Summary
Brain swelling is the major predictor of mortality in pediatric cerebral malaria (CM). However, the mechanisms leading to swelling remain poorly defined. Here, we combined neuroimaging, parasite transcript profiling, and laboratory blood profiles to develop machine learning models of malarial retinopathy and brain swelling. We found that parasite var transcripts encoding endothelial protein C receptor (EPCR) binding domains, in combination with high parasite biomass and low platelet levels, are strong indicators of CM cases with malarial retinopathy. Swelling cases presented low platelet levels and increased transcript abundance of parasite PfEMP1 DC8 and group A EPCR-binding domains. Remarkably, the dominant transcript in 50% of swelling cases encoded PfEMP1 group A CIDRα1.7 domains. Furthermore, a recombinant CIDRα1.7 domain from a pediatric CM brain autopsy inhibited the barrier protective properties of EPCR in human brain endothelial cells in vitro. Together, these findings suggest a detrimental role for EPCR-binding CIDRα1 domains in brain swelling.
Keywords: Cerebral malaria, brain swelling, EPCR, PfEMP1, var, Plasmodium falciparum
eTOC blurb
Brain swelling is associated with cerebral malaria mortality, but the parasite and host factors responsible for development of brain swelling are unknown. Kessler et al demonstrate an association of low platelet count and EPCR-binding PfEMP1 with brain swelling in children with cerebral malaria.
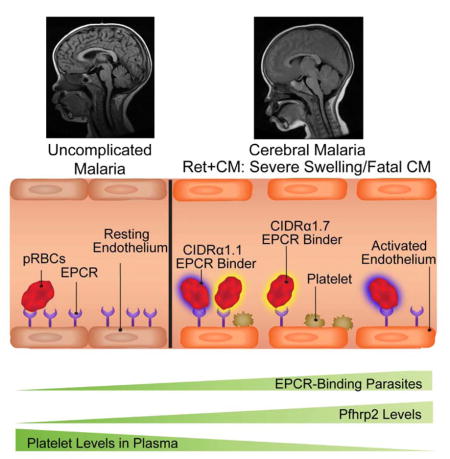
Introduction
Cerebral malaria (CM) is associated with sequestration of Plasmodium falciparum parasitized red blood cells (pRBCs) in cerebral vasculature. While parenteral artemesinins have dramatically improved patient survival (Dondorp et al., 2010), they do not specifically target the blood-brain barrier (BBB) disruption and brain swelling associated with pediatric CM fatality (Brown et al., 2001; Dorovini-Zis et al., 2011; Seydel et al., 2015). A better understanding of CM pathophysiology may inform new adjunctive treatment strategies to reduce mortality and improve the neurological outcomes of survivors (Birbeck et al., 2010).
P. falciparum sequestration is mediated by erythrocyte membrane protein 1 (PfEMP1), a protein family that can be classified into three major groups (A, B, C) on the basis of upstream sequence (UpsA/B/C) and chromosome location (Lavstsen et al., 2003). PfEMP1 contain multiple extracellular binding domains referred to as Duffy binding-like (DBL) or cysteine-rich interdomain regions (CIDR). Sequence classification has revealed conserved tandem arrangements of domains known as domain cassettes (DC) (Rask et al., 2010; Smith et al., 2000) and provided molecular insight into protein diversification. The N-terminal DBL-CIDR head structure has diverged between var groups, such that group B and C PfEMP1 encode CD36 binding properties (CIDRα2–6 domains) (Hsieh et al., 2016; Robinson et al., 2003) whereas group A proteins have diversified into those that bind endothelial protein C receptor (EPCR) (CIDRα1 domains) or non-EPCR binders (CIDRβ/γ/δ domains) (Lau et al., 2015; Turner et al., 2013). The latter group A subset is less well characterized, although some mediate rosetting of pRBCs with unparasitized RBCs (Ghumra et al., 2012). The PfEMP1 repertoire also includes a B/A chimera var gene that encodes an EPCR-binding CIDRα1 domain characterized by the DC8 cassette (DBLα2-CIDRα1.1/8-DBLβ12-DBLγ4/6) (Turner et al., 2013).
Prior var expression studies have implicated DC8 and group A EPCR binding PfEMP1 in severe malaria (Abdi et al., 2015; Jensen et al., 2004; Jespersen et al., 2016; Lavstsen et al., 2012). As EPCR plays a central role in vascular homeostasis by facilitating the activation of the plasma protease protein C to activated protein C (APC) (Mosnier et al., 2007), it has been hypothesized that parasites may potentiate cerebral vascular dysfunction by inhibiting the protective APC-EPCR interaction (Gillrie et al., 2015; Petersen et al., 2015; Turner et al., 2013). Indeed, pediatric CM brain autopsies have revealed alterations in coagulation (fibrin deposits and platelet infiltrates) (Dorovini-Zis et al., 2011; Grau et al., 2003; Hochman et al., 2015; Moxon et al., 2015) and increased BBB permeability (Brown et al., 2001; Dorovini-Zis et al., 2011). However, the specific contribution of the CIDRα1-EPCR interaction to late stage disease mechanisms important for CM pathogenesis or fatality has been largely unaddressed.
A challenge of identifying CM disease mechanisms is that up to one-quarter of children presenting with WHO-defined CM have another non-malaria cause of death at autopsy (Hochman et al., 2015; Taylor et al., 2004). Malarial retinopathy, a set of ocular abnormalities characterized by patchy retinal whitening, vessel discoloration, and retinal hemorrhages detected by funduscopic exam, is the most accurate clinical sign of histologically confirmed pediatric CM and improves case definition (Lewallen et al., 1996; Seydel et al., 2015).
Our patient population is a rigorously characterized pediatric CM cohort that underwent funduscopy to determine malarial retinopathy status (Ret+CM or Ret−CM) and MRI for evaluation of brain swelling during acute infection. We employed machine learning models to investigate parasite and host risk factors associated with retinopathy and severe brain swelling and conducted a functional characterization of a PfEMP1 variant associated with fatal malaria in our autopsy cohort. Our findings provide evidence for a role of EPCR-binding domains in CM brain swelling.
Results
Study Population Characteristics
During 2015–16, 75 children with WHO-defined P. falciparum CM and 38 children with P. falciparum uncomplicated malaria (UM) were studied (Table S1; Figure S1). Of 75 CM cases (Figure 1A), 57 children had malarial retinopathy (Ret+CM) and 18 were retinopathy negative (Ret−CM) on funduscopic exam. Eight Ret+CM cases had insufficient parasite material for var typing (Figure S1; Tables S1 and S2). Subsequent analyses included 49 Ret+CM, 18 Ret−CM, and 38 UM patients.
Figure 1. Case definitions of P. falciparum severe malaria syndromes in children.
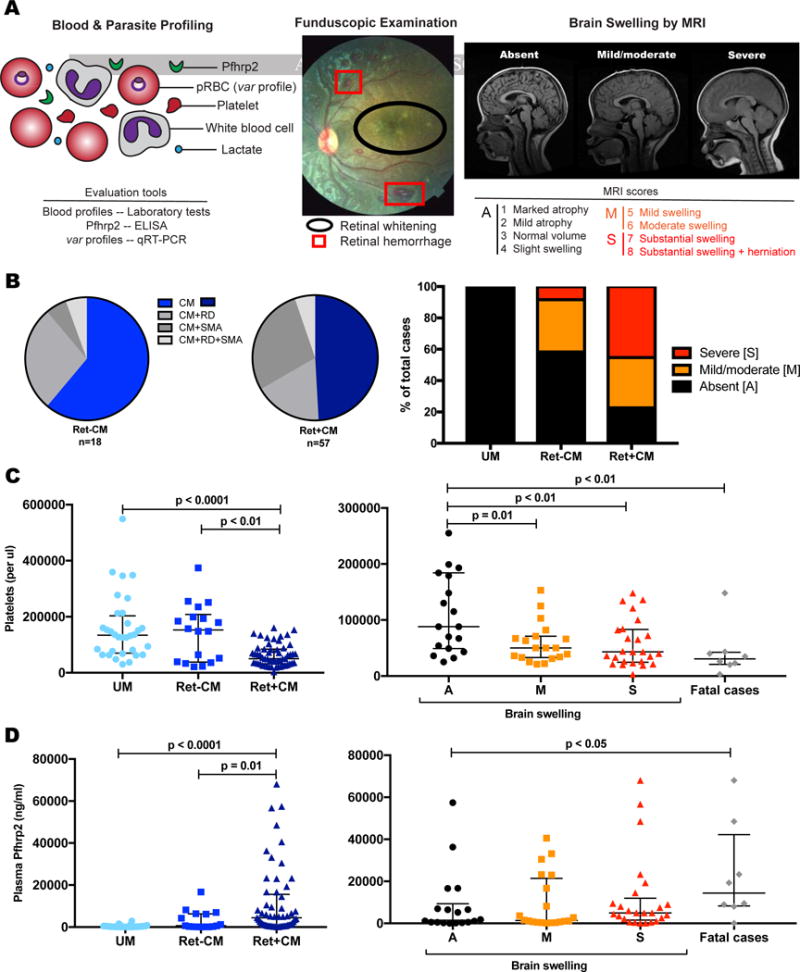
(A) Cases underwent blood and parasite profiling (UM, CM), funduscopic examination (CM), and MRI for brain swelling evaluation (CM). Signs of malarial retinopathy are indicated.
(B) Ret+CM and Ret−CM cases have overlapping but differential malaria-associated disease symptoms, including the presence and severity of brain swelling.
(C) Ret+CM, brain swelling, and fatality are inversely associated with circulating platelet levels. Median and IQR are represented by horizontal lines.
(D) Ret+CM and fatal cases are characterized by higher levels of Pfhrp2. Median and IQR are represented by horizontal lines.
See also Figure S1 and Tables S1 and S2.
Severe childhood malaria consists of severe malarial anemia, respiratory distress, and CM that occurs alone or in overlapping syndromes. Children with Ret+ or Ret−CM had lower hemoglobin levels relative to UM cases (Median [IQR]: 7.6 [6.4, 9.0] g/dL and 7.4 [6.7, 10.0] g/dL vs. 9.9 [8.5, 11.7] g/dL; p<0.0001) (Table 1). Some children had severe malarial anemia (Hgb <5g/dL or PCV<15) and/or respiratory distress (Figure 1B, Table S1). Platelet levels were significantly lower in the Ret+CM cases relative to both Ret−CM cases (p=0.004) and UM cases (p<0.0001) (Table 1, Figure 1C). Notably, Ret+CM cases had higher levels of severe brain swelling (MRI scores >6): 24 of 53 Ret+CM cases and 1 of 12 Ret−CM cases (Figure 1B; Table S1). All mortality was in children with severe brain swelling (Table S1). Mortality rates for UM, Ret−CM, and Ret+CM were 0%, 5.6%, and 12.3%, respectively.
Table 1.
Clinical characteristics of patient groups
| Patient Characteristic | Ret+CM (n = 57) | Ret−CM (n = 18) | p* | UM (n = 38) | p* |
|---|---|---|---|---|---|
| Age (yr), median [IQR] | 4.0 [3.0, 5.7] | 3.7 [1.9, 5.7] | 0.49 | 5.5 [2.0, 7.0] | 0.33 |
| Male, n (%) | 35 (61.4) | 9 (50.0) | 0.39 | 22 (57.9) | 0.73 |
| Smear score, median [IQR] | 3 [4] | 4 [4] | 0.05 | 4 [4] | <0.0001 |
| Parasitemia (103/ul), median [IQR] | 4.7 [0.5, 96.3] | 61.1 [29.2, 229.8] | 0.04 | NA | |
| Pfhrp2 (ng/ml), median [IQR] | 4466 [14476] | 609 [6251] | 0.01 | 190 [461] | <0.0001 |
| Hgb (g/dL), median [IQR] | 7.6 [6.4, 9.0] | 7.4 [6.7, 10.0] | 0.44 | 9.9 [8.5, 11.7] | <0.0001 |
| Platelets (103/pl), median [IQR] | 50 [84] | 152.5 [199] | 0.004 | 134 [72.5, 194] | <0.0001 |
| Total WBC (103/pl), median [IQR] | 8.5 [7.0, 11.6] | 8.6 [4.9, 12.8] | 0.58 | 7.5 [6.0, 9.4] | 0.09 |
| Lactate (mmol/L), median [IQR] | 3.4 [2.1, 5.6] | 3.3 [2.3, 4.0] | 0.49 | NA | |
| Blantyre coma score, median [IQR] | 2 [2] | 2 [2] | 0.73 | UC | |
| Severe malarial anemia, n (%) | 19(33.3) | 2 (11.1) | 0.07 | 0(0) | <0.0001 |
| Respiratory distress, n (%) | 13 (22.8) | 6 (33.3) | 0.37 | 0(0) | 0.002 |
| Jaundice, n (%) | 5 (8.8) | 0(0) | 0.19 | 0(0) | 0.06 |
| Brain swelling°, n (%) | 41 (77.4) | 5 (41.7) | 0.01 | UC | |
| MRI scores (scale 1–8) | |||||
| 1–4 – Absence of swelling | 12 (22.6) | 7 (58.3) | |||
| 5–6 – Mild/moderate swelling | 17(32.1) | 4 (33.3) | |||
| 7–8 – Severe swelling | 24 (45.3) | 1 (8.3) | |||
| Mortality, n (%) | 7(12.3) | 1 (5.6) | 0.42 | 0(0) | 0.025 |
IQR, interquartile range; UC, uncomplicated and by definition BCS=5, no swelling; NA, not available.
Ret+CM (n=53), Ret−CM (n=12); Brain swelling = MRI score > 4.
p-values correspond to Wilcoxon rank-sum or chi-squared test between indicated group and Ret+CM. See also Tables SI and S2
Ret+CM is associated with higher Pfhrp2 levels and increased transcript levels of EPCR-binding domains
Parasite biomass (circulating and sequestered pRBCs) was estimated using plasma Pfhrp2 levels. Consistent with previous studies (Dondorp et al., 2005; Fox et al., 2013; Seydel et al., 2012), we found elevated levels of Pfhrp2 in Ret+CM cases relative to both UM (p<0.0001) and Ret−CM cases (p=0.01) (Figure 1D).
To investigate parasite cytoadhesion we performed var transcript profiling by qRT-PCR on circulating ring stage pRBCs using var primers that target groups A, B, C or specific adhesion domains (Table S3) (Lavstsen et al., 2012; Rottmann et al., 2006). While all three var groups were detected in most patients, group A var transcripts were expressed at the highest levels in all patients (Table S4). Group A transcript level was significantly greater in Ret+CM cases compared to UM cases (Table S4, p<0.001), and Ret−CM cases had intermediate levels.
Domain primers give additional insight into expression of PfEMP1 variants that have diversified in binding properties (Figure 2A). Transcripts linked to EPCR-binding, including both DC8 and group A EPCR-binding variants, were significantly increased in Ret+CM compared to UM (Figures 2B, 2C, and Table S4). Additionally, a group A head structure linked to rosetting/unknown binding properties was enriched in Ret+CM cases (DBLα1.5/6a, DBLα1.5/6b, and CIDRδ of DC16), as was a C-terminal domain linked to PECAM-1 binding (DBLγ and DBLβ7/9 of DC5) (Figures 2D, 2E, and Table S4) (Berger et al., 2013) The application of second generation primers (Mkumbaye et al., 2017; Petersen et al., 2016) that can distinguish between the six subtypes of EPCR-binding CIDRα1 domains confirmed that both DC8 (CIDRα1.1) and group A (CIDRα1.4–1.7) transcript levels were significantly increased in Ret+CM cases compared to UM, whether considered individually or as the sum of all CIDRα1 transcripts (Figure S2 and Table S4).
Figure 2. Specific PfEMP1 domains/variants are overexpressed in Ret+CM cases.
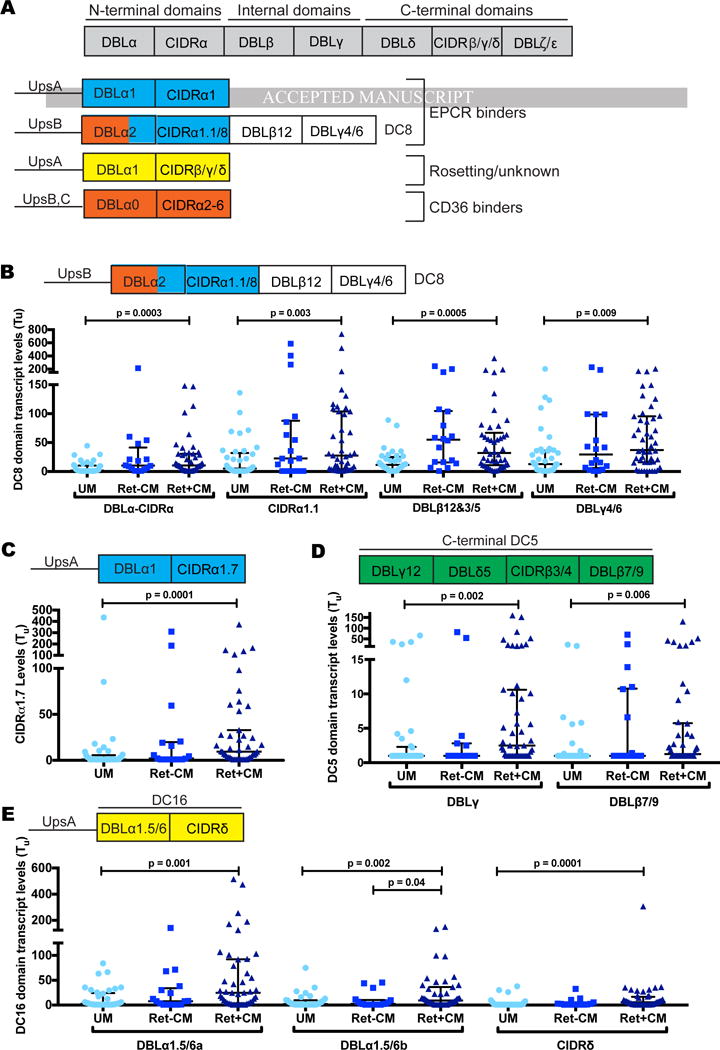
(A) PfEMP1 schematic illustrating binding domains and corresponding phenotypes.
(B–E) Scatter dot plot showing var transcript levels across patient groups, Medians and IQRs are represented by horizontal lines.
See also Figure S2 and Table S3 and S4.
By comparison, while DC8 CIDRα1.1 and group A CIDRα1.5 and CIDRα1.7-containing var transcripts tended to be higher in Ret+CM cases (Figure 3B and 3C), none of the six EPCR-binding CIDRα1 subtypes differed significantly by retinopathy status (Figure S2 and Table S4). Transcript abundance of a C-terminal domain (DBLε2) and of the DC16 subset was increased in Ret+CM cases, but did not achieve statistical significance when corrected for multiple comparisons (Figure 2E; Table S4).
Figure 3. DC8 CIDRα1.1 and group A CIDRα1.7 EPCR-binding transcripts are enriched in CM cases with severe swelling and fatal outcomes.
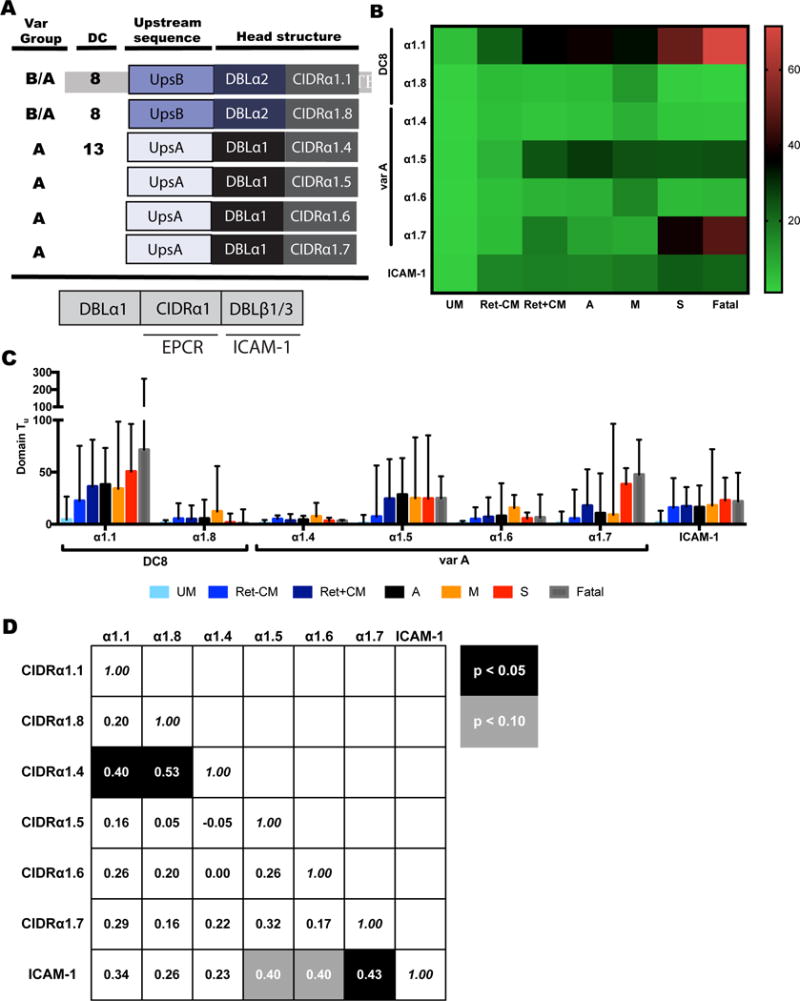
(A) Schematic diagrams of EPCR-binding head structures by CIDRα1 sub-classification and the domain arrangement for dual EPCR and ICAM-1-binding (CIDRα1-DBLβ1/3) sequences.
(B) Heat map of median values of CIDRα1 subtype and ICAM-1-binding motif expression (dual EPCR and ICAM-1 binders).
(C) Bar graphs showing transcript levels of CIDRα1 subtypes and ICAM-1-binding motif. Median and IQR are indicated.
(D) Spearman correlations of transcript levels for individual EPCR-binding CIDRα1 subtypes and ICAM-1-binding motif amongst CM cases. Rho and statistical significance (correcting for multiple comparisons) are indicated.
See also Figure S2 and Tables S3–5.
Recent work revealed that a subset of group A PfEMP1 with dual binding properties to EPCR and intercellular adhesion molecule 1 (ICAM-1) may be associated with pediatric CM (Lennartz et al., 2017). Using this motif-specific primer, we found higher levels of transcripts in CM cases than UM cases (Table S4, p<0.0001), but no difference in transcript level by retinopathy status (Figure 3A–C). Since the EPCR and ICAM-1 binding domains act synergistically in parasite adhesion (Avril et al., 2016; Lennartz et al., 2017), we also examined the correlation of expression of dual ICAM-1 and EPCR binders with different EPCR binding domains in all CM cases (Figure 3D). CIDRα1.5/6/7 domains, often associated with ICAM-1 binding motifs (Lennartz et al., 2017), had the strongest correlation with ICAM-1 binding motifs in CM, but when corrected for multiple comparisons, only CIDRα1.7 showed significant correlation with ICAM-1 binding motif expression (Figure 3D).
PfEMP1 expression in CM cases with severe brain swelling
Increased brain volume (swelling) is a major risk factor for death in pediatric CM (Seydel et al., 2015). Brain volume is scored on a scale of 1–8, with brain swelling defined as a score >4, and severe swelling as a score >6 (Figure 1A). There was an inverse relationship between Pfhrp2 levels and circulating platelet levels in severe swelling and fatal cases (Figure 1C and 1D).
To investigate cytoadhesion and brain swelling, we compared var expression profiles of CM cases with no swelling (absent n=18) to CM cases with evidence of swelling (mild/moderate n=17, severe n=22). While none of the PfEMP1 domains were uniquely associated with swelling (Figure 3B and 3C, Table S5), both the DC8 CIDRα1.1 and the group A CIDRα1.7 EPCR-binding domains showed a trend toward higher expression in CM cases with severe swelling or a fatal outcome (Figures 3B, 3C, and S2).
Machine learning models of malarial retinopathy
To better understand the interplay between parasite and host factors important for severe disease, we generated machine learning models using random forests (RF) (Liaw and Wiener, 2002). We used RF models to discriminate patient groups with various clinical classifications (Figures 4A–C, S3), using var transcript, platelets, and Pfhrp2 levels as features. We also incorporated white blood cell (WBC) counts and lactate levels, markers of CM severity (Seydel et al., 2015), into the models. We applied these models to measure the mean decrease in classifier accuracy (MDCA) for each included feature (e.g. var domain) upon removal of the corresponding feature. For statistical rigor, we applied the mProbes algorithm to determine the significance of features ranked in the RF models while controlling for multiple comparisons (Finney et al., 2014). To identify var domains whose expression appeared to solely reflect parasite biomass, we applied a Pfhrp2 conditional mutual information (CMI)-based filter to each var primer (Wyner, 1978). PfEMP1 domains that did not provide additional information beyond Pfhrp2 levels (CMI p <0.20), were filtered from the model.
Figure 4. Machine learning models of retinopathy and swelling.
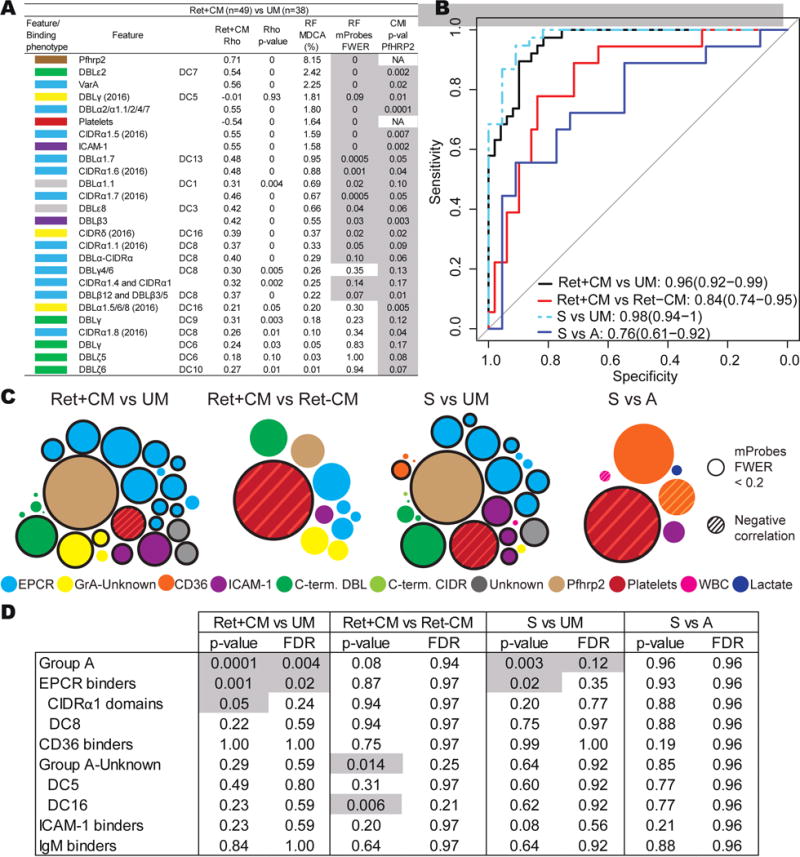
(A) Host and parasite features that discriminate Ret+CM patients from UM cases after Pfhrp2 filtration. Other retinopathy and swelling models are found in Figure S3.
(B) Receiver operating characteristic curve of the RF models. Area under the curve (95% confidence intervals).
(C) Bubble graphs that summarize RF models. The area of the bubble represents the MDCA. Significant features are represented by outlined bubbles.
(D) Primers targeting different var domains were grouped according to domain classification or predicted binding phenotype to create set enrichments. Statistical significance and false discovery rate (FDR) are indicated.
See also Table S6.
In the RF model that compares Ret+CM to UM, ten var primers targeting DC8 and group A-EPCR binding PfEMP1 presented a significant, positive association with retinopathy (Rho>0.30, p ≤0.05, familywise error rate (FWER) ≤0.20) (Figure 4A). Of all features, Pfhrp2 levels had the strongest predictive score for retinopathy (MDCA=8.15%, mProbes FWER = 0), and low platelet level was the sixth ranked feature (Figure 4A and 4C). In addition to the strong signal of EPCR-binding variants, abundance of a C-terminal domain, DBLε2, was the second most important feature (MDCA 2.42%, FWER=0) (Figure 4A and 4C). The model showed strong predictive performance, as measured by the receiver operating characteristics curve [area under the curve (AUC) =0.96] (Figure 4B).
Many of the primers that target EPCR-binding PfEMP1 variants can amplify different regions within the same PfEMP1, and act as proxies for one another, artificially decreasing the MDCA. To overcome this limitation, we built set enrichment models that compare FWER values of grouped primers that share the same architecture/phenotype versus other primers (Table S6). The set enrichment of the Ret+CM vs. UM model confirmed the importance of parasite-EPCR interactions in retinopathy patients, as grouped primers that target EPCR-binding PfEMP1, were significantly enriched (FDR FWER ≤0.20) (Figure 4D).
In a second retinopathy model comparing Ret+CM vs. Ret−CM (AUC=0.84), Pfhrp2 levels, EPCR-binding var domains, and the DBLε2 domain assumed positions of lesser importance, while low platelet levels (thrombocytopenia) became the most important feature (Figure 4C and S3). Taken together, these models suggest that Ret+ and Ret−CM patients are infected with parasites displaying closely related adhesion types. Overall, the RF models suggest that high parasite biomass, low platelet levels, and high parasite transcript levels of EPCR-binding var genes contribute to the development of retinal pathology.
Machine learning models of swelling
Our machine learning models accurately discriminated CM cases with severe swelling from uncomplicated malaria (S vs. UM; AUC 0.98, Figure 4B). The model shares features observed to be important for the Ret+CM vs. UM model. The main differences were a slight decrease in the predictive power of Pfhrp2 and an increase in the MDCA of low platelet levels (Figures 4C and S3). Seven features targeting EPCR-binding proteins significantly predicted brain swelling (Figures 4C and S3). High transcript levels of group A, including EPCR-binding domains, was the sole significant feature in the set enrichment model after correcting for multiple comparisons (Figure 4D).
Conversely, the model that compares patients with and without swelling (S vs. A: AUC of 0.76) did not show differential expression of EPCR-binding domains (Figure 4C and S3), even though EPCR binding domains CIDRα1.1/7 tended to be increased in severe swelling and fatal patients (Figure 3B and 3C). The only factor that had a significant MDCA in this swelling model was thrombocytopenia (Figure 4C and S3). Additionally, new var domains CIDRα2.3/5/6/7/9/10 (CD36 binder) and DBLβ5 (CD36-ICAM-1 binder) were present in the model, but did not have a significant MDCA. None of the var groups remained significant in the set enrichment model (Figure 4D). Taken together, both univariate and machine learning analyses suggest that high parasite biomass and expression of EPCR-binding variants are important features in CM patients, however, none of the EPCR-binding subtypes was uniquely associated with swelling. Thrombocytopenia is the feature most strongly associated with brain swelling.
CIDRα1.7 transcripts and brain swelling
As DC8 CIDRα1.1 and group A CIDRα1.7 transcript levels were increased in peripheral blood of severe swelling and fatal cases (Figures 3B and 3C) we sought to connect these observations to our var typing of brains from autopsies of pediatric CM cases from 1999–2003 (Tembo et al., 2014). Most of the var sequences present in a patient can be captured by amplifying DBLα “tags” using degenerate primers targeting semi-conserved motifs in the N-terminal DBLα domain (Figure 5A)(Lavstsen et al., 2012).We previously found two common DBLα tags amplified directly from brain tissue of fatal pediatric CM cases: var62B1-1 (5 of 20, 25% CM cases; GenBank: KC678109) and var28B1-1 (4 of 20, 20% of CM cases; GenBank: KC678110) (Tembo et al., 2014). Downstream sequencing revealed that var62B1-1 is a group A CIDRα1.7 domain followed by a predicted ICAM-1 binding motif (DBLα1.1-CIDRα1.7-DBLβ3) (Tembo et al., 2014). BLAST analysis of the var28B1-1 tag shows it has highest homology to DC8 variants (Jespersen et al., 2016) (Table S7), suggesting that both DC8 and group A EPCR-binding CIDRα1 are associated with fatal CM.
Figure 5. Diversity and similarity in CIDRα1.7 sequences from swelling cases.
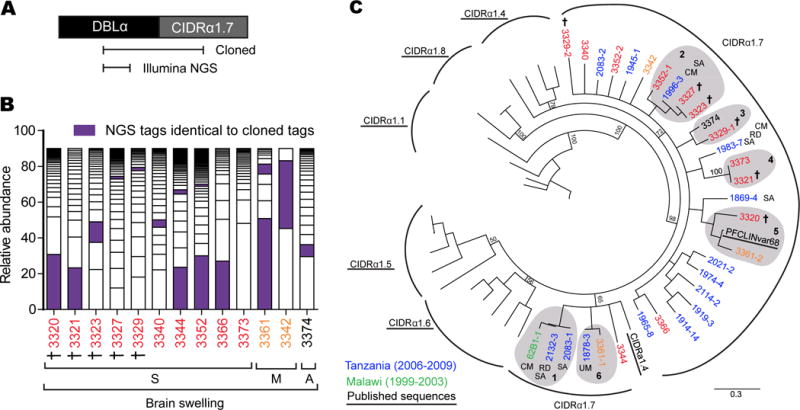
(A) Schematic illustrating the position of the cloned DBLα-CIDRα1.7 fragment and the DBLα tag covered by Illumina sequencing.
(B) Representation of the proportion of unique DBLα tags from Illumina sequencing that made up 90% of the var repertoire in each patient. DBLα tags that matched those from the cloned DBLα-CIDRα1.7 fragment are highlighted (purple). (See Figure S4; Tables S7–S9).
(C) A neighbor-joining tree of CIDRα1.7 sequences amplified from children with Ret+CM in Malawi compared to published sequences from Tanzania (Jespersen et al., 2016) and representative CIDRα1 sequences (Rask et al., 2010), which were included to provide outgroup comparisons. Clusters of identical or highly related sequences are highlighted in grey shading and labeled 1–6. Only patient 3373 did not have a matching DBLα tag-CIDRα1.7. †: Fatal cases, CM: Cerebral malaria, RD: Respiratory distress, SA: Severe anemia.
To investigate var genes associated with severe swelling, we amplified DBLα tags and performed Illumina sequencing from a subset of thirteen patients with high CIDRα1.7 transcript levels (Figure S4A–B). Using a 96% sequence identity cut-off, we found 338 unique tags among CM cases with a mean of 28 ± 14 DBLα tags per patient (range: 3 – 52 tags accounting for the top 90% of sequences) (Figure 5B and Table S8), similar to the combined number of DBLα tags amplified from brain, heart, and gut of the pediatric CM autopsy cohort (Tembo et al., 2014). None of the top 90% of DBLα tags from brain swelling cases in 2015–16 was identical to var62B1-1 or var28B1-1. Furthermore, only 8 tags were shared at 96% identity among the cohorts in 1999–2003 and 2015–16 (Table S9). This genetic diversity extended to the 2015–16 cohort, where there were only 15 tags (4%) shared between patients at the 96% identity cut-off (Table S9). To classify DBLα tags, we performed BLAST searches against a custom library of 521 PfEMP1 sequences from (Rask et al., 2010) and (Jespersen et al., 2016) (Figure S4C). Although this approach may misclassify some tags, the top 40–90% of DBLα tags in either patients were predicted to be from EPCR-binding variants (Figure S4D), highlighting the dominance of this binding phenotype in Ret+CM and brain swelling cases (Figure 3).
While amplification of DBLα tags provides an assessment of the expressed var repertoire in each patient, the tags do not include the downstream CIDR domain. To further investigate the relationship between CIDRα1.7 domains and swelling, we performed targeted cloning of cDNA fragments extending from the DBLα forward primer to the CIDRα1.7 domain (Figure 5A). As expected, all amplified sequences clustered with other annotated CIDRα1.7 sequences (Figure 5C). None of the 2015–16 CIDRα1.7 sequences were closely related to the 62B1-1 CM autopsy variant, but 62B1-1 was nearly identical to sequences from two severe anemia cases from Tanzania, neither of whom had CM (cluster 1, Figure 5C) (Jespersen et al., 2016). Many of the 2015–16 swelling cases were identical or highly related (clusters 2–6, Figure 5C). Strikingly, CIDRα1.7 sequences were prominent var tags in the majority of the severe swelling cases, and the dominant tag in six cases (Figure 5B), suggesting they were major parasite variants in the peripheral blood. Given that the CIDRα1.7 variant represents ~2.25% of the typical var repertoire (Rask et al., 2010), the probability that at least 6 of 13 CM cases would have CIDRα1.7 as the top variant by random chance is extremely low (binomial p: 1.93e-07). Even in patients with less dominant CIDRα1.7 tags (e.g. fatal case 3327) (Figure 5B), the amplified sequence was identical to other fatal or severe swelling cases (Figure 5C). Although CIDRα1.7 sequences from severe swelling and fatal cases were highly related over a short time period, there appears to be rapid turn-over in the parasite population, as the Tembo et al. sequences (1999–2003) were not found in 2015–16 in Malawi.
A brain autopsy CIDRα1.7 variant binds to EPCR and interferes with APC-EPCR function
We expressed CM brain variant 62B1-1 CIDRα1.7 domain as a recombinant protein to characterize its functional properties (Figure 6A). 62B1-1-CIDRα1.7 binds to EPCR with picomolar affinity (KD of 0.6 nM, kon of 6.18E+04 M−1s−1 and koff of 3.71E−05 s−1) comparable to other CIDRα1 domains (Figure 6B) (Lau et al., 2015). We observed dose-dependent and saturated binding of 62B1-1-CIDRα1.7 to CHO745-EPCR cells, but not to CHO745 cells or CHO745-CD36 cells (Figure S5A). IT4var31-CIDRα4, a control CD36-binding domain, bound to CHO745-CD36 cells, but not to CHO745-EPCR cells or CHO745 cells (Figure S5B).
Figure 6. Inhibition of the APC-EPCR interaction by a common brain-specific 62B1-1-CIDRα1.7 domain isolated from a fatal pediatric CM case.
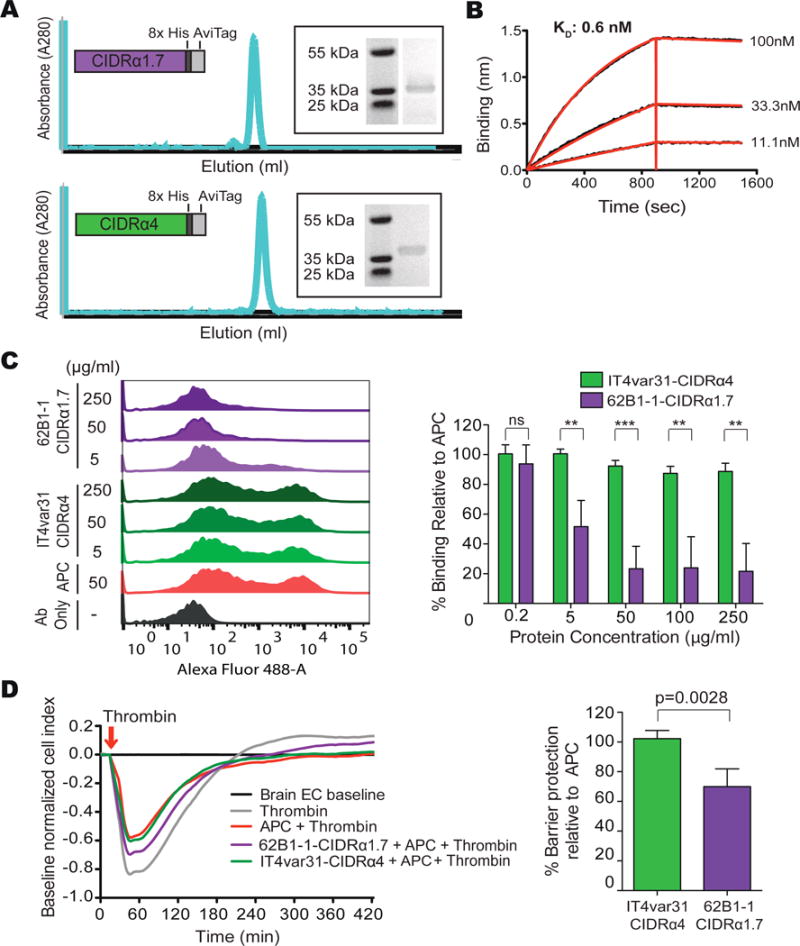
(A) Schematic of the expressed recombinant proteins, size exclusion chromatography trace and SDS-PAGE.
(B) 62B1-1-CIDRα1.7 binds to EPCR with picomolar affinity. A representative graph from n = 3 independent experiments.
(C) 62B1-1-CIDRα1.7 blocks binding of APC to EPCR. Left: representative flow cytometry histograms show binding of APC to CHO745-EPCR. Right: percentage of APC binding to CHO745-EPCR cells in the presence of varying concentrations of the CIDR domains, relative to APC binding alone. Mean ± SD of n = 3–5 independent experiments. Ab: Antibody. See Figure S5.
(D) 62B1-1-CIDRα1.7 partially blocks APC-mediated protection from thrombin-induced barrier disruption of primary human brain endothelial cells (HBMECs). Left: graph showing the cell index over time. Right: graph showing the level of APC protection from thrombin-induced barrier disruption in the presence of 62B1-1-CIDRα1.7 and IT4var31-CIDRα4, relative to APC alone. Mean ± SD of n = 4 independent experiments **p<0.01, ***p<0.001 by the Student’s unpaired, two-tailed t-test.
Whereas CIDRα1 domains block binding of APC to EPCR to varying extents (Bernabeu et al., 2016; Sampath et al., 2015), it is unknown whether CIDRα1 domains associated with severe brain swelling or fatal malaria cases have similar activity. 62B1-1-CIDRα1.7 inhibits binding of APC to EPCR in a dose-dependent fashion, while there was no inhibition by the CD36-binding IT4var31-CIDRα4 domain (Figure 6C). One physiologic function of the APC-EPCR interaction is to protect against disruption of the endothelial cell layer induced by the clotting factor thrombin. To determine if 62B1-1-CIDRα1.7 could interfere with this function, we tested the ability of APC to protect against thrombin-induced permeability of a primary human brain endothelial cell monolayer in the presence of 62B1-1-CIDRα1.7. Whereas APC diminished thrombin-induced barrier disruption by 30% and led a more rapid recovery to baseline values (Figure 6D), 62B1-1-CIDRα1.7 partially blocked the APC protective effect by ~30% and there was an increased lag of recovery (Figure 6D). Conversely, there was no inhibition by the CD36-binding IT4var31-CIDRα4 domain. Taken together, these results suggest that 62B1-1-CIDRα1.7 interferes with the EPCR-APC interaction, and suggest a detrimental role for EPCR binding domains in brain swelling.
Discussion
In this study, we assessed factors and mechanisms for CM pathogenesis and brain swelling by combining clinical measures of disease severity and molecular typing of var expression in peripheral blood from a stringently defined clinical cohort. Machine learning models highlight the importance of plasma Pfhrp2 levels, low platelet levels, and specific PfEMP1 variants in CM pathogenesis (summarized in Figure S6).
Pfhrp2, a surrogate of parasite biomass, was a top parasite factor in both a malarial retinopathy model (Ret+CM vs. UM) and a swelling model (CM with severe swelling vs. UM), consistent with previous findings showing that Pfhrp2 concentrations distinguish retinopathy-positive CM and predict progression to CM in Malawian children (Fox et al., 2013; Seydel et al., 2012). Collectively, these observations suggest that higher parasite biomass is a key factor in CM (Figure S6). Indeed, secreted Pfhrp2 can activate the inflammasome pathway in endothelial cells (Pal et al., 2016), and high parasite biomass may directly contribute to CM pathogenesis, as elevated in vitro concentrations of ruptured pRBCs induce endothelial barrier disruption (Gallego-Delgado et al., 2016; Gillrie et al., 2012).
Low circulating platelet levels emerge as another important factor associated with brain swelling. Although thrombocytopenia (<150,000 platelets per ȝl) is a common finding in severe P. falciparum malaria, it has not been consistently associated with an increased risk of death in children (Chimalizeni et al., 2010; Ladhani et al., 2002). Thrombocytopenia may be a biomarker reflecting widespread endothelial activation and/or dysfunction, which is implicated in disease severity. Additionally, platelet accumulation in cerebral microvasculature (Grau et al., 2003; Hochman et al., 2015; Milner et al., 2014) may potentiate endothelial damage caused by sequestered pRBCs and facilitate the recruitment of more pRBCs and monocytes through direct binding interactions and the release of microparticles (Faille et al., 2009; Pain et al., 2001). Pediatric CM is associated with alterations in coagulation, including increased plasma levels of fibrin degradation products and soluble thrombomodulin in Ret+CM and fatal cases (Moxon et al., 2015) and the loss of EPCR and fibrin deposits in cerebral microvasculature (Dorovini-Zis et al., 2011; Moxon et al., 2013). Taken together, these data argue that coagulation dysfunction plays a central role in pediatric CM and involves platelet recruitment/sequestration and disruption of the protein C pathway two — events that are likely interdependent.
Pro-coagulant and barrier disruptive pathways may converge on parasite cytoadhesion properties. Our data suggest that pediatric CM infections are accompanied by the expansion of parasites expressing EPCR-binding domains, as well as group A head structures with rosetting/unknown binding properties. A higher rosetting rate has been linked to disease severity (Carlson et al., 1990; Ghumra et al., 2012), but was not directly assessed here. Additionally, a C-terminal DBLε2 domain was a top var feature in the retinopathy and swelling models, but it was not elevated in all patients. Notably, the DBLε2 domain binds to human IgM (Jeppesen et al., 2015), an adhesion property that can strengthen pRBC binding affinity (Akhouri et al., 2016; Stevenson et al., 2015).
While EPCR-binding var transcripts have consistently been associated with a variety of severe malaria disease syndromes (Abdi et al., 2015; Bernabeu et al., 2016; Bertin et al., 2013; Jensen et al., 2004; Jespersen et al., 2016; Lavstsen et al., 2012; Rottmann et al., 2006; Turner et al., 2013; Warimwe et al., 2012), the relationship to brain swelling has not been studied. Our study extends previous work suggesting higher transcript levels of DC8 and group A EPCR-binding variants in severe pediatric malaria (Jespersen et al., 2016; Mkumbaye et al., 2017) and the presence of both parasite subsets in Ret+ and Ret−CM cases (Abdi et al., 2015). Strikingly, we found that a CIDRα1.7 sequence was the dominant var tag in 50% of swelling cases. Recent findings suggest that a subset of group A variants with dual EPCR and ICAM-1 binding properties (Avril et al., 2016; Bengtsson et al., 2013; Lennartz et al., 2017; Oleinikov et al., 2009) play a role in pediatric CM. We found that dual EPCR and ICAM-1 binding sequences are enriched in CM cases, but do not distinguish by retinopathy or swelling status. Overall, our study of children in 2015–16 implicates a role for both CIDRα1.7 (linked to ICAM-1 motifs) and DC8 (not linked to ICAM-1 motifs) in stringently characterized CM and brain swelling and extends prior studies that identified a common CIDRα1.7 and a DC8-like sequence from multiple brain autopsies performed from 1999–2003 (Tembo et al., 2014). Collectively, our data, spanning nearly two decades of CM patients in Malawi, argue for the contribution of both EPCR binding types in cerebral pathology.
We further functionally characterized a CIDRα1.7 domain that was specifically amplified from brain autopsy material (Tembo et al., 2014). The 62B1-1-CIDRα1.7 domain impaired the barrier-protective functions of the protein C-EPCR pathway in human brain endothelial cells and has picomolar affinity for EPCR compared to nanomolar affinity for APC binding to EPCR (Fukudome et al., 1996), suggesting that it may act as an effective competitor for binding. Coupled with loss of EPCR in the brains of children with CM (Moxon CA et al. 2013), the binding nature of 62B1-1-CIDRα1.7 and similar variants may be sufficiently damaging to compromise the BBB properties in pediatric CM.
There is limited understanding of the longevity of dangerous parasite binding variants in the parasite population. While the 62B1-1-CIDRα1.7 variant was common in the 1999–2003 autopsy series (Tembo et al., 2014), it was replaced by other CIDRα1.7 transcripts in severe swelling or fatal cases in 2015–16. This finding supports previous serological observations suggesting that there is periodic turnover of the immunodominant epitopes of PfEMP1 associated with severe malaria (Warimwe et al., 2016). The 62B1-1-CIDRα1.7 variant was nearly identical to var transcripts isolated from two patients with severe malaria anemia in Tanzania (Jespersen et al., 2016), reinforcing the possibility that an element of clinical protective immunity may be directed at cross-reactive epitopes (Lau et al., 2015), shared by parasites causing different malaria syndromes.
In conclusion, our findings highlight the interaction of high parasite biomass, low platelet levels, and EPCR-binding parasite variants in CM. We provide evidence that parasites expressing CIDRα1 domains have expanded in children with severe or fatal brain swelling and may impair EPCR function. These findings have important implications for vaccine design against severe malaria and strengthen the rationale of therapeutics that counteract endothelial barrier breakdown by restoring normal function of the APC-EPCR cytoprotective signaling pathway in children with CM.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Kami Kim (kami.kim@einstein.yu.edu; after November 1, 2017 kamikim@health.usf.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human subjects and enrollment criteria
During the 2015 and 2016 malaria seasons (January-June), 38 UM and 75 CM patients were recruited to the study at Queen Elizabeth Central Hospital (QECH) in Blantyre, Malawi. The parents and/or guardians of UM cases were approached for enrollment by a recruitment nurse in the Accident and Emergency (A&E) Department at QECH. Inclusion criteria included for UM cases were defined as 1–12 years old, (history of) fever, Blantyre coma score of 5, peripheral P. falciparum parasitemia, and no overt signs of compromised health, malnutrition, or progression to severe malaria. Approximately 25% of UM cases approached were not enrolled due to refused consent or progression to severe disease. The UM children enrolled had a median age of 5.5 years, and 57.9% were male (see Table 1 for details). All children admitted to the Pediatric Malaria Research Ward with WHO-defined cerebral malaria (P. falciparum parasitemia, Blantyre coma score fl2 that remains following treatment of hypoglycemia and/or seizures, and exclusion of other identifiable causes of coma) that were 6 months to 12 years old met inclusion criteria for the study. Consistent with previous years on the research ward, approximately 15% of the CM parents/guardians denied consent. For all cases, 5 ml blood was drawn into citrate anti-coagulant tubes at the time of study enrollment. All blood samples were collected by trained clinicians and processed immediately following blood draw by laboratory staff. At time of admission, all CM cases underwent (direct and indirect) dilated ophthalmological funduscopic examination. Children with retinal findings, including retinal whitening, hemorrhages, and/or abnormal vessels were classified as retinopathy-positive (Ret+CM) and children with normal ocular fundi as retinopathy-negative (Ret−CM). While WHO-CM clinical criteria are sensitive for true CM, addition of retinal exam improves specificity. Ret−CM are more likely to have parasitemia with a non-malaria etiology of coma, but the Ret−CM group is clinically heterogeneous, so some individuals may have CM. Altogether, children with Ret+CM had a median age of 4 years where 61.4% were male. Similarly, children enrolled with Ret−CM had a median age of 3.7 years, and the gender distribution was 50:50 (see table 1 for more details). CM cases that did not meet the predefined criteria for parasitized red blood cell sample analysis (see var expression by qRT PCR) were excluded from subsequent analyses. The study was approved by the institutional review boards at the University of Malawi College of Medicine, Michigan State University, and the Albert Einstein College of Medicine. Informed consent was obtained from the parent/guardian of all study participants prior to enrollment.
Sample size estimation
The final sample sizes of n=57 Ret+CM cases and n=38 UM cases achieved greater than 80% power to detect a difference of ≥0.52 SD in Pfhrp2 and platelets levels between the two groups with a significance level (alpha) of 0.05 using a one-sided two-sample t-test.
Cell lines
The primary human brain microvascular endothelial cells (ACBRI 376 HBMECs) were obtained from Cell Systems at passage 4. They were grown and used within 14 passages for permeability assays. CHO745-EPCR cells (Sampath et al., 2015) were previously described. Information regarding the sex of the cells used is unavailable for the primary brain endothelial cells and transfected CHO745 cells.
METHOD DETAILS
Plasma Pfhrp2 quantification
Plasma samples were analyzed for Pfhrp2 by enzyme-linked immunosorbent assay (ELISA) according to validated methods (Fox et al., 2013) by personnel blinded to clinical diagnosis, retinal findings, and brain swelling severity. Briefly, samples were diluted 1:100 or 1:500 in phosphate-buffered saline (PBS) and plated in duplicate onto plates coated with anti-hrp2 antibody (Cellabs, Brookvale, Australia). Recombinant hrp2 was plated at multiple dilutions to generate a standard curve for accurate quantification of Pfhrp2 levels in the experimental samples. Minor modifications were made to the manufacturer’s protocol, including all incubations carried out at 37°C in a humidified chamber. Upon optical density (OD) measurement, samples that did not lie in the linear range of the generated standard curve were re-diluted (1:10 or 1:1000) and re-analyzed.
MRI scans and brain swelling scores
Scans were performed onsite at QECH using a 0.35-T Signa Ovation Excite MRI scanner (General Electric). MRI results were interpreted independently by two radiologists blinded to the patient’s clinical and retinopathy status. Brain volume was scored on the appearance of the cerebral hemispheres using a scale of 1–8 (1 marked atrophy, 2 mild atrophy, 3 normal volume, 4 slight swelling, 5 mild swelling, 6 moderate selling, 7 substantial swelling with sulcal and cisternal effacement, and 8 substantial swelling with sulcal and cisternal effacement and herniation). Scores of 1–4 were considered absent of swelling, while scores of 5–6 and 7–8 were defined as mild/moderate swelling and severe swelling, respectively. MRI scan/swelling data was available for CM cases that were considered medically stable and did not wake up from malarial coma prior to scan administration.
var expression by qRT-PCR
Parasitized red blood cells were stored in TRIzol at −80° C. Upon thawing, RNA was extracted using a standard phenol-chloroform method. Genomic DNA was removed (TURBO DNA-free, Ambion), and cDNA was synthesized using random hexamers and SuperScript III reverse transcriptase (Invitrogen). Var transcript levels were determined by qRT-PCR using published amplification conditions (Lavstsen et al., 2012; Mkumbaye et al., 2017; Rottmann et al., 2006), SYBR green, and an AB7300 real-time system. Complete removal of DNA using the stated protocol was previously validated by comparing expression levels in conditional var knockdown parasites (DC-J; Deitsch) to 3D7 expression levels in vitro. Experimental var expression levels were determined by relative quantification of the average expression of two housekeeping genes: fructose bis-phosphate aldolase and seryl-tRNA synthetase (ΔCt = Ctvar – CtAvgControls). Only samples with a CtAvgControls ≤30 were included for analysis. Final expression levels are presented as Transcript units (Tu = 2(5−ΔCt)). All qRT-PCR data was analyzed blinded to retinopathy and brain swelling classification.
Library preparation for Illumina MiSeq
DBLα tags were PCR-amplified from cDNA from a selection of patients with severe, moderate, or no brain swelling. 20–40 cycle PCR amplification was done in either one step, using the previously described varF_dg2 and brlong2 primers (Lavstsen et al., 2012) ligated to 5′ and 3′ MiSeq adaptor sequences (5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGCAMGMAGTTTYGCNGATATWGG and 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGTCTTCDSYCCATTCVTCRAACCA) or in 2 steps with varF_dg2 and brlong2 primers first, followed by MiSeq adaptor-DBLα primers (sequences shown above). PCR amplification was carried out as follows using Phusion High-Fidelity DNA polymerase (New England Biolabs) and final primer concentrations of 0.5 μM: 98°C for 45 s, followed by 20–40 cycles of 98°C for 10 s, 50°C for 20 s, 68°C for 20 s and final extension at 72°C for 7 min. Amplicons were gel-purified and served as the template for second-round amplification where P5 and P7 index sequences were added in 10 cycles using Nextera primers (Illumina) and the aforementioned PCR conditions. Amplicons were purified using FlashGels (Lonza) and quantified using the KAPA library quantification kit (Kapa Biosystems) by qPCR performed on an Applied Biosystems 7500 Fast real-time PCR machine (Applied Biosystems). Libraries were combined at 500 pM and denatured using 0.2N NaOH according to the Illumina MiSeq protocol. The denatured library was diluted in HT1 hybridization buffer (Illumina) to 10 pM and a 12.5 pM PhiX Control library added to a final concentration of 40% to compensate for the AT nucleotide bias of P. falciparum. The library was loaded into a 600-cycle V3 cartridge (Illumina) for sequencing.
DBLα tag sequencing and data processing
The initial processing of FASTQ sequence data was performed as previously described (Vigdorovich et al., 2016) with some modifications and additional steps. Briefly, forward and reverse reads were assembled to reconstruct amplicon sequences using FLASH (Magoc and Salzberg, 2011). Reads that did not overlap were discarded. Using cutadapt (Martin, 2011), assembled sequences were trimmed at the 5′ and 3′ ends to remove primer and adaptor sequences. Any reads that lacked primer sequences were discarded. Sequences with low-confidence base calls (N’s) were also filtered out using FASTX-toolkit (http://hannonlab.cshl.edu/fastx_toolkit). Clustering of reads from each patient was done using USEARCH version v8.0.1623 (Edgar, 2010) (command line parameters: “-sizein -sizeout -id 0.96 -centroids filename.fasta -sort size –quiet”). Sequences were considered identical if they had ≥96% nucleotide sequence identity. Reads with <5 identical sequences were excluded from downstream analyses, as these likely resulted from amplification or sequencing errors. A consensus sequence for each cluster was generated from alignments made using centroid to group sequences. Each cluster contained sequences at a given identity cut-off (≥96% identity) and the cluster size represents the relative abundance of those sequence reads.
BLAST analysis of DBLα tags
BLAST searches of the DBLα tags from NGS were performed against a custom library of 521 mostly full-length and annotated var genes [361 PfEMP1 sequences from (Rask et al., 2010) and 160 sequences from (Jespersen et al., 2016)]. BLAST searches were performed using the BLAST function in Geneious version 8.1.8. Predictions of the PfEMP1 type associated with each DBLα tag were made based on the top 5 hits of each BLAST search. Predictions were considered high confidence when ≥4 of the top 5 hits were of the same type, reasonable confidence when 3 of the top 5 hits were of the same type and ambiguous when <3 were of the same type.
Cloning of DBLα-CIDRα1.7 fragments
We used a hemi-nested strategy to clone a 1.2 kb fragment extending from the DBLα-tag to the CIDRα1.7 domain in 13 patients with severe, moderate or no swelling. In the first step, a PCR product was amplified from parasite cDNA using primers varF_dg2 and CIDRα1.7 reverse primer 5′- CTTTTTGTTTAACCCATYTGTCAAAAC (Lavstsen et al. 2012) according to the following PCR conditions: 98°C for 5 min, followed by 20–40 cycles of 98°C for 30 s, 50°C for 30 s, 68°C for 3 min and final extension at 68°C for 7 min. Amplicons were gel-purified and used as the template for the second amplification step, which employed the varF_dg2 primer and the reverse sequence of the CIDRα1.7 forward primer 5′-TTATCTTTCCACGTTATAGTTTCCG. Resulting amplicons were gel-purified and cloned into the PCR-Blunt II-TOPO plasmid (Life Technologies). Bacterial clones were analyzed by sequencing.
Database accessibility of sequences
All cloned var and NGS sequences have been submitted to GenBank and the Sequence Reads Archive (SRA). GenBank and SRA accession numbers for DBLα tags are in Table S7. DBLα-CIDRα1.7 GenBank IDs: 3320: MF069418; 3321: MF069419; 3323: MF069420; 3327: MF069421; 3329-1: MF069422; 3329-2: MF069423; 3340: MF069424; 3342: MF069425; 3344: MF069426; 3352-1: MF069427; 3352-2: MF069428; 3361-1: MF069429; 3361-2: MF069430; 3366: MF069431; 3373: MF069432; 3374: MF069433.
Recombinant protein expression
Biotinylated recombinant EPCR was produced as previously described (Sampath et al., 2015). Briefly, the extracellular region of human EPCR was cloned into pCEP4 (Life Technologies) with a tPA leader sequence and a C-terminal His tag and AviTag™, and expressed in HEK293F cells using linear polyethylenimine (Polysciences) as a transfection reagent. EPCR recombinant protein was recovered from the culture supernatant by NiNTA column chromatography followed by size- exclusion chromatography. Biotinylation was achieved in vitro using recombinant BirA ligase. Biotinylated-EPCR was then purified by size-exclusion chromatography with final resuspension in PBS.
The 62B1-1-CIDRα1.7 domain from a fatal cerebral malaria case (accession number KC678109) and IT4var31-CIDRα4 were codon-optimized (CIDRα1.7 – GeneArt, CIDRα4 – IDT) and synthesized (gblocks; IDT), then cloned into pcDNA3_4 with a tPA leader sequence and a C-terminal 8x-His tag and AviTag™. Expression in HEK293F cells and purification by size-exclusion chromatography was done as described above for EPCR. Final resuspension of purified CIDRα1.7 and CIDRα4 was in 2 mM EDTA (in HBS).
CIDR-EPCR binding by biolayer interferometry
The kinetics of binding of CIDR domains to EPCR was determined on an Octet Qke instrument (ForteBio, Inc.) as previously described (Sampath et al., 2015). Briefly, 50 μg/ml of biotinylated EPCR was immobilized on streptavidin biosensors (ForteBio) for 3.3 min and then equilibrated in kinetics buffer (PBS-0.02% Tween, 0.05% sodium azide, 0.1 mg/ml BSA) for 60 s. The EPCR-coupled biosensors were then immersed in 250 μg/ml biotin to ensure that any free streptavidin sites were occupied, followed by washing in PBS and then immersion in CIDR domains. The association phase was monitored for 900 s and the dissociation phase, for 600 s in kinetics buffer. Using the data analysis software (ForteBio, Inc.), the data were double-reference subtracted and fitted to a 1:1 langmuir binding model to determine the mean kon, koff and apparent KD values. BLI experiments were performed three times.
Cell-based CIDR and APC-EPCR binding assay
CHO745 cell binding and APC competition assays were performed as previously described (Sampath et al., 2015) with slight modifications. Briefly, adherent cells were lifted with 8 mM EDTA (in PBS) and washed and resuspended in complete Hank’s buffered salt solution (HBSS with 3 mM CaCl2, 0.6 mM MgCl2, 1% BSA, pH 7.4). 1 × 105 cells were incubated with varying concentrations of CIDR domains (binding assay) or a mixture of CIDR domains and 50 μg/ml APC [(Sigma Aldrich) competition assay] for 30 mins at 4°C. Cells were washed twice. For the binding assay, CIDR binding was detected with an anti-His mAb (1:100, Life Technologies), followed by an anti-mouse Alexa Fluor 488 antibody. For the competition assay, APC binding was detected by a goat anti-APC mAb (1:100, Affinity Biologicals), followed by a rabbit anti-goat Alexa Fluor 488 F(ab’)2 probe (1:200, Life Technologies). Cells were then washed twice in complete HBSS and analyzed by flow cytometry (LSRII, Becton Dickinson). Data were analyzed using FlowJo version 10 (Tree Star). Binding of CIDR domains was determined, as well as binding of APC alone relative to APC binding in the presence of CIDR domains. CIDR binding titration experiments were performed three times in duplicate or triplicate, while APC-competition assays were performed three to five times in triplicate.
HBMEC barrier permeability assay
Barrier permeability assays were carried out using a real-time cell analyzer (xCELLigence system, ACEA Biosciences) as previously described (Bernabeu et al., 2016). Briefly, a 96-well plate fitted with gold microelectrodes (ACEA Biosciences) was seeded with ~10,000 human brain microvascular endothelial cells per well. Cell proliferation in EBM-2 media (Lonza) was monitored for 72 h under standard culture conditions in the xCELLigence system. On the day of the assay, 50 μg/ml recombinant CIDR domains were added to cells for 30 min, followed by 100 nM APC (Haematologic Technologies) for 2 h. Barrier integrity of the cell monolayer was perturbed by addition of 5 nM Thrombin (Sigma-Aldrich). Electrical impedance across the cell monolayer was monitored after thrombin challenge every minute for 120 min, then every 5 min for 245 min and then every 30 minutes thereafter. The cell index (a measure of cell impedance) of treated cells was normalized to untreated cells at the time point immediately preceding thrombin challenge. The protection afforded by APC against thrombin-induced barrier disruption was set to 100% and compared to the level of protection of APC in the presence of CIDR domains. Barrier permeability assays were done four times in triplicate.
QUANTIFICATION AND STATISTICAL ANALYSIS
Univariate analysis
Sample size estimation was estimated using PASS. Univariate analyses were performed using Stata 12.1 or Prism 7.0 for Macintosh. The distribution of the data was examined using graphical approaches. The normality assumption was tested using the Shapiro-Wilk test. For continuous variables, statistical differences were determined by Wilcoxon rank-sum, Kruskal-Wallis, or Spearman correlation. The data is reported as median [IQR] or rho. Differences in categorical variables were tested using chi-squared test, and the data is represented as n (% of group). For discrete variables (Figure 6), statistically significant differences were determined by the Student’s unpaired, two-tailed t-test (**p<0.01, ***p<0.001), where n represents independent experiments (details can be found in the legend of Figure 6). Other graphically depicted data represents ‘n’ individual pediatric malaria cases (e.g. Figures 1–3 and S2). All univariate statistics were conducted using an α level <0.05, and adjustments for multiple comparisons were performed using Simes Method or Dunn’s correction.
Machine learning models
To further evaluate the parasite factors responsible for the development of Ret+CM and/or brain swelling and identify var domains that are contributing to disease pathology independently of or in addition to increasing sequestered parasite biomass, we utilized machine learning models as previously described (Bernabeu et al., 2016). Random Forest (RF) machine learning classifiers were developed to identify primers that could make accurate binary classifications of patients in different comparisons: Ret+CM (n=49) vs. UM (n=38); Ret+CM vs. Ret−CM (n=18); CM with Severe Swelling (S, n=22) vs. UM; and CM with Severe Swelling (S) vs. CM with Absent Swelling (A, n=18). Classifiers were fit to primer Tu values, along with Pfhrp2, platelet levels and white blood cell counts using the R randomForest package (Liaw and Wiener, 2002), with 1,000,000 trees sampled to train each model. Lactate levels were also included as a potential feature for the S vs. A comparison.
To ensure that each var primer did not merely represent a surrogate of Pfhrp2 levels, a cumulative mutual information (CMI)-based filtering approach was applied to select primers. Prior to model training, the CMI of var primer expression values related to the patient outcome group (UM/Ret+CM/Ret−CM/A/S) given Pfhrp2 measurements was calculated using the condinformation() function from the R infotheo package (Meyer, 2014). Pfhrp2 and var primer expression were quantized using log(n samples) bins for the CMI calculation. CMI significance was calculated by randomly resampling individual expression values from the var expression matrix to create synthetic primers which were CMI evaluated as above. Only var primers showing higher CMI than 80% of the synthetic primers were included in the RF classifiers.
The resulting RF models were used to estimate the mean decrease in classifier accuracy (MDCA) when a particular parasite factor, such as a single var domain, is removed from the model. To control for multiple comparisons in the RF analysis, a FWER was estimated for each feature using the mProbes algorithm (Huynh-Thu et al., 2012), which compares selected primers to new synthetic features generated by randomly shuffling model var primers. Receiver operating characteristic (ROC) analysis was used to assess the predictive ability of the RF classifiers using the R pROC package (Robin et al., 2011). ROC curves were constructed based on consensus out-of-bag sample predictions generated during RF training.
To help elucidate the complex and redundant nature of our var targets, we applied a set enrichment analysis to determine the contribution of related var ‘sets’. Sets were developed that combine individual domains/targets with related structures or in related functional categories (e.g. domain cassettes; group A/B/C; binding phenotypes; adhesion traits) (Table S6). To determine set enrichment, primers were ranked in order of MDCA values from the RF analysis, and a Wilcoxon rank-sum was calculated to test for a significant increase in rank for each defined set compared with all other var targets not included in the set.
DATA AND SOFTWARE AVAILABILITY
Deposited sequences
The PfEMP1 CIDRα1.7 partial sequences and the DBLα tag assembled sequences were deposited in GenBank under accession ID codes MF069418-MF069433 and MF100158-MF100510, respectively (https://www.ncbi.nlm.nih.gov/nuccore/?term=Linking EPCR-binding PfEMP1 to brain swelling in pediatric cerebral malaria). The DBLα tag raw data was deposited in the SRA under study ID code SRP106913 (https://www.ncbi.nlm.nih.gov/sra/?term=SRP106913).
Supplementary Material
Table S7. Results of DBLα tag BLAST searches with SRA and GenBank Accession Numbers, Related to Figure 5
Highlights.
Stringently defined cerebral malaria cases profiled for PfEMP1/var expression
Low platelet count linked to malarial retinopathy and brain swelling
EPCR-binding PfEMP1 enriched in malarial retinopathy and brain swelling
Parasite PfEMP1 CIDRα1.7 domain from brain autopsies counteracts EPCR protection
Acknowledgments
We thank the patients and their families that made this study possible. We thank Patricia Phula and the Blantyre Malaria Project (BMP) laboratory staff for help with sample collection and processing, Alice Liomba (BMP) for organizing data, Jack Gormley for the 2016 ophthalmoscopy, Matt Levy for sharing equipment, and Adriana Lippy for illustration. The study was made possible by NIH grants K08MH089848 and T32 AI 070117 (SH), R01AI34969 (TT), R01AI114766 and R01HL130488 (JS), P41GM109824 (JA), NIH National Center for Advancing Translational Science (NCATS) Einstein-Montefiore CTSA TL1TR001072 (AK); the Burroughs Wellcome Fund (AK), grant 1061993 from the National Health and Medical Research Council of Australia (SR), and support from the Michigan State University College of Osteopathic Medicine (TT). Data in this paper are from a thesis submitted in partial fulfillment of the requirements for the Degree of Doctor of Philosophy in the Graduate Division of Medical Sciences at Albert Einstein College of Medicine – Yeshiva University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Conceptualization, AK, SD, MB, JS, KS, and KK; Machine learning analysis, FD and SAD; Formal analysis, AK, SD, and MB; MRI interpretation, SK, MP; Retinopathy evaluation, IM; Investigation, AK, SD, MB, VH, and ND. Oversight of severe malaria and mild malaria cohorts, TT, WM, SR, and KS; Next-generation sequence analysis, SD, VV, and BO. Recombinant protein production, SD and ND; Binding and barrier assays, SD; Writing – Original Draft, AK, SD, and MB; Writing – Review & Editing, AK, SD, MB, JS, and KK with input from all authors; Supervision, DS, JA, WBM, KS, JS, and KK.
The authors report no conflicts of interest.
References
- Abdi AI, Kariuki SM, Muthui MK, Kivisi CA, Fegan G, Gitau E, Newton CR, Bull PC. Differential Plasmodium falciparum surface antigen expression among children with Malarial Retinopathy. Sci Rep. 2015;5:18034. doi: 10.1038/srep18034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhouri RR, Goel S, Furusho H, Skoglund U, Wahlgren M. Architecture of Human IgM in Complex with P. falciparum Erythrocyte Membrane Protein 1. Cell Rep. 2016;14:723–736. doi: 10.1016/j.celrep.2015.12.067. [DOI] [PubMed] [Google Scholar]
- Avril M, Bernabeu M, Benjamin M, Brazier AJ, Smith JD. Interaction between Endothelial Protein C Receptor and Intercellular Adhesion Molecule 1 to Mediate Binding of Plasmodium falciparum- Infected Erythrocytes to Endothelial Cells. MBio. 2016;7 doi: 10.1128/mBio.00615-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson A, Joergensen L, Rask TS, Olsen RW, Andersen MA, Turner L, Theander TG, Hviid L, Higgins MK, Craig A, et al. A novel domain cassette identifies Plasmodium falciparum PfEMP1 proteins binding ICAM-1 and is a target of cross-reactive, adhesion-inhibitory antibodies. J Immunol. 2013;190:240–249. doi: 10.4049/jimmunol.1202578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SS, Turner L, Wang CW, Petersen JE, Kraft M, Lusingu JP, Mmbando B, Marquard AM, Bengtsson DB, Hviid L, et al. Plasmodium falciparum expressing domain cassette 5 type PfEMP1 (DC5-PfEMP1) bind PECAM1. PloS one. 2013;8:e69117. doi: 10.1371/journal.pone.0069117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernabeu M, Danziger SA, Avril M, Vaz M, Babar PH, Brazier AJ, Herricks T, Maki JN, Pereira L, Mascarenhas A, et al. Severe adult malaria is associated with specific PfEMP1 adhesion types and high parasite biomass. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E3270–3279. doi: 10.1073/pnas.1524294113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin GI, Lavstsen T, Guillonneau F, Doritchamou J, Wang CW, Jespersen JS, Ezimegnon S, Fievet N, Alao MJ, Lalya F, et al. Expression of the domain cassette 8 Plasmodium falciparum erythrocyte membrane protein 1 is associated with cerebral malaria in Benin. PloS one. 2013;8:e68368. doi: 10.1371/journal.pone.0068368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birbeck GL, Molyneux ME, Kaplan PW, Seydel KB, Chimalizeni YF, Kawaza K, Taylor TE. Blantyre Malaria Project Epilepsy Study (BMPES) of neurological outcomes in retinopathy-positive paediatric cerebral malaria survivors: a prospective cohort study. Lancet Neurol. 2010;9:1173–1181. doi: 10.1016/S1474-4422(10)70270-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown H, Rogerson S, Taylor T, Tembo M, Mwenechanya J, Molyneux M, Turner G. Blood-brain barrier function in cerebral malaria in Malawian children. Am J Trop Med Hyg. 2001;64:207–213. doi: 10.4269/ajtmh.2001.64.207. [DOI] [PubMed] [Google Scholar]
- Carlson J, Helmby H, Hill AV, Brewster D, Greenwood BM, Wahlgren M. Human cerebral malaria: association with erythrocyte rosetting and lack of anti-rosetting antibodies. Lancet. 1990;336:1457–1460. doi: 10.1016/0140-6736(90)93174-n. [DOI] [PubMed] [Google Scholar]
- Chimalizeni Y, Kawaza K, Taylor T, Molyneux M. The platelet count in cerebral malaria, is it useful to the clinician? Am J Trop Med Hyg. 2010;83:48–50. doi: 10.4269/ajtmh.2010.09-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondorp AM, Desakorn V, Pongtavornpinyo W, Sahassananda D, Silamut K, Chotivanich K, Newton PN, Pitisuttithum P, Smithyman AM, White NJ, et al. Estimation of the total parasite biomass in acute falciparum malaria from plasma PfHRP2. PLoS Med. 2005;2:e204. doi: 10.1371/journal.pmed.0020204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondorp AM, Fanello CI, Hendriksen IC, Gomes E, Seni A, Chhaganlal KD, Bojang K, Olaosebikan R, Anunobi N, Maitland K, et al. Artesunate versus quinine in the treatment of severe falciparum malaria in African children (AQUAMAT): an open-label, randomised trial. Lancet. 2010;376:1647–1657. doi: 10.1016/S0140-6736(10)61924-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorovini-Zis K, Schmidt K, Huynh H, Fu W, Whitten RO, Milner D, Kamiza S, Molyneux M, Taylor TE. The neuropathology of fatal cerebral malaria in malawian children. Am J Pathol. 2011;178:2146–2158. doi: 10.1016/j.ajpath.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Faille D, Combes V, Mitchell AJ, Fontaine A, Juhan-Vague I, Alessi MC, Chimini G, Fusai T, Grau GE. Platelet microparticles: a new player in malaria parasite cytoadherence to human brain endothelium. FASEB J. 2009;23:3449–3458. doi: 10.1096/fj.09-135822. [DOI] [PubMed] [Google Scholar]
- Finney OC, Danziger SA, Molina DM, Vignali M, Takagi A, Ji M, Stanisic DI, Siba PM, Liang X, Aitchison JD, et al. Predicting antidisease immunity using proteome arrays and sera from children naturally exposed to malaria. Mol Cell Proteomics. 2014;13:2646–2660. doi: 10.1074/mcp.M113.036632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox LL, Taylor TE, Pensulo P, Liomba A, Mpakiza A, Varela A, Glover SJ, Reeves MJ, Seydel KB. Histidine-rich protein 2 plasma levels predict progression to cerebral malaria in Malawian children with Plasmodium falciparum infection. The Journal of infectious diseases. 2013;208:500–503. doi: 10.1093/infdis/jit176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukudome K, Kurosawa S, Stearns-Kurosawa DJ, He X, Rezaie AR, Esmon CT. The endothelial cell protein C receptor. Cell surface expression and direct ligand binding by the soluble receptor. J Biol Chem. 1996;271:17491–17498. doi: 10.1074/jbc.271.29.17491. [DOI] [PubMed] [Google Scholar]
- Gallego-Delgado J, Basu-Roy U, Ty M, Alique M, Fernandez-Arias C, Movila A, Gomes P, Weinstock A, Xu W, Edagha I, et al. Angiotensin receptors and beta-catenin regulate brain endothelial integrity in malaria. J Clin Invest. 2016;126:4016–4029. doi: 10.1172/JCI87306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghumra A, Semblat JP, Ataide R, Kifude C, Adams Y, Claessens A, Anong DN, Bull PC, Fennell C, Arman M, et al. Induction of strain-transcending antibodies against Group A PfEMP1 surface antigens from virulent malaria parasites. PLoS Pathog. 2012;8:e1002665. doi: 10.1371/journal.ppat.1002665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillrie MR, Avril M, Brazier AJ, Davis SP, Stins MF, Smith JD, Ho M. Diverse functional outcomes of Plasmodium falciparum ligation of EPCR: potential implications for malarial pathogenesis. Cellular microbiology. 2015;17:1883–1899. doi: 10.1111/cmi.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillrie MR, Lee K, Gowda DC, Davis SP, Monestier M, Cui L, Hien TT, Day NP, Ho M. Plasmodium falciparum histones induce endothelial proinflammatory response and barrier dysfunction. Am J Pathol. 2012;180:1028–1039. doi: 10.1016/j.ajpath.2011.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau GE, Mackenzie CD, Carr RA, Redard M, Pizzolato G, Allasia C, Cataldo C, Taylor TE, Molyneux ME. Platelet accumulation in brain microvessels in fatal pediatric cerebral malaria. The Journal of infectious diseases. 2003;187:461–466. doi: 10.1086/367960. [DOI] [PubMed] [Google Scholar]
- Hochman SE, Madaline TF, Wassmer SC, Mbale E, Choi N, Seydel KB, Whitten RO, Varughese J, Grau GE, Kamiza S, et al. Fatal Pediatric Cerebral Malaria Is Associated with Intravascular Monocytes and Platelets That Are Increased with HIV Coinfection. MBio. 2015;6:e01390–01315. doi: 10.1128/mBio.01390-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh FL, Turner L, Bolla JR, Robinson CV, Lavstsen T, Higgins MK. The structural basis for CD36 binding by the malaria parasite. Nat Commun. 2016;7:12837. doi: 10.1038/ncomms12837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh-Thu VA, Saeys Y, Wehenkel L, Geurts P. Statistical interpretation of machine learning-based feature importance scores for biomarker discovery. Bioinformatics. 2012;28:1766–1774. doi: 10.1093/bioinformatics/bts238. [DOI] [PubMed] [Google Scholar]
- Jensen AT, Magistrado P, Sharp S, Joergensen L, Lavstsen T, Chiucchiuini A, Salanti A, Vestergaard LS, Lusingu JP, Hermsen R, et al. Plasmodium falciparum associated with severe childhood malaria preferentially expresses PfEMP1 encoded by group A var genes. J Exp Med. 2004;199:1179–1190. doi: 10.1084/jem.20040274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen A, Ditlev SB, Soroka V, Stevenson L, Turner L, Dzikowski R, Hviid L, Barfod L. Multiple Plasmodium falciparum Erythrocyte Membrane Protein 1 Variants per Genome Can Bind IgM via Its Fc Fragment Fcmu. Infection and immunity. 2015;83:3972–3981. doi: 10.1128/IAI.00337-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespersen JS, Wang CW, Mkumbaye SI, Minja DT, Petersen B, Turner L, Petersen JE, Lusingu JP, Theander TG, Lavstsen T. Plasmodium falciparum var genes expressed in children with severe malaria encode CIDRalpha1 domains. EMBO Mol Med. 2016;8:839–850. doi: 10.15252/emmm.201606188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladhani S, Lowe B, Cole AO, Kowuondo K, Newton CR. Changes in white blood cells and platelets in children with falciparum malaria: relationship to disease outcome. British journal of haematology. 2002;119:839–847. doi: 10.1046/j.1365-2141.2002.03904.x. [DOI] [PubMed] [Google Scholar]
- Lau CK, Turner L, Jespersen JS, Lowe ED, Petersen B, Wang CW, Petersen JE, Lusingu J, Theander TG, Lavstsen T, et al. Structural conservation despite huge sequence diversity allows EPCR binding by the PfEMP1 family implicated in severe childhood malaria. Cell Host Microbe. 2015;17:118–129. doi: 10.1016/j.chom.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavstsen T, Salanti A, Jensen AT, Arnot DE, Theander TG. Sub-grouping of Plasmodium falciparum 3D7 var genes based on sequence analysis of coding and non-coding regions. Malaria journal. 2003;2:27. doi: 10.1186/1475-2875-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavstsen T, Turner L, Saguti F, Magistrado P, Rask TS, Jespersen JS, Wang CW, Berger SS, Baraka V, Marquard AM, et al. Plasmodium falciparum erythrocyte membrane protein 1 domain cassettes 8 and 13 are associated with severe malaria in children. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E1791–1800. doi: 10.1073/pnas.1120455109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennartz F, Adams Y, Bengtsson A, Olsen RW, Turner L, Ndam NT, Ecklu-Mensah G, Moussiliou A, Ofori MF, Gamain B, et al. Structure-Guided Identification of a Family of Dual Receptor- Binding PfEMP1 that Is Associated with Cerebral Malaria. Cell Host Microbe. 2017;21:403–414. doi: 10.1016/j.chom.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewallen S, Bakker H, Taylor TE, Wills BA, Courtright P, Molyneux ME. Retinal findings predictive of outcome in cerebral malaria. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1996;90:144–146. doi: 10.1016/s0035-9203(96)90116-9. [DOI] [PubMed] [Google Scholar]
- Liaw A, Wiener M. Classification and regression by randomForest. R News. 2002;2:18–22. [Google Scholar]
- Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 2011 [Google Scholar]
- Meyer EP. infotheo: Information-Theoretic Measures 2014 [Google Scholar]
- Milner DA, Jr, Whitten RO, Kamiza S, Carr R, Liomba G, Dzamalala C, Seydel KB, Molyneux ME, Taylor TE. The systemic pathology of cerebral malaria in African children. Front Cell Infect Microbiol. 2014;4:104. doi: 10.3389/fcimb.2014.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mkumbaye SI, Wang CW, Lyimo E, Jespersen JS, Manjurano A, Mosha J, Kavishe RA, Mwakalinga SB, Minja DT, Lusingu JP, et al. The Severity of Plasmodium falciparum Infection Is Associated with Transcript Levels of var Genes Encoding Endothelial Protein C Receptor-Binding P. falciparum Erythrocyte Membrane Protein 1. Infection and immunity. 2017;85 doi: 10.1128/IAI.00841-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosnier LO, Zlokovic BV, Griffin JH. The cytoprotective protein C pathway. Blood. 2007;109:3161–3172. doi: 10.1182/blood-2006-09-003004. [DOI] [PubMed] [Google Scholar]
- Moxon CA, Chisala NV, Mzikamanda R, MacCormick I, Harding S, Downey C, Molyneux M, Seydel KB, Taylor TE, Heyderman RS, et al. Laboratory evidence of disseminated intravascular coagulation is associated with a fatal outcome in children with cerebral malaria despite an absence of clinically evident thrombosis or bleeding. J Thromb Haemost. 2015;13:1653–1664. doi: 10.1111/jth.13060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxon CA, Wassmer SC, Milner DA, Jr, Chisala NV, Taylor TE, Seydel KB, Molyneux ME, Faragher B, Esmon CT, Downey C, et al. Loss of endothelial protein C receptors links coagulation and inflammation to parasite sequestration in cerebral malaria in African children. Blood. 2013;122:842–851. doi: 10.1182/blood-2013-03-490219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleinikov AV, Amos E, Frye IT, Rossnagle E, Mutabingwa TK, Fried M, Duffy PE. High throughput functional assays of the variant antigen PfEMP1 reveal a single domain in the 3D7 Plasmodium falciparum genome that binds ICAM1 with high affinity and is targeted by naturally acquired neutralizing antibodies. PLoS pathogens. 2009;5:e1000386. doi: 10.1371/journal.ppat.1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pain A, Ferguson DJ, Kai O, Urban BC, Lowe B, Marsh K, Roberts DJ. Platelet- mediated clumping of Plasmodium falciparum-infected erythrocytes is a common adhesive phenotype and is associated with severe malaria. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:1805–1810. doi: 10.1073/pnas.98.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal P, Daniels BP, Oskman A, Diamond MS, Klein RS, Goldberg DE. Plasmodium falciparum Histidine-Rich Protein II Compromises Brain Endothelial Barriers and May Promote Cerebral Malaria Pathogenesis. MBio. 2016;7 doi: 10.1128/mBio.00617-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen JE, Bouwens EA, Tamayo I, Turner L, Wang CW, Stins M, Theander TG, Hermida J, Mosnier LO, Lavstsen T. Protein C system defects inflicted by the malaria parasite protein PfEMP1 can be overcome by a soluble EPCR variant. Thromb Haemost. 2015;114:1038–1048. doi: 10.1160/TH15-01-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen JE, Mkumbaye SI, Vaaben AV, Manjurano A, Lyimo E, Kavishe RA, Mwakalinga SB, Mosha J, Minja DT, Lusingu JP, et al. Plasma Ang2 and ADAM17 levels are elevated during clinical malaria; Ang2 level correlates with severity and expression of EPCR-binding PfEMP1. Sci Rep. 2016;6:35950. doi: 10.1038/srep35950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rask TS, Hansen DA, Theander TG, Gorm Pedersen A, Lavstsen T. Plasmodium falciparum erythrocyte membrane protein 1 diversity in seven genomes–divide and conquer. PLoS Comput Biol. 2010;6 doi: 10.1371/journal.pcbi.1000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, Muller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson BA, Welch TL, Smith JD. Widespread functional specialization of Plasmodium falciparum erythrocyte membrane protein 1 family members to bind CD36 analysed across a parasite genome. Molecular microbiology. 2003;47:1265–1278. doi: 10.1046/j.1365-2958.2003.03378.x. [DOI] [PubMed] [Google Scholar]
- Rottmann M, Lavstsen T, Mugasa JP, Kaestli M, Jensen AT, Muller D, Theander T, Beck HP. Differential expression of var gene groups is associated with morbidity caused by Plasmodium falciparum infection in Tanzanian children. Infection and immunity. 2006;74:3904–3911. doi: 10.1128/IAI.02073-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath S, Brazier AJ, Avril M, Bernabeu M, Vigdorovich V, Mascarenhas A, Gomes E, Sather DN, Esmon CT, Smith JD. Plasmodium falciparum adhesion domains linked to severe malaria differ in blockade of endothelial protein C receptor. Cellular microbiology. 2015;17:1868–1882. doi: 10.1111/cmi.12478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydel KB, Fox LL, Glover SJ, Reeves MJ, Pensulo P, Muiruri A, Mpakiza A, Molyneux ME, Taylor TE. Plasma concentrations of parasite histidine-rich protein 2 distinguish between retinopathy-positive and retinopathy-negative cerebral malaria in Malawian children. The Journal of infectious diseases. 2012;206:309–318. doi: 10.1093/infdis/jis371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydel KB, Kampondeni SD, Valim C, Potchen MJ, Milner DA, Muwalo FW, Birbeck GL, Bradley WG, Fox LL, Glover SJ, et al. Brain swelling and death in children with cerebral malaria. N Engl J Med. 2015;372:1126–1137. doi: 10.1056/NEJMoa1400116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JD, Subramanian G, Gamain B, Baruch DI, Miller LH. Classification of adhesive domains in the Plasmodium falciparum erythrocyte membrane protein 1 family. Molecular and biochemical parasitology. 2000;110:293–310. doi: 10.1016/s0166-6851(00)00279-6. [DOI] [PubMed] [Google Scholar]
- Stevenson L, Huda P, Jeppesen A, Laursen E, Rowe JA, Craig A, Streicher W, Barfod L, Hviid L. Investigating the function of Fc-specific binding of IgM to Plasmodium falciparum erythrocyte membrane protein 1 mediating erythrocyte rosetting. Cellular microbiology. 2015;17:819–831. doi: 10.1111/cmi.12403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor TE, Fu WJ, Carr RA, Whitten RO, Mueller JS, Fosiko NG, Lewallen S, Liomba NG, Molyneux ME. Differentiating the pathologies of cerebral malaria by postmortem parasite counts. Nat Med. 2004;10:143–145. doi: 10.1038/nm986. [DOI] [PubMed] [Google Scholar]
- Tembo DL, Nyoni B, Murikoli RV, Mukaka M, Milner DA, Berriman M, Rogerson SJ, Taylor TE, Molyneux ME, Mandala WL, et al. Differential PfEMP1 Expression Is Associated with Cerebral Malaria Pathology. PLoS pathogens. 2014;10:e1004537. doi: 10.1371/journal.ppat.1004537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner L, Lavstsen T, Berger SS, Wang CW, Petersen JE, Avril M, Brazier AJ, Freeth J, Jespersen JS, Nielsen MA, et al. Severe malaria is associated with parasite binding to endothelial protein C receptor. Nature. 2013;498:502–505. doi: 10.1038/nature12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigdorovich V, Oliver BG, Carbonetti S, Dambrauskas N, Lange MD, Yacoob C, Leahy W, Callahan J, Stamatatos L, Sather DN. Repertoire comparison of the B-cell receptor-encoding loci in humans and rhesus macaques by next-generation sequencing. Clin Transl Immunology. 2016;5:e93. doi: 10.1038/cti.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warimwe GM, Abdi AI, Muthui M, Fegan G, Musyoki JN, Marsh K, Bull PC. Serological Conservation of Parasite-Infected Erythrocytes Predicts Plasmodium falciparum Erythrocyte Membrane Protein 1 Gene Expression but Not Severity of Childhood Malaria. Infection and immunity. 2016;84:1331–1335. doi: 10.1128/IAI.00772-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warimwe GM, Fegan G, Musyoki JN, Newton CR, Opiyo M, Githinji G, Andisi C, Menza F, Kitsao B, Marsh K, et al. Prognostic indicators of life-threatening malaria are associated with distinct parasite variant antigen profiles. Sci Transl Med. 2012;4:129ra145. doi: 10.1126/scitranslmed.3003247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyner AD. A definition of conditional mutual information for arbitrary ensembles. Inf Control. 1978;38:51–59. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S7. Results of DBLα tag BLAST searches with SRA and GenBank Accession Numbers, Related to Figure 5


