Cantonwine D, Lamadrid-Figueroa H, Schnaas L, et al. 2011. Prenatal lead exposure and weight of 0- to 5-year-old children in Mexico City. Environ Health Perspect 119(10):1436–1441, PMID: 21715242, 10.1289/ehp.1003184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, et al. 2014. Minfi: A flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30(10):1363–1369, PMID: 24478339, 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakulski KM, Feinberg JI, Andrews SV, Yang J, Brown S, McKenney SL, et al. 2016. DNA methylation of cord blood cell types: Applications for mixed cell birth studies. Epigenetics 11(5):354–362, PMID: 27019159, 10.1080/15592294.2016.1161875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher O, Muckle G, Jacobson JL, Carter RC, Kaplan-Estrin M, Ayotte P, et al. 2014. Domain-specific effects of prenatal exposure to PCBs, mercury, and lead on infant cognition: Results from the environmental contaminants and child development study in nunavik. Environ Health Perspect 122(3):310–316, PMID: 24441767, 10.1289/ehp.1206323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunn TL, Parsons PJ, Kao E, Dietert RR. 2001. Exposure to lead during critical windows of embryonic development: Differential immunotoxic outcome based on stage of exposure and gender. Toxicol Sci 64(1):57–66, PMID: 11606801, 10.1093/toxsci/64.1.57. [DOI] [PubMed] [Google Scholar]
- Chen YA, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, et al. 2013. Discovery of cross-reactive probes and polymorphic cpgs in the illumina infinium humanmethylation450 microarray. Epigenetics 8(2):203–209, PMID: 23314698, 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiesa ME, Rosenberg CE, Fink NE, Salibián A. 2006. Serum protein profile and blood cell counts in adult toads Bufo arenarum (Amphibia: Anura: Bufonidae): Effects of sublethal lead acetate. Arch Environ Contam Toxicol 50(3):384–391, PMID: 16446996, 10.1007/s00244-004-0252-4. [DOI] [PubMed] [Google Scholar]
- Chuang HY, Schwartz J, Gonzales-Cossio T, Lugo MC, Palazuelos E, Aro A, et al. 2001. Interrelations of lead levels in bone, venous blood, and umbilical cord blood with exogenous lead exposure through maternal plasma lead in peripartum women. Environ Health Perspect 109(5):527–532, PMID: 11401766, 10.1289/ehp.01109527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, et al. 2010. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics 11:587, PMID: 21118553, 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström K, Rydbeck F, Kippler M, Wojdacz TK, Arifeen S, Vahter M, et al. 2015. Prenatal lead exposure is associated with decreased cord blood DNA methylation of the glycoprotein VI gene involved in platelet activation and thrombus formation. Environ Epigenet 1(1):dvv007, 10.1093/eep/dvv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulk C, Barks A, Liu K, Goodrich JM, Dolinoy DC. 2013. Early-life lead exposure results in dose- and sex-specific effects on weight and epigenetic gene regulation in weanling mice. Epigenomics 5(5):487–500, PMID: 24059796, 10.2217/epi.13.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillman MW, Rich-Edwards JW, Rifas-Shiman SL, Lieberman ES, Kleinman KP, Lipshultz SE. 2004. Maternal age and other predictors of newborn blood pressure. J Pediatr 144(2):240–245, PMID: 14760269, 10.1016/j.jpeds.2003.10.064. [DOI] [PubMed] [Google Scholar]
- Hannon E, Lunnon K, Schalkwyk L, Mill J. 2015. Interindividual methylomic variation across blood, cortex, and cerebellum: Implications for epigenetic studies of neurological and neuropsychiatric phenotypes. Epigenetics 10(11):1024–1032, PMID: 26457534, 10.1080/15592294.2015.1100786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka A, Yano Ki K, Kagami N, Takeshige K, Mio H, Anazawa H, et al. 2001. Stem cell growth factor: In situ hybridization analysis on the gene expression, molecular characterization and in vitro proliferative activity of a recombinant preparation on primitive hematopoietic progenitor cells. Hematol J 2(5):307–315, PMID: 11920266, 10.1038/sj/thj/6200118. [DOI] [PubMed] [Google Scholar]
- Jacob B, Ritz B, Heinrich J, Hoelscher B, Wichmann H-E. 2000. The effect of low-level blood lead on hematologic parameters in children. Environ Res 82(2):150–159, PMID: 10662529, 10.1006/enrs.1999.4011. [DOI] [PubMed] [Google Scholar]
- Jaffe AE, Murakami P, Lee H, Leek JT, Fallin MD, Feinberg AP, et al. 2012. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int J Epidemiol 41(1):200–209, PMID: 22422453, 10.1093/ije/dyr238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelliffe-Pawlowski LL, Miles SQ, Courtney JG, Materna B, Charlton V. 2006. Effect of magnitude and timing of maternal pregnancy blood lead (Pb) levels on birth outcomes. J Perinatol 26(3):154–162, PMID: 16453008, 10.1038/sj.jp.7211453. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. 2007. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8(1):118–127, PMID: 16632515, 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Jones L, Parker JD, Mendola P. 2010. Blood lead and mercury levels in pregnant women in the United States, 2003–2008. NCHS Data Brief 52:1–8, PMID: 21211167. [PubMed] [Google Scholar]
- Joubert BR, Felix JF, Yousefi P, Bakulski KM, Just AC, Breton C, et al. 2016. DNA methylation in newborns and maternal smoking in pregnancy: Genome-wide consortium meta-analysis. Am J Hum Genet 98(4):680–696, PMID: 27040690, 10.1016/j.ajhg.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggett RW. 1993. An age-specific kinetic model of lead metabolism in humans. Environ Health Perspect 101(7):598, PMID: 8143593, 10.1289/ehp.93101598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Chen Y, Gao D, Jing J, Hu Q. 2014. Prenatal and postnatal lead exposure and cognitive development of infants followed over the first three years of life: a prospective birth study in the Pearl River Delta region, China. Neurotoxicology 44:326–334, PMID: 25124739, 10.1016/j.neuro.2014.07.001. [DOI] [PubMed] [Google Scholar]
- Liu J, Morgan M, Hutchison K, Calhoun VD. 2010. A study of the influence of sex on genome wide methylation. PloS One 5(4):e10028, PMID: 20386599, 10.1371/journal.pone.0010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mio H, Kagami N, Yokokawa S, Kawai H, Nakagawa S, Takeuchi K, et al. 1998. Isolation and characterization of a cDNA for human, mouse, and rat full-length stem cell growth factor, a new member of C-type lectin superfamily. Biochem Biophys Res Commun 249(1):124–130, PMID: 9705843, 10.1006/bbrc.1998.9073. [DOI] [PubMed] [Google Scholar]
- Papanikolaou NC, Hatzidaki EG, Belivanis S, Tzanakakis GN, Tsatsakis AM. 2005. Lead toxicity update. A brief review. Med Sci Monit 11(10):RA329–RA336, PMID: 16192916. [PubMed] [Google Scholar]
- Perkins M, Wright RO, Amarasiriwardena CJ, Jayawardene I, Rifas-Shiman SL, Oken E. 2014. Very low maternal lead level in pregnancy and birth outcomes in an eastern Massachusetts population. Ann Epidemiol 24(12):915–919, PMID: 25444892, 10.1016/j.annepidem.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese SE, Zhao S, Wu MC, Joubert BR, Parr CL, Håberg SE, et al. 2017. DNA methylation score as a biomarker in newborns for sustained maternal smoking during pregnancy. Environ Health Perspect 125(4):760–766, PMID: 27323799, 10.1289/EHP333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlén SE, Greco D, et al. 2012. Differential DNA methylation in purified human blood cells: Implications for cell lineage and studies on disease susceptibility. PloS One 7(7):e41361, PMID: 22848472, 10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez OF, Lee J, Yu King Hing N, Kim SE, Freeman JL, Yuan C. 2017. Lead (Pb) exposure reduces global DNA methylation level by non-competitive inhibition and alteration of dnmt expression. Metallomics 9(2):149–160, PMID: 27934997, 10.1039/C6MT00198J. [DOI] [PubMed] [Google Scholar]
- Schiavo G, Greensmith L, Hafezparast M, Fisher EM. 2013. Cytoplasmic dynein heavy chain: The servant of many masters. Trends Neurosci 36(11):641–651, PMID: 24035135, 10.1016/j.tins.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JS, Kidd SK, Anderson DW. 2013. Influence of developmental lead exposure on expression of DNA methyltransferases and methyl cytosine-binding proteins in hippocampus. Toxicol Lett 217(1):75–81, PMID: 23246732, 10.1016/j.toxlet.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JS, Mettil W, Anderson DW. 2012. Differential effect of postnatal lead exposure on gene expression in the hippocampus and frontal cortex. J Mol Neurosci 47(1):76–88, PMID: 22160880, 10.1007/s12031-011-9686-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senut MC, Sen A, Cingolani P, Shaik A, Land SJ, Ruden DM. 2014. Lead exposure disrupts global DNA methylation in human embryonic stem cells and alters their neuronal differentiation. Toxicol Sci 139(1):142–161, PMID: 24519525, 10.1093/toxsci/kfu028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrani RE, Gioia IA, Corchs JL. 1997. Lead effects on structural and functional cellular parameters in human red cells from a prenatal hematopoiesis stage. Biometals 10(4):331–335, PMID: 9353882. [DOI] [PubMed] [Google Scholar]
- Sun X, Xie Y, Wu L, Zhu W, Hu J, Lu R, et al. 2012. Lead acetate reduces the ability of human umbilical cord mesenchymal stem cells to support hematopoiesis in vitro. Mol Med Rep 6(4):827–832, PMID: 22858795, 10.3892/mmr.2012.1014. [DOI] [PubMed] [Google Scholar]
- Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, et al. 2013. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 29(2):189–196, PMID: 23175756, 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triche TJ, Weisenberger DJ, Van Den Berg D, Laird PW, Siegmund KD. 2013. Low-level processing of Illumina Infinium DNA Methylation BeadArrays. Nucleic Acids Res 41(7):e90–e90, PMID: 23476028, 10.1093/nar/gkt090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgolini MB, Rossi-George A, Weston D, Cory-Slechta DA. 2008. Influence of low level maternal Pb exposure and prenatal stress on offspring stress challenge responsivity. Neurotoxicology 29(6):928–939, PMID: 18951918, 10.1016/j.neuro.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (World Health Organization). 2015. Lead poisoning and health. http://www.who.int/mediacentre/factsheets/fs379/en/ [accessed 16 March 2016].
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.