Abstract
Plants and algae have developed various light-harvesting mechanisms for optimal delivery of excitation energy to the photosystems. Cryptophyte algae have evolved a novel soluble light-harvesting antenna utilizing phycobilin pigments to complement the membrane-intrinsic Chl a/c-binding LHC antenna. This new antenna consists of the plastid-encoded β-subunit, a relic of the ancestral phycobilisome, and a novel nuclear-encoded α-subunit unique to cryptophytes. Together, these proteins form the active α1β·α2β-tetramer. In all cryptophyte algae investigated so far, the α-subunits have duplicated and diversified into a large gene family. Although there is transcriptional evidence for expression of all these genes, the X-ray structures determined to date suggest that only two of the α-subunit genes might be significantly expressed at the protein level. Using proteomics, we show that in phycoerythrin 545 (PE545) of Guillardia theta, the only cryptophyte with a sequenced genome, all 20 α-subunits are expressed when the algae grow under white light. The expression level of each protein depends on the intensity of the growth light, but there is no evidence for a specific light-dependent regulation of individual members of the α-subunit family under the growth conditions applied. GtcpeA10 seems to be a special member of the α-subunit family, because it consists of two similar N- and C-terminal domains, which likely are the result of a partial tandem gene duplication. The proteomics data of this study have been deposited to the ProteomeXchange Consortium and have the dataset identifiers PXD006301 and 10.6019/PXD006301.
Keywords: Cryptophyta, Phycobilin, Phycobiliprotein, Translation, TAT-pathway, Proteomics
Introduction
Photosynthesis is the process that powers all life on Earth, generates renewable energy and food, and counteracts the greenhouse effect. Algae and cyanobacteria are without doubt the most productive photosynthetic organisms on Earth. The first step of photosynthesis is harvesting of sunlight by designated pigment-binding antenna complexes, which in the photosynthetic light reaction rapidly transfer the absorbed light energy to a reaction center (Blankenship 2014; Mirkovic et al. 2016). While the reaction centers of Photosystem II (PSII) and Photosystem I (PSI) remained highly conserved during evolution, various antenna systems have evolved in photosynthetic organisms. Prokaryotic cyanobacteria and eukaryotic red algae contain phycobilisomes as their major antennae-rods of stacked phycobiliproteins, to which the linear tetrapyrrole phycobilin pigments are covalently bound (Adir 2005; Watanabe and Ikeuchi 2013). These structures are extrinsically associated with the stromal side of the thylakoid membrane. In higher plants and green algae, the most abundant antenna is the chlorophyll a/b-binding light-harvesting complex (referred to as LHC), which is inserted into the thylakoid membrane (Neilson and Durnford 2009). As well as the phycobilisome, red algae also have a related LHC antenna, which binds only Chl a and is mainly associated with PSI (Gantt et al. 2003).
In addition to these photosynthetic organisms, several major algal groups acquired their chloroplasts by secondary endosymbiogenesis from a red algal endosymbiont (Gibbs 1981). A particularly interesting example is the cryptophyte algae, which are unique in having retained a remnant of the red algal nucleus, called the nucleomorph. The nucleomorph is located in the periplastid space, next to the chloroplast envelope, and is surrounded by two additional membranes derived from the red algal plasma membrane and the host endomembrane system (Gould et al. 2008). Cryptophytes use two different light-harvesting systems: phycobiliproteins and the chlorophyll Chl a/c-binding proteins (MacPherson and Hiller 2003; Broughton et al. 2006). The Chl a/c-binding proteins are homologous to the LHCs of the red algae and the Chl a/b-LHCs of the green lineage (Green and Durnford 1996; Durnford et al. 1999), and like these they are integrated in the thylakoid membrane. In contrast, the phycobiliprotein antenna of cryptophytes is unique (Glazer and Wedemayer 1995; MacColl et al. 1999). Rather than the complex multiprotein phycobilisome stucture, the cryptophyte phycobiliproteins are small proteins located in the thylakoid lumen (Spear-Bernstein and Miller 1989). They are compact tetrameric complexes made of two identical copies of a 18–20 kDa β-subunit (related to a phycobilisome β-subunit) and two small (8–10 kDa) subunits which were named “α-subunits,” but they have no relatedness to the phycobilisome α-subunits or to any other protein in sequence databases (Wilk et al. 1999; Doust et al. 2004). High resolution crystal structures have been determined for Rhodomonas sp. phycoerythrin (PE) 545 (Wilk et al. 1999) (Fig. 1) as well as several other cryptophyte phycobiliproteins (Doust et al. 2004; Harrop et al. 2014; Arpin et al. 2015).
Fig. 1.
Crystal structure of Rhodomonas sp. PE545 (PDB 1xg0, Wilk et al. 1999). The α-1 subunit is displayed in yellow and the α-2 subunit in purple. The β-subunits are shown in blue
In general, very little is known about light-harvesting mechanisms in cryptophytes. Many of these algae are adapted to low light and are suggested to take advantage of quantum coherence to improve the efficiency of energy transfer (Collini et al. 2010; Harrop et al. 2014). The photoprotective mechanism of two cryptophyte species has been studied in Rhodomonas salina (Kaňa et al. 2012) and Guillardia theta (Funk et al. 2011; Cheregi et al. 2015), and it does not appear to involve the phycobiliprotein antenna. G. theta has PE545, like Rhodomonas, and its nuclear genome has a surprisingly large number of genes encoding α-subunits (Gould et al. 2007; Curtis et al. 2012) compared to the six genes isolated by standard cloning techniques in Rhodomonas CS24 (Broughton et al. 2006). To find out if all the G. theta genes are expressed at the protein level, we used proteomics to test and optimize the gene models, and we found that all 20 α-subunit genes are indeed expressed into protein. Our data also show that the expression of the α-subunits depends on the intensity of the growth light, but there is little evidence for a specific light regulation of individual members of the α-subunit family under our experimental conditions.
Materials and methods
Culturing and cell counting
Guillardia theta cells (CCMP2712) were obtained from the Provasoli-Guillard National Center for Culture of Marine Phytoplankton. Cultures were grown in Fernbach culture flasks in h/2 media (Guillard 1975) under white light at 20 °C with light–dark cycle of 12:12 h, and shaken at 120 rpm. Light intensities were low light (7.5 µmol m−2 s−1, LL), standard light (30 µmol m−2 s−1, SL), and high light (150 µmol m−2 s−1, HL). Cultures of 1 l were started with the same number of cells (~105). Every day cell number and size were determined using a calibrated Coulter Counter (Beckman Multisizer III) equipped with a 70 µm aperture. Samples were measured in triplicates. Samples were harvested after 4 days (HL), or 7 days (SL) at a cell number of 1.6 × 106 cells/ml, which corresponds to the late exponential phase. Cells grown at LL were harvested after 14 days, when they had reached a density of about 1 × 106 cells/ml.
Pigment determination
Chlorophyll a and c concentrations in the cells were determined by absorption using an UV/VIS spectrophotometer (Unicam UV 550, Thermo Spectronic, UK) and calculated according to the equations of Jeffrey and Humphrey (1975). Triplicates of 5 ml of the algal suspension were filtered onto Whatman GF/F filters; the pigments were extracted by 90% acetone for 24 h at 4 °C in darkness.
Absorption and fluorescence spectra
For absorption spectra, cells were collected on nitrocellulose membrane filters (Pragochema, Czech Republic), and the filters were then positioned in the integrating sphere of a Unicam UV550 spectrophotometer (Thermo Spectronic, UK). Absorbance was measured between 400 and 800 nm, with a bandwidth of 4 nm.
77 K fluorescence emission spectra were measured using a Fluorolog-3 spectrofluorometer (Horiba jobin Yvon, Japan). One ml of culture, diluted to the same cell number/ml (5 × 105) were used in each measurement. Fluorescence was excited at 435 or 545 nm and measured from 550 to 800 nm with a bandwidth of 1 nm.
Fluorescence yield quenching (NPQ) and maximum photochemical efficiency of PSII (Fv/Fm) were measured using an AquaPen-C AP-C 100 device (Photon Systems Instruments, Czech Republic).
Gene models and protein sequences
Gene models for 21 nuclear-encoded α-subunits were identified during the annotation of the draft genome of G. theta (Curtis et al. 2012). Where possible, model numbers were based on those assigned to the corresponding ESTs in Gould et al. (2008). In preparation for proteomic analysis, each gene model was carefully reexamined with respect to transcript support, intron splice sites, and model completeness, including the targeting sequences. CpeA7 and CpeA11 mapped to the same position on the genome, so CpeA7 has been deleted from the genome annotation, leaving 20 complete gene models. Alternative models were generated for several genes and, in every case, tested using the peptide sequences identified by mass spectrometry. A fasta file with the latest versions of the gene models is available at ProteomeXchange in the dataset PXD006301.
Sequence analysis
Protein sequence alignments were generated with MAFFT version 7 (http://mafft.cbrc.jp/alignment/server/index.html) (Katoh and Standley 2013), using the default settings, and refined with BioEdit ver. 5.0.9 (Hall 1999). The analysis of the targeting sequences was performed according to Gould et al. (2008), C-terminal transit peptide cleavage sites were predicted according to Huesgen et al. (2013), and cleavage of the lumenal targeting domain by the presence of an AXA motif.
Design of the sample set and preparation of protein extracts for mass spectrometry analysis
The sample set included five biological replicates of extracts containing soluble proteins from G. theta grown at LL, SL and HL. The extracts were prepared by harvesting the cells in late exponential phase: after 14 days of growth in low light, 7 days of growth in standard light, or 4 days of growth in high light. A variable volume of culture containing the same number of cells (4.5 × 108) was harvested for each biological replicate and each culture condition. The pelleted cells were resuspended in 1 ml precooled breaking buffer (0.25 M Sorbitol, 20 mM HEPES, 0.4 mM Na2EDTA, pH ~7.5), transferred to 2 ml screw cap tubes containing 1 ml glass beads, frozen in liquid nitrogen, and kept at −80 °C until further analysis. Cells were thawed then broken on ice in darkness in a Bead beater (Glen Mills) in three cycles of 1 min with pauses of 3 min in between. Glass beads and whole cell debris were removed by centrifugation at ~600×g for 3 min. The green supernatant was centrifuged again at 21,000×g for 10–15 min to remove fragments of thylakoid membranes, giving a clear dark pink supernatant containing the phycobiliproteins as one of the major components.
The sample preparation included reduction of cysteine residues using 5 mM DTT for 20 min at 56 °C and subsequent alkylation of the thiol groups in the presence of 15 mM fresh iodoacetamide for 15 min in the dark. Next, the proteins were desalted using Zeba spin 0.5 ml columns (Thermo Fisher Scientific, Stockholm, Sweden) that were equilibrated using 50 mM Hepes pH 8.0, and the protein concentration was determined according to the method of Lowry as described by Peterson (1977). To minimize disturbance through the absorbance of the phycoerythrins, protein concentrations were measured at 750 nm.
The sample set for the preparation of in-solution digests included three technical replicates for each of the 15 biological samples. Of each sample, an aliquot containing 50 µg protein in 100 µl of 50 mM Hepes pH 8.0 was prepared, and the proteins were digested for 3 h at 37 °C in the presence of 18 ng/µl of sequencing grade trypsin (Promega Biotech AB, Nacka, Sweden). The digestion was stopped by adding 10% formic acid to a final concentration of 0.5%, and the samples were stored in −80 °C.
Mass spectrometry analysis and bioinformatics
Automated Data Dependent Acquisition (DDA) spectra were acquired using a Synapt G2-Si mass spectrometer linked on-line to an ACQUITY UPLC M-Class System (Waters AB, Sollentuna, Sweden). The data acquisition was performed in the positive ion mode using continuum data format and lock mass calibration. In the MS mode, spectra were acquired over the range of 350–2000, and in the MS/MS mode, spectra acquisition was performed over the range of 50–2000 using charge state recognition up to four charges and eight MS/MS channels. In both the MS and MS/MS modes, a scan time of 0.4 s and an interscan scan time of 0.015 s were used. The settings for the cone voltage was 40 V. Fragmentation in the MS/MS mode was performed using MS Trap collision energy profiles ranging from 20 to 25 V in the low-mass range and from 30 to 45 V in the high-mass range. Data were acquired in the time window from 10 to 68 min.
Nanoliquid chromatography separation of peptides was performed at a flow rate of 280 nl/min and 35 °C using a combination of a Trap V/M Symmetry C18 column (100 Å, 5 µm, 180 µm × 20 mm) and HSS T3 C18 analytical column (1.8 µm, 75 µm × 250 mm) (Waters AB, Sollentuna, Sweden). The gradient was generated using 75% acetonitrile, 25% isopropanol in 0.1% formic acid (solvent B) and included the following steps: 0.5 min, 5% B; 1 min 5% B, 37 min, 41% B, 41 min, 95% B, 53 min 95% B, 57 min, 5% B. The total run time of the LC method was 69 min.
Processing of the DDA data was performed using the ProteinLynx Global Server 3.0 software (Waters AB, Sollentuna, Sweden) and the recommended settings for High Definition Data Direct Analysis (HD-DDA), including lockspray calibration and fast deisotoping in both the MS and MS/MS mode. Database searches using the peak lists of the processed mass spectra were performed using the Mascot search engine (version 2.5) in a set of databases, which included a homemade database of the gene models of the phycoerythrins of G. theta, a database of the JGI gene models of G. theta without the phycoerythrins, a database of contaminants and Glu-1-fibrinopeptide B. The search parameters permitted a mass error of 5 ppm (MS mode) and 0.05 Da, respectively (MS/MS mode) and tryptic cleavage with one missed cleavage site. Modifications included variable oxidation of methionine, and variable deamidation of asparagine and glutamine, and fixed modification of cysteine residues by carbamidomethylation. For a given database search for a biological sample, the peak-lists of the three technical replicates were merged. The Percolator of the Mascot search engine was on, and no cutoffs were applied to the percolated Mascot scores.
The use of the Percolator improved the sensitivity of the searches and allowed the detection of the unique tryptic peptides distinguishing GtcpeA1 from GtcpeA21, and GtcpeA9 from GtcpeA19. To detect N-terminal peptides, the databases searches were also performed using semitryptic cleavage, and matched spectra were inspected manually. A semiquantitative analysis of the relative expression of the α- and β-subunits was performed according to Dowle et al. (2016). The proteomics work was performed at the KBC Proteomics Core Facility at the Umeå University and the Swedish University of Agricultural Sciences, and the mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaino et al. 2014; Vizcaíno et al. 2016) partner repository with the dataset identifiers as PXD006301 and 10.6019/PXD006301.
Prediction of structure models
A prediction of the structures of the α- and β-phycoerythrins was performed by threading using the Phyre2 web server (Kelley et al. 2015), and the predicted structures were assessed by structural alignments to the structure templates using FATCAT (Ye and Godzik 2003, 2004a) on the public FATCAT server (Ye and Godzik 2004b; Li et al. 2006). The images of the structural alignments of the subunits of phycoerythrin 545 (PDB 1xg0) with the predicted folds of phycobiliproteins of G. theta were created using Chimera (Pettersen et al. 2004).
Results and discussion
Growth and light acclimation of G. theta
To investigate the growth of G. theta under different light intensities, cultures were grown under low light (7.5 µmol m−2 s−1, LL), standard light (30 µmol m−2 s−1, SL), and high light (150 µmol m−2 s−1, HL) in a light:dark regime of 12:12 h at 20 °C and cells counted daily. Within 4 days of culturing the high-light, resp cultures reached a cell number of 1.6 × 106 cells/ml, while under standard conditions the same number of cells was reached after 7 days culturing. The low-light cells grew very slowly and after 14 days in culture had only reached ~1 × 106 cells/ml. All cultures were harvested in exponential stage for pigment- and biophysical analyses.
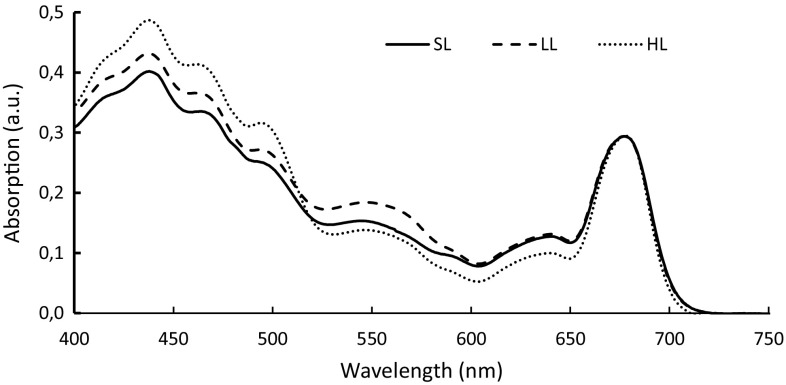
Besides the membrane integral chlorophyll-containing antenna, G. theta contains phycoerythrin 545 (PE545), the red pigment–protein complex located in the thylakoid lumen (Wilk et al. 1999; Broughton et al. 2006). The pigmentation of G. theta cells grown at different light intensities varied noticeably: cells grown under LL conditions were deep red in color, cells grown under SL conditions had a reddish/brownish color, while cells grown under HL displayed a yellowish-green color (not shown). Absorption spectra of suspensions with similar cell density (5 × 105 cells/ml) of these cultures showed absorption maxima corresponding to Chl a (436 and 680 nm), Chl c (465 nm), carotenoids (495 nm), and phycoerythrin (550 nm) (Fig. 2). The spectra were normalized at 678 nm, the maximum of PSII absorption. Absorbance at 550 nm was much higher in LL-grown cells than in HL-grown cells, reflecting their higher PE545/Chl levels as reported in earlier studies (Faust and Gantt 1973; Lichtlé 1979). Conversely, carotenoid absorption was highest in HL, resp cells (Fig. 2).
Fig. 2.
Room temperature absorption spectra of whole cells of G. theta grown under three light intensities: standard-light (SL) (30 µmol m−2 s−1), low-light (LL) (7.5 µmol m−2 s−1), or high-light (HL) (150 µmol m−2 s−1) conditions. Spectra were normalized at 678 nm, the maximum of PSII absorption. The same number of cells was used for each measurement. SL and HL cells were measured in late exponential phase; the slow-growing LL cells were probably in mid-exponential phase
Chls a and c were estimated in acetone extracts (Table 1). Cells grown under SL had about three times the amount of Chl per cell compared to HL grown cells (Table 1), while the Chl/cell content in LL grown cells was about 60% of the one in SL cells, probably because the low level of light limited not only cell growth, but also pigment biosynthesis. The Chl a/Chl c ratio increased from 3:1 in LL, resp cells to 6:1 in HL-grown cells, indicating the Chl c containing antenna to be downregulated in high light. The maximal efficiency of PSII photochemistry (Fv/Fm) measured in the harvested cells demonstrated optimal PSII efficiency (higher than 0.7, the maximum reported in the literature) in all cultures, independent of the light intensity. This indicates that the cells were acclimated to their growth-light levels and are not suffering photoinhibition. This is reinforced by that fact that the non-photochemical quenching (NPQ) capacities of HL cells were similar to that of SL cells (0.59 and 0.57, respectively), while LL cells had only developed half of the protective capacity (NPQ of 0.3) (Table 1).
Table 1.
Photosynthetic parameters of cells grown under different light intensities
| Low light (LL) | Standard light (SL) | High light (HL) | |
|---|---|---|---|
| Chl/cell (pg/cell) | 0.776 (±0.016) | 1.300 (±0.07) | 0.441 (±0.007) |
| Chl a/Chl c | 3.56 (±0.17) | 4.4 (±0.63) | 6.2 (±0.3) |
| Fv/Fm | 0.77(±0) | 0.77(±0.01) | 0.72 (±0.01) |
| NPQ | 0.30 (±0.04) | 0.59 (±0.03) | 0.57 (±0.02) |
Standard- and high-light cells were harvested in the late exponential phase at 1.6 × 106 cells/ml. Low-light cells were in the mid-exponential phase when harvested at 1.0 × 106 cells/ml. Average values and standard deviations are calculated from 3 to 5 biological replicates
Low-temperature (77 K) fluorescence emission spectra of intact cells are shown in Fig. 3. Exciting Chl a at 435 nm, two PSII specific emission peaks were noticed with maxima at 686 and 696 nm (Fig. 3a). The main contributor to the 695 nm maximum most likely is a low-energy chlorophyll that appears to be associated with His-114 of CP47, as in the cyanobacterium Synechocystis sp. PCC 6803 (Shen and Vermaas 1994). The other chlorophylls of the PSII-core complex together are represented by the 685 nm peak (Shen et al. 1993). While in cells grown at SL and HL the ratio of A686/A696 was lower than 1, in cells grown at LL the 696 nm peak was dominant. The rise of the A696 nm peak in LL cells was also seen in the 77 K spectra when PE545 was excited at 545 nm (Fig. 3b). The PE pool, which is accumulating in LL cells, seems energetically better connected to PSII. The 77 K spectra of SL- and LL cultures further displayed a third emission maxima at 578 nm, which is attributed to a pool of PE present only during the logarithmic growth phase (Cheregi et al. 2015). In HL-grown cells, this emission peak was completely absent. This pool of PE therefore either is energetically more strongly coupled to the photosystems during HL or it is degraded.
Fig. 3.
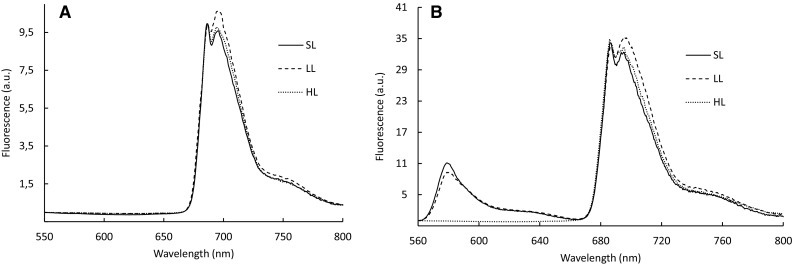
Low-temperature (77 K) fluorescence spectra of G. theta cells grown under standard-light (SL) (30 µmol m−2 s−1), low-light (LL) (7.5 µmol m−2 s−1) or high-light (HL) (150 µmol m−2 s−1) conditions. Chlorophyll a was excited at 435 nm (a), and phycoerythrin PE545 was excited at 545 nm (b). Spectra were normalized to PSII fluorescence at 687 nm. The same number of cells was used for each measurement
Gene models and expression of the α-subunits
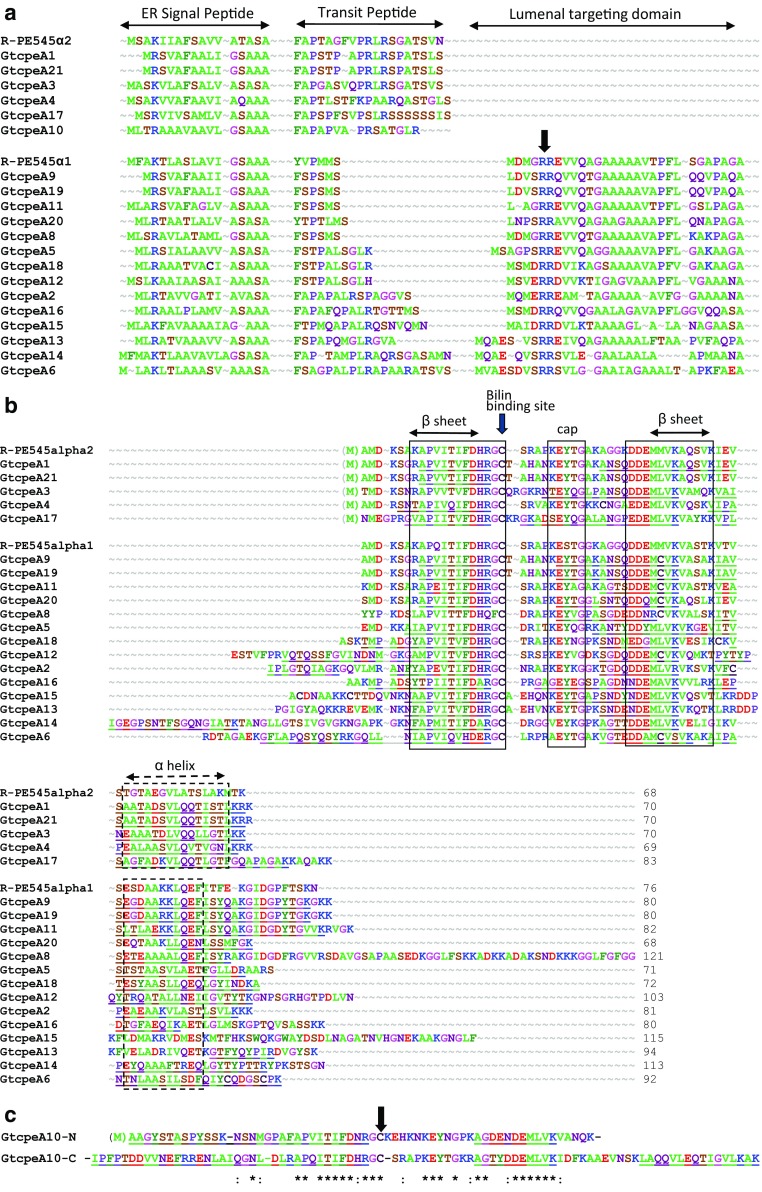
The nuclear genome of G. theta contains 20 different genes encoding PE545 α-subunits, annotated GtcpeA1-6 and GtcpeA8-21 (Curtis et al. 2012). In our reexamination of the gene models, the incomplete GtcpeA7 model mapped to the same position on the genome as GtcpeA11, and it was therefore deleted from the genome annotation. The 20 genes are scattered across a number of scaffolds, with the exception of two pairs of neighboring genes: GtcpeA1-A9 on scaffold 57 and GtcpeA21-A19 on scaffold 117. GtcpeA9 and GtcpeA19 encode almost identical protein sequences, as do GtcpeA1 and GtcpeA21. These closely related pairs are clearly the result of a duplication of the whole gene pair followed by transposition. Like the three pairs of α-subunit genes in Rhodomonas sp. CS24 (Broughton et al. 2006), the two genes are divergently transcribed. One member of each pair encodes a slightly longer protein precursor with the full tripartite targeting sequence including the lumenal TAT targeting domain (α1-type), whereas the other has only the ER signal peptide plus the transit peptide (α2-type) (Fig. 4). Since the plastid-encoded β-subunit also lacks the lumenal targeting domain (LTD), this suggested that the tetramer is assembled in the plastid stroma and then targeted across the thylakoid membrane by the α1-type protein (Broughton et al. 2006; Gould et al. 2008). The G. theta α-subunit sequences are closely related to those of Rhodomonas, so it is expected that the holoprotein will have the same three-dimensional structure as the PE545 of Rhodomonas sp. CS24 (Wilk et al. 1999).
Fig. 4.
Multiple sequence alignment of the G. theta α-subunit sequences colored according to chemical similarity. a Precursor targeting sequences. All precursors start with an ER signal peptide, cleaved from the following chloroplast transit peptide at the conserved motif AXA_(F/Y) (Gruber et al. 2007). The C-terminal cleavage site of the chloroplast transit peptide is predicted according to Huesgen et al. (2013) with a conserved Leu or Met at the -2 position. Where present, the lumenal targeting domain has a twin arginine motif near the N-terminus (arrow) and a hydrophobic core, typical of proteins targeted to the thylakoid lumen via the TAT import machinery, and a C-terminal AXA cleavage site to release the mature protein. b Mature proteins aligned with the α1- and α2-sequences from the crystal structure of PE545 from Rhodomonas sp. CS24 (Wilk et al. 1999). Peptides identified by mass spectrometry are underlined. N-terminal Met are in parenthesis to indicate the high probability that they are removed by Met aminopeptidase (Huesgen et al. 2013; reviewed by Giglione and Meinnel 2001). Highly conserved regions corresponding to secondary structural elements of the Rhodomonas structure are boxed. The EYxG motif forms a cap on the end of one of the β-subunit helices. Arrow, bilin-binding site. c Internal duplication of GtcpeA10. The mature sequence was split into N- and C-terminal halves and aligned to show sequence relatedness. Both halves a bilin-binding site. Peptides identified by mass spectrometry are underlined. Parenthesis, removed N-terminal Met; arrow, bilin-binding site
The rest of the α-subunit genes do not fall neatly into pairs with and without LTDs (Fig. 4a). There are 14 genes encoding a complete tripartite targeting sequence that includes an LTD, and 6 that have no LTD. They all have a canonical endoplasmic reticulum signal peptide (SP), with predicted SP-transit peptide cleavage site AXA-FXP as determined for diatoms (Gruber et al. 2007; Huesgen et al. 2013). The transit peptides are varied in length but have the same overall composition as diatom and plant transit peptides, being enriched in hydroxylated amino acids (S and T) and with at least one positive charge except for the shortest ones. C-terminal cleavage sites were predicted according to Huesgen et al. (2013), with a conserved Leu or Met at the -2 position. All LTDs have the conserved twin arginine motif followed by a hydrophobic domain characteristic of the TAT-transport system and an AXA-motif at the C-terminal cleavage site, with the exception of GtcpeA13 which most likely has the C-terminal cleavage site VFA.
In Fig. 4b, the predicted mature protein sequences are aligned with the α1- and α2-sequences from the crystal structure of Rhodomonas PE545 (Wilk et al. 1999; Harrop et al. 2014). All sequences share two blocks of high sequence similarity, which correspond to the two β-strands seen in the crystal structure, and the conserved FDxRGC motif, where the Cys residue is covalently linked to dihydrobiliverdin. They also share a somewhat conserved block predicted to form the characteristic C-terminal alpha helix. These structural elements are so conserved across all six cryptophyte phycobiliproteins structures, which have been determined at high resolution (Harrop et al. 2014; Arpin et al. 2015), that it is safe to conclude that the G. theta α-subunits will also have a similar fold when assembled with their β-subunit partners.
A number of the sequences encoded by genes with LTDs have long N- and/or C-terminal extensions. The N-terminal extensions of GtcpeA2 and GtcpeA14 are supported by peptide sequences (Fig. 4b, underlined), and probably represent the true mature N-termini. There is also peptide support for the extensions of GtcpeA6 and GtcpeA12. At the C-terminal end, the distinction between α1 type (with LTD) and the α2 type (shorter, no LTD) breaks down, with both long and short C-termini, many with peptide support as detailed in the next section about the proteomics analysis.
A most interesting gene model is GtcpeA10, which has a second FDxRGC motif as the result of a partial internal duplication. Figure 4c shows that the N- and C-terminal parts of this gene model have a higher sequence similarity to one another than to the other α-subunits of this group. Both parts have high peptide coverage. We have found homologs to this sequence in transcriptomes of two other cryptophytes with PE545 and one with PE566 (data not shown) showing that this duplicated sequence arose in the common ancestor of the clades with PE and has not been eliminated by selection or degenerated into a pseudogene. This implies that this gene model represents a functional protein, but it raises the question of how it would interact with the other α- and β-subunits.
Proteomics analysis
To test the gene models and their expression, our proteomic analysis included G. theta cultures grown under white light of three different intensities: 30 µmol m−2 s−1 (SL), 7.5 µmols m−2 s−1 (LL), and 150 µmols m−2 s−1 (HL). For each light condition, five biological replicates were grown, of which in turn three technical replicates were analyzed by mass spectrometry. In total, our dataset consists of 45 LC-MS/MS runs and the processed data contain 69615 spectra, of which 5950 match tryptic peptides of the phycobiliproteins. The datasets from the LC-MS/MS analysis were first used for database searches with the Mascot search engine, to verify and optimize the gene models. Table 2 gives the percolated Mascot scores, which have higher sensitivity than standard Mascot scores [for explanation. see, e.g., (Brosch et al. 2009)]. Table 2 shows that in the samples from algae that were grown under SL and LL conditions, all 20 α-subunits were detected. For most of the α-subunits, the identified peptides have two or more unique sequences (Table 3), and together they cover large parts of the predicted mature proteins, which are underlined in Fig. 4b, c.
Table 2.
Phycoerythrins of G. theta expressed under different light growth conditions
| Gene model | Low-light growth conditions | Standard-light growth conditions | High-light growth conditions | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | |
| GtcpeA1 | 988 | 1298 | 478 | 700 | 697 | 1421 | 667 | 730 | 1390 | 860 | 971 | 733 | 495 | 802 | 855 |
| GtcpeA2 | 102 | 130 | 106 | 47 | 42 | 204 | 181 | 68 | 280 | 276 | 26 | ||||
| GtcpeA3 | 511 | 453 | 254 | 274 | 369 | 983 | 423 | 423 | 669 | 394 | 86 | 453 | 172 | 313 | 183 |
| GtcpeA4 | 208 | 256 | 83 | 67 | 116 | 387 | 226 | 172 | 205 | 89 | |||||
| GtcpeA5 | 138 | 223 | 169 | 138 | 184 | 466 | 223 | 297 | 378 | 285 | 64 | 62 | 176 | 48 | |
| GtcpeA6 | 342 | 673 | 299 | 563 | 379 | 653 | 256 | 439 | 753 | 810 | 332 | 423 | 216 | 581 | 377 |
| GtcpeA8 | 158 | 270 | 136 | 18 | 200 | 327 | 262 | 239 | 368 | 365 | 42 | ||||
| GtcpeA9 | 909 | 381 | |||||||||||||
| GtcpeA10 | 858 | 1037 | 384 | 537 | 539 | 1421 | 621 | 554 | 1553 | 876 | 81 | 509 | 194 | 220 | 178 |
| GtcpeA11 | 365 | 411 | 134 | 236 | 191 | 612 | 324 | 223 | 716 | 535 | 240 | 323 | 202 | 302 | 244 |
| GtcpeA12 | 576 | 825 | 325 | 500 | 301 | 613 | 298 | 425 | 728 | 426 | 133 | 412 | 237 | 560 | 583 |
| GtcpeA13 | 31 | 89 | 25 | 172 | 102 | 93 | 92 | 93 | 50 | 35 | 42 | 18 | |||
| GtcpeA14 | 381 | 440 | 213 | 332 | 290 | 750 | 283 | 292 | 812 | 646 | 162 | 189 | 224 | 367 | 152 |
| GtcpeA15 | 355 | 477 | 205 | 333 | 281 | 484 | 210 | 332 | 581 | 441 | 231 | 194 | 130 | 278 | 318 |
| GtcpeA16 | 774 | 1679 | 810 | 700 | 1019 | 1913 | 779 | 793 | 1461 | 996 | 1026 | 925 | 889 | 1592 | 909 |
| GtcpeA17 | 78 | 359 | 165 | 98 | 159 | 567 | 479 | 217 | 253 | 147 | |||||
| GtcpeA18 | 516 | 968 | 381 | 589 | 610 | 1448 | 953 | 440 | 529 | 507 | 263 | 301 | 460 | 811 | 438 |
| GtcpeA19 | 975 | 269 | 404 | 370 | 1136 | 852 | 404 | 375 | 278 | ||||||
| GtcpeA9 or GtcpeA19 | 323* | 754* | 477* | ||||||||||||
| GtcpeA20 | 543 | 1031 | 375 | 675 | 615 | 1282 | 573 | 605 | 1514 | 900 | 638 | 447 | 405 | 558 | 608 |
| GtcpeA21 | 973 | 1241 | 469 | 681 | 677 | 1283 | 660 | 702 | 1320 | 816 | 883 | 730 | 463 | 767 | 848 |
| Gt-Beta | 2308 | 3619 | 2077 | 2400 | 1500 | 2869 | 1151 | 1437 | 4407 | 4133 | 1443 | 1304 | 1139 | 1768 | 1581 |
Percolated Mascot scores of the antenna gene models identified in fifteen biological replicates
The database searches were performed with the Mascot 2.5 search engine, and the search results have ≥95% confidence and a false discovery rate of ≤1%. Each search includes the peak list files of three technical replicates, and the table summarizes the results from 45 LC-MS/MS runs. If a search did not give a significant score for a gene model, the cell is empty
*Identified peptides could not be destinguished between GtcpeA9 and GtcpeA19 in database search
Table 3.
Experimentally detected unique peptides of the phycobiliproteins of G. theta
| Protein | Start and end in precursor | Start and end in predicted mature protein | Unique tryptic peptide | Highest peptide score | Lowest peptide expectation value | Spectra with expectation values <0.05 |
|---|---|---|---|---|---|---|
| Beta | 1–7 | MLDAFSR (N-terminus) | 47.64 | 1.70E−05 | 14 | |
| 16–28 | AAYVGGADLQALK | 117.63 | 1.70E−12 | 90 | ||
| 16–29 | AAYVGGADLQALKK | 90.98 | 8.10E−10 | 147 | ||
| 85–91 | DGEIILR | 39.35 | 0.00012 | 6 | ||
| 92–108 | YVSYALLSGDSSVLEDR | 155.02 | 3.20E−16 | 289 | ||
| 115–129 | ETYSSLGVPANSNAR | 120 | 1.00E−12 | 230 | ||
| 130–149 | AVSIMKACAVAFINNTASQR | 145.61 | 2.80E−15 | 30 | ||
| 136–149 | ACAVAFINNTASQR | 155.02 | 3.20E−16 | 160 | ||
| 150–171 | KLSTPQGDCSGLASECASYFDK | 114.82 | 3.30E−12 | 31 | ||
| 151–171 | LSTPQGDCSGLASECASYFDK | 155.02 | 3.20E−16 | 26 | ||
| GtcpeA1 | 42–51 | 9–18 | APVITVFDHR | 73.05 | 5.00E−08 | 181 |
| GtcpeA2 | 59–68 | 1–10 | IPLGTQIAGK (N-terminus) | 25.67 | 0.0027 | 30 |
| 75–88 | 17–30 | ANFYAPEVTIFDHR | 76.29 | 2.40E−08 | 6 | |
| 102–112 | 44–54 | TGDQDDEMLVR | 89.35 | 1.20E−09 | 10 | |
| 118–127 | 60–69 | VFCPEAEAAK | 13.82 | 0.041 | 1 | |
| 128–137 | 70–79 | VLASTLSVLK | 77.34 | 1.80E−08 | 39 | |
| GtcpeA3 | 45–54 | 9–19 | APVVTVFDHR | 91.3 | 7.40E−10 | 35 |
| 62–80 | 26–44 | NTEYQGLPANSQDDEMLVK | 155.02 | 3.20E−16 | 90 | |
| 61–80 | 25–44 | RNTEYQGLPANSQDDEMLVK | 18.77 | 0.013 | 1 | |
| 86–105 | 50–69 | VAINEAAATDLVQQLLGTLK | 120.64 | 8.60E−13 | 16 | |
| 86–106 | 50–70 | VAINEAAATDLVQQLLGTLKK (C-terminus) | 106.65 | 2.20E−11 | 13 | |
| GtcpeA4 | 43–55 | 6–18 | SNTAPIVQIFDHR | 92.01 | 6.30E−10 | 35 |
| 84–104 | 47–67 | VIPAPEALAASVLQVTVGNLK | 93.35 | 4.60E−10 | 12 | |
| GtcpeA5 | 63–74 | 6–17 | AIAPVITIFDHR | 112.95 | 5.10E−12 | 20 |
| 88–98 | 31–41 | ANTYDDYMLVK | 58.6 | 1.40E−06 | 16 | |
| 87–98 | 30–41 | KANTYDDYMLVK | 58.06 | 1.60E−06 | 8 | |
| 101–124 | 44–52 | GEVITVSTSTAASVLAETFGLLDR | 37.6 | 0.00017 | 1 | |
| 99–124 | 42–67 | VKGEVITVSTSTAASVLAETFGLLDR | 141.73 | 6.70E−15 | 32 | |
| GtcpeA6 | 78–89 | 9–20 | GFLAPQSYQSYR | 91.23 | 7.50E−10 | 120 |
| 90–106 | 21–37 | KGQLLNIAPVIQVHDER | 55.2 | 3.00E−06 | 16 | |
| 91–106 | 22–37 | GQLLNIAPVIQVHDER | 97.83 | 1.60E−10 | 16 | |
| 120–132 | 51–63 | VGTEDDAMCVSVK | 132.58 | 5.50E−14 | 174 | |
| 135–161 | 66–92 | AIPANTNLAASILSDFQIYCQDGSCPK (C-terminus) | 126.81 | 2.10E−13 | 129 | |
| GtcpeA8 | 77–90 | 25–38 | EYVGPASGDEDDNR | 76.52 | 2.20E−08 | 6 |
| 99–117 | 47–65 | ITVSETEAAAALQEFISYR | 154.66 | 3.40E−16 | 41 | |
| GtcpeA9 | 98–107 | 48–57 | IAVSEGDAAK | 26.6 | 0.0022 | 3 |
| GtcpeA10 | 34–46 | 2–14 | AAGYSTASPYSSK (N-terminus) | 28.85 | 0.0013 | 3 |
| 47–64 | 15–32 | NSNMGPAFAPVITIFDNR | 155.02 | 3.20E−16 | 71 | |
| 79–89 | 47–57 | AGDENDEMLVK | 28.24 | 0.0015 | 10 | |
| 95–107 | 63–75 | IPFPTDDVVNEFR | 145.05 | 3.10E−15 | 50 | |
| 95–108 | 63–76 | IPFPTDDVVNEFRR | 25.22 | 0.003 | 7 | |
| 109–120 | 77–88 | ENLAIQGNLDLR | 67.25 | 1.90E−07 | 18 | |
| 109–130 | 77–98 | ENLAIQGNLDLRAPQITIFDHR | 26.91 | 0.002 | 3 | |
| 121–130 | 89–98 | APQITIFDHR | 125.77 | 2.60E−13 | 79 | |
| 144–154 | 112–122 | AGTYDDEMLVK | 32.63 | 0.00055 | 29 | |
| 166–179 | 134–147 | LAQQVLEQTIGVLK | 84.23 | 3.80E−09 | 29 | |
| 166–181 | 134–149 | LAQQVLEQTIGVLKAK (C-terminus) | 31.42 | 0.00072 | 1 | |
| GtcpeA11 | 59–68 | 8–17 | APEITIFDHR | 89.96 | 1.00E−09 | 61 |
| 82–92 | 31–41 | AGTSDDEMCVK | 42.6 | 5.50E−05 | 10 | |
| 98–107 | 47–56 | VEASLTLAEK | 27.29 | 0.0019 | 25 | |
| 119–129 | 68–78 | GIDGDYTGVVK | 70.2 | 9.50E−08 | 55 | |
| GtcpeA12 | 64–79 | 8–23 | VQTQSSFGVINDNMGK | 129.58 | 1.10E−13 | 51 |
| 80–91 | 24–35 | GAMPVITVFDHR | 67.79 | 1.70E−07 | 14 | |
| 105–115 | 49–59 | SGDQDDEMCVK | 32.64 | 0.00054 | 6 | |
| 120–129 | 64–73 | TPYTYPQYTR | 52.09 | 6.20E−06 | 52 | |
| 130–145 | 74–89 | QATALLNEIIGVTYTK | 155.02 | 3.20E−16 | 50 | |
| GtcpeA13 | 79–90 | 18–29 | NFAPVITVFDHR | 66.65 | 2.20E−07 | 11 |
| 105–115 | 44–54 | SNDENDEMLVK | 18.13 | 0.015 | 2 | |
| 141–149 | 80–88 | GTFYQYPIR | 64.48 | 3.60E−07 | 32 | |
| GtcpeA14 | 67–84 | 1–18 | IGEGPSNTFSGQNGIATK (N-terminus) | 131.28 | 7.50E−14 | 65 |
| 85–99 | 19–33 | TANGLLGTSIVGVGK | 146.89 | 2.00E−15 | 46 | |
| 107–118 | 41–52 | NFAPMITIFDAR | 95.41 | 2.90E−10 | 43 | |
| 132–142 | 66–76 | AGTTDDEMLVK | 28.18 | 0.0015 | 1 | |
| 150–160 | 84–94 | VPEYQAAAFTR | 73.4 | 4.60E−08 | 51 | |
| 161–171 | 95–105 | EQLGYTYPTTR | 57.5 | 1.80E−06 | 55 | |
| GtcpeA15 | 78–89 | 17–28 | NAAPVITIFDHR | 96.09 | 2.50E−10 | 55 |
| 98–114 | 37–53 | EYTGAPSNDYNDEMLVK | 155.02 | 3.20E−16 | 60 | |
| GtcpeA16 | 66–81 | 4–19 | MPADSYTPIITIFDAR | 155.02 | 3.20E−16 | 96 |
| 86–105 | 24–43 | GAGEYEGSPAGDNNDEMAVK | 155.81 | 2.60E−16 | 186 | |
| 111–122 | 49–60 | LEPDTGFAEQIK | 57.2 | 1.90E−06 | 7 | |
| 110–122 | 48–60 | KLEPDTGFAEQIK | 88.18 | 1.50E−09 | 31 | |
| 111–131 | 49–69 | LEPDTGFAEQIKAETLGLMSK | 19.97 | 0.01 | 1 | |
| GtcpeA17 | 44–55 | 8–19 | GVAPIITVFDHR | 44.81 | 3.30E−05 | 3 |
| 60–81 | 24–45 | GKADSEYQGALANGPEDEMLVK | 28.48 | 0.0014 | 3 | |
| 62–81 | 26–45 | ADSEYQGALANGPEDEMLVK | 154.68 | 3.40E−16 | 31 | |
| 87–96 | 51–60 | VPLSAGFADK | 35.26 | 0.0003 | 3 | |
| 97–113 | 61–77 | VLQQTLGTFGQAPAGAK | 155.02 | 3.20E−16 | 21 | |
| GtcpeA18 | 59–75 | 4–20 | TMPADGYAPVITVFDHR | 113.61 | 4.40E−12 | 81 |
| 89–99 | 34–44 | SNDMEDGMLVK | 54.99 | 3.20E−06 | 54 | |
| 107–126 | 52–71 | VTESYAASLLQEQLGYINDK | 153.61 | 4.40E−16 | 60 | |
| 107–127 | 52–72 | VTESYAASLLQEQLGYINDKA (C-terminus) | 154.66 | 3.40E−16 | 64 | |
| GtcpeA19 | 98–107 | 48–57 | IAVSEGDAAR | 51.02 | 7.90E−06 | 15 |
| GtcpeA20 | 77–93 | 25–41 | EYTGGLSNTQDDQMCVK | 155.02 | 3.20E−16 | 84 |
| 109–120 | 57–68 | LLQENLSSMFGK (C-terminus) | 153.03 | 5.00E−16 | 56 | |
| GtcpeA21 | 42–51 | 9–18 | APVVTIFDHR | 66.35 | 2.30E−07 | 176 |
The table shows the tryptic peptides with unique sequences of the phycobiliproteins of G. theta that are detected in the Mascot database searches. The search parameters permitted a mass error of 5 ppm and one missed cleavage site. The expectation value corresponds to the E-value of a Blast search result, and expectation values <0.05 have a confidence of >95%
For GtcpeA8 and GtcpeA15, the sequence coverage was lower, but for each of these proteins, two unique peptides were identified, which provides sufficient confidence for their identification. In the samples of algae that were grown under HL conditions, GtcpeA4 and GtcpeA17 were not detectable, which is probably due to their amount being below the detection threshold of our assay. In addition, GtcpeA8 was barely detectable under HL conditions.
The sequences of the GtcpeA1 and GtcpeA21 precursor proteins differ by only three amino acid residues, and those of GtcpeA9 and GtcpeA19 by only two (Fig. 4). The mature proteins of GtcpeA1 and GtcpeA21 are distinguished from each other by only one tryptic peptide, which is APVITVFDHR in GtcpeA1 and APVVTIFDHR in GtcpeA21. However, both peptides were detected by more than 100 spectra (Table 3) and in all of the 15 biological samples analyzed (Table 2), which supports the expression of these proteins. GtcpeA9 has the unique tryptic peptide IAVSEGDAAK and GtcpeA19 the unique tryptic peptide IAVSEGDAAR, neither of which is found in any other protein. The peptide IAVSEGDAAR of GtcpeA19 was detected in 9 of the 15 biological samples analyzed, and the peptide IAVSEGDAAK of GtcpeA9 in two samples, which supports the expression of these proteins. In summary, our data support the expression of all 20 α-subunit proteins, and they are also consistent with the presence of 225 EST sequences for the α-subunit proteins in Genbank, which include at least one high-quality EST sequence for each α-subunit.
The experimentally identified peptides also allow an assignment of some of the N- and C-terminal sites of the α1-type (with LTD) and α2-type (no LTD) α-subunits. The long N-terminal extensions of the α1-type GtcpeA2 and GtcpeA14 are supported by the peptides IPLGTQIAGK and IGEGPSNTFSGQNGIATK (Table 3; Fig. 4b, underlined), which probably represent the mature N-termini of these proteins. At the C-terminal end, the peptides AIPANTNLAASILSDFQIYCQDGSCPK and VTESYAASLLQEQLGYINDKA provide support for the predicted C-termini of the gene models of GtcpeA6 and GtcpeA18, and the peptide LLQENLSSMFGK for the one of GtcpeA20. For many of the other α1-subunits, the support of the C-terminal parts of their gene models through identified peptides is good, although no peptides were found to support the very long GtcpeA8 and GtcpeA15 tails. Since the long 3′-ends of these genes have not been experimentally verified with 3′-RACE, it is possible that they are the result of sequencing errors.
As for the α2-type GtcpeA10, the peptide AAGYSTASPYSSK matches the predicted N-terminus. The N-terminal Met of the putative mature GtcpeA10, which precedes the sequence AAGYSTASPYSSK, is likely cleaved off after the import into the chloroplast stroma (Huesgen et al. 2013). In addition, the peptide LAQQVLEQTIGVLKAK supports the C-terminal end of this protein. Finally, there is also support for the C-terminal end of GtcpeA3 by the peptide VAINEAAATDLVQQLLGTLKK. As for the other α2-type α-subunits, no peptides that match the predicted C-termini of their gene models were detected, but the coverage of their C-terminal parts is good (Fig. 4b, c).
The low level or the absence of peptides for some proteins under HL (Table 2) suggested that some subunits might have a control function that is regulated by the intensity of the growth light. Due to ion suppression in electro spray-ionization, quantization by direct comparison of peptides from different proteins is not possible. Usually, relative quantization methods are applied, in which the relative levels of individual proteins under different conditions are measured. In this study, we used a semiquantitative evaluation by peptide counting to test if individual α-subunits are regulated by the different intensities of the white growth light, under which the algae were grown. The goal of this approach was to reject the working hypothesis that no individual α-subunit is regulated by light under the experimental conditions of this study.
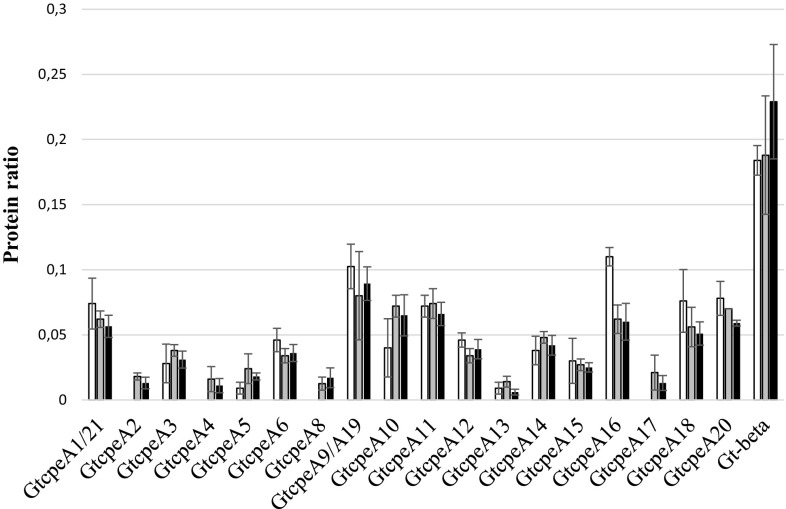
Our electrospray ionization mass spectrometry method allows no absolute quantization but only the relative comparison of unique peptides of the individual phycobiliproteins (e.g., the presence of GtcpeA14 at different light intensities). Figure 5 shows for each light condition the relative ratios of the individual α- and β-subunits to the entire pool of the phycobiliprotein peptides. The result of this evaluation is that the relative ratios of each α- and β-subunit do not change significantly under LL, SL or HL conditions within the limits of the standard deviations of our quantization. There is one outlier for GtcpeA16 under HL conditions, but it is not strong enough to reject the hypothesis that the relative ratios of the α- and β-subunits do not change significantly, when the algae are grown under white light of different intensities. It therefore appears that the PE545 antennas function as a pool that is regulated by light intensity. As long as the spectral composition of the growth light does not change, there seems to be no need for the algae to change the composition of the subunit pool, and it is enough if the algae adjust the pool size to adapt to the light intensity in the surrounding environment. Studies of subunit expression under different growth conditions and a more accurate quantization than our peptide counting experiment might reveal more details, which might change this picture, but that is beyond the scope of this study, and at this point, there is no evidence for the differential light regulation of the functions of individual α- and β-subunits under white growth light.
Fig. 5.
Relative expression of the individual phycobiliproteins of G. theta relative to the pool of all phycobiliproteins. G. theta cells were grown either at low light (LL, black bars), standard light (SL, gray bars), or high light (HL, white bars). For each light condition, the ratios of the individual phycobiliproteins to the entire phycobiliprotein pool were determined by semiquantitative peptide counting according to Dowle et al. (2016). GtcpeA1 and GtcpeA21, and GtcpeA9 and GtcpeA19 were quantified as groups and not as individual proteins, because of their high sequence similarity. The data are based on five biological replicates with three technical replicates each. The standard deviations were calculated from the ratios of the phycobiliproteins obtained for each biological replicate and included in the graph as error bars
Structure prediction
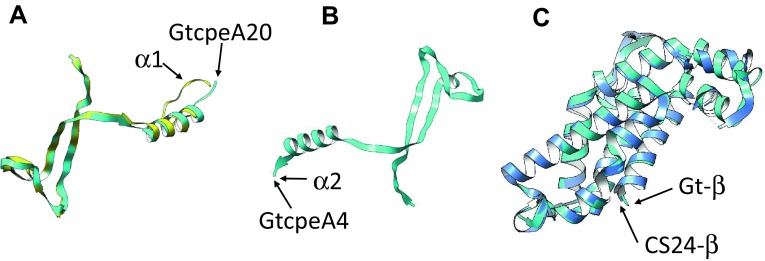
To address the question of whether the gene models of the α- and β-subunits of G. theta PE545 are likely to have the same fold as the subunits of PE545 of Rhodomonas sp. CS24 (Doust et al. 2004) and related antenna complexes, we searched for suitable fold models using the Phyre 2 server (Kelley et al. 2015) (Fig. 6). We found that all sequences of the mature α-subunits aligned with 99.9% confidence or higher to the folds of the α-1 and α-2 subunits of Rhodomonas sp. CS24 (d1xg0a_ and d1xg0b_) (Doust et al. 2004) or to the fold of the α-1 subunit of phycocyanin PC645 of Chroomonas sp. CCMP270 (c4lmsA_) (Harrop et al. 2014). Both folds are very similar and can be aligned to one another with high confidence, with the exception of the N-terminal extensions and C-terminal tails. This was not surprising, since Harrop et al. (2014) showed that the three-dimensional structures of four different cryptophyte phycobiliproteins (PE545, PC645, PC612, and PE555) could be superimposed. A critical assessment of these alignments shows, however, that 13 of the 20 α-subunits of G. theta are predicted to have more than 50% disorder, which means that their structure predictions have low confidence. Nevertheless, good alignments with a sequence coverage of 84–99% were obtained for GtcpeA4, GtcpeA5, GtcpeA11, GtcpeA16 and GtcpeA20. As an example, the alignments of GtcpeA4 and GtcpeA20 to the fold of the corresponding Rhodomonas sp. CS24 α-subunits are shown in Fig. 6a, b. The central parts of GtcpeA13 and GtcpeA15 also give good alignments with a sequence coverage of 69 and 57%, but their N- and C-terminal tails do not match the model fold. Searches for structure models for the β-phycoerythrin of G. theta resulted in an excellent match to the β-phycoerythrin of Rhodomonas sp. CS24 (Doust et al. 2004) (Fig. 6c), which is expected since their primary sequences are almost identical. In summary, our modeling work suggests that the phycobiliprotein complexes of G. theta have similar three-dimensional structures as that of Rhodomonas sp. CS24, even though some differences in the N- and C-terminal extensions of some α-subunits might exist.
Fig. 6.
Structure predictions of the α-subunits. The folds of GtcpeA4, GtcpeA20, and the β-subunit of G. theta PE545 were predicted using Phyre 2 (Kelley et al. 2015) and aligned using FATCAT (Ye and Godzik 2003) to the experimental folds of their models in the PE545 of Rhodomonas sp. CS24 (Doust et al. 2004). a Alignment of GtcpeA20 (cyan) to the α1-subunit (yellow) of the Rhodomonas PE545 tetramer (Fig. 1). The alignment has sequence coverage of 99% and a confidence of 100%. b Alignment of GtcpeA4 (cyan) to the α2-subunit (yellow) of the Rhodomonas PE545 tetramer. The alignment has sequence coverage of 99% and a confidence of 100%. The similarity between these folds is so high that the color of the Rhodomonas chain is not visible. c. Alignment of the G. theta β-subunit (Gt-β, cyan) to that of Rhodomonas PE545 (CS24-β, blue). The alignment has sequence coverage of 100% and a confidence of 100%
The expressions of 6 different α1-type and 14 different α2-type proteins raise the intriguing possibility that the G. theta chloroplast has multiple types of tetramers with different combinations of α- and β-subunits. In support of this, the pioneering work of Hiller and Martin (1987) on purified PE545 from Rhodomonas sp. CS24 (then called Chroomonas) resolved four fractions with distinctive isoelectric points using a chromatofocusing column. The major fraction (pI 6.24) contained both α1- and α2-type bands, while a minor fraction (pI 7.16) contained only the lighter α2-type, and the other two fractions only the heavier α1-type. This suggested the existence of α1β α1β and α2β α2β as well as α1β α2β tetramers. In the case of G. theta, there could be an even greater variety of tetramers.
This brings up another complication. In light of our current knowledge of targeting to secondary plastids: the α2β α2β tetramers could not be transported into the thylakoid lumen because neither α-subunit would have a LTD. However, if they escaped degradation by stromal proteases, there is no reason why they could not associate with the stromal surface of the thylakoid membrane and transfer energy to the photosystems, since there appears to be no preferred orientation or packing arrangement for the tetramers within the lumen. It would then be possible to envisage redox or other control being exerted on at least a small fraction of the phycobiliprotein population.
Conclusions
Our proteomics analysis shows that all the 20 genome-predicted α-subunits of G. theta PE545 are expressed at the protein level, and suggests that the PE545 α-subunits operate as a pool that is regulated up and down depending on the light intensity. The similarity of these sequences to those of the published crystal structure of Rhodomonas sp. CS24, as well as the very high similarity of the β-subunit sequences in these two species suggest that all the PE545s have similar three-dimensional structures. GtcpeA10 with its internal duplication might be able to link two partial tetramers or to form oligomers.
Acknowledgements
The authors would like to thank Drs. Cheryl A. Kerfeld and Matthew R. Melnicki, the University of California, Berkely, USA, for hosting O.C. in their lab and their kind introduction to protein modeling. The support for this work provided by the KBC Proteomics Facility at Umeå University and the Swedish University of Agricultural Sciences is greatly appreciated.
Funding
This work was supported by the Swedish Energy Agency (2012-005889) (to C.F.), the Umeå University, and the Artificial Leaf and Solar Fuel Project (KAW 2011-0055) (to C.F.). The work was initiated when BRG was a visiting Professor in Umeå financed by the sabbatical program of Umeå University.
Footnotes
Thomas Kieselbach, Otilia Cheregi and Beverley R. Green have contributed equally to the work.
Contributor Information
Thomas Kieselbach, Email: Thomas.Kieselbach@umu.se.
Otilia Cheregi, Email: Otilia.cheregi@gmail.se.
Beverley R. Green, Email: brgreen@mail.ubc.ca
Christiane Funk, Phone: +46-90-786 7633, Email: Christiane.Funk@umu.se.
References
- Adir N. Elucidation of the molecular structures of components of the phycobilisome: reconstructing a giant. Photosynth Res. 2005;85(1):15–32. doi: 10.1007/s11120-004-2143-y. [DOI] [PubMed] [Google Scholar]
- Arpin PC, Turner DB, McClure SD, Jumper CC, Mirkovic T, Challa JR, Lee J, Teng CY, Green BR, Wilk KE, Curmi PMG, Hoef-Emden K, McCamant DW, Scholes GD. Spectroscopic studies of cryptophyte light harvesting proteins: vibrations and coherent oscillations. J Phys Chem B. 2015;119(31):10025–10034. doi: 10.1021/acs.jpcb.5b04704. [DOI] [PubMed] [Google Scholar]
- Blankenship RE. Molecular mechanisms of photosynthesis. 2. Oxford: Wiley; 2014. [Google Scholar]
- Brosch M, Yu L, Hubbard T, Choudhary J. Accurate and sensitive peptide identification with Mascot percolator. J Proteome Res. 2009;8(6):3176–3181. doi: 10.1021/pr800982s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton MJ, Howe CJ, Hiller RG. Distinctive organization of genes for light-harvesting proteins in the cryptophyte alga Rhodomonas. Gene. 2006;369(0):72–79. doi: 10.1016/j.gene.2005.10.026. [DOI] [PubMed] [Google Scholar]
- Cheregi O, Kotabová E, Prášil O, Schröder WP, Kaňa R, Funk C. Presence of state transitions in the cryptophyte alga Guillardia theta. J Exp Bot. 2015;66(20):6461–6470. doi: 10.1093/jxb/erv362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collini E, Wong CY, Wilk KE, Curmi PMG, Brumer P, Scholes GD (2010) Coherently wired light-harvesting in photosynthetic marine algae at ambient temperature. Nature 463(7281):644–647. http://www.nature.com/nature/journal/v463/n7281/suppinfo/nature08811_S1.html [DOI] [PubMed]
- Curtis BA, Tanifuji G, Burki F, Gruber A, Irimia M, Maruyama S, Arias MC, Ball SG, Gile GH, Hirakawa Y, Hopkins JF, Kuo A, Rensing SA, Schmutz J, Symeonidi A, Elias M, Eveleigh RJM, Herman EK, Klute MJ, Nakayama T, Obornik M, Reyes-Prieto A, Armbrust EV, Aves SJ, Beiko RG, Coutinho P, Dacks JB, Durnford DG, Fast NM, Green BR, Grisdale CJ, Hempel F, Henrissat B, Hoppner MP, Ishida K-I, Kim E, Koreny L, Kroth PG, Liu Y, Malik S-B, Maier UG, McRose D, Mock T, Neilson JAD, Onodera NT, Poole AM, Pritham EJ, Richards TA, Rocap G, Roy SW, Sarai C, Schaack S, Shirato S, Slamovits CH, Spencer DF, Suzuki S, Worden AZ, Zauner S, Barry K, Bell C, Bharti AK, Crow JA, Grimwood J, Kramer R, Lindquist E, Lucas S, Salamov A, McFadden GI, Lane CE, Keeling PJ, Gray MW, Grigoriev IV, Archibald JM (2012) Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature 492(7427):59–65. http://www.nature.com/nature/journal/v492/n7427/abs/nature11681.html#supplementary-information [DOI] [PubMed]
- Doust AB, Marai CNJ, Harrop SJ, Wilk KE, Curmi PMG, Scholes GD. Developing a structure–function model for the cryptophyte phycoerythrin 545 using ultrahigh resolution crystallography and ultrafast laser spectroscopy. J Mol Biol. 2004;344(1):135–153. doi: 10.1016/j.jmb.2004.09.044. [DOI] [PubMed] [Google Scholar]
- Dowle AA, Wilson J, Thomas JR. Comparing the diagnostic classification accuracy of iTRAQ, peak-area, spectral-counting, and emPAI methods for relative quantification in expression proteomics. J Proteome Res. 2016;15(10):3550–3562. doi: 10.1021/acs.jproteome.6b00308. [DOI] [PubMed] [Google Scholar]
- Durnford DG, Deane JA, Tan S, McFadden GI, Gantt E, Green BR. A phylogenetic assessment of the eukaryotic light-harvesting antenna proteins, with implications for plastid evolution. J Mol Evol. 1999;48(1):59–68. doi: 10.1007/PL00006445. [DOI] [PubMed] [Google Scholar]
- Faust MA, Gantt E. Effect of light intensity and glycerol on the growth, pigment composition and ultrastructure of Chroomonas sp. J Phycol. 1973;9(4):489–495. [Google Scholar]
- Funk C, Alami M, Tibiletti T, Green BR. High light stress and the one-helix LHC-like proteins of the cryptophyte Guillardia theta. Biochim Biophys Acta (BBA) 2011;1807(7):841–846. doi: 10.1016/j.bbabio.2011.03.011. [DOI] [PubMed] [Google Scholar]
- Gantt E, Grabowski B, Cunningham FX. Light-harvesting antennas in photosynthesis. Netherlands: Kluwer Academics; 2003. Antenna systems of red algae: phycobilisomes with photosystem II and chlorophyll complexes with photosystem I; pp. 307–322. [Google Scholar]
- Gibbs SP. The chloroplast of some algal groups may have evolved from endosymbiotic eukaryotic algae. Ann N Y Acad Sci. 1981;361(FEB):193–208. doi: 10.1111/j.1749-6632.1981.tb46519.x. [DOI] [PubMed] [Google Scholar]
- Giglione C, Meinnel T (2001) Organellar peptide deformylases: universality of the N-terminal methionine cleavage mechanism. Trends Plant Sci 6(12):566–572 [DOI] [PubMed]
- Glazer AN, Wedemayer GJ. Cryptomonad biliproteins: an evolutionary perspective. Photosynth Res. 1995;46(1):93–105. doi: 10.1007/BF00020420. [DOI] [PubMed] [Google Scholar]
- Gould SB, Fan E, Hempel F, Maier U-G, Klösgen RB. Translocation of a phycoerythrin α subunit across five biological membranes. J Biol Chem. 2007;282(41):30295–30302. doi: 10.1074/jbc.M701869200. [DOI] [PubMed] [Google Scholar]
- Gould SB, Waller RF, McFadden GI. Plastid evolution. Annu Rev Plant Biol. 2008;59(1):491–517. doi: 10.1146/annurev.arplant.59.032607.092915. [DOI] [PubMed] [Google Scholar]
- Green BR, Durnford DG. The chlorophyll-carotenoid proteins of oxygenic photosynthesis. Ann Rev Plant Physiol Plant Mol Biol. 1996;47:685–714. doi: 10.1146/annurev.arplant.47.1.685. [DOI] [PubMed] [Google Scholar]
- Gruber A, Vugrinec S, Hempel F, Gould SB, Maier U-G, Kroth PG. Protein targeting into complex diatom plastids: functional characterisation of a specific targeting motif. Plant Mol Biol. 2007;64(5):519–530. doi: 10.1007/s11103-007-9171-x. [DOI] [PubMed] [Google Scholar]
- Guillard RL. Culture of phytoplankton for feeding marine invertebrates. In: Smith W, Chanley M, editors. Culture of marine invertebrate animals. New York: Springer; 1975. pp. 29–60. [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Harrop SJ, Wilk KE, Dinshaw R, Collini E, Mirkovic T, Teng CY, Oblinsky DG, Green BR, Hoef-Emden K, Hiller RG, Scholes GD, Curmi PMG. Single-residue insertion switches the quaternary structure and exciton states of cryptophyte light-harvesting proteins. Proc Natl Acad Sci. 2014;111(26):E2666–E2675. doi: 10.1073/pnas.1402538111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller RG, Martin CD. Multiple forms of a type I phycoerythrin from a Chroomonus sp. (Cryptophyceae) varying in subunit composition. Biochim Biophys Acta. 1987;923:198–202. [Google Scholar]
- Huesgen PF, Alami M, Lange PF, Foster LJ, Schröder WP, Overall CM, Green BR. Proteomic amino-termini profiling reveals targeting information for protein import into complex plastids. PLoS ONE. 2013;8(9):e74483. doi: 10.1371/journal.pone.0074483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey S, Humphrey New spectrophotometric equations for determining chlorophylls a1, b1, c1 and c2 in higher plants, algae and natural phytoplankton. Biochem Physiol Pflanz. 1975;167:191–194. doi: 10.1016/S0015-3796(17)30778-3. [DOI] [Google Scholar]
- Kaňa R, Kotabová E, Sobotka R, Prášil O. Non-photochemical quenching in cryptophyte alga Rhodomonas salina is located in chlorophyll a/c antennae. PLoS ONE. 2012;7(1):e29700. doi: 10.1371/journal.pone.0029700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protocols. 2015;10(6):845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Ye Y, Godzik A. Flexible structural neighborhood: a database of protein structural similarities and alignments. Nucleic Acids Res. 2006;34(Database issue):D277–D280. doi: 10.1093/nar/gkj124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtlé C. Effects of nitrogen deficiency and light of high intensity on Cryptomonas rufescens (Cryptophyceae) Protoplasma. 1979;101(4):283–299. doi: 10.1007/BF01276969. [DOI] [Google Scholar]
- MacColl R, Eisele LE, Dhar M, Ecuyer J-P, Hopkins S, Marrone J, Barnard R, Malak H, Lewitus AJ. Bilin organization in cryptomonad biliproteins. BioChemistry. 1999;38(13):4097–4105. doi: 10.1021/bi982059c. [DOI] [PubMed] [Google Scholar]
- MacPherson AN, Hiller RG. Light-harvesting antennas in photosynthesis. Netherlands: Kluwer Academics; 2003. Light-harvesting systems in chlorophyll c-containing algae; pp. 307–322. [Google Scholar]
- Mirkovic T, Ostroumov EE, Anna JM, van Grondelle R, Govindjee, Scholes GD. Light absorption and energy transfer in the antenna complexes of photosynthetic organisms. Chem Rev. 2016 doi: 10.1021/acs.chemrev.6b00002. [DOI] [PubMed] [Google Scholar]
- Neilson JAD, Durnford DG. Evolutionary distribution of light-harvesting complex-like proteins in photosynthetic eukaryotes. Genome. 2009;53(1):68–78. doi: 10.1139/G09-081. [DOI] [PubMed] [Google Scholar]
- Peterson GL. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal Biochem. 1977;83(2):346–356. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF chimera: a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Shen G, Vermaas WFJ. Mutation of chlorophyll ligands in the chlorophyll-binding CP47 protein as studied in a Synechocystis sp. PCC 6803 photosystem I-less background. BioChemistry. 1994;33(23):7379–7388. doi: 10.1021/bi00189a044. [DOI] [PubMed] [Google Scholar]
- Shen G, Eaton-Rye JJ, Vermaas WFJ. Mutation of histidine residues in CP47 leads to destabilization of the photosystem II complex and to impairment of light energy transfer. BioChemistry. 1993;18(32):5109–5115. doi: 10.1021/bi00070a019. [DOI] [PubMed] [Google Scholar]
- Spear-Bernstein L, Miller KR. Unique location of the phycobiliprotein light-harvesting pigment in the Cryptophyceae. J Phycol. 1989;25(3):412–419. doi: 10.1111/j.1529-8817.1989.tb00245.x. [DOI] [Google Scholar]
- Vizcaino JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Rios D, Dianes JA, Sun Z, Farrah T, Bandeira N, Binz P-A, Xenarios I, Eisenacher M, Mayer G, Gatto L, Campos A, Chalkley RJ, Kraus H-J, Albar JP, Martinez-Bartolome S, Apweiler R, Omenn GS, Martens L, Jones AR, Hermjakob H (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotech 32(3):223–226. doi:10.1038/nbt.2839http://www.nature.com/nbt/journal/v32/n3/abs/nbt.2839.html#supplementary-information [DOI] [PMC free article] [PubMed]
- Vizcaíno JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu Q-W, Wang R, Hermjakob H. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44(D1):D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Ikeuchi M. Phycobilisome: architecture of a light-harvesting supercomplex. Photosynth Res. 2013;116(2):265–276. doi: 10.1007/s11120-013-9905-3. [DOI] [PubMed] [Google Scholar]
- Wilk KE, Harrop SJ, Jankova L, Edler D, Keenan G, Sharples F, Hiller RG, Curmi PMG. Evolution of a light-harvesting protein by addition of new subunits and rearrangement of conserved elements: crystal structure of a cryptophyte phycoerythrin at 1.63-Å resolution. Proc Natl Acad Sci. 1999;96(16):8901–8906. doi: 10.1073/pnas.96.16.8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Godzik A. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics. 2003;19(suppl_2):ii246–ii255. doi: 10.1093/bioinformatics/btg1086. [DOI] [PubMed] [Google Scholar]
- Ye Y, Godzik A. Database searching by flexible protein structure alignment. Protein Sci. 2004;13(7):1841–1850. doi: 10.1110/ps.03602304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Godzik A. FATCAT: a web server for flexible structure comparison and structure similarity searching. Nucleic Acids Res. 2004;32(Web Server issue):W582–W585. doi: 10.1093/nar/gkh430. [DOI] [PMC free article] [PubMed] [Google Scholar]