Abstract
Recent advances in human neuroimaging research have revealed that white‐matter connectivity can be described in terms of an integrated network, which is the basis of the human connectome. However, the developmental changes of this connectome in childhood are not well understood. This study made use of two independent longitudinal diffusion‐weighted imaging data sets to characterize developmental changes in the connectome by estimating age‐related changes in fractional anisotropy (FA) for reconstructed fibers (edges) between 68 cortical regions. The first sample included 237 diffusion‐weighted scans of 146 typically developing children (4–13 years old, 74 females) derived from the Pediatric Longitudinal Imaging, Neurocognition, and Genetics (PLING) study. The second sample included 141 scans of 97 individuals (8–13 years old, 62 females) derived from the BrainTime project. In both data sets, we compared edges that had the most substantial age‐related change in FA to edges that showed little change in FA. This allowed us to investigate if developmental changes in white matter reorganize network topology. We observed substantial increases in edges connecting peripheral and a set of highly connected hub regions, referred to as the rich club. Together with the observed topological differences between regions connecting to edges showing the smallest and largest changes in FA, this indicates that changes in white matter affect network organization, such that highly connected regions become even more strongly imbedded in the network. These findings suggest that an important process in brain development involves organizing patterns of inter‐regional interactions. Hum Brain Mapp 39:157–170, 2018. © 2017 Wiley Periodicals, Inc.
Keywords: brain development, DWI, graph theory, MRI, brain network
INTRODUCTION
In recent years, researchers have unraveled the macroscale network of projections that form the basis for interactions between disparate brain regions, also referred to as the human connectome [Sporns, 2011]. Neuroimaging techniques, including diffusion weighted imaging (DWI), have permitted researchers to map bundles of axons, which have shown to be relatively compatible with major fiber tracts. Herewith, the macroscale layout of the human connectome can be estimated in vivo, which has resulted in the characterization of individual anatomical connectivity with predictive power for cognitive abilities [Kim et al., 2016; Koenis et al., 2015; Li et al., 2009; Sporns, 2011] and mental health outcomes [Kaufmann et al, 2017; Collin et al., 2015]. In the last years, there have been strong advances in studying the adult human connectome, but less is known about how this connectome changes during childhood, even though it is well known that this is a time period of substantial changes in brain development and white‐matter volume [Brouwer et al., 2012; Giedd et al., 2015; Hagmann et al., 2010]. During primary school age years, children show large changes in cognitive abilities, and researchers have speculated that these arise from the interaction of disparate brain regions [Johnson, 2011].
Prior research showed that childhood is characterized by widespread changes in FA and white matter volume [Brouwer et al., 2012; Giedd et al., 2015; Hagmann et al., 2010; Muetzel et al. 2015]. In addition, different maturation rates across various brain regions have been observed [Lebel et al., 2008; Tamnes et al., 2010; Brouwer et al., 2012; Simmonds et al., 2014; Krogsrud et al., 2016; Pohl et al., 2016]. However, it is currently not well understood if these white matter changes in childhood reflect substantial reorganization of network topology, or whether they merely contribute to a global increase in network efficiency.
Grey matter development studies showed specific regional change during child development [Gogtay et al., 2004; Giedd et al., 1999; Sowell et al., 2001; Wierenga et al., 2014]. Functional neuroimaging studies in primary school children suggest that refinement of the brain network is driven by a dual process of integration (increased neural synchrony as modeled by increased functional correlation strength) and segregation (decreased functional correlation strength) [Brown et al., 2005 ; Fair et al., 2009; Schlaggar et al., 2002 ; Supekar et al., 2009]. These functional connectivity studies measure activity in the cortex and assume its connections based on correlated activity. In contrast, DWI assesses tissue properties of the white‐matter connections directly, which may add important information on developmental changes in the brain network. The present study was designed to test these issues by examining developmental changes in the organization of DWI derived network reconstructions.
Previous studies applied graph theoretical analyses to study developmental changes in network topology [van den Heuvel et al., 2015; Wierenga et al., 2015; Dennis et al., 2013]. Initial evidence that there may be systematic regional developmental changes in the human connectome comes from studies focusing on adolescents. In adolescence, white‐matter connections showed substantial changes among a set of densely connected hub regions [Baker et al., 2015; Kaufmann, 2017], which have been called the “rich club” [van den Heuvel & Sporns, 2011]. This study extends these initial studies by testing human connectome development in childhood, which is the time period presumably characterized by changes in interactive specialization [Johnson, 2011]. Moreover, important advances can be made by using longitudinal data sets which have proven to (1) have more power to detect changes by including within person change, (2) reduce cohort effects and thereby noise, and (3) tests for change rather than cross‐sectional differences [Mills and Tamnes, 2014]. This study capitalizes on these methods by applying complex network analysis to two independent sets of DWI data including a total of 243 children aged 4–13.5 years old including serial data resulting in a total of 378 scans.
The first question addressed in this study concerns the spatial pattern of age effects in macro‐scale structural connectivity. We hypothesize that this age period is characterized by a selective regional specific pattern of development, which would appear in different anatomical network layouts and topology for different developmental rates. To assess if there is indeed a regional specific maturational pattern of connectome development, we examined the anatomical layout and network topology of connections (edges) that show the most substantial change in FA. We tested if this set of edges differentiated from edges that did not show substantial developmental changes. To do so, we compared edges showing the largest level of change to edges showing the smallest level of change in FA. We hypothesize that large changes favor a specific set of edges between rich‐club regions, as previously observed in adolescence [Baker et al., 2015]. This is thought to support the developmental change in functional integration that marks the change in patterns of activity from adolescence into adulthood [Fair et al., 2009].
The second question that is addressed concerns the underlying biology that gives rise to the patterns of change. We hypothesized that long fibers show the largest change in FA, as previously reported in a smaller cross‐sectional dataset [Hagmann et al., 2010]. This hypothesis is in line with findings of resting‐state functional imaging studies, where functional correlations showed age related decreases in short range connectivity and age related increases in long‐range connectivity [Supekar, Musen, & Menon, 2009; but see Power et al., 2012]. Finally, we explored the link between developmental changes in network topology and the age‐dependent change in fibers traced from their corresponding cortical region [Jeon et al., 2015]. We test for consistency and replicability of our results by analyzing two independent datasets [Poldrack et al., 2017; see Tamnes et al. 2017 and van den heuvel et al., 2013].
MATERIALS AND METHODS
The Human Research Protections Program and institutional review board at the University of California, San Diego (UCSD) approved the research protocol to collect and share the data of the Pediatric Longitudinal Imaging, Neurocognition, and Genetics (PLING) study. The BrainTime study was approved by the Institutional Review Board at Leiden University Medical Center.
Participants
This PLING sample included 237 scans of 146 typically developing children between 4 and 13.5 years of age (74 females) recruited in the greater San Diego area by the Center for Human Development at UCSD (see Table 1 for demographics). Participants were recruited through local postings, outreach activities, and through school and community contacts in San Diego, CA, USA. Participants had no diagnosis of neurological disorders; history of head trauma; preterm birth (<36 weeks); diagnosis of autism spectrum disorder, bipolar disorder, schizophrenia, mental retardation, or contraindications for MRI (such as dental braces, metallic or electronic implants, or claustrophobia). Data acquisition procedures for the PLING study were similar to the PING study, which has been described in detail [Jernigan et al., 2016]. Written parental informed consent was obtained from all participants, in addition child assent was obtained for all participants older than 7 years.
Table 1.
Participant characteristics
| PLING | BrainTime | |||||
|---|---|---|---|---|---|---|
| Characteristics | Males | Females | Total | Males | Females | Total |
| Number of individuals, n | 79 | 67 | 146 | 35 | 62 | 97 |
| Total number of scans, n | 125 | 112 | 237 | 56 | 85 | 141 |
| Number of scans by wave, n | ||||||
| Wave 1 (age range) | 79 (4.2–12.9) | 67 (4.5–12.4) | 146 | 35 (8.2–13.2) | 62 (8.2–13.4) | 97 |
| Wave 2 (age range) | 35 (5.8–10.8) | 34 (5.5–11.3) | 69 | 16 (10.5–13.1) | 18 (10.2–13.5) | 34 |
| Wave 3 (age range) | 9 (6.9–10.8) | 8 (6.6–10.8) | 17 | 5 (12.5–13.5) | 5 (13.5–13.4) | 10 |
| Wave 4 (age range) | 2 (8.5–9.6) | 3 (8.8–9.0) | 5 | |||
The BrainTime sample included 141 scans of 97 individuals (62 females) aged between 8 and 13.5 years old. This is a subsample of a large accelerated longitudinal research project as described previously [Braams et al., 2015, Achterberg et al., 2016; Peper et al., 2015] (see Table 1 for demographics). Subjects were recruited through schools in Leiden, the Netherlands. Self‐report questionnaires were administered to confirm the absence of psychopathology, neurological, or mental health problems or the use of psychotropic medication. Written informed consent was obtained from all participants and their parents.
Data Acquisition
To reduce data loss and maximize subject comfort and compliance, a variety of procedures were developed. These procedures were applied depending on the subject's needs and included exposure and habituation to the scanner, parent, or technician accompaniment in the scanning environment and extra rests in between scans.
For the PLING dataset, all participants had an MRI scan on a GE 3T Signa HDx 3T Discovery 750× scanner (GE Healthcare, Waukesha, WI) using an eight‐channel phased array head coil. The imaging sequences included a high‐resolution 3D T1‐volume and a set of diffusions‐weighted scans. The T1‐weighted volume was optimized for maximum grey/white matter contrast and acquired using prospective motion correction (PROMO) and a magnetization prepared rapid gradient echo sequence (flip angle = 8°; receiver bandwith = ±31.25 kHz, freq = 256, phase = 192, slice thickness = 1.2 mm, FoV = 24 cm; TE = 3.5 ms; TR = 8.1 ms; TI =640 ms).
Furthermore, a set of axial diffusion‐weighted scans were collected with integrated B0 distortion correction (DISCO) (30 directions, b value = 1,000 s/mm2) together with two sets of diffusion‐unweighted scans (b value = 0 s/mm2, flip angle 90°, FOV 24 × 24 cm, freq = 96, phase = 96, slice thickness = 2.5 mm, TE = 83 ms; TR = 13,600 ms) were acquired. Standardized quality control procedures were followed including computer algorithms and visual inspection ratings by trained imaging technicians at the Center for Human Development. Subjects that showed excessive head movement were excluded.
For the BrainTime dataset, MRI scans were acquired on a 3 T Philips Achieva scanner, using a six‐element SENSE receiver head coil (Philips, Best, The Netherlands) at Leiden University Medical Centre. The image sequence included a high‐resolution 3D T1‐weighted volume (flip angle = 8°, slice thickness = 1.2 mm, FoV = 24 cm; TE = 4.6 ms; TR = 9.8 ms) and two transverse diffusion‐weighted scans (30 directions, b value = 1,000 s/mm2) and 5 sets of diffusion‐unweighted scans (b value = 0 s/mm2, flip angle 90°, FOV 240 × 240 mm, freq = 96, phase = 96, slice thickness = 2 mm, TE = 69 ms; TR = 7,315 ms). Visual inspection ratings by trained imaging technicians at Leiden University were performed, and subjects that showed excessive movement were excluded.
Data Preprocessing; T1 Data
Within each individual dataset, we used the T1 images for anatomical reference and the selection of brain network nodes. Tissue classification and anatomical labeling was performed on the basis of the T1‐weighted MR image using the well‐validated and well‐documented Freesurfer v5.3.0 software (http://surfer.nmr.mgh.harvard.edu/). Technical details of the automated reconstruction scheme are described elsewhere [Dale, Fischl, & Sereno, 1999].
DWI Data Preprocessing
The full protocol for the DWI preprocessing pipeline has been described elsewhere [Romme, 2017]. In short, it involved the following steps: First, both DWI sets were realigned and corrected for common distortions [Andersson & Skare, 2002]. Second, diffusion images were corrected for eddy‐current distortions and realigned to the b = 0 image. Third, we fitted a diffusion profile within each voxel using the two sets of 30 weighted images and the average b = 0 image from each subject. From the resulting tensor, the main diffusion direction in each voxel was selected as the principal eigenvector resulting from the eigenvalue decomposition of the fitted tensor, marking the preferred diffusion direction in each voxel. For each voxel in the brain mask, the FA values were computed, indicating the level of anisotropic diffusion [Basser & Pierpaoli, 1996; Beaulieu & Allen, 1994]. Fourth, information on the preferred diffusion direction was extracted within each voxel in the brain mask to reconstruct streamlines based on deterministic fiber tracking using the FACT algorithm (fiber assignment by continuous tracking) [Mori & van Zijl, 2002; Mori et al., 1999; van den Heuvel et al., 2009]. DTI has shown relative high specificity of connection reconstruction as compared to other, more advanced algorithms for the estimation of the diffusion profile, which is indicated to be important when studying the topological structure of networks [Zalesky et al., 2016; Van den Heuvel et al. 2017]. Streamlines were reconstructed by starting eight seeds in each voxel; these seeds were evenly distributed across the volume of the voxel. A streamline was started from each seed following the main diffusion direction (selected as the principal eigenvector) until the streamline entered a voxel with a low level of diffusion preference (FA < 0.1), made an unexpected sharp angular turn (angle > 45°), or left the brain mask.
Network Reconstruction
Each reconstructed network per time point of each individual was represented as a graph, matrix M. This matrix includes 68 cortical brain regions defined as the nodes of the graph and the edges between nodes and (i.e., brain region and region ). These edges are based on the streamlines that were reconstructed as described above, and an example of a representative subject can be observed in Figure 1.
Figure 1.
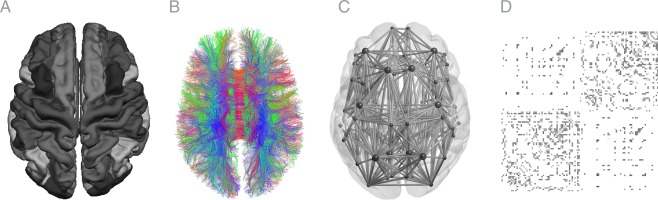
Representation of reconstructed anatomical parcellation of freesurfer (A), DWI streamlines (B), and graph representation (C) for one representative subject of the BrainTime dataset. D shows a single‐subject connectome as a connectivity matrices with rows and columns depicting source ( ) and target regions ( ). Pathways are grouped by hemisphere colors represent mean FA values ranging from 0.1 (grey) to 0.7 (black). [Color figure can be viewed at http://wileyonlinelibrary.com]
Network Node Definition
The 68 nodes of the networks were represented by brain regions that were automatically segmented with Freesurfer v5.3.0 software using the Desikan–Killiany atlas [Desikan et al., 2006]. Individual T1‐weighted images were co‐registered to the b = 0 images (rigid transformations using mutual information). This procedure has been previously described [van den Heuvel et al., 2008, 2009].
To assess the robustness of results, we additionally examined the impact of an alternative network node definition using higher resolution parcellation of the brain from the Lausanne atlas (219 cortical nodes) [Cammoun et al., 2012].
Network Edge Definition
The FACT algorithm is sensitive to image resolution and noise and hence there is the possibility of tracing pseudo‐streamlines (e.g., false positives). To minimize the number of false positives, we included edges if two regions were connected by at least three fiber streamlines [Li et al., 2009; Lo et al., 2010; Shu et al., 2009]. This cutoff did not affect our measure of interest (mean FA) as the mean number of streamlines was not related to mean FA (PLING: value = 0.891; BrainTime P value = 0.597).
The connection strength of each edge was assessed by the mean FA of values in each voxel along the streamlines [van den Heuvel et al., 2008, 2009]. Variation in FA has been associated with physical properties of the fiber bundles, such as packing density, myelination, and axon diameter [Beaulieu, 2002]. Although FA is affected by either one or a composite of these measures, they have in common that they reflect tissue characteristics that have functional relevance [Wolff & Balaban, 1994]. Therefore, our measure of interest was mean FA, and the weights of each edge were set as such.
There was variability in edge topology between subjects; hence, we estimated age‐related change only for those edges that were present in a minimum number of participants. This variability was not related to age. We set this threshold at 60%, as this has been shown to minimalize the balance between false positives and false negatives [de Reus & van den Heuvel, 2013].
Network Analysis
For graph theoretical analysis, we used the Brain Connectivity Toolbox [Rubinov & Sporns, 2010]. Each matrix M was used to compute the measures described below:
Number of edges, the total count of edges included in each un‐thresholded binary matrix for each individual and time point.
Node degree, the number of edges connected to each node in matrix .
Node strength, the sum of weights (mean FA) of all edges to each node in matrix .
Path length, the minimum number of binary steps to get from one node to another node , averaged over all nodes .
Normalized clustering coefficient gamma ( ), the ratio of number of edges between direct neighbors of a given node and the total number of possible edges between these neighbors [Watts & Strogatz, 1998]. Because this measure is highly related to the degree of each node, we compared the cluster coefficient of node to the average cluster coefficients of its corresponding node in 500 random graphs. These random graphs were constructed by redistributing edge weights of each matrix while keeping the degree distribution intact [Maslov and Sneppen, 2002; Rubinov and Sporns, 2010]. Next, normalized clustering gamma ( ) is estimated as the cluster coefficient relative to clustering in the random networks.
Rich club classification, nodes with a high node degree that display more connectivity than one would expect on the basis of their degree alone are said to form a rich club core. This study used rich club classification based on previous studies in children, adults and clinical groups [Baker et al., 2015; Ball et al., 2014; Daianu et al., 2015; Grayson et al., 2014; van den Heuvel and Sporns, 2011;van den Heuvel et al., 2012]. This a priori definition of rich club regions is an unbiased method and permits comparison with the literature. Regions included bilateral superior frontal gyrus, precuneus, superior parietal gyrus, and insula. Nodes in the network were classified as rich club or peripheral nodes. Next, edges were categorized into rich club edges (between rich club nodes), feeder edges (nonrich club to rich club nodes), and local edges (between nonrich club nodes).
Functional modules, each network node was assigned a priori to one of the functional network modules defined by Yeo et al. [2011]. Baum et al. [2017] showed that even though these module partitions were defined using functional imaging, the modularity quality of the functional partition showed to fit structural connectivity data.
Statistical Analysis
Age‐related change in total number of edges, total number of streamlines, global mean FA, streamline length, and each edge in matrix M were assessed using linear mixed modeling. This model accounts for irregular intervals between measures, missing data and within person dependence, and was therefore particularly suited for our datasets. More formally, each dependent measure of the th individual and th time point was modeled as follows:
Here, denotes the age of the th individual at the th time of his/her scan. The dependent variable is modeled as a function age ( ) plus a random person effect ( ) plus error ( ). Intercept and age, were fixed effects, while within person dependence ( ) was modeled as a random effect. Nonlinear models including cubic and quadratic age terms did not improve model fit and were therefore not used in further analyses. We did not include gender in our model as it did not improve model fit. This is in line with previous observations of no to limited gender effects on DWI measures [Wierenga et al., 2015; Krogsrud et al., 2015].
For assessment of the anatomical distribution of edges and anatomical layout of nodes, a χ 2 analysis was used. Furthermore, for the analysis of network topology, ANOVA analysis was performed.
Selection of Edges
We estimated age‐related changes in FA ( ) for each edge included in our group level analyses with the statistical model described above. We were interested in edges showing either substantial or little change in FA, as an increase in FA for all edges would not per se result in changes in network topology and organization but could merely result in a global change in network efficiency. Rather a spatial heterogeneous pattern of change would affect network topology and organization. To address this pattern, we classified edges into different categories: edges that showed the largest change in FA and edges that showed the smallest change in FA. Edges were defined as largest or smallest change in FA when they had a of one above or below the average , respectively.
To improve power, we selected only those edges within the edge categories that formed an interconnected structure, that is, connected component. This method is comparable to cluster‐extent‐based methods that are often used in functional neuroimaging studies to account for the multiple comparison problem [Bullmore, Suckling, & Overmeyer, 1999]. Rather than selecting a (unknown) threshold for component size we only included the largest connected‐components observed in the sets of largest and smallest change.
RESULTS
Global Changes in Number of Edges, Streamlines, and Mean FA
First, we addressed the question if the structural layout is stable across the age range. For this goal, we assessed the total number of edges for each matrix and observed that indeed this did not change with age in both data sets, see results in Table 2. Furthermore, there were no significant developmental changes in the mean number of streamlines in both data sets.
Table 2.
Global age‐related changes
| PLING | BrainTime | BrainTime parc 219 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β age | df | t | P value | β age | df | t | P value | β age | df | t | P value | |
| Total number of edges | 1.887 | 90 | 1.164 | 0.247 | 3.266 | 37 | 1.450 | 0.155 | −3.378 | 37 | −0.307 | 0.761 |
| Mean NOS | 0.916 | 90 | 1.806 | 0.074 | −0.046 | 37 | −0.065 | 0.948 | 0.034 | 37 | 0.105 | 0.917 |
| Mean FA | 0.002 | 90 | 4.506 | <0.001 | 0.004 | 37 | 6.398 | <0.001 | 0.004 | 37 | 7.157 | <0.001 |
| Mean FA + ICV | 0.002 | 90 | 4.387 | <0.001 | 0.004 | 37 | 6.396 | <0.001 | 0.004 | 37 | 7.154 | <0.001 |
Next, we examined the global developmental change in FA. To do so, we estimated mean FA for each network at each time point (l) of each individual (m) and observed that mean FA increased with age in both data sets (PLING: < 0.001; BrainTime: < 0.001). Mean FA was not mediated by intracranial volume (PLING: value < 0.001; BrainTime: value < 0.001). Similar results were found when using the Lausanne 219 cortical parcellation schedule ( value < 0.001).
Anatomical Distribution of Edges
The anatomical layout of the connected components including edges showing the largest change in FA and smallest changes in FA are displayed in Figure 2. First, the overlap in anatomical layout of the largest component of the two datasets were compared to the overlap in 10 000 random networks. This analysis confirmed the stability of these components across the datasets where the overlap showed to be significant (large change component: value < 0.0001; small change component: value = 0.0058).
Figure 2.
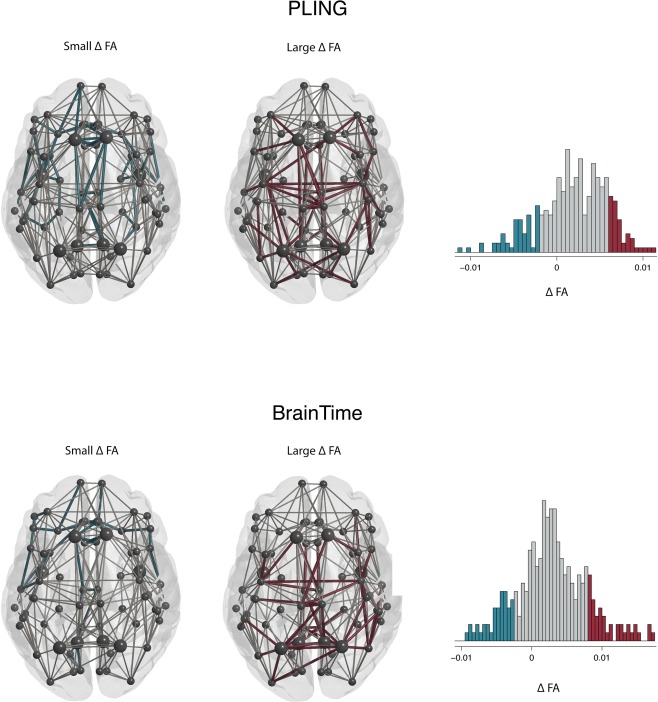
Connected components of largest (red) and smallest changes (blue) in FA frontal lobe at the top of the image. The PLING dataset is represented in the top row and the BrainTime dataset in the bottom row. Nodes (circles) and edges (lines) are displayed for the reconstructed thresholded group averaged brain networks. The histogram shows values for all edges in the group network. Edges that showed an age‐related change of 1 SD smaller than the mean change in FA are displayed in blue (small FA). Connections larger than 1 above mean change in FA are displayed in red (large FA). The largest two connected components were selected for both sets of edges. Rich‐club nodes are represented by large circles. [Color figure can be viewed at http://wileyonlinelibrary.com]
We next addressed the question if large changes in FA favor a specific set of edges. As shown in Figure 3 and Table 3, the large change component included a greater number of feeder edges than the small change component, which reached significance in the BrainTime dataset (P value = 0.001), but not the PLING dataset ( value = 0.223). As only 8 of 68 nodes were labeled as rich club regions, the number of possible rich club and feeder edges was smaller than the possible number of peripheral edges. To control for this, we compared the observed number of edges in the largest and smallest change components to the total possible number of edges. A χ 2 analysis indicated that there was a significant effect for edges with large change compared to all possible connections (PLING: value < 0.001; BrainTime: value < 0.001) but not for edges with the smallest change in FA in the BrainTime dataset (BrainTime; P value = 0.812). The PLING dataset did show a significant difference between the small change component and the total possible number of edges (PLING: value < 0.001). The Lausanne parcellation in the BrainTime dataset confirmed the pattern in the Desikan–Killiany parcellation.
Figure 3.
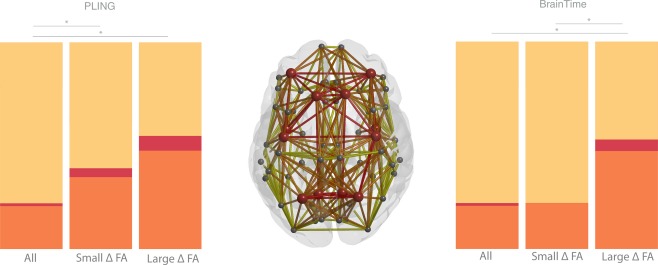
Percentage of anatomical distribution of rich club, feeder, and local edges in red, orange, and yellow, respectively. The middle graph shows a schematic representation of the group averaged reconstructed brain network. Nodes (circles) represent brain regions where rich club nodes are indicated by red circles. The bar graphs on the left (PLING set) and right (BrainTime set) indicate that a larger proportion of feeder and hub edges and a smaller proportion of peripheral edges were observed in the edges showing large age‐related changes in FA (large FA) compared to the distribution of edges showing the smallest change in FA (small FA) and compared to the distribution of edge categories for all possible edges. [Color figure can be viewed at http://wileyonlinelibrary.com]
Table 3.
Anatomical distribution of edges
| PLING | BrainTime | BrainTime parc 219 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| χ 2 | df | N | P value | χ 2 | df | N | P value | χ 2 | df | N | P value | |
| Hub/feeder/rich club | ||||||||||||
| Large Δ FA vs small Δ FA | 4.982 | 2 | 88 | 0.223 | 16.216 | 2 | 73 | 0.001 | 6.100 | 2 | 384 | 0.047 |
| Large Δ FA vs total | 55.13 | 2 | 1198 | <0.001 | 49.202 | 2 | 1198 | <0.001 | 25.739 | 2 | 23072 | <0.001 |
| Small Δ FA vs total | 16.66 | 2 | 1202 | <0.001 | 0.956 | 2 | 1187 | 0.812 | 0.0414 | 2 | 22968 | 0.813 |
Streamline Length
The next questions we addressed concerns the underlying biology associated with these patterns of change. Edge categories showed to differ in mean streamline length (see results Table 4): edges that showed large changes in FA were longer (PLING: = 51.979, = 32.150; BrainTime: = 51.939, = 35.658) than edges that showed small changes in FA (PLING: = 25.377, = 23.289; BrainTime: = 16.104, = 13.619), this difference was significant in both datasets (PLING: value < 0.001; BrainTime: value <0.001) and the Lausanne 219 parcellation ( value <0.001). This effect remained significant when we accounted for average FA for each edge. Together, these findings demonstrate particularly strengthening of long‐range connectivity.
Table 4.
Streamline length
| PLING | BrainTime | BrainTime parc 219 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| F | df | P value | F | df | P value | F | df | P value | |
| Large Δ FA > small Δ FA | 40.00 | 174 | <0.001T | 49.82 | 144 | <0.001T | 101.7 | 447 | <0.001T |
T = significant after covarying for mean FA.
Anatomical Layout
In the following section, we mapped the anatomical layout of cortical regions and age‐dependent change in fibers traced from these regions. To do so, we identified three sets of nodes: (i) nodes connecting to edges that showed large change, (ii) nodes connecting to edges that showed small change, and (iii) nodes connecting to both edge categories.
In the BrainTime dataset, the anatomical layout across the four lobes significantly differed between nodes connected to edges showing large age‐related change in FA (i) and nodes that connected to edges showing small changes in FA, as shown by a χ 2 analysis ( value = 0.015). Large change was observed for a relative large number of nodes in the parietal and frontal lobes, while small changes were observed for a relative large number of nodes in the occipital lobe. This difference was not significant in the PLING dataset ( value = 0.905). Table 5 shows the relative contribution by the number of nodes identified in each node category divided by the number of nodes in each lobe.
Table 5.
Anatomical distribution of nodes connecting to edges with the smallest and largest changes in FA
| PLING | BrainTime | BrainTime parc 219 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Lobe | Large Δ FA | Small Δ FA | Both | Large Δ FA | Small Δ FA | Both | Large Δ FA | Small Δ FA | Both |
| Frontal, % | 27 | 35 | 31 | 12 | 19 | 35 | 34 | 26 | 33 |
| Parietal, % | 29 | 43 | 29 | 50 | 0 | 50 | 23 | 30 | 37 |
| Temporal, % | 25 | 30 | 20 | 20 | 20 | 5 | 17 | 28 | 37 |
| Occipital, % | 12 | 38 | 25 | 0 | 38 | 12 | 13 | 48 | 30 |
%= percentage of nodes connected to edges categories; Δ = age related change.
We next investigated the anatomical layout of edges in functional networks: the limbic system showed mostly small changes in FA while visual regions showed predominantly large changes in FA (Fig. 4). Furthermore, regions in somatomotor and sensory systems (insula, pre‐ and postcentral gyrus, paropercularis, paracentral region, superior temporal and transverse temporal region), and default mode network (inferior parietal cortex, middle temporal cortex, cingulate cortex, precuneus and superior frontal cortex) showed a larger proportion of edges with the largest change compared to edges with the smallest change in FA, this pattern was more pronounced in the BrainTime dataset. The visual system showed a similar number of edges showing large and small changes.
Figure 4.
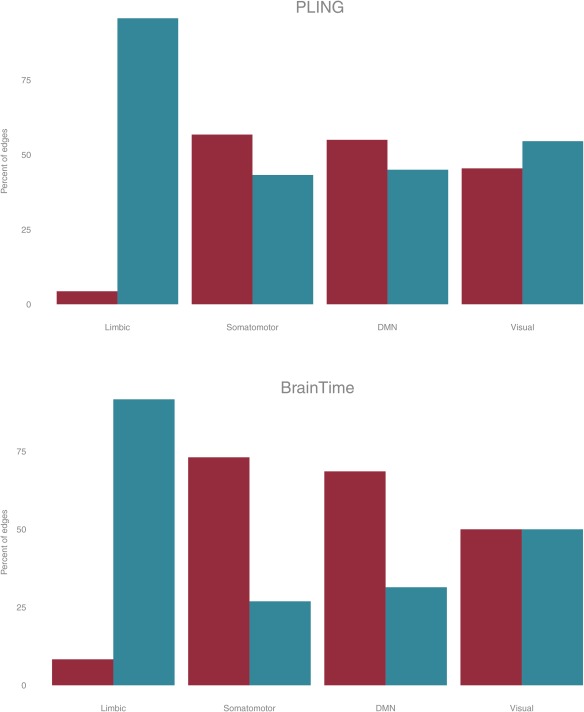
Subnetworks for the PLING (top) and BrainTime (bottom) datasets showing percentage of edges that had large change (dark grey/red) and small changes (light grey/blue) in FA connecting to nodes in one of the following subnetwork: cortical limbic network, somato‐motor network, default mode network, and visual network (left to right). [Color figure can be viewed at http://wileyonlinelibrary.com]
Network Topology
We next compared the topological properties of nodes that were uniquely connected to either large (i) or small (ii) changing edges or to both edge categories (iii) to further address the question if development of the connectome is region specific (see results in Table 6). In the BrainTime data set, it was observed that nodes that were connected to edges with large changes in FA (i) had greater average node strength ( value = 0.002) and shorter path length ( value < 0.001) compared to nodes solely connected to edges showing small change in FA (ii). Gamma also significantly differed between node categories, where larger gamma was observed for nodes connecting to large changing edges ( value = 0.003). This effect was not replicated in the PLING dataset. The Lausanne parcellation showed significant effects of node strength and gamma, but not path length.
Table 6.
Topology of nodes across node groups
| PLING | Brain Time | Brain Time parc 219 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| F | df | P value | F | df | P value | F | df | P value | |
| Average node strength | 0.298 | 56 | 0.744 | 7.149 | 41 | 0.002 | 4.894 | 192 | 0.008 |
| Path length | 0.611 | 56 | 0.546 | 10.446 | 41 | <0.001 | 0.210 | 192 | 0.811 |
| Gamma | 1.068 | 56 | 0.351 | 6.939 | 41 | 0.003 | 3.381 | 192 | 0.036 |
| Change in node strength | 0.117 | 56 | 0.890 | 1.667 | 41 | 0.201 | 1.361 | 192 | 0.259 |
| Change in path length | 0.323 | 56 | 0.725 | 1.661 | 41 | 0.203 | 0.366 | 192 | 0.694 |
| Change in gamma | 0.441 | 56 | 0.645 | 2.429 | 41 | 0.101 | 0.997 | 192 | 0.371 |
Furthermore, we tested how the two components (large and small changes in FA) related to developmental changes in the topological organization of the network. Age‐related changes in node strength, path length, and gamma did not show significant differences between node groups ( value > 0.05).
DISCUSSION
This study investigated the spatial pattern of human connectome development in two independent longitudinal datasets of 4–13‐year‐old children. These white‐matter connections may represent potential routes of information flow between pairs of brain regions and herewith may contribute to information processing and synchronization patterns between distant regions [Singer, 1993]. We observed that there is a selective regional specific pattern of network development, as edges that show substantial change across age differ in anatomical layout and network topology from edges that show little change. We furthermore confirmed previous findings that long fibers show larger changes in FA than short fibers [Hagmann et al., 2010]. Together these findings are interpreted to suggest that developmental changes in white‐matter connections may promote developmental changes in the brain network that are marked by simultaneous progressive and regressive neurobiological changes [Rubinov & Sporns, 2010].
The primary aim of this study was to test if the connectivity network follows global or specific developmental changes. A robust finding across the two samples, was that the brain network follows a heterogeneous pattern of development in childhood and herewith extends previous findings that show different rates of development in major white matter tracts [Dean et al., 2015; Lebel et al., 2008]. The observations in this study contrast findings in previous studies that suggested a rather global pattern of white matter development [Brouwer et al., 2012; Giedd et al., 2015; Hagmann et al., 2010; Muetzel et al. 2015]. This study made use of advanced analysis methods and longitudinal data sets and therefore had increased power to detect region specific and heterogeneous change, consistent with the hypothesis that developmental progressions occur through interactive specialization [Johnson et al., 2011].
Interestingly, edges that showed large and small changes in FA had different anatomical layouts. The frontal and parietal lobes had a substantial large number of edges that showed large increases in FA values. In addition, edges within the cortical limbic network predominantly showed small changes in FA values. This spatial heterogeneous developmental pattern is in line with findings of connectome development in a smaller cross‐sectional sample of toddlers where efficiency in medial nodes significantly increased with age, while in lateral located nodes decreases were observed [Huang et al., 2015]. Furthermore, the pattern of development observed in this study indicates that a considerably large number of feeder edges, connecting peripheral to rich club regions, showed the largest changes in FA. Although speculative, this may indicate that childhood development is characterized by changes in the ability to integrate complex information, as rich club regions are known to process information from multiple functional modalities [van den Heuvel and Sporns, 2011]. Developmental changes in connectivity between peripheral and rich club regions may also be reflected in cognitive changes during this age period, such as changes in attention and working memory processes [Diamond et al., 2013], as these processes are dependent on the efficient integration of information from multiple regions [Braun et al., 2015]. An outstanding question for future research concerns how development of white‐matter connectivity correlates with developmental advances in cognitive control.
The second question we addressed concerned the underlying biology that gives rise to the patterns of change. We extend previous findings by showing that longer fibers had larger changes in network topology than shorter fibers [Hagmann et al., 2010]. This finding may indicate that long‐range connectivity shows a delayed maturational pattern compared to short‐range connectivity, as previously reported in functional imaging studies [Supekar, Musen, & Menon, 2009, but see Power et al., 2012; van Dijk et al., 2012]. Together these findings suggest that childhood is marked by both progressive and regressive neurobiological processes that lead to some cortical regions being more strongly embedded in the brain network while other regions are subtracted from the brain network through the course of development.
The last question we addressed concerns how these changes in FA relate to network topology. We observed in the BrainTime dataset that network topology differed between edges showing large and small changes in FA, where edges showing substantial changes connected to regions with higher node strength, path length and gamma than edges showing little change. Note that these results could not be replicated across the two datasets and should therefore be interpreted with care.
A strength of this study is the use of two large independent longitudinal samples, that allow for unambiguous replication and the assessment of within subject changes [Poldrack et al., 2017]. Several findings showed to be robust across the two datasets and parcellation of cortical nodes. These results included; mean FA showed age‐related increases in both datasets, as expected. Also, no age‐effects in global number of edges and mean number of streamlines was observed. This result supports previous observations that the majority of tracts are already present early in development [Ball et al., 2014; van den Heuvel et al., 2015; Wierenga et al., 2015]. Furthermore, the anatomical distribution of edges that showed the largest and smallest change in FA significantly overlapped between the datasets, and both datasets revealed that edges showing large changes had larger streamline length than edges showing small changes. In addition, both datasets showed that cortical limbic structures connected to a larger number of small change edges. Furthermore, the BrainTime dataset showed a pattern where large changing edges included a larger number of feeder and hub edges, this effect was not significant in the PLING dataset. Also, the observation that the large and small change components showed significant differences in anatomical distribution between lobes and differences in network topology was significant in the BrainTime but not PLING dataset; hence, these results should be interpreted with caution.
Several limitations should be considered when interpreting the results. First, DWI is known to be sensitive to motion artifacts [Yendiki et al., 2013]. This was partially tackled with integrated distortion correction algorithms in the PLING dataset and the quality assessment on all scans, where subjects that showed excessive movement were excluded. Second, the spatial resolution of our imaging technique makes it currently unfeasible to trace short cortical U‐fibers. Thus, our fiber‐tracing technique may identify only a fraction of the actual neural interactions involved [Sporns 2011]. Third, it should be kept in mind that the main communication path between two regions can also occur via a third region; this may best be detected with functional correlation analysis. Therefore, the integration between distant regions through synchronization cannot be apparently manifested solely based on DWI‐based networks used in this study.
An interesting question for future research will be to unravel the underlying biological conditions that are related to the age‐related variability in FA. Although FA has one of the highest correlations with myelin water fraction, in comparison with other diffusion measures [Mädler et al., 2008], a number of other biological conditions may effect FA, including physical properties of the fiber bundles (e.g., changes in axonal diameter and packing density) or their environment (e.g., angiogenesis). Moreover, a composite of these factors that differ per region may be at play. However, some of these conditions may be more plausible in relation to the developmental changes observed in this study than others. For example, changes in the axon packing density is an unlikely candidate for the observed developmental changes in this study as an increase in the number of axons in this age range is not in line with findings in animal models [Price et al., 2006]. Alternative measures, such as T1‐weighted T2‐weighted ratio, might provide additional information on changes in myelin deposition [Ganzetti et al., 2014; Glasser and Van Essen, 2011]. Combining these measures into multimodal approaches is recommended for future studies.
In summary, we showed that refinement of the brain network in childhood is a selective systematic region specific process, supporting the notion that this developmental period is characterized by large changes in interactive specialization [Johnson, 2011]. This was supported by the finding that substantial differences in anatomical layout and topological organization were observed between edges that showed the largest increase in FA compared to edges that showed the smallest change in FA. Mapping the specific developmental pattern of the connectome may help us better understand the processes essential for childhood cognitive development [Diamond et al., 2013] and may ultimately predict vulnerability and guide interventions.
ACKNOWLEDGMENTS
The authors thank all subjects and their parents for participating in this study. PLING data are disseminated by the Center for Human Development, University of California, San Diego.
Data used in preparation of this article were obtained from the Pediatric Longitudinal Imaging, Neurocognition, and Genetics Study (PLING) (http://www.chd.ucsd.edu/research/pling.html). As such, the PLING investigators contributed to the design and implementation of PLING and/or collected and provided data but did not participate in the analysis or writing of this report. A complete listing of PLING investigators can be found at http://www.chd.ucsd.edu/research/pling.html.
REFERENCES
- Achterberg M, Peper JS, van Duijvenvoorde ACK, Mandl RCW, Crone EA (2016): Frontostriatal white matter integrity predicts development of delay of gratification: A longitudinal study. J Neurosci 36:1954–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson JLR, Skare S (2002): A model‐based method for retrospective correction of geometric distortions in diffusion‐weighted EPI. NeuroImage 16:177–199. [DOI] [PubMed] [Google Scholar]
- Baker STE, Lubman DI, Yucel M, Allen NB, Whittle S, Fulcher BD, et al. (2015): Developmental changes in brain network hub connectivity in late adolescence. J Neurosci 35:9078–9087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball G, Aljabar P, Zebari S, Tusor N, Arichi T, Merchant N, et al. (2014): Rich‐club organization of the newborn human brain. Proc Natl Acad Sci 111:7456–7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basser PJ, Pierpaoli C (1996): Microstructural and physiological features of tissues elucidated by quantitative‐diffusion‐tensor MRI. J Magn Reson B 111:209–219. [DOI] [PubMed] [Google Scholar]
- Baum GL, Ciric R, Roalf DR, Betzel RF, Moore TM, Shinohara RT, et al. (2017): Modular segregation of structural brain networks supports the development of executive function in youth. Curr Biol 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu C, Allen PS (1994): Water diffusion in the giant axon of the squid: Implications for diffusion‐weighted MRI of the nervous system. Magn Reson Med 32:579–583. [DOI] [PubMed] [Google Scholar]
- Beaulieu C (2002): The basis of anisotropic water diffusion in the nervous system ‐ a technical review. NMR Biomed 15:435–455. [DOI] [PubMed] [Google Scholar]
- Braams BR, van Duijvenvoorde ACK, Peper JS, Crone EA (2015): Longitudinal changes in adolescent risk‐taking: A comprehensive study of neural responses to rewards, pubertal development, and risk‐taking behavior. J Neurosci 35:7226–7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun U, Schäfer A, Walter H, Erk S, Romanczuk‐Seiferth N, Haddad L, et al. (2015): Dynamic reconfiguration of frontal brain networks during executive cognition in humans. Proc Natl Acad Sci 112:11678–11683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer RM, Mandl RCW, Schnack HG, van Soelen ILC, van Baal GC, Peper JS, et al. (2012): White matter development in early puberty: A longitudinal volumetric and diffusion tensor imaging twin study. PLoS One 7:e32316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TT, Lugar HM, Coalson RS, Miezin FM, Petersen SE, Schlaggar BL (2005): Developmental changes in human cerebral functional organization for word generation. Cereb Cortex (New York, N.Y.: 1991) 15:275–290. [DOI] [PubMed] [Google Scholar]
- Bullmore ET, Suckling J, Overmeyer S (1999): IEEE Xplore Abstract ‐ Global, voxel, and cluster tests, by theory and permutation, for a difference between two groups of structural MR images of the brain. IEEE Trans Med Imaging 18. [DOI] [PubMed] [Google Scholar]
- Cammoun L, Gigandet X, Meskaldji D, Thiran JP, Sporns O, Do KQ, Maeder P, Meuli R, Hagmann P (2012): Mapping the human connectome at multiple scales with diffusion spectrum MRI. J Neurosci Methods 203:386–397. [DOI] [PubMed] [Google Scholar]
- Collin G, de Nijs J, Hulshoff Pol HE, Cahn W, van den Heuvel MP (2015): Connectome organization is related to longitudinal changes in general functioning, symptoms and IQ in chronic schizophrenia. Schizophrenia Res. [DOI] [PubMed] [Google Scholar]
- Daianu M, Jahanshad N, Nir TM, Jack CR, Weiner MW, Bernstein MA, et al. (2015): Rich club analysis in the Alzheimer's disease connectome reveals a relatively undisturbed structural core network. Hum Brain Mapp 36:3087–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI (1999): Cortical surface‐based analysis. I. Segmentation and surface reconstruction. NeuroImage 9:179–194. [DOI] [PubMed] [Google Scholar]
- Dean DC, O'Muircheartaigh J, Dirks H, Waskiewicz N, Walker L, Doernberg E, et al. (2015): Characterizing longitudinal white matter development during early childhood. Brain Struct Funct 220:1921–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis EL, Jahanshad N, McMahon KL, de Zubicaray GI, Martin NG, Hickie IB, et al. (2013): Development of brain structural connectivity between ages 12 and 30: A 4‐Tesla diffusion imaging study in 439 adolescents and adults. NeuroImage 64:671–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, et al. (2006): An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage 31:968–980. [DOI] [PubMed] [Google Scholar]
- Diamond A (2013): Executive functions. Ann Rev Psychol. 64:135–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk KRA, Sabuncu MR, Buckner RL (2012): The influence of head motion on intrinsic functional connectivity MRI. NeuroImage 59:431–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair DA, Cohen AL, Power JD, Dosenbach NUF, Church JA, Miezin FM, et al. (2009): Functional brain networks develop from a “local to distributed” organization. PLoS Comput Biol 5:e1000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganzetti M, Wenderoth N, Mantini D (2014): Whole brain myelin mapping using T1‐ and T2‐weighted MR imaging data. Front Hum Neurosci 8:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedd JN, Blumenthal J, Jeffries NO, Castellanos FX, Liu H, Zijdenbos A, Paus T, Evans AC, Rapoport JL (1999): Brain development during childhood and adolescence: A longitudinal MRI study. Nat Neurosci 2:861–863. [DOI] [PubMed] [Google Scholar]
- Giedd JN, Raznahan A, Alexander‐Bloch A, Schmitt E, Gogtay N, Rapoport JL (2015): Child psychiatry branch of the National Institute of Mental Health longitudinal structural magnetic resonance imaging study of human brain development. Neuropsychopharmacology 40:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Van Essen DC (2011): Mapping human cortical areas in vivo based on myelin content as revealed by T1‐ and T2‐weighted MRI. J Neurosci 31:11597–11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogtay N, Giedd JN, Lusk L, Hayashi KM, Greenstein D, Vaituzis AC, et al. (2004): Dynamic mapping of human cortical development during childhood through early adulthood. Proc Natl Acad Sci 101:8174–8179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson DS, Ray S, Carpenter S, Iyer S, Dias TGC, Stevens C, et al. (2014): Structural and functional rich club organization of the brain in children and adults. PLoS One 9:e88297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann P, Sporns O, Madan N, Cammoun L, Pienaar R, Wedeen VJ, et al. (2010): White matter maturation reshapes structural connectivity in the late developing human brain. Proc Natl Acad Sci 107:19067–19072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Shu N, Mishra V, Jeon T, Chalak L, Wang ZJ, et al. (2015): Development of human brain structural networks through infancy and childhood. Cerebral Cortex 25:1389–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon T, Mishra V, Ouyang M, Chen M, Huang H (2015): Synchronous changes of cortical thickness and corresponding white matter microstructure during brain development accessed by diffusion MRI tractography from parcellated cortex. Front Neuroanat 9: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernigan TL, Brown TT, Hagler DJ, Akshoomoff N, Bartsch H, Newman E, et al. (2016): The Pediatric Imaging, Neurocognition, and Genetics (PING) data repository. NeuroImage 124:1149–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MH (2011): Interactive specialization: A domain‐general framework for human functional brain development? Dev Cogn Neurosci 1:7–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann T, Alnæs D, Doan NT, Brandt CL (2017): Delayed stabilization and individualization in connectome development are related to psychiatric disorders. Nature. doi: 10.1016/j.neuroimage.2010.08.063 [DOI] [PubMed] [Google Scholar]
- Kim D‐J, Davis EP, Sandman CA, Sporns O, O'Donnell BF, Buss C, Hetrick WP (2016): Children's intellectual ability is associated with structural network integrity. NeuroImage 124:550–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenis MMG, Brouwer RM, van den Heuvel MP, Mandl RCW, van Soelen ILC, Kahn RS, et al. (2015): Development of the brain's structural network efficiency in early adolescence: A longitudinal DTI twin study. Hum Brain Mapp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogsrud SK, Fjell AM, Tamnes CK, Grydeland H, Mork L, Due‐Tønnessen P, et al. (2016): Changes in white matter microstructure in the developing brain—A longitudinal diffusion tensor imaging study of children from 4 to 11years of age. NeuroImage 124:473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel C, Walker L, Leemans A, Phillips L, Beaulieu C (2008): Microstructural maturation of the human brain from childhood to adulthood. NeuroImage 40:1044–1055. [DOI] [PubMed] [Google Scholar]
- Li Y, Liu Y, Li J, Qin W, Li K, Yu C, Jiang T (2009): Brain anatomical network and intelligence. PLoS Comput Biol 5:e1000395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo CY, Wang PN, Chou KH, Wang J, He Y, Lin CP (2010): Diffusion tensor tractography reveals abnormal topological organization in structural cortical networks in Alzheimer's disease. J Neurosci 30:16876–16885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mädler B, Drabycz SA, Kolind SH, Whittall KP, MacKay AL (2008): Is diffusion anisotropy an accurate monitor of myelination? Correlation of multicomponent T2 relaxation and diffusion tensor anisotropy in human brain. Magn Reson Imaging 26:874–888. [DOI] [PubMed] [Google Scholar]
- Maslov S, Sneppen K (2002): Specificity and stability in topology of protein networks. Science (New York, NY) doi: 10.1126/science.1065103 [DOI] [PubMed] [Google Scholar]
- Mills KL, Tamnes CK (2014): Methods and considerations for longitudinal structural brain imaging analysis across development. Dev Cogn Neurosci 9:172–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, van Zijl PC (2002): Fiber tracking: Principles and strategies—a tech‐ nical review. NMR Biomed 15:468–480. [DOI] [PubMed] [Google Scholar]
- Mori S, Crain BJ, Chacko VP, van Zijl PC (1999): Three‐dimensional track‐ ing of axonal projections in the brain by magnetic resonance imaging. Ann Neurol 45:265–269. [DOI] [PubMed] [Google Scholar]
- Muetzel RL, Mous SE, van der Ende J, Blanken LME, van der Lugt A, Jaddoe VWV, et al. (2015): White matter integrity and cognitive performance in school‐age children: A population‐based neuroimaging study. NeuroImage 119:119–128. [DOI] [PubMed] [Google Scholar]
- Peper JS, de Reus MA, van den Heuvel MP, Schutter DJLG (2015): Short fused? Associations between white matter connections, sex steroids, and aggression across adolescence. Hum Brain Mapp 36:1043–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl KM, Sullivan EV, Rohlfing T, Chu W, Kwon D, Nichols BN, et al. (2016): Harmonizing DTI measurements across scanners to examine the development of white matter microstructure in 803 adolescents of the NCANDA study. NeuroImage 130:194–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldrack RA, Baker CI, Durnez J, Gorgolewski KJ, Matthews PM, Munafò MR, et al. (2017): Scanning the horizon: Towards transparent and reproducible neuroimaging research. Nat Rev Neurosci 18:115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE (2012): Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. NeuroImage 59:2142–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DJ, Kennedy H, Dehay C, Zhou L, Mercier M, Jossin Y, et al. (2006): The development of cortical connections. Eur J Neurosci 23:910–920. [DOI] [PubMed] [Google Scholar]
- de Reus MA, van den Heuvel MP (2013): The parcellation‐based connectome: Limitations and extensions. NeuroImage 80:397–404. [DOI] [PubMed] [Google Scholar]
- Romme IAC, de Reus MA, Ophoff RA, Kahn RS, van den Heuvel MP (2017): Connectome disconnectivity and cortical gene expression in patients with schizophrenia. Biol Psychiatry 81:495–502. [DOI] [PubMed] [Google Scholar]
- Rubinov M, Sporns O (2010): Complex network measures of brain connectivity: Uses and interpretations. NeuroImage [DOI] [PubMed] [Google Scholar]
- Schlaggar BL, Brown TT, Lugar HM, Visscher KM, Miezin FM, Petersen SE (2002): Functional neuroanatomical differences between adults and school‐age children in the processing of single words. Science (New York, N.Y.) 296:1476–1479. [DOI] [PubMed] [Google Scholar]
- Shu N, Liu Y, Li J, Li Y, Yu C, Jiang T (2009): Altered anatomical network in early blindness revealed by diffusion tensor tractography. PLoS ONE 4:e7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds DJ, Hallquist MN, Asato M, Luna B (2014): Developmental stages and sex differences of white matter and behavioral development through adolescence: A longitudinal diffusion tensor imaging (DTI) study. NeuroImage 92:356–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer W (1993): Synchronization of cortical activity and its putative role in information processing and learning. Ann Rev Psychol 55:349–374. [DOI] [PubMed] [Google Scholar]
- Sowell ER, Thompson PM, Tessner KD, Toga AW (2001): Mapping continued brain growth and gray matter density reduction in dorsal frontal cortex: Inverse relationships during postadolescent brain maturation. J Neurosci 21:8819–8829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O (2011): The human connectome: A complex network. Ann N Y Acad Sci. [DOI] [PubMed] [Google Scholar]
- Supekar K, Musen M, Menon V (2009): Development of large‐scale functional brain networks in children. PLoS Biol 7:e1000157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamnes CK, Østby Y, Fjell AM, Westlye LT, Due‐Tønnessen P, Walhovd KB (2010): Brain maturation in adolescence and young adulthood: Regional age‐related changes in cortical thickness and white matter volume and microstructure. Cereb Cortex 20:534–548. [DOI] [PubMed] [Google Scholar]
- Tamnes CK, Herting MM, Goddings AL, Meuwese R, Blakemore SJ, Dahl RE, et al. (2017): Development of the cerebral cortex across adolescence: A multisample study of interrelated longitudinal changes in cortical volume, surface area and thickness. J Neurosci 3302–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel M, Mandl R, Luigjes J, Hulshoff Pol H (2008): Microstruc‐ tural organization of the cingulum tract and the level of default mode functional connectivity. J Neurosci 28:10844–10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Mandl RCW, Kahn RS, Hulshoff Pol HE (2009): Functionally linked resting‐state networks reflect the underlying structural connectivity architecture of the human brain. Hum Brain Mapp 30:3127–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Sporns O (2011): Rich‐club organization of the human connectome. J Neurosci 31:15775–15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Kahn RS, Goni J, Sporns O (2012): High‐cost, high‐capacity backbone for global brain communication. Proc Natl Acad Sci 109:11372–11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Kersbergen KJ, de Reus MA, Keunen K, Kahn RS, Groenendaal F, et al. (2015): The neonatal connectome during preterm brain development. Cereb Cortex 25:3000–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Sporns O, Collin G (2013): Abnormal rich club organization and functional brain dynamics in schizophrenia. Jama. [DOI] [PubMed] [Google Scholar]
- van den Heuvel MP, de Lange SC, Zalesky A (2017): Proportional thresholding in resting‐state fMRI functional connectivity networks and consequences for patient‐control connectome studies: Issues and recommendations. NeuroImage 152:437–449. [DOI] [PubMed] [Google Scholar]
- Watts DJ, Strogatz SH (1998): Collective dynamics of ‘small‐world’ networks. Nature 393:440–442. [DOI] [PubMed] [Google Scholar]
- Wierenga LM, Langen M, Oranje B, Durston S (2014): Unique developmental trajectories of cortical thickness and surface area. NeuroImage 87:120–126. [DOI] [PubMed] [Google Scholar]
- Wierenga LM, van den Heuvel MP, van Dijk S, Rijks Y, de Reus MA, Durston S (2015): The development of brain network architecture. Hum Brain Mapp 37:717–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff SD, Balaban RS (1994): Magnetization transfer imaging: Practical as‐ pects and clinical applications. Radiology 192:593–599. [DOI] [PubMed] [Google Scholar]
- Yendiki A, Koldewyn K, Kakunoori S, Kanwisher N, Fischl B (2013): Spurious group differences due to head motion in a diffusion MRI study. NeuroImage 88:C, 79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BTT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, et al. (2011): The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol 106:1125–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalesky A, Fornito A, Cocchi L, Gollo LL (2016): Connectome sensitivity or specificity: Which is more important? NeuroImage 142:407–420. [DOI] [PubMed] [Google Scholar]


