Abstract
Background:
Mercury is a global pollutant, and prenatal exposure is associated with adverse health effects. To date, no studies have evaluated the association between prenatal mercury exposure and DNA hydroxymethylation, an epigenetic modification important for tissue differentiation and embryonic development.
Objectives:
We sought to evaluate the association between prenatal mercury exposure and offspring global DNA methylation and hydroxymethylation at birth and test for persistence of the association in childhood.
Methods:
Within Project Viva, a U.S. prebirth cohort, we examined associations of maternal second trimester red blood cell mercury (RBC-Hg) concentrations with global 5-hydroxymethylcytosine () and 5-methylcytosine () DNA content in blood collected at birth (), early childhood (; 2.9 to 4.9 y), and midchildhood (; 6.7 to 10.5 y).
Results:
Median prenatal RBC-Hg concentration was []. At birth, median cord blood , , and their ratio were 4.95%, 0.22%, and 24.37, respectively. The mean adjusted difference [95% confidence interval (CI)] of blood for a doubling in prenatal RBC-Hg concentration was (, 0.002), (, ), and 0.005% (, 0.018) at birth, early, and midchildhood, respectively. The corresponding relative adjusted change in the genomic ratio of to for a doubling in prenatal RBC-Hg concentration was 4.70% (0.04, 9.58), 22.42% (7.73, 39.11), and 0.73% (, 5.88) at birth, early, and midchildhood, respectively. No associations were present between prenatal maternal RBC-Hg and at any time point.
Conclusions:
Prenatal mercury exposure was associated with lower genomic content and a corresponding increase in the ratio of to in cord blood. This association was persistent in early but not midchildhood blood. Our results demonstrate the potential malleability of epigenetic modifications associated with mercury exposure in utero. https://doi.org/10.1289/EHP1467
Introduction
Mercury is a global contaminant that bioaccumulates in the environment. Coal-burning emissions and industrial waste are among the major anthropogenic sources of mercury, and ocean mercury levels have tripled since the industrial revolution (Lamborg et al. 2014). Nonoccupational human exposures predominately occur through the consumption of fish and other seafood contaminated with methylmercury that has been biomagnified up the food chain (Driscoll et al. 2013). Methylmercury readily crosses the placenta and blood–brain barrier, exposing the fetus during critical windows of development (Stern and Smith 2003). Prenatal exposure to mercury, typical of regular fish consumption with elevated mercury content, has been associated with lower cognitive test scores and may also hinder infant growth in the first few years of life (Karagas et al. 2012). The specific mechanism of mercury toxicity remains poorly characterized, but several mechanisms, including oxidative stress, disruption of calcium homeostasis, and alterations of neurotransmitters, among others, are hypothesized to be involved (Castoldi et al. 2003). Recently, epidemiological studies have shown that prenatal mercury exposure is associated with DNA methylation at specific genomic regions (Bakulski et al. 2015; Cardenas et al. 2015) that could serve as a mechanism linking exposure and infant neurobehavioral outcomes (Maccani et al. 2015).
Epigenetic modifications during fetal development play a critical role during embryogenesis regulating cell lineage commitment, chromosome silencing, retrotransposon repression, and genetic imprinting (Cheng et al. 2015). One of the most widely studied epigenetic modifications is DNA methylation at the 5’ position of cytosine (C) nucleotides, also referred to as 5-methylcytosine (5mC) (Relton et al. 2015). More recently, it has been demonstrated that 5mC can undergo oxidation by the ten-eleven translocation (TET) family of enzymes to form 5-hydroxymethylcytosine (5hmC) (Dao et al. 2014). Regulation of gene expression by 5hmC has been shown to be independent of 5mC and plays a crucial role during embryogenesis, particularly in neurogenesis (Etchegaray et al. 2015; Hahn et al. 2013). Similar to 5mC, genomic 5hmC is cell type–specific, and it is most abundant in neurons, embryonic stem cells, and pluripotent cells (Ruzov et al. 2011). While there is emerging literature on mercury exposure with DNA methylation and its potential role in neurodevelopment and fetal programming, we are not aware of any studies that have examined associations with 5hmC genomic content. Epigenetic measurements associated with prenatal mercury exposure might help elucidate mechanisms associated with toxicity, particularly for exposures occurring in the developing fetus during critical periods of epigenomic remodeling (Waterland and Michels 2007). Alternatively, unique epigenetic signatures could serve as biomarkers of exposure and be used as biosensors even years after the exposure window (Ladd-Acosta 2015). Identifying if epigenomic changes at birth and sustained throughout childhood are associated with prenatal exposure to mercury will help elucidate their potential role as biomarkers of exposure or disease susceptibility.
In the present analysis, we examined the association of prenatal maternal second trimester mercury exposure with global DNA 5hmC and 5mC content in cord blood. We tested for persistence of the association among blood DNA samples collected from children during early and midchildhood. We hypothesized that prenatal maternal mercury concentration would be associated with both global DNA methylation and hydroxymethylation, and the observed association would persist into early and midchildhood, reflecting persistent epigenetic reprogramming events occurring in utero.
Methods
Study Population
Mother–child pairs were participants in Project Viva, a prospective prebirth cohort study (Oken et al. 2014). This cohort was recruited between 1999 and 2002 during the mothers’ first prenatal visits at Atrius Harvard Vanguard Medical Associates, a multispecialty medical group practice in Massachusetts, United States. Eligibility criteria included fluency in English, gestational age less than 22 wk at the first prenatal visit, and singleton pregnancy. Of the total 2,128 live births, cord blood was collected from 1,018 born at one of the two study hospitals. We measured global genomic 5hmC and 5mC content in 481 cord blood samples for mothers who provided consent for genetic analysis. Of these, 306 samples had complete covariate information and maternal prenatal blood mercury concentrations. We evaluated persistence of cord blood associations in 68 children with available blood epigenetic measurements in early childhood (age: 2.9 to 4.9 y) and 260 children with blood epigenetic measurements from midchildhood (age: 6.7 to 10.5 y) with complete exposure and covariate information. Not all participants were measured at all three time points: of the 306 infants eligible for analyses in cord blood, 63 provided early childhood blood, and 144 provided samples for midchildhood analyses after restricting on complete covariate information. We did not restrict our analyses to participants with repeated epigenomic measurements at all three time points in order to maximize the sample size at each time point, but instead performed a sensitivity analyses among repeated measurements (sample flow diagram is depicted in Figure S1). Mothers provided written informed consent at recruitment and at postpartum follow-up visits. The Institutional Review Board of Harvard Pilgrim Health Care reviewed and approved all study protocols.
Maternal Prenatal Red Blood Cell Mercury
Sample collection and exposure assessment for prenatal mercury exposure in this cohort has been previously described (Oken et al. 2016). Briefly, at the second trimester visit, we obtained a maternal blood sample that was centrifuged to separate plasma from blood erythrocytes. A separate aliquot of erythrocytes was provided for mercury analysis. Aliquots were stored at and analyzed for total mercury using the Direct Mercury Analyzer 80 (Milestone Inc.). Results are reported as mercury concentration in the original red blood cell sample in ng/g. The detection limit was of sample, and the percentage recovery for standards ranged from 90% to 110%. A total of 32 samples were below the limit of detection (LOD), and we used the instrument estimated red blood cell mercury (RBC-Hg) concentration below the LOD for analyses. During pregnancy, participants also completed a semiquantitative food frequency questionnaire (FFQ) that we previously calibrated against erythrocyte levels of elongated fatty acids (Fawzi et al. 2004). Participants self-reported their consumption of fish with six frequency response options ranging from “never/less than 1 per month” to “1 or more servings per day.” We combined responses to estimate average total fish intake as average servings per week for the first and the second trimesters.
DNA Methylation and Hydroxymethylation
We isolated of DNA from buffy coat and enzymatically hydrolyzed it to individual deoxyribonucleosides by a simple, one-step DNA hydrolysis procedure consisting of a digest mix prepared by adding phosphodiesterase I, alkaline phosphatase, and benzonase Nuclease to Tris-HCl buffer (Sigma-Aldrich, Diegem, Belgium). We spiked extracted DNA with an internal standard mixture, dried it, and then hydrolyzed at 37°C for at least 8 h with digest mixture. After hydrolysis, we added ACN (acetonitrile): (90:8, v/v) to each sample. We prepared stock solutions of 5mC, 5hmC, and cytosine (C) by dissolving commercial standards (Sigma-Aldrich) in 22 high-performance liquid chromatography–grade to prepare the calibration standards. Finally, we measured global genomic DNA methylation and hydroxymethylation of cytosine nucleotides simultaneously for each sample using ultrapressure liquid-chromatography combined with tandem mass spectrometry, as previously described (Godderis et al. 2015). The lower limits of detection were , , and for 5mC, 5hmC, and C, respectively. The accuracy for the concentrations tested ranged from 87% to 114.8% of the target, and the precision expressed as relative standard errors ranged from 0.8% to 12.1%. Global DNA methylation is reported as a percentage of 5mC over the sum of 5mC, 5hmC, and C [], while global DNA hydroxymethylation is expressed as a percentage of 5hmC versus the sum of 5mC, 5hmC, and C []. A ratio of to is also reported (). All samples analyzed were above the LOD. However, a total of 24 samples, 11 in cord blood, 4 in early, and 9 in midchildhood were below the limit of quantification and therefore excluded from analyses. For further details on the methodology, see the supplementary materials on the methods that include analytical conditions, calibration curves, and validation parameters for our method (Tables S3–S4 and Figure S7).
Covariates
At study enrollment, we obtained information on maternal race/ethnicity, education, prepregnancy body mass index, smoking during pregnancy, marital status, parity, age, and household income. Infant sex, birth weight, and type of delivery were abstracted from medical records. We determined birth weight for gestational age and sex z-score from a U.S. national reference (Oken et al. 2003). Infant race/ethnicity was collected by interview. We obtained data on micronutrient intake using self-administered semiquantitative FFQ during the first and second trimesters that were previously validated and modified to be used in pregnancy (Fawzi et al. 2004). We evaluated the effect of adjusting for intake of four methyl donors (vitamin B-12, folate, betaine, and choline) estimated from FFQs in the first or second trimester of pregnancy, given that one carbon metabolism plays a central role in DNA methylation and prenatal maternal methyl donor intake might play a role in fetal programming of the epigenome (Pauwels et al. 2016).
Statistical Analyses
We estimated medians and interquartile ranges (IQRs) for , , and their ratio in cord blood across demographic and prenatal nutritional information for the study population among all mother–child pairs available with epigenomic measurements at birth without restricting on prenatal mercury exposure (). We tested unadjusted associations for , , and their ratio with participant characteristics with two levels using a nonparametric Wilcoxon-rank sum test, and use the Kruskal-Wallis test for characteristics with more than two levels. We used Spearman correlation coefficients to estimate the association of and in cord blood with levels in blood during early or midchildhood. We also evaluated changes in , , and their ratio among subjects with repeated measurements from cord blood and in early () or cord blood and midchildhood () using Wilcoxon signed-rank test for paired samples.
Both RBC-Hg concentrations and the ratio of and were right skewed. Therefore, we them to approximate a normal distribution. We used separate linear regression models to estimate associations between prenatal maternal RBC-Hg and , , and the ratio of to in cord blood as well as in blood collected during early and midchildhood. The ratio of to was modeled separately as a potential target for DNA demethylation.
We estimated unadjusted as well as adjusted associations using two multivariate linear regression models: Model 1, a linear regression model adjusted for covariates selected a priori that were nominally associated with the outcome in univariate analyses () or that change the effect estimate for mercury by at least 10% regardless of statistical significance. Model 1 was adjusted for maternal education, age at enrollment, marital status, first trimester vitamin B-12 intake, and second trimester fish consumption, as well as child race/ethnicity, sex, gestational age, and birth weight for gestational age z-score. The model was further adjusted for first trimester folate intake, while the model was adjusted by first trimester betaine intake. The model for the ratio of to was adjusted for both first trimester folate and betaine intake. Model 2 was adjusted for the same covariates as Model 1, but also included estimated cell type proportions. Cell type composition was estimated from genome-wide DNA methylation arrays (HumanMethylation450 BeadChip; Illumina) available in a subset of samples and estimated using the minfi package of R (version 3.3.0, R Core Team) (Aryee et al. 2014). We used an adult reference DNA methylation panel to estimate leukocyte composition in early and midchildhood and a cord blood DNA methylation reference panel, which includes nucleated red blood cells (nRBCs), to estimate nucleated cell types at birth (Bakulski et al. 2016). Associations between cell type composition in cord blood and both epigenomic measures are summarized in Table S1. We report the relative percent change in the ratio of 5mC to 5hmC per doubling in exposure, as regression coefficients were estimated from linear models where RBC-Hg and the ratio of and were both (log–log adjusted models) to meet model assumptions. Unadjusted and fully adjusted models were restricted to complete covariate information used in fully adjusted models in order to compare models.
We evaluated persistence by testing associations between prenatal maternal RBC-Hg concentration and epigenetic measurements among all participants who provided blood samples during early or midchildhood. Estimates from unadjusted and adjusted models are reported. Early and midchildhood adjusted linear regression models were similar to Model 2 for cord blood with the addition of child age in days at blood collection and replacing cell type composition with the estimated leukocyte distribution obtained from genome-wide DNA methylation arrays measured during early or midchildhood. We evaluated effect modification by testing the interaction between sex and RBC-Hg for all epigenomic measures across time points. Model fit and assumptions were examined using scatterplots of the standardized residuals vs. the fitted values, residuals vs. the leverage, and quantile-quantile plots of the standardized residuals. All analyses were performed using R 3.3.0 (www.r-project.org).
Results
Sociodemographic and Unadjusted Associations
A total of 481 participants with measurements of cord blood DNA and were available for bivariate analyses. The median content for the entire sample was 0.22% () (min–max: 0.024-0.87; skewness: 1.04) and the median was 4.95% () (min–max: 2.24–10.81; skewness: 0.94). The median ratio of to in cord blood was 24.37 (). In unadjusted analyses, median and in cord blood varied by participant characteristics (Table 1).
Table 1.
Sample characteristics of 481 mother–child pairs in the Project Viva cohort and distribution of cord blood DNA 5-hydroxymethylcytosine (5hmC) and 5-methylcytosine (5mC) by these characteristics.
| Characteristics | % | [median (IQR)] | [median (IQR)] | 5mC to 5hmC ratio [median (IQR)] | |||
|---|---|---|---|---|---|---|---|
| p-Value | p-Value | p-Value | |||||
| Overall | 0.22% (0.19) | — | 4.95% (2.52) | — | 24.37 (14.89) | — | |
| Maternal race/ethnicity | |||||||
| White | 71.9 | 0.22 (0.19) | 0.62 | 4.95 (2.49) | 24.8 (15.4) | 0.91 | |
| Black | 11.9 | 0.23 (0.15) | 4.94 (2.06) | 0.09 | 23.5 (15.5) | ||
| Hispanic | 6.9 | 0.24 (0.19) | 6.31 (3.02) | 23.5 (13.2) | |||
| Other | 9.4 | 0.21 (0.15) | 4.58 (1.99) | 22.7 (10.2) | |||
| Prepregnancy BMI () | |||||||
| Underweight () | 3.5 | 0.19 (0.19) | 0.91 | 5.37 (3.16) | 0.63 | 28.5 (18.3) | 0.89 |
| Normal () | 60.2 | 0.21 (0.19) | 4.88 (2.64) | 24.2 (14.8) | |||
| Overweight () | 22.1 | 0.23 (0.20) | 5.15 (2.56) | 24.6 (14.4) | |||
| Obese () | 14.2 | 0.23 (0.14) | 4.88 (2.11) | 23.1 (14.2) | |||
| Missing | — | — | — | ||||
| College graduate | |||||||
| No | 33.5 | 0.24 (0.22) | 0.03 | 5.14 (2.59) | 0.03 | 23.3 (14.1) | 0.13 |
| Yes | 66.5 | 0.20 (0.18) | 4.82 (2.41) | 24.7 (15.1) | |||
| Self-reported Smoking | |||||||
| Never | 68.6 | 0.23 (0.18) | 0.68 | 5.00 (2.49) | 0.30 | 23.7 (15.0) | 0.89 |
| Former | 20.8 | 0.20 (0.21) | 4.70 (2.78) | 25.4 (14.2) | |||
| During pregnancy | 10.6 | 0.20 (0.17) | 4.95 (1.80) | 23.9 (14.7) | |||
| Nulliparous | |||||||
| No | 53.8 | 0.23 (0.19) | 0.71 | 4.82 (2.40) | 0.87 | 24.1 (14.8) | 0.72 |
| Yes | 46.2 | 0.21 (0.18) | 5.07 (2.55) | 24.5 (14.6) | |||
| Any alcohol intake (first trimester) | |||||||
| No | 25.2 | 0.24 (0.19) | 0.38 | 5.11 (2.32) | 0.84 | 24.4 (13.0) | 0.45 |
| Yes | 74.8 | 0.22 (0.19) | 4.95 (2.54) | 24.5 (15.5) | |||
| Missing | — | — | — | ||||
| Maternal age at enrollment (y) | |||||||
| 15.7–29.6 | 25.2 | 0.23 (0.20) | 0.75 | 5.31 (2.49) | 0.56 | 23.8 (13.9) | 0.65 |
| 24.5 | 0.20 (0.19) | 4.87 (2.46) | 24.6 (14.5) | ||||
| 25.2 | 0.23 (0.18) | 4.71 (2.37) | 23.7 (12.2) | ||||
| 25.2 | 0.22 (0.19) | 4.94 (2.68) | 24.4 (17.8) | ||||
| Married or cohabitating | |||||||
| No | 9.1 | 0.27 (0.26) | 0.03 | 5.84 (3.61) | 0.08 | 22.6 (15.3) | 0.24 |
| Yes | 90.9 | 0.22 (0.18) | 4.88 (2.43) | 24.5 (15.0) | |||
| Household income | |||||||
| No | 40 | 0.23 (0.22) | 0.83 | 4.90 (2.54) | 0.65 | 24.8 (17.1) | 0.95 |
| Yes | 60 | 0.22 (0.17) | 5.04 (2.54) | 24.2 (13.5) | |||
| Missing | — | — | — | ||||
| First trimester fish (serving/wk) | |||||||
| 0 | 10.9 | 0.25 (0.16) | 0.10 | 5.08 (2.13) | 0.71 | 22.4 (12.6) | 0.03 |
| 75.7 | 0.23 (0.20) | 4.97 (2.50) | 24.0 (15.4) | ||||
| 13.4 | 0.19 (0.12) | 5.98 (2.40) | 28.0 (12.5) | ||||
| Missing | — | — | — | ||||
| Second trimester fish (serving/wk) | |||||||
| 0 | 13.2 | 0.24 (0.21) | 0.06 | 5.03 (2.42) | 0.94 | 22.6 (13.6) | 0.01 |
| 75.6 | 0.23 (0.20) | 4.95 (2.46) | 24.2 (14.2) | ||||
| 11.2 | 0.17 (0.14) | 4.88 (3.66) | 30.7 (19.2) | ||||
| Missing | — | — | — | ||||
| 2nd trimester RBC-Hg (ng/g) | |||||||
| 0.15–1.65 | 25.2 | 0.20 (0.18) | 0.030 | 4.71 (2.37) | 0.88 | 23.7 (15.3) | 0.008 |
| 24.2 | 0.25 (0.23) | 5.27 (2.77) | 23.1 (14.3) | ||||
| 24.2 | 0.20 (0.22) | 4.72 (2.84) | 23.9 (14.4) | ||||
| 25.2 | 0.18 (0.15) | 5.00 (2.43) | 27.8 (16.2) | ||||
| Missing | — | — | — | ||||
| Child sex | |||||||
| Male | 53.4 | 0.22 (0.18) | 0.61 | 4.83 (2.46) | 0.42 | 24.5 (15.3) | 0.74 |
| Female | 46.6 | 0.22 (0.19) | 5.05 (2.49) | 23.7 (14.3) | |||
| Child race/ethnicity | |||||||
| White | 67.8 | 0.22 (0.19) | 0.84 | 4.94 (2.46) | 0.03 | 24.6 (15.5) | 0.59 |
| Black | 12.9 | 0.23 (0.16) | 4.83 (2.06) | 23.1 (14.7) | |||
| Hispanic | 4.8 | 0.24 (0.19) | 6.84 (3.91) | 26.0 (18.6) | |||
| Other | 14.6 | 0.22 (0.17) | 4.71 (2.50) | 24.2 (11.2) | |||
| C-section birth | |||||||
| No | 83.4 | 0.22 (0.18) | 0.41 | 4.95 (2.41) | 0.49 | 24.1 (13.7) | 0.31 |
| Yes | 16.6 | 0.20 (0.20) | 4.72 (2.84) | 26.8 (15.3) | |||
| Gestation length | |||||||
| No | 95.6 | 0.22 (0.19) | 0.69 | 4.93 (2.43) | 0.37 | 24.3 (15.2) | 0.93 |
| Yes | 4.4 | 0.23 (0.21) | 6.38 (3.39) | 25.1 (7.5) | |||
| BW/GA category | |||||||
| Small for gestational age (SGA) | 4.4 | 0.18 (0.06) | 0.10 | 4.62 (2.07) | 0.53 | 29.5 (11.8) | 0.05 |
| Appropriate for gestational age 10–90 | 80.9 | 0.23 (0.19) | 5.00 (2.56) | 23.7 (13.9) | |||
| Large for gestational age (LGA) | 14.8 | 0.20 (0.16) | 4.85 (2.57) | 26.0 (16.0) | |||
Note: BMI, body mass index; BW, body weight; IQR, interquartile range. p-Value calculated from a Kruskal-Wallis test for groups with more than two levels and a Wilcoxon-rank sum test for groups with only two levels.
In unadjusted analyses, prenatal RBC-Hg concentration measured during the second trimester of pregnancy was associated with global content measured in cord blood (Table 1). Namely, the lowest cord blood 5hmC content was observed among participants in the highest prenatal exposure quartile (0.18%) compared to those in the third (0.20%), second (0.25%), or first (0.20%) quartiles of exposure. The ratio of 5mC to 5hmC was higher in the highest prenatal mercury exposure quartile (27.9) compared to the third (23.9), second (23.1), or first (23.7) quartiles of exposure.
Correlations among Epigenomic Measurements and Longitudinal Changes
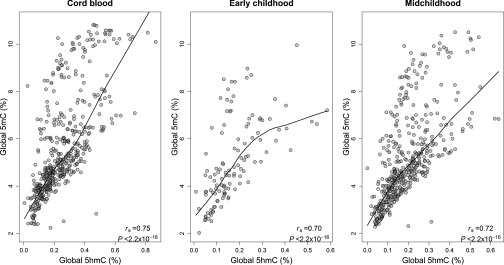
Among all participants with epigenomic measurements, global and were strongly correlated in cord blood (; ), as well as in blood collected during early (; ) and in midchildhood (; ), illustrated in Figure 1.
Figure 1.
*Scatter plots, Spearman correlations coefficients and locally weighted scatterplot smoothing lines for 5-hydroxymethylcytosine (5hmC) and 5-methylcytosine (5mC) at three sample collection periods: at birth (cord blood, ), early childhood (), and midchildhood blood ().
Note: All participants with blood epigenomic measurements and not restricted to prenatal mercury exposure.
Among participants with repeated epigenomic measurements (), median cord blood was higher (0.22%) compared to median measured in blood during early childhood (0.15%; ). Similarly, median was higher in cord blood (5.06%) compared to the median measured during early childhood (4.63%; ). Correspondingly, among participants with repeated epigenomic measurements in cord blood and midchildhood (), median cord blood was higher (0.24%) compared to median levels of measured in blood during midchildhood (0.16%; ), as well as levels of in cord blood (5.52%) compared to measurements in midchildhood blood (5.31%; ).
However, median levels of measured during early (0.14%) and midchildhood (0.13%) (; ) and the corresponding levels of genomic content (4.57% and 4.42%, ) were similar. A similar trend of elevated and in cord blood compared to early or midchildhood blood was observed among all participants (Figure S2).
Association of 5-Hydroxymethylcytosine and 5-Methylcytosine with Prenatal Red Blood Cell Mercury Concentrations
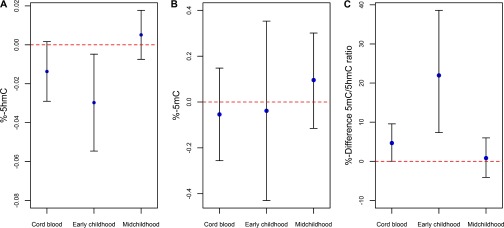
In unadjusted linear regression analyses, a doubling in prenatal mercury exposure was associated with lower global [; 95% confidence interval (CI): , 0.000; ] and an increase in the ratio of 5mC to 5hmC (; 95% CI: 1.60, 9.95; ) in cord blood. A doubling in RBC-Hg concentration was associated with a 0.015% mean decrease in global 5hmC content (95% CI: , 0.0003; ) after adjusting for maternal education, age at enrollment, marital status, first trimester vitamin B-12, first trimester betaine intake, second trimester fish consumption, and child race/ethnicity, sex, gestational age, and birth weight for gestational age z-scores (Table 2). After further adjustment for the estimated cell type composition, this association was slightly attenuated (; 95% CI: , 0.002; ), Table 2. Correspondingly, a doubling in prenatal mercury exposure was associated with a 4.80% increase in the ratio of to (95% CI: 0.18, 9.69; ) after adjusting for potential confounders. This association remained consistent after further adjusting for the estimated cell type composition of cord blood (; 95% CI: 0.04, 9.58; ). In this sample, no associations were observed with in cord blood and prenatal mercury exposure (Table 2).
Table 2.
Percent change in global 5-hydroxymethylcytosine (5hmC), global 5-methylcytosine (5mC), and their ratio in cord blood per doubling in prenatal maternal red blood cell mercury (RBC-Hg) concentrations.
| Model | Change in global (95% CI) | p-Value | Change in global (95% CI) | p-Value | %-change in the ratio of 5mC to 5hmC (95% CI) | p-Value | |
|---|---|---|---|---|---|---|---|
| Unadjusted | 306 | (, 0.000) | 0.05 | (, 0.149) | 0.74 | 5.70% (1.60, 9.95) | 0.006 |
| *Model 1 | 306 | (, 0.000) | 0.06 | (, 0.128) | 0.47 | 4.80% (0.18, 9.69) | 0.04 |
| †Model 2 | 306 | (, 0.002) | 0.09 | (, 0.151) | 0.63 | 4.70% (0.04, 9.58) | 0.05 |
Model 1 adjusted for maternal education, age at enrollment, marital status, first trimester vitamin B-12 intake, second trimester fish consumption, and child race/ethnicity, sex, gestational age, and birth weight for gestational age z-scores. Additionally, the 5-hydroxymethylcytosine (%-5hmC) model was further adjusted for first trimester betaine intake, while the 5-methylcytosine (%-5mC) model was further adjusted for first trimester folate intake. The model for the ratio of 5mC to 5hmC was adjusted for both betaine and folate intake during the first trimester.
Model 2: [( T cells, T cells, Natural killer cells, monocytes, granulocytes, and nucleated red blood cells (nRBCs)].
In early childhood (age: 2.9 to 4.9 y), a doubling in RBC-Hg concentration was associated with a 0.031% mean decrease in global measured in blood (; 95% CI: , ; ) after adjusting for covariates (Table 3). Correspondingly, in fully adjusted models, a doubling in prenatal RBC-Hg concentration was associated with a 22.42% increase in the ratio of to in early childhood (95% CI: 7.73, 39.11; ). Prenatal RBC-Hg concentration was not associated with in early childhood. No associations were observed between prenatal RBC-Hg concentration and global , , or their ratio measured during midchildhood in unadjusted or fully adjusted models (Table 3). Fully adjusted estimates of the associations of prenatal RBC-Hg concentration with epigenomic measures at all measured time points are shown in Figure 2. In sensitivity analyses restricted to participants with repeated epigenomic measurements available in cord blood and in early childhood () or cord blood and midchildhood () yielded consistent results (Figure S3). Analyses stratified by gender suggested that the association might be stronger for females. However, in fully adjusted models, there was no evidence of a significant interaction by sex for the association between prenatal mercury exposure and global at birth () or in early childhood (). Similarly, the association between prenatal mercury exposure and the ratio of 5mC to 5hmC at birth was not significantly different by gender () or in early childhood (). Adjustment for maternal smoking, either sustained during pregnancy or any smoking during pregnancy, did not influence the results. Furthermore, we observed similar results when samples below the LOD were replaced by the (results not shown).
Table 3.
Percent change in global 5-hydroxymethylcytosine (5hmC), global 5-methylcytosine (5mC), and their ratio in blood collected early and midchildhood per doubling in prenatal maternal red blood cell mercury (RBC-Hg) concentrations.
| Childhood time point | Change in global (95% CI) | p-Value | Change in global (95% CI) | p-Value | % change in the ratio of 5mC to 5hmC (95% CI) | p-Value | |
|---|---|---|---|---|---|---|---|
| Early childhood (2.9 to 4.9 y) | |||||||
| Unadjusted | 68 | (, 0.005) | 0.15 | (, 0.300) | 0.89 | 10.69% (0.10, 22.39) | 0.05 |
| Adjusteda | 68 | (, ) | 0.02 | (, 0.337) | 0.77 | 22.42% (7.73, 39.11) | |
| Midchildhood (6.7 to 10.5 y) | |||||||
| Unadjusted | 260 | 0.002% (, 0.013) | 0.77 | 0.010% (, 0.192) | 0.91 | 1.00% (, 5.51) | 0.65 |
| Adjusteda | 260 | 0.005% (, 0.018) | 0.41 | 0.094% (, 0.301) | 0.37 | 0.73% (, 5.88) | 0.78 |
Linear regression models adjusted for maternal education, age at enrollment, marital status, first trimester vitamin B-12 intake, second trimester fish consumption, and child race/ethnicity, sex, gestational age, birth weight for gestational age z-scores, child age at blood collection, and estimated leukocyte composition in early or midchildhood ( T cells, T cells, Natural killer cells, monocytes, and granulocytes). Additionally, the %-5hmC model was further adjusted for first trimester betaine intake, while the %-5mC model was further adjusted for first trimester folate intake. The model for the ratio of 5mC to 5hmC was adjusted for both betaine and folate intake during the first trimester.
Figure 2.
Fully adjusted association for A) 5-hydroxymethylcytosine (%-5hmC); B) 5-methylcytosine (%-5mC;) and C) %-difference in the ratio of 5mC to 5hmC per doubling in prenatal maternal mercury concentrations.
Discussion
In this U.S. prebirth cohort, higher prenatal maternal mercury exposure measured during the second trimester of pregnancy was associated with lower global DNA content in cord blood after adjusting for potential confounders. This association was persistent in early (2.9 to 4.9 y), but not in midchildhood (6.7 to 10.5 y). Correspondingly, prenatal mercury exposure was associated with an increase in the ratio of to in cord blood, also persisting into early but not midchildhood. No associations between prenatal maternal red blood cell mercury concentrations and global DNA content were evident. Global 5hmC and 5mC were higher in cord blood compared to childhood blood samples. Additionally, both 5hmC and 5mC were positively correlated with each other in cord blood in early and midchildhood blood. Our study highlights the potential toxicity of prenatal mercury exposure during fetal epigenetic reprogramming and suggests that the association might be reversible during childhood.
Although 5hmC is an epigenetic modification known to be important during embryogenesis, little is known about its physiological role and predictors. The distribution of 5hmC has been shown to be a relatively stable epigenetic mark in vitro and in animal models, and not just present as a transient demethylation product, likely playing an independent regulatory function regulation (Bachman et al. 2014; Iurlaro et al. 2013; Spruijt et al. 2013). For example, stem cell models have shown that enhancer regions in the genome have greater abundance of 5hmC having a repressive distal role on transcription (Choi et al. 2014; Yu et al. 2012). In embryonic stem cells, 5hmC plays a dual role by maintaining chromatin structure for both upregulated genes upon differentiation and for polycomb group repressed genes (Pastor et al. 2011; Wu et al. 2011). Disruption of 5hmC levels by knocking down TET2 function has been shown to affect hematopoietic cell differentiation, a potential contributor to myeloid malignancies, highlighting the important role of 5hmC levels in blood (Pronier et al. 2011).
Few studies have examined fetal 5hmC disruption during human development. For example, among cases of preeclampsia and gestational diabetes, both conditions known to affect the intrauterine environment, 5hmC content and TET2 expression in umbilical vein tissue were observed to be altered compared to controls, suggesting that an adverse intrauterine environment can influence 5hmC during development (Sun et al. 2016). In this same study, it was shown with cell cultures that hypoxia is a potential mechanism for the observed alteration in 5hmC levels. Furthermore, oxidative stress has been shown to alter the distribution of 5hmC at several genomic regions in both cell cultures and in vivo. For example, inducing oxidative stress led to a global decrease in hydroxymethylation in vitro and in vivo in the epithelium of a double-knockout animal model deficient in glutathione peroxidases 1 and 2, major antioxidant enzymes (Delatte et al. 2015). Consistent with these models, we also observed a decrease in global measurements of 5hmC in the cord blood associated with increasing levels of prenatal mercury exposure. Interestingly, the leading hypothesized mechanisms for mercury-associated toxicity include the generation of reactive oxygen species and increased oxidative stress (Farina et al. 2011a; Farina et al. 2011b). Moreover, metal-induced oxidative stress is also thought to be a unifying process for epigenetic disruption (Baccarelli and Bollati 2009; Hou et al. 2012). Oxidative stress or antioxidant depletion might be involved in the observed association between prenatal mercury exposure and lower 5hmC content. Future experimental studies could test this hypothesis.
A few studies have shown that 5hmC in blood might be sensitive to environmental exposures. In a cross-sectional study, in adult participants from the Strong Heart Study (American Indians from Arizona, Oklahoma, North Dakota, and South Dakota; 45 to 74 y old), higher cadmium and arsenic exposure were each associated with higher of blood DNA (Tellez-Plaza et al. 2014). This study also reported moderate correlations (: 0.32 to 0.54) for the relationship between 5hmC and 5mC. Likewise, another study of adults reported similar Pearson correlations between 5hmC and 5mC measured in blood at different time points ( to 0.22) and an increase in with elevated particulate matter () exposure (Sanchez-Guerra et al. 2015). Although these two studies reported lower correlations among epigenomic measures (5hmC and 5mC) compared to our data, their target populations were adults, and both studies used an enzyme-linked immunosorbent assay commercial method for the quantification of 5mC and 5hmC that differs from our mass spectrometry methodology. In these two previous studies, arsenic and exposure were positively associated with content, but not . We observed an inverse association between prenatal mercury exposure and , but the association with was null, suggesting that 5hmC might be a more sensitive biomarker of environmental exposures. Moreover, the observed increase in the ratio of 5mC to 5hmC paired with the null associations for 5mC suggests that prenatal mercury exposure does not influence the demethylation pathway of 5mC to the 5hmC product. Instead, our results suggest that the 5hmC association with prenatal maternal mercury exposure is independent of 5mC levels in the genome. The health implications of the associations between metals and in both children and adults are currently unknown. Prospective epidemiological and molecular studies with metal exposure assessment, epigenetic measures that distinguish 5hmC from 5mC, and health end points over the follow-up are needed.
Several studies have examined the association between prenatal mercury exposure and DNA methylation across the genome. Namely, a previous epigenome-wide association study of mother–infant pairs documented hypermethylation of top CpG sites along with shifts in cell type composition of cord blood relative to maternal methylmercury concentrations in a population exposed to low levels of mercury mainly through the diet (Cardenas et al. 2015). Another study of mother–infant pairs likely exposed through dietary sources documented methylation changes of a genomic region associated with both total prenatal mercury and methylmercury exposure, but the association was attenuated after adjusting for cell type composition (Bakulski et al. 2015). In a study of dental health professionals occupationally exposed to mercury, a cross-sectional relationship was observed between methylmercury concentration and hypomethylation of a candidate gene (SEPP1), but no relationship with global DNA methylation of repetitive elements such as Long Interspersed Element (LINE-1) (Goodrich et al. 2013). In an epigenome-wide association study of our cohort, we observed that prenatal maternal RBC-Hg concentration was associated with regional DNA methylation of the PON1 gene in cord blood for males, an association that persisted in early childhood but was attenuated in midchildhood, supporting our findings of the potential malleability of the epigenetic modifications early in life (Cardenas et al. 2017). None of the gene-specific CpGs previously reported to be associated with prenatal mercury exposure in our cohort were significantly correlated with global measures of DNA methylation or DNA hydroxymethylation (Figure S4). Although PON1 cord blood DNA methylation was associated with child cognitive function in early childhood in our previous Epigenome Wide Association Study (EWAS) (Cardenas et al. 2017), we did not observe an association for global and measured in cord blood or early childhood with child cognitive function (Table S2). Additionally, we tested whether DNA methylation measurements from the Illumina Infinium HumanMethylation450 BeadChip or 450K array were associated with our global measurements of and among the same samples using the Global Analysis of Methylation Profiles package of R (Zhao et al. 2015). Our global measures of DNA were not associated with the cumulative density function () or the density distribution of the 450K measurements (). Furthermore, our global measurements of DNA was not significantly associated with either the cumulative density function () or the density distribution of the 450K measurements (). As sensitivity analyses, we also examined if any individual CpG site from the 450K array was associated with global and evaluated whether those CpGs were also associated with prenatal mercury exposure. After adjusting for covariates and multiple comparisons, we did not observe any significant associations between CpGs correlated with global and prenatal mercury exposure (Figure S5). However, these results must be interpreted with caution, as the genomic inflation for the EWAS of was relatively large (), which that could lead to the overestimation of statistical significance (van Iterson et al. 2017). Additionally, a more appropriate study design is needed to address this question, for example, implementing oxidative bisulfite treatment of samples to distinguish individuals CpGs that are hydroxymethylated. Processing and technical batch adjustment for the 450K data have been previously described (Cardenas et al. 2017).
Although these studies measured localized rather than global genomic DNA methylation, they provide evidence that prenatal exposure could potentially affect DNA methylation levels of specific genomic regions. Furthermore, in two animal studies conducted in brain tissue of polar bears (Pilsner et al. 2010) and whole blood from alligators (Nilsen et al. 2016), mercury levels were negatively associated with global DNA methylation of each specific tissue, suggesting that mercury might lead to demethylation of the genome independent of tissue type. In sensitivity analyses, we investigated whether cord blood global or were associated with other measures of DNA methylation, such as LINE-1 DNA methylation previously measured in this cohort (Boeke et al. 2012). We did not see any significant or strong correlations between our global epigenetic measures and LINE-1 DNA methylation. Furthermore, prenatal mercury exposure was not associated with LINE-1 cord blood DNA methylation (Figure S6).
The adjusted associations of prenatal mercury exposure with , along with the corresponding increase in the ratio of to , were persistent into early childhood but absent in midchildhood, even though our sample size was greater in the latter. This relationship suggests that the 5hmC association with prenatal mercury exposure might be malleable and potentially reversible throughout childhood. This is also supported by our sensitivity analyses, showing that among individuals with repeated epigenomic measurements in cord blood and early childhood (), the association persists, but among individuals with repeated epigenomic measurements in cord blood and midchildhood (), the association does not persist. Global measurements of both 5hmC and 5mC have been shown to decline with age, consistent with our observation of higher levels for both epigenomic measures in cord blood compared to early or midchildhood blood (Buscarlet et al. 2015). However, both epigenomic measurements were not significantly different in early and midchildhood.
Measuring global 5hmC and 5mC content in the genome is innovative in epidemiological studies. Liquid chromatography combined with tandem mass spectrometry has been shown to be among the top two best-performing assays for global DNA epigenomic measurements in a multicenter benchmarking study and observed to most accurately reflect expected differences in global DNA methylation (BLUEPRINT Consortium et al. 2016). Also, our global epigenomic measurements in cord blood might differ from previous studies, as both the proportion of 5hmC and 5mC represent the estimated amount of modified cytosine in the entire genome as opposed to other global assays that only target repetitive DNA elements and are a proxy for global epigenomic modifications. For example, we did not see any significant associations between our global measurement of 5hmC and 5mC with either LINE-1 cord blood DNA methylation or the global distribution of CpG methylation measured with the array. This is not unexpected, as both LINE-1 and the array provide methylation measurements at very distinct genomic positions and have limited coverage of all CpGs present in the human genome. In contrast, our liquid chromatography method has a true global genomic coverage.
However, an important limitation of this method is the lack of information on localized methylation or hydroxymethylation. Given the previously characterized dual role of 5hmC in stem cells, it would be beneficial to measure 5hmC with greater resolution in order to characterize its role in transcriptional regulation and response to environmental exposures at specific genomic coordinates within DNA samples. Another important limitation of this study is that both epigenomic measurements were taken from cord blood and blood DNA, and not isolated from a target organ, such as the brain. Given the cell type specificity of 5hmC and 5mC, our measurement represents an aggregate of different nucleated blood cell types. However, we did adjust for major cell type distributions, including nRBCs found in cord blood, estimated from DNA methylation arrays shown to have good correlations with laboratory measurements (Cardenas et al. 2016). Our cell type adjustment minimizes the chance that our findings are due to cell type heterogeneity. Our study has several strengths, including the relatively large number of samples, the prospective assessment of both epigenomic measurements at birth and at two time points during childhood, and the use of an objective biomarker of prenatal mercury exposure at a sensitive window of fetal development during pregnancy. We used second trimester maternal RBC-Hg concentrations as an unbiased biomarker of exposure with an estimated half-life of 72 d (Mahaffey et al. 2004), and 70-95% of the red blood cell mercury content is estimated to be methylmercury (Mortensen et al. 2014). In our cohort, we also expect that the major source of exposure is dietary, mainly from fish, although we cannot rule out other sources. Although this is an observational study and confounding cannot be ruled out, we adjusted for many different confounders, including important prenatal maternal nutritional information, that have been characterized and validated in our cohort.
Conclusion
Our study is the first to show that prenatal mercury exposure, a global persistent environmental contaminant, is associated with lower 5hmC levels in cord blood DNA, and this association persists in early childhood blood, but likely not beyond that time period. Levels of 5hmC play a critical role in embryogenesis and cell lineage commitment, and are now appreciated as an independent epigenetic control mechanism of transcriptional regulation. The prenatal toxicity of mercury might be mediated by the observed alteration in 5hmC content serving as biomarker of exposure or disease susceptibility. Furthermore, our results highlight the potential role of prenatal mercury exposure during fetal epigenetic programming and suggest that this association is reversible in childhood.
Supplemental Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01 NR013945, R01 ES021357, R01 HD034568, K24 HD069408, R01 ES016314, R01 HL 111108, and P30 ES009089).
References
- Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, et al. 2014. Minfi: a flexible and comprehensive bioconductor package for the analysis of infinium DNA methylation microarrays. Bioinformatics 30(10):1363–1369, PMID: 24478339, 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A, Bollati V. 2009. Epigenetics and environmental chemicals. Curr Opin Pediatr 21(2):243, PMID: 19663042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman M, Uribe-Lewis S, Yang X, Williams M, Murrell A, Balasubramanian S. 2014. 5-hydroxymethylcytosine is a predominantly stable DNA modification. Nat Chem 6(12):1049–1055, PMID: 25411882, 10.1038/nchem.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakulski KM, Feinberg JI, Andrews SV, Yang J, Brown S, McKenney SL, et al. 2016. DNA methylation of cord blood cell types: Applications for mixed cell birth studies. Epigenetics 11(5):354–362, PMID: 27019159, 10.1080/15592294.2016.1161875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakulski KM, Lee H, Feinberg JI, Wells EM, Brown S, Herbstman JB, et al. 2015. Prenatal mercury concentration is associated with changes in DNA methylation at TCEANC2 in newborns. Int J Epidemiol 44(4):1249–1262, PMID: 25906783, 10.1093/ije/dyv032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BLUEPRINT Consortium, Bock C, Halbritter F, Carmona FJ, Tierling S, Datlinger P, et al. 2016. Quantitative comparison of DNA methylation assays for biomarker development and clinical applications. Nat Biotechnol 34(7):726–737. PMID: 27347756, 10.1038/nbt.3605. [DOI] [PubMed] [Google Scholar]
- Boeke CE, Baccarelli A, Kleinman KP, Burris HH, Litonjua AA, Rifas-Shiman SL, et al. 2012. Gestational intake of methyl donors and global LINE-1 DNA methylation in maternal and cord blood: prospective results from a folate-replete population. Epigenetics 7(3):253–260, PMID: 22430801, 10.4161/epi.7.3.19082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscarlet M, Tessier A, Provost S, Mollica L, Busque L. 2015. Human blood cell level of 5-hydroxymethylcytosine (5hmC) declines steadily during aging and is multifactorial. Exp Hematol 44(11):1072–1084, PMID: 27475703, 10.1016/j.exphem.2016.07.009. [DOI] [PubMed] [Google Scholar]
- Cardenas A, Allard C, Doyon M, Houseman EA, Bakulski KM, Perron P, et al. 2016. Validation of a DNA methylation reference panel for the estimation of nucleated cells types in cord blood. Epigenetics 11(11):773–779, PMID: 27668573, 10.1080/15592294.2016.1233091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas A, Koestler DC, Houseman EA, Jackson BP, Kile ML, Karagas MR, et al. 2015. Differential DNA methylation in umbilical cord blood of infants exposed to mercury and arsenic in utero. Epigenetics 10(6):508–515, PMID: 25923418, 10.1080/15592294.2015.1046026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas A, Rifas-Shiman SL, Agha G, Hivert MF, Litonjua AA, DeMeo DL, et al. 2017. Persistent DNA methylation changes associated with prenatal mercury exposure and cognitive performance during childhood. Sci Rep 7(1):288, PMID: 28325913, 10.1038/s41598-017-00384-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoldi AF, Coccini T, Manzo L. 2003. Neurotoxic and molecular effects of methylmercury in humans. Rev Environ Health 18(1):19–32, PMID: 12875509, 10.1515/REVEH.2003.18.1.19. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Xie N, Jin P, Wang T. 2015. DNA methylation and hydroxymethylation in stem cells. Cell Biochem Funct 33(4):161–173, PMID: 25776144, 10.1002/cbf.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi I, Kim R, Lim HW, Kaestner KH, Won KJ. 2014. 5-hydroxymethylcytosine represses the activity of enhancers in embryonic stem cells: a new epigenetic signature for gene regulation. BMC Genomics 15(1):670, PMID: 25106691, 10.1186/1471-2164-15-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao T, Cheng R, Revelo M, Mitzner W, Tang W. 2014. Hydroxymethylation as a novel environmental biosensor. Curr Envir Health Rep 1(1):1–10, PMID: 24860723, 10.1007/s40572-013-0005-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatte B, Jeschke J, Defrance M, Bachman M, Creppe C, Calonne E, et al. 2015. Genome-wide hydroxymethylcytosine pattern changes in response to oxidative stress. Sci Rep 5:12714, PMID: 26239807, 10.1038/srep12714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll CT, Mason RP, Chan HM, Jacob DJ, Pirrone N. 2013. Mercury as a global pollutant: sources, pathways, and effects. Environ Sci Technol 47(10):4967–4983, PMID: 23590191, 10.1021/es305071v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchegaray JP, Chavez L, Huang Y, Ross KN, Choi J, Martinez-Pastor B, et al. 2015. The histone deacetylase SIRT6 controls embryonic stem cell fate via TET-mediated production of 5-hydroxymethylcytosine. Nat Cell Biol 17(5):545–557, PMID: 25915124, 10.1038/ncb3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina M, Aschner M, Rocha JB. 2011a. Oxidative stress in MeHg-induced neurotoxicity. Toxicol Appl Pharmacol 256(3):405–417, PMID: 21601588, 10.1016/j.taap.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina M, Rocha JB, Aschner M. 2011b. Mechanisms of methylmercury-induced neurotoxicity: Evidence from experimental studies. Life Sci 89(15-16):555–563, PMID: 21683713, 10.1016/j.lfs.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawzi WW, Rifas-Shiman SL, Rich-Edwards JW, Willett WC, Gillman MW. 2004. Calibration of a semi-quantitative food frequency questionnaire in early pregnancy. Ann Epidemiol 14(10):754–762, PMID: 15519898, 10.1016/j.annepidem.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Godderis L, Schouteden C, Tabish A, Poels K, Hoet P, Baccarelli AA, et al. 2015. Global methylation and hydroxymethylation in DNA from blood and saliva in healthy volunteers. Biomed Res Int 2015:845041, PMID: 26090450, 10.1155/2015/845041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JM, Basu N, Franzblau A, Dolinoy DC. 2013. Mercury biomarkers and DNA methylation among Michigan dental professionals. Environ Mol Mutagen 54(3):195–203, PMID: 23444121, 10.1002/em.21763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MA, Qiu R, Wu X, Li AX, Zhang H, Wang J, et al. 2013. Dynamics of 5-hydroxymethylcytosine and chromatin marks in Mammalian neurogenesis. Cell Rep 3(2):291–300, PMID: 23403289, 10.1016/j.celrep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Zhang X, Wang D, Baccarelli A. 2012. Environmental chemical exposures and human epigenetics. Int J Epidemiol 41(1):79–105, 10.1093/ije/dyr154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iurlaro M, Ficz G, Oxley D, Raiber EA, Bachman M, Booth MJ, et al. 2013. A screen for hydroxymethylcytosine and formylcytosine binding proteins suggests functions in transcription and chromatin regulation. Genome Biol 14(10):R119, PMID: 24156278, 10.1186/gb-2013-14-10-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagas MR, Choi AL, Oken E, Horvat M, Schoeny R, Kamai E, et al. 2012. Evidence on the human health effects of low-level methylmercury exposure. Environ Health Perspect 120(6):799–806, PMID: 22275730, 10.1289/ehp.1104494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd-Acosta C. 2015. Epigenetic signatures as biomarkers of exposure. Curr Environ Health Rep 2(2):117–125, PMID: 26231361, 10.1007/s40572-015-0051-2. [DOI] [PubMed] [Google Scholar]
- Lamborg CH, Hammerschmidt CR, Bowman KL, Swarr GJ, Munson KM, Ohnemus DC, et al. 2014. A global ocean inventory of anthropogenic mercury based on water column measurements. Nature 512(7512):65–68, PMID: 25100482, 10.1038/nature13563. [DOI] [PubMed] [Google Scholar]
- Maccani JZ, Koestler DC, Lester B, Houseman EA, Armstrong DA, Kelsey KT, et al. 2015. Placental DNA methylation related to both infant toenail mercury and adverse neurobehavioral outcomes. Environ Health Perspect 123(7):723–729, PMID: 25748564, 10.1289/ehp.1408561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahaffey KR, Clickner RP, Bodurow CC. 2004. Blood organic mercury and dietary mercury intake: National Health and Nutrition Examination Survey, 1999 and 2000. Environ Health Perspect 112(5):562–570, PMID: 15064162, 10.1289/ehp.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen ME, Caudill SP, Caldwell KL, Ward CD, Jones RL. 2014. Total and methyl mercury in whole blood measured for the first time in the US population: NHANES 2011–2012. Environ Res 134:257–264, PMID: 25173092, 10.1016/j.envres.2014.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen FM, Parrott BB, Bowden JA, Kassim BL, Somerville SE, Bryan TA, et al. 2016. Global DNA methylation loss associated with mercury contamination and aging in the American alligator (Alligator mississippiensis). Sci Total Environ 545–546:389–397, PMID: 26748003, 10.1016/j.scitotenv.2015.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oken E, Baccarelli AA, Gold DR, Kleinman KP, Litonjua AA, De Meo D, et al. 2014. Cohort profile: project viva. Int J Epidemiol 44(1):37–48, PMID: 24639442, 10.1093/ije/dyu008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oken E, Kleinman KP, Rich-Edwards J, Gillman MW. 2003. A nearly continuous measure of birth weight for gestational age using a United States national reference. BMC Pediatr 3:6, PMID: 12848901, 10.1186/1471-2431-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oken E, Rifas-Shiman SL, Amarasiriwardena C, Jayawardene I, Bellinger DC, Hibbeln JR, et al. 2016. Maternal prenatal fish consumption and cognition in mid childhood: Mercury, fatty acids, and selenium. Neurotoxicol Teratol 57:71–78, PMID: 27381635, 10.1016/j.ntt.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, et al. 2011. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature 473(7347):394–397, PMID: 21552279, 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels S, Duca RC, Devlieger R, Freson K, Straetmans D, Van Herck E, et al. 2016. Maternal methyl-group donor intake and global DNA (hydroxy) methylation before and during pregnancy. Nutrients 8(8):474, PMID: 27509522, 10.3390/nu8080474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilsner RJ, Lazarus AL, Nam DH, Letcher RJ, Sonne C, Dietz R, et al. 2010. Mercury‐associated DNA hypomethylation in polar bear brains via the luminometric methylation assay: A sensitive method to study epigenetics in wildlife. Molecular Ecology 19(2):307–314, PMID: 20002585, 10.1111/j.1365-294X.2009.04452.x. [DOI] [PubMed] [Google Scholar]
- Pronier E, Almire C, Mokrani H, Vasanthakumar A, Simon A, da Costa Reis Monte Mor B, et al. 2011. Inhibition of TET2-mediated conversion of 5-methylcytosine to 5-hydroxymethylcytosine disturbs erythroid and granulomonocytic differentiation of human hematopoietic progenitors. Blood 118(9):2551–2555, PMID: 21734233, 10.1182/blood-2010-12-324707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relton CL, Hartwig FP, Smith G. 2015. From stem cells to the law courts: DNA methylation, the forensic epigenome and the possibility of a biosocial archive. Int J Epidemiol 44(4):1083–1093, PMID: 26424516, 10.1093/ije/dyv198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzov A, Tsenkina Y, Serio A, Dudnakova T, Fletcher J, Bai Y, et al. 2011. Lineage-specific distribution of high levels of genomic 5-hydroxymethylcytosine in mammalian development. Cell Res 21(9):1332–1342, PMID: 21747414, 10.1038/cr.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Guerra M, Zheng Y, Osorio-Yanez C, Zhong J, Chervona Y, Wang S, et al. 2015. Effects of particulate matter exposure on blood 5-hydroxymethylation: Results from the Beijing truck driver air pollution study. Epigenetics 10(7):633–642, PMID: 25970091, 10.1080/15592294.2015.1050174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, et al. 2013. Dynamic readers for 5-(hydroxy) methylcytosine and its oxidized derivatives. Cell 152(5):1146–1159, PMID: 23434322, 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- Stern AH, Smith AE. 2003. An assessment of the cord blood: maternal blood methylmercury ratio: implications for risk assessment. Environ Health Perspect 111(12):1465–1470, PMID: 12948885, 10.1289/ehp.6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Song MM, Wei B, Gao Q, Li L, Yao B, et al. 2016. 5-Hydroxymethylcytosine-mediated alteration of transposon activity associated with the exposure to adverse in utero environments in human. Hum Mol Genet 25(11):2208–2219, PMID: 27005421, 10.1093/hmg/ddw089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellez-Plaza M, Tang WY, Shang Y, Umans JG, Francesconi KA, Goessler W, et al. 2014. Association of global DNA methylation and global DNA hydroxymethylation with metals and other exposures in human blood DNA samples. Environ Health Perspect 122(9):946–954, PMID: 24769358, 10.1289/ehp.1306674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Iterson M, van Zwet EW, BIOS Consortium, Heijmans BT. 2017. Controlling bias and inflation in epigenome-and transcriptome-wide association studies using the empirical null distribution. Genome Biol 18(1):19, PMID: 28129774, 10.1186/s13059-016-1131-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterland RA, Michels KB. 2007. Epigenetic epidemiology of the developmental origins hypothesis. Annu Rev Nutr 27:363–388, PMID: 17465856, 10.1146/annurev.nutr.27.061406.093705. [DOI] [PubMed] [Google Scholar]
- Wu H, D'Alessio AC, Ito S, Wang Z, Cui K, Zhao K, et al. 2011. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev 25(7):679–684, PMID: 21460036, 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, et al. 2012. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149(6):1368–1380, PMID: 22608086, 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N, Bell DA, Maity A, Staicu AM, Joubert BR, London SJ, et al. 2015. Global analysis of methylation profiles from high resolution CpG data. Genet Epidemiol 39(2):53–64, PMID: 25537884, 10.1002/gepi.21874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.