Abstract
Cassava (Manihot esculenta Crantz) is an important security crop that faces severe yield loses due to cassava brown streak disease (CBSD). Motivated by the slow progress of conventional breeding, genetic improvement of cassava is undergoing rapid change due to the implementation of quantitative trait loci mapping, Genome-wide association mapping (GWAS), and genomic selection (GS). In this study, two breeding panels were genotyped for SNP markers using genotyping by sequencing and phenotyped for foliar and CBSD root symptoms at five locations in Uganda. Our GWAS study found two regions associated to CBSD, one on chromosome 4 which co-localizes with a Manihot glaziovii introgression segment and one on chromosome 11, which contains a cluster of nucleotide-binding site-leucine-rich repeat (NBS-LRR) genes. We evaluated the potential of GS to improve CBSD resistance by assessing the accuracy of seven prediction models. Predictive accuracy values varied between CBSD foliar severity traits at 3 months after planting (MAP) (0.27–0.32), 6 MAP (0.40–0.42) and root severity (0.31–0.42). For all traits, Random Forest and reproducing kernel Hilbert spaces regression showed the highest predictive accuracies. Our results provide an insight into the genetics of CBSD resistance to guide CBSD marker-assisted breeding and highlight the potential of GS to improve cassava breeding.
Introduction
Cassava (Manihot esculenta Crantz) is a primary source of income and dietary calories for millions of people, and the high starch content of its storage roots is exploited in industry1. Although cassava is a resilient crop, its production in East Africa is often constrained by viral diseases including cassava brown streak virus disease (CBSD) which causes significant yield loses2–4. This disease is caused by two virus species of the genus Ipomovirus, family Potyviridae: cassava brown streak virus (CBSV) and Ugandan cassava brown streak virus (UCBSV)5–7. Both cassava brown streak viruses have successfully colonized a broad altitudinal range in South, East, and Central Africa and the steady spread of the disease is a threat to cassava production in West Africa3,6. In the field, CBSVs are transmitted by the whitefly (Bemisia tabaci) in a semi-persistent manner and through the exchange of infected cassava cuttings among farmers8,9. In susceptible clones, the viruses cause a myriad of symptoms including yellow chlorotic patterns along minor veins of leaves, necrotic streaks on the stems and brown or grey corky root necrosis10–15.
Breeding for durable CBSD resistance, through the development and propagation of CBSD-resistant varieties, has been the standard strategy to restrict disease spread. To date varieties with immunity to CBSVs from conventional breeding have not been reported. However, resistance in the form of restricted virus accumulation has been demonstrated, as has reduced symptom expression and recovery after clonal propagation7,16,17.
Numerous factors have hindered the rate of genetic progress in CBSD resistance breeding using conventional breeding approaches. These factors include the availability of suitable levels of resistance, the lack of well-characterized CBSD resistant varieties, genotype by environment interaction18–20, inconsistent year-to-year symptom expression, as well as reduced flowering, length of the breeding cycle, limited genetic diversity and slow rate of multiplication of planting materials21,22. Conventional cassava breeding can take three to six years from seedling germination to multi-location yield trials and additional years are required for evaluation of promising genotypes before superior clones are released as varieties23.
So far, few studies have used genomic-enabled approaches to increase the understanding of the mechanism of resistance and identify candidate genes and/or molecular markers associated with CBSD resistance or tolerance19,20,24,25. Recently, a transcriptome analysis of two cassava varieties, Namikonga (resistant) and Albert (susceptible), has facilitated the identification of a set of candidate genes that collectively may confer resistance to CBSD in Namikonga25. QTL mapping in biparental crosses successfully identified a set of QTL and candidate genes associated with resistance to CBSD induced root necrosis and CBSD foliar symptoms19,20. Notably, the characterization of QTL regions associated with CBSD resistance in the Tanzanian local cultivar Namikonga and the Tanzanian landrace Kiroba, suggest that some of those regions were introgressed from Manihot glaziovii19,20.
Crop biotechnology approaches such as RNAi technology have also been proposed to accelerate the integration of CBSD resistance into farmer-preferred cultivars26–28. Evaluation in the greenhouse and the field of transgenic lines, has shown efficient, durable and stable siRNA-derived resistance to CBSD across agro-ecological regions28.
Currently, genetic improvement of cassava is undergoing rapid change through the implementation of genomic-enabled tools by breeding programs in Africa (www.nextgencassava.org). Preliminary studies suggest that Genome-wide association studies (GWAS) and genomic selection (GS) are effective approaches for cassava breeding. Genome-wide association mapping studies in cassava have led to the identification of QTL regions associated with cassava mosaic disease resistance (CMD)29, beta-carotene content30 and dry matter content31. Genomic selection is particularly promising as an alternative method to marker-assisted selection (MAS) and conventional phenotypic selection because it can accelerate genetic gains due to the selection of parental genotypes with superior breeding values at the seedling stage based on genotypes alone32–34. Indeed, the evaluation of the performance of genomic prediction models using phenotypic and genotyping by sequencing (GBS) datasets from three African cassava breeding programs highlight the potential of GS as a breeding tool for some traits23,35,36.
In the present study, we followed a GWAS approach in combination with genomic prediction to unravel the genetic architecture of CBSD in two Ugandan breeding populations. The objectives of this study were: (1) to identify sources of resistance to CBSD in the GWAS panel, (2) to assess the current predictive accuracy for CBSD (3) to identify genomic prediction models that account for CBSD genetic architecture including the evaluation of a synergistic implementation of GWAS and GS and (4) to identify significant polymorphisms to guide CBSD marker-assisted breeding to improve cassava breeding in the face of increasing disease threats to agricultural production.
Results
Phenotypic variability
Foliar and root disease scoring was performed according to a standard CBSD scoring scale that ranges from 1 to 5 (Supplementary Fig. S1). The distribution of CBSD de-regressed BLUPs is presented in Supplementary Figs S2 and S3. Both GWAS panels exhibited differential response foliar symptom response to CBSVs at 3, 6 and 9 months after planting (MAP) and root severity (CBSDRS) at 12 MAP as demonstrated by the variability of the de-regressed BLUPs. Interestingly, clones which displayed an intermediate response were more abundant than clones with a susceptible or resistance response.
Phenotypic correlations were calculated within panels and within and across locations for foliar symptom severity scores at 3 MAP and 6 MAP and CBSDRS. Correlations across locations for each panel are given in Supplementary Figure S4 and Supplementary Tables S2 and S3. Clear differences were observed in CBSD severity scores. For Panel 1, the lowest correlation value corresponded to CBSDRS and was found between the locations Ngetta and Kasese (0.09) while the highest correlation value, which also corresponded to CBSDRS, was found between the locations Namulonge and Kasese (0.60) (Supplementary Table S2A). For Panel 2, correlation values ranged across locations between −0.08 at 9 MAP (Namulonge-Kamuli) and 0.51 at 3 MAP (Kamuli-Serere) (Supplementary Table S2B).
Across traits within locations, the highest correlation values were found in Panel 1 for foliar scorings 3 MAP and 6 MAP (r2 > 0.5) (Supplementary Table S3A). For Panel 2, correlation across traits varied depending on the location. Nonetheless, correlations across foliar traits were higher than those found between foliar and root symptom severity scores (Supplementary Table S3B).
Broad-sense and SNP heritability
For both panels and across locations the heritability estimates for CBSD symptom severity scorings at 3 MAP, 6 MAP, 9 MAP, and CBDRS were low to intermediate. Broad-sense heritability (H2) estimates across panels and locations spanned a wide range of values from 0.11 for 3 MAP at Namulonge (hotspot CBSD location) (Panel 1) to 0.75 for scorings 9 MAP at Kamuli (moderate CBSD prevalence) (Panel 2) (Table 1). Specifically, for GWAS Panel 1, broad-sense heritability (H2) estimates ranged between 0.11 for 3 MAP at Namulonge and 0.73 for CBSDRS at Ngetta (low CBSD pressure). For GWAS Panel 2, H2 estimates ranged from 0.24 for scorings 3 MAP to 0.75 at Kamuli for 9 MAP.
Table 1.
Broad-sense heritability (H2) and SNP heritability (h2) of foliar and root CBSD severity. For each panel, the heritability values were estimated per location and by combining locations (see methods).
| Trait | H2 | h2 | LOCATION-YEAR | Panel |
|---|---|---|---|---|
| CBSD 3 MAP | 0.11 | 0.32 | NAMULONGE | 1 |
| CBSD 6 MAP | 0.31 | 0.39 | NAMULONGE | 1 |
| CBSDRS | 0.55 | 0.59 | NAMULONGE | 1 |
| CBSD 3 MAP | 0.43 | 0.48 | NGETTA | 1 |
| CBSD 6 MAP | 0.51 | 0.53 | NGETTA | 1 |
| CBSDRS | 0.73 | 0.72 | NGETTA | 1 |
| CBSD 3 MAP | 0.27 | 0.29 | KASESE | 1 |
| CBSD 6 MAP | 0.21 | 0.27 | KASESE | 1 |
| CBSDRS | 0.39 | 0.47 | KASESE | 1 |
| CBSD 3 MAP | 0.61 | 0.17 | MULTI-LOCATION | 1 |
| CBSD 6 MAP | 0.35 | 0.31 | MULTI-LOCATION | 1 |
| CBSDRS | 0.37 | 0.34 | MULTI-LOCATION | 1 |
| CBSD 3 MAP | 0.60 | 0.37 | NAMULONGE | 2 |
| CBSD 6 MAP | 0.60 | 0.32 | NAMULONGE | 2 |
| CBSD 9 MAP | 0.68 | 0.34 | NAMULONGE | 2 |
| CBSDRS | 0.24 | 0.53 | NAMULONGE | 2 |
| CBSD 3 MAP | 0.63 | 0.28 | SERERE | 2 |
| CBSD 6 MAP | 0.60 | 0.28 | SERERE | 2 |
| CBSD 9 MAP | 0.73 | 0.34 | SERERE | 2 |
| CBSDRS | 0.15 | 0.48 | SERERE | 2 |
| CBSD 3 MAP | 0.56 | 0.27 | KAMULI | 2 |
| CBSD 6 MAP | 0.62 | 0.29 | KAMULI | 2 |
| CBSD 9 MAP | 0.75 | 0.34 | KAMULI | 2 |
| CBSDRS | 0.28 | 0.44 | KAMULI | 2 |
| CBSD 3 MAP | 0.42 | 0.28 | MULTI-LOCATION | 2 |
| CBSD 6 MAP | 0.47 | 0.34 | MULTI-LOCATION | 2 |
| CBSD 9 MAP | 0.56 | 0.38 | MULTI-LOCATION | 2 |
| CBSDRS | 0.25 | 0.33 | MULTI-LOCATION | 2 |
CBSD: Cassava brown streak disease, MAP: months after planting, CBSDRS cassava brown streak root severity.
Further, we combined genotypic and phenotypic data and estimated SNP heritability values using variance components that were obtained as a result of fitting a one-step model for each panel, each location, and multi-location datasets. For Panel 1, H2 and SNP heritability values were approximately the same across locations except for Namulonge (3 MAP) that displayed an H2 value of 0.11 and for the multi-location dataset (3 MAP), which showed the most significant difference between H2 and SNP heritability estimates. For Panel 2, H2 estimates were consistently higher than SNP heritability estimates except for Namulonge, Serere, Kamuli and the multi-location dataset for CBSDRS.
Genome-wide association study
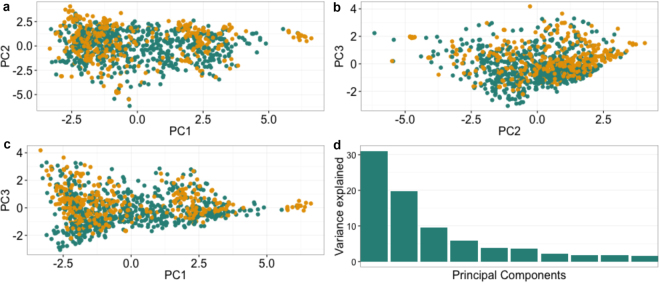
The genetic structure of the GWAS panels was estimated by using principal components analysis (PCA). Overall, the first three principal components (PCs) accounted for 60% of the genetic variation observed in the data (Fig. 1), the first PC accounted for 30% of the observed variation while the second and third PCs contributed 20% and 10% respectively. Most clones showed no clear separation, which indicates the presence of only one cluster and low stratification across panels.
Figure 1.
Plot of the first three principal components (PCs). Panel 1 and Panel 2 clones were used in the PC analysis. The top panels and the lower left panel display the distribution of clones in PC1-PC3. The lower right panel shows the variance explained by the first ten principal components. Colours correspond to members of Panel 1 (green) and Panel 2 (orange) clones.
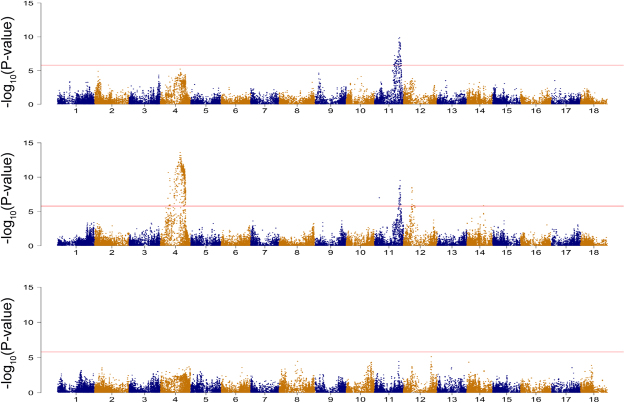
Manhattan plots of the genotype-phenotype associations at 3 MAP, 6 MAP, and CBDRS based on the combination of ~1000 clone multi-location de-regressed BLUPs are presented in Fig. 2. Additional GWAS analyses performed on each panel and location are given in Supplementary Tables S4 and S5 and Supplementary Figs S5–S12.
Figure 2.
Manhattan plots of three CBSD severity scorings in leaves and roots. The GWAS results presented correspond to the combined dataset. Association tests were performed for CBSD symptom severity on leaves at (a) 3 and (b) 6 month after planting (MAP) and (c) on roots (CBSDRS). The horizontal line indicates the genome-wide significance level (−log10(P-value) = 5.9).
SNP markers with a −log10(P-value) which exceeded the Bonferroni threshold >5.9 were considered to be statistically significant and were further annotated into coding regions (genes) of the cassava genome (Supplementary Table S4). Using the multi-location dataset, 83 significant SNP markers were identified as being associated with foliar symptoms at 3 MAP. All the significant markers were located on chromosome 11 and from these, 61 were annotated within genic regions (Supplementary Table S4). The top SNP −log10(P-value) = 9.38 within the QTL on chromosome 11 explained 6% of the observed phenotypic variance (Supplementary Table S5). For foliar severity scores at 6 MAP, we identified SNP associations on chromosome 11, chromosome 4 and chromosome 12. On chromosome 11, 33 SNPs passed the Bonferroni threshold, and from these markers, 27 were annotated within genic regions. This QTL was in the same chromosomal location as that found for 3 MAP foliar CBSD QTL and explained 5% of the observed phenotypic variance (Fig. 2). Although several of the SNPs on chromosome 11 associated with 6 MAP CBSD exceeded the Bonferroni threshold, only six SNPs were in linkage disequilibrium (r2 > 0.6) with the highest −log10(P-value) SNP hit.
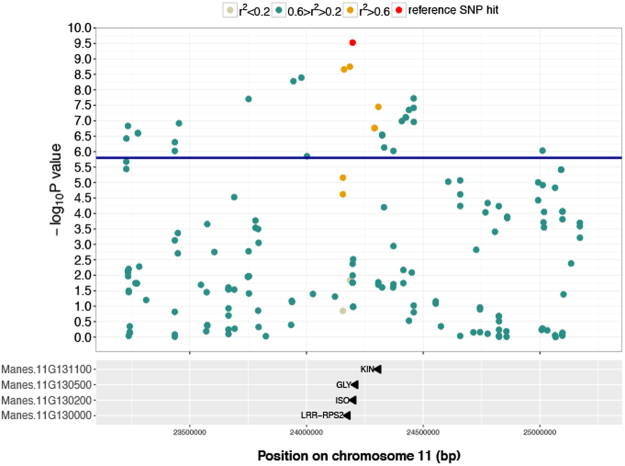
Genes on chromosome 11 containing SNP markers, with a correlation value of r2 > 0.2 with the highest −log10(P-value) marker, were annotated and classified as candidate genes. The genes identified were Manes.11G130500, Manes.11G130000, Manes.11G130200 and Manes.11G131100. Manes.11G130500 is known to encode glycine-rich protein. Manes.11G130000 encodes a Leucine-rich repeat (LRR) containing protein. Manes.11G130200 encodes the trigger factor chaperone and peptidyl-prolyl trans. Finally, Manes.11G131100 encodes a protein kinase (Fig. 3).
Figure 3.
Local Manhattan plot surrounding the peak on chromosome 11. The plot spans a 2 Mb region on chromosome 11. At the top, the SNP indicated in red is the SNP with the highest −log10(P-value) = 9.38 on that chromosome for CBSD symptom severity at 3 MAP. Colours indicate the Pearson’s correlation coefficient (r2) between the top significant GWAS SNP hit on this chromosome and neighboring markers in the given window. Markers with r2 > 0.2 in annotated genes are indicated in the panel below.
On chromosome 4 the significant SNPs defining the QTL were in high linkage disequilibrium (Fig. 4) and co-localized with an introgression segment from a wild relative of cassava (M. glaziovii)37,38. We further confirmed the presence and segregation of the introgressed genome segment in both panels using a set of diagnostic markers from M. glaziovii (Supplementary Fig. S13). Because of the high level of linkage disequilibrium at the QTL location, we do not highlight a single locus or loci as candidate gene(s) associated with CBSD foliar severity.
Figure 4.
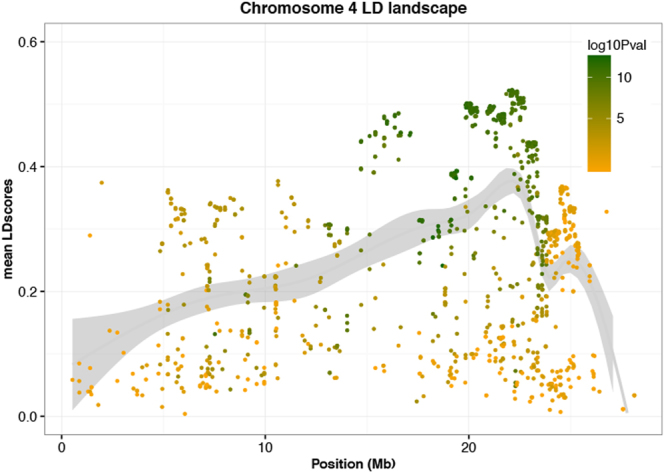
Local Manhattan plot of Chromosome 4. LD score values (r2) for each marker on chromosome 4 plotted against physical distances between markers. The smooth line represents a relative measure of the local LD in chromosome 4. Dot colors depend on the −log10(P-value) obtained for CBSD symptom severity at 6 MAP.
The significant QTL on chromosome 12 associated with foliar severity at 6 MAP had previously been found to be associated with CMD resistance in cassava29. To confirm the association of that QTL with foliar severity scores at 6 MAP we re-fit the first-step mixed-models used to obtain de-regressed BLUPs, this time including CMD severity as a fixed-effect covariate. After the corrected de-regressed BLUPs were included in the GWAS analysis, the QTL on chromosome 12 was no longer significant, and only the QTL on chromosomes 4 and 11 remained (Supplementary Fig. S14).
SNPs surpassing the Bonferroni threshold could not be identified for CBSDRS across panels. However, analysis of the multi-location data for Panel 1 identified significant regions of CBSDRS association on chromosomes 5, 11 and 18 (−log10(P-value) > 6.5), which explained 8, 6 and 10% of the phenotypic variance respectively.
Genome-wide prediction
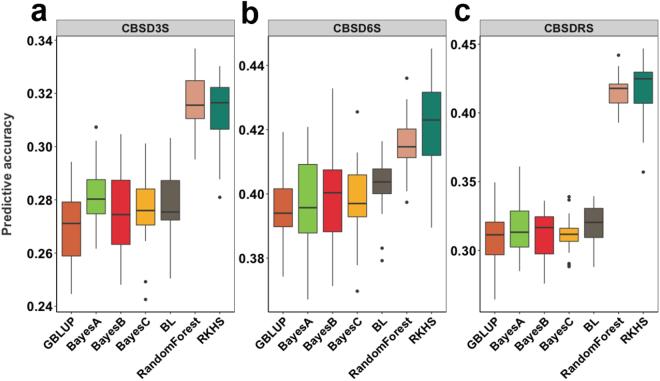
Using the combined dataset, we compared the performance of seven genomic prediction models with different assumptions on trait genetic architecture. Some model predictions represent genomic estimated breeding values (GEBV) in that they are sums of additive effects of markers, while other model predictions represent genomic estimated total genetic values (GETGV) because they include non-additive effects. Prediction accuracy for CBSD related traits had mean values across methods of 0.29 (3 MAP), 0.40 (6 MAP) and 0.34 (CBSDRS) (Fig. 5 and Supplementary Table S6A).
Figure 5.
Plot of cross-generation prediction accuracies for CBSD severity symptoms. Seven genomic prediction methods (colors, x-axis within panels) were tested for CBSD symptom severity predictive accuracy (y-axis within panels) on leaves at (a) 3 and (b) 6 month after planting (MAP) and on roots (c) 12 MAP.
These accuracies varied from 0.27 (BayesB and GBLUP) to 0.32 (RF) for 3 MAP, from 0.40 (most methods) to 0.41 (RF) and 0.42 (RKHS) for 6 MAP and from 0.31 (BayesA, B, C and GBLUP) to 0.42 (RF and RKHS) for CBSDRS. It is clear from the results that higher predictive accuracies were consistently achieved when using RF and RKHS for the prediction of both foliar and root CBSD resistance traits. For foliar symptoms, the increase in predictive accuracy using RF and RKHS was modest in comparison to GBLUP, whereas for CBSDRS the predictive accuracy increased from 0.31 using GBLUP to 0.42 using RF and RKHS models.
GWAS-guided genomic prediction
Based on the genome-wide association results, we identified for foliar 3 MAP and 6 MAP and CBSDRS the strongest marker associations on chromosomes 4 and 11. To test if GWAS results can help to improve genomic prediction accuracy markers from chromosomes 4, 11 and markers on other chromosomes were used independently to construct covariance matrices that were fitted together in a multikernel GBLUP model (Supplementary Fig. S15). For all CBSD traits, the mean predictive accuracy results from the single-kernel GBLUP model were similar to the mean total predictive accuracy following the multi-kernel approach (Supplementary Table S6B). However, the contribution of the individual kernels to the total predictive accuracies was different. For example, in the multikernel GBLUP model for CBSD 3 MAP (0.27), the highest contribution to the total predictive accuracy came from chromosome 11 and the rest of the genome (0.19). In contrast, the multikernel GBLUP model for 6 MAP gave the highest predictive accuracy (0.40) with the majority of prediction coming from chromosome 4 (0.29). Finally, the multikernel GBLUP approach for CBSDRS had a total predictive accuracy of 0.30 with the rest of the genome (0.29) contributing most to the total predictive accuracy (Supplementary Fig. S15).
Discussion
Efforts to understand CBSD resistance have focused on population development and QTL mapping, viral strain characterization, development of transgenic lines and evaluation of local and elite cassava genotypes to identify possible sources of resistance10,19,20,39,40. Although QTL and candidate genes associated with resistance to CBSD have been identified through bi-parental mapping, further transcriptional studies are needed to validate results and uncover the genetics of resistance to CBSD19,20,25.
We evaluated two GWAS panels from the NaCRRI breeding program in Uganda for CBSD severity symptoms in leaves and roots at locations with different CBSD disease pressure and environmental conditions. In both GWAS panels, the frequency distribution of scores within locations and the range of correlation values across locations (−0.08 to 0.60) reflect the differences in CBSD pressure at these locations. For example, Kasese and Namulonge are both hotspots for CBSD and showed a higher correlation in comparison to Kasese and Ngetta, which differ on their CBSD prevalence. The high variability observed within and across GWAS panels reflects differences in population composition, trait genetic architecture, field design and environmental effects including differences in virus strains or species which may contribute to poor correlations across locations.
In addition to scoring variability within and across locations, both panels showed variation in foliar CBSD symptom expression at 3, 6 and 9 MAP and root necrosis CBSDRS. These results are consistent with the observation that symptom expression changed with the age of the plant, with the loss of lower symptomatic leaves or symptoms being obscured in older plants16,41. Nonetheless, the correlation among foliar CBSD severities, in both panels and across locations, were consistently higher than the correlation between foliar and root severities. Similar to other studies20,21, our results demonstrate variability of symptom expression across environments and suggest that mechanisms of CBSD resistance operate somewhat independently in leaves and storage roots17, or are under different genetic control.
Given the availability of multi-location phenotypic data and GBS genotyping datasets, we assessed the potential of GWAS to identify QTL associated with CBSD resistance in leaves and storage roots. For all traits, several factors played a role in the identification of significantly associated SNPs including differences in panel size, age of the plant at scoring, CBSD prevalence and environmental condition at the different locations. Based on preliminary GWAS results in individual panels, we decided to increase the study power and resolution of our GWAS by combining multi-location scores from both panels. GWAS in the combined population detected SNPs significantly associated (P-value > 5.9) with foliar CBSD symptoms at 3 and 6 MAP on chromosomes 4 and 11. In contrast, no significant SNPs associated with CBSDRS could be detected, possibly due genotype-by-environment (GxE) interaction with CBSD and polygenic control or mechanisms of resistance under different genetic control contributing to CBSDRS19. However, when Panel 1 was analyzed independently, significant regions associated with CBSDRS were identified on chromosomes 5, 11 and 18. Similarly, Nzuki et al.19 in an analysis of a biparental population identified a region on chromosome 5 associated with root necrosis and a region on chromosome 11 associated with both root necrosis and foliar symptoms. Moreover, Masumba et al.20 found chromosomes 11 and 18 associated with CBSD root necrosis. Both QTL mapping studies were developed from crosses between Tanzanian clones and support the idea that much of the CBSD resistance present in the Ugandan breeding population has its origin in Tanzania.
Genomic annotation of the foliar GWAS results showed that a cluster of genes underlies the significant QTL associated with foliar disease resistance on chromosome 11; candidate genes that were identified for further study are Manes.11G131100, Manes.11G130500, Manes.11G130200 and Manes.11G130000. Lozano et al.42 previously reported Manes.11G130000 when studying the distribution of NBS-LRR gene family in the cassava genome. In addition,Manes.11G130000 was among the differentially expressed genes during early transcriptome response to brown streak virus infection in the susceptible line 60444 from the ETH cassava germplasm collection40. The QTL on chromosome 11 is particularly unstable across locations, which may be related to NBS-LRR genes conferring resistance to a particular viral strain3,7.
Throughout the 1940s and 1950s at the Amani Research Station in Tanzania, M. glaziovii and cassava varieties of Brazilian origin were successfully used for crosses to obtain varieties which showed high levels of field resistance to CBSD43. Similar to Bredeson et al.38, in our study, we identified widespread evidence for interspecific hybridization in the Ugandan cassava breeding panels. One of these introgression regions is located on chromosome 4, which in our study was associated with foliar severity. However, the degree of linkage disequilibrium in that region of chromosome 4 was a major constraint for the identification of a gene or genes responsible for CBSD resistance38. Interestingly, this region was not detected in the QTL mapping population involving the inter-specific hybrid, Namikonga21, but was detected in a cross with Kiroba20, another inter-specific hybrid presumably originating from the Amani breeding program.
Genomic selection (GS) can accelerate genetic gains through the use of phenotypic and genotypic data from a training population for early selection of seedlings to develop superior varieties32–34. We applied genomic prediction models that have different underlying assumptions: the GBLUP model assumes an infinitesimal genetic architecture, Bayesian methods such as BayesA and BayesB relax the assumption of common variance across marker effects44–46 and RKHS and RF can model epistasis. For all traits, using the combined dataset, we found moderate predictive accuracies in the 0.27–0.42 range; in general, predictive accuracies were in accordance with broad-sense heritability estimates, which ranged for Panel 1 between 0.37–0.61 and Panel 2 between 0.25–0.46. In general, we observed a superiority of the RF and RKHS models to predict both CBSD foliar symptoms and root necrosis, though the increase in predictive accuracy was more prominent for root necrosis. Similar to our results, previous studies that contrasted the performance of various prediction methods in cassava showed that RF and RKHS displayed higher predictive accuracy, particularly with phenotypes known to have a significant amount of non-additive genetic variation such as yield-related traits23. Based our findings, non-additive effects are likely to play a role shaping CBSD resistance in cassava in particular for root necrosis.
Even though a priori knowledge of the loci associated with a trait is not needed for GS, we tested a multiple kernel approach by fitting three kernels with genomic relationship matrices constructed with SNP markers from chromosomes 4 (Gchr4), 11 (Gchr11) and SNPs from other chromosomes (Gallchr-[4,11]). While the use of a multiple kernel approach did not increase predictive accuracies, the highest contribution of each kernel to the total predictive accuracy modeled the genetic architecture of CBSD traits. Based on our results, we conclude that genomic selection is a promising breeding tool for selecting for CBSD resistance. Based on our results, we conclude that genomic selection is a promising breeding tool for selecting for CBSD resistance and that the use of prediction models that consider both additive and non-additive effects could be advantageous.
Cassava brown streak disease is devastating cassava production in regions already affected by CBSVs and poses a high risk to countries in Central and West Africa where CBSD is not currently present. The GWAS and GS results presented in this study support previous findings which indicate that resistance to CBSD is polygenic39 and unstable across environments, which is an indication of quantitative resistance47. Although we were able to identify a candidate NBS-LRR gene on chromosome 11, the function of this gene in CBSD resistance requires further validation and more importantly, there is the possibility that this gene might not be a source of durable resistance to CBSVs. Further work will require screening large diversity panels, possibly including wild relatives, in multiple environments, and efforts to identify QTL specific to certain viral strains to further support the development of CBSD resistance.
Materials and Methods
Plant material and phenotyping
We assembled two cassava GWAS panels composed of 429 and 872 clones, respectively. These clones represented germplasm diversity derived from the International Institute for Tropical Agriculture (IITA) and the International Center for Tropical Agriculture (CIAT) (Supplementary Table 1).
GWAS Panel 1 clones were evaluated in replicated two-row 5 m plots in an alpha lattice field design from 2012 through 2014 at Namulonge, Ngetta, and Kasese in Uganda. While GWAS Panel 2 clones were evaluated in single row 10 m plots in an augmented design48,49 with 6 checks randomized in each incomplete block of 25 clones during the 2014 to 2015 season at Namulonge, Kamuli, and Serere.
Foliar disease severity was visually scored following the standard field phenotyping protocol for CBSD at 3, 6, and 9 months after planting (MAP) on a 1–5 scale13,50,51. Additional method description can be found in the supplementary methods section and Supplementary Fig. S1.
Two-stage genomic analyses
The first stage in data analysis involved accounting for trial design using a linear mixed model to obtained de-regressed BLUPs, and the second stage involved the use of de-regressed BLUPs in GWAS and Genomic prediction.
For Panel 1 we fit the model:
| 1 |
In this model, β included a fixed effect for the population mean and location. The incidence matrix Zclone and the vector c represent a random effect for clone and represents the identity matrix. The range variable, which is the row or column along which plots are arrayed, is nested in location-rep and is represented by the incidence matrix Zrange(loc.) and random effects vector . Block effects were nested in ranges and incorporated as random effects with incidence matrix Zblock(range) and effects vector . Residuals ε were distributed .
For Panel 2 we fit the model
| 2 |
Where y was the vector of raw phenotypes, β included fixed effects for the population mean, the location and finally an effect for checks. The incidence matrix Zclone and the vector c are the same as the model above, and the blocks were also modeled with incidence matrix and b represents the random effect for block. The best linear predictors (BLUPs) of the clone effect (ĉ) were extracted as de-regressed BLUPS following the formula52:
| 3 |
Where PEV is the prediction error variance of the BLUP and is the clonal variance component. We used the lmer function from the lme4 R package53 to fit the models described above.
Genotyping by sequencing (GBS)
Total genomic DNA was extracted from young leaves according to standard procedures using the DNAeasy plant mini extraction kit54. GBS libraries were constructed using the ApeKI restriction enzyme as previously described55. Marker genotypes were called using TASSEL GBS pipeline V456 after aligning the reads to the Cassava v6 reference genome57,58. Markers with more than 60% missing calls were removed.
The resulting marker dataset consisted of 173,647 bi-allelic SNP markers and was imputed using Beagle 4.159. After imputation, 63,016 SNPs had an AR2 (Estimated Allelic r-squared) higher than 0.3 and were kept for downstream analysis. From the remaining imputed markers, 41,530 had a minor allele frequency (MAF) higher than 0.01.
Genetic correlations and heritability estimates
Correlation across CBSD traits and within each location was estimated using using the de-regressed BLUP values obtained after fitting the aforementioned linear mixed model. Broad-sense heritability values on an entry-mean basis were calculated using the variance components estimated using the mixed-models described above, which represent the first-step of the two-step genomic analysis.
Finally, SNP-based heritability was calculated for each GWAS panel by fitting a single-step mixed-effects model using the emmreml function from the EMMREML R package60. The random effect was modeled as having a co-variance proportional to the kinship matrix, which was calculated using the A.mat function from the rrBLUP R package61.
Genome-wide association analysis for CBSV severity
A principal component analysis (PCA) was performed across panels to identify any population stratification between the two GWAS panels. We used the imputed dataset of 63,016 SNP markers to calculate the PCs with the function princomp in R62.
With the imputed dataset of 63,016 SNP markers and 986 individuals, GWAS was performed using a mixed linear model association analysis (MLMA) accounting for kinship and filtering by MAF > 0.05 as implemented in GCTA (v 1.26.0)63. We followed a leave one chromosome out approach in which the chromosome with the tested candidate SNP markers is excluded from the genomic relationship (GRM) calculation. Manhattan plots were generated using R package qqman with a Bonferroni threshold of 5.964.
Genomic prediction models
GBLUP
In this prediction model, the GEBVs are obtained after fitting a linear mixed model where the genomic realized relationship matrix is based on SNP marker dosages. Accordingly, the genomic relationship matrix was constructed using the function A.mat in the R package rrBLUP61. GBLUP predictions were made with the function emmreml in the R package EMMREML60.
Multi-kernel GBLUP
Because the most significant QTLs for foliar CBSD severity at 3 and 6 MAP were mapped on chromosomes 4 and 11 (this paper) we followed a multi-kernel approach by fitting three kernels with genomic relationship matrices constructed with SNP markers from chromosomes 4 (Gchr4), 11 (Gchr11) and SNPs from the other chromosomes (Gallchr-[4,11]). Multi-kernel GBLUP predictions were made with the function emmremlMultiKernel in the R package EMMREML60.
Reproducing kernel Hilbert spaces (RKHS)
We use a Gaussian kernel function:
| 4 |
where Kij is the measured relationship between two individuals, dij is their Euclidean genetic distance based on marker dosages, and θ is a tuning (“bandwidth”) parameter that determines the rate of decay of correlation among individuals. This function is nonlinear, and the kernels used for RKHS can capture non-additive as well as additive genetic variation65. To fit a multiple-kernel model with six covariance matrices, we used the emmremlMultiKernel function in the EMMREML package, with the following bandwidth parameters: 0.0000005, 0.00005, 0.0005, 0.005, 0.01, 0.05 (Multi-kernel RKHS) and allowed REML to find optimal weights for each kernel.
Bayesian maker regressions
We tested four Bayesian prediction models: BayesCpi46, the Bayesian LASSO66, BayesA, and BayesB32. The Bayesian models we tested allow for alternative genetic architecture by way of differential shrinkage of marker effects. We performed Bayesian predictions with the R package BGLR67.
Random Forest
Random Forest (RF) is a machine learning method used for regression and classification68–70. Random Forest regression with marker data has been shown to capture epistatic effects and has been successfully used for prediction70–74. We implemented RF using the Random Forest package in R75 with the parameter, ntree set to 500 and the number of variables sampled at each split (mtry) equal to 300.
Introgression Segment Detection
To identify introgression segments in the two GWAS panels, we followed the approach described in Bredeson et al.38. We used the M. glaziovii diagnostic markers identified in Supplementary Dataset 2 of Bredeson et al.38. These ancestry informative (AI) SNPs were identified as being fixed for different alleles in a sample of two pure M. esculenta (Albert and CM33064) and two pure M. glaziovii.
Out of 173,647 SNP in our imputed dataset, 12,502 matched published AI SNPs. For these AI SNPs, we divided each chromosome into non-overlapping windows of 20 SNP. Within each window, for each individual, we calculated the proportion of SNPs that were M. glaziovii homozygous (G/G), M. esculenta homozygous (E/E), or heterozygous (G/E). We assigned G/G, G/E or E/E ancestry to each window, for each individual only when the proportion of the most common genotype in that window was at least twice the proportion of the second most common genotype. If this was not the case, we assigned windows a “No Call” status.
Linkage disequilibrium plots
To confirm the presence of a haplotype on chromosome 4, we calculated LD scores for every SNP marker on chromosome 4 in a 1 Mb window using GCTA63. Briefly, the LD score for a given marker is calculated as the sum of R2 adjusted between the index marker and all markers within a specified window:
| 5 |
where “n” is the population size and R2 is the usual estimator of the squared Pearson’s correlation76. The resulting LD scores were then plotted against log10(P-value) from GWAS of every marker on chromosome 4.
To highlight the importance of the associated markers on chromosome 11 we calculated pairwise squared Pearson’s correlation coefficient (r2) between the top significant GWAS SNP hit on this chromosome and neighboring markers in a window of 2 Mb (1 Mb upstream and 1 Mb downstream)77,78.
Candidate gene identification
We used the mlma GCTA output to filter out SNP markers based on −log10(P-value) > 5.9. The resulting significant SNP markers were then mapped onto genes using the SNP location and gene description from the M.esculenta_305_v6.1.gene.gff3 available in Phytozome 1158 for Manihot esculenta v6.1 using bedtools79.
Data availability
The phenotypic and genotypic data generated and analyzed during this study are available in the Cassavabase repository, https://www.cassavabase.org/.
Electronic supplementary material
Acknowledgements
This work was generously funded by the “Next Generation cassava breeding project” through funds from the Bill and Melinda Gates Foundation and the Department for International development of the United Kingdom (UKaid), Grant 760 1048542. We give special thanks to John Francis Osingada for DNA processing, and the entire NaCRRI root crops team for assistance in field establishment, maintenance and data collection. We also acknowledge support from the Cassavabase team, Lukas Mueller, Guillaume Bauchet and Mukisa Rachael for phenotypic data curation, uploads to Cassavabase, SNP processing and imputation.
Author Contributions
I.S.K., and D.P.D.C. Conducted all analyses, wrote the original manuscript. I.S.K., A.O., and R.K. Conducted field trials, performed phenotyping and curated the data. R.L., and M.W. Imputed GBS data and gave editorial input into the manuscript. R.K., Y.B., V.G., J.L.J., and S.O. Conceived and designed the study. Editorial input into the analyses and the manuscript. M.F. Conceptual and editorial input into the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Siraj Ismail Kayondo and Dunia Pino Del Carpio contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-19696-1.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pérez JC, et al. Genetic variability of root peel thickness and its influence in extractable starch from cassava (Manihot esculenta Crantz) roots. Plant Breed. 2011;130:688–693. doi: 10.1111/j.1439-0523.2011.01873.x. [DOI] [Google Scholar]
- 2.ASARECA: ASARECA Annual Report 2012: Transforming Agriculture for Economic Growth in Eastern and Central Africa (2013).
- 3.Ndunguru J, et al. Analyses of twelve new whole genome sequences of cassava brown streak viruses and ugandan cassava brown streak viruses from East Africa: Diversity, supercomputing and evidence for further speciation. PLoS One. 2015;10:e0139321. doi: 10.1371/journal.pone.0139321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patil BL, Legg JP, Kanju E, Fauquet CM. Cassava brown streak disease: A threat to food security in Africa. J. Gen. Virol. 2015;96:956–968. doi: 10.1099/vir.0.000014. [DOI] [PubMed] [Google Scholar]
- 5.Mbanzibwa D, et al. Genetically distinct strains of Cassava brown streak virus in the Lake Victoria basin and the Indian Ocean coastal area of East Africa. Arch. Virol. 2009;154:353–359. doi: 10.1007/s00705-008-0301-9. [DOI] [PubMed] [Google Scholar]
- 6.Winter S, et al. Analysis of cassava brown streak viruses reveals the presence of distinct virus species causing cassava brown streak disease in East Africa. J. Gen. Virol. 2010;91:1365–1372. doi: 10.1099/vir.0.014688-0. [DOI] [PubMed] [Google Scholar]
- 7.Alicai, T. et al. Cassava brown streak virus has a rapidly evolving genome: implications for virus speciation, variability, diagnosis and host resistance. Sci. Rep. 6, 10.1038/srep36164 (2016). [DOI] [PMC free article] [PubMed]
- 8.Legg JP, et al. Spatio-temporal patterns of genetic change amongst populations of cassava Bemisia tabaci whiteflies driving virus pandemics in East and Central Africa. Virus Res. 2014;186:61–75. doi: 10.1016/j.virusres.2013.11.018. [DOI] [PubMed] [Google Scholar]
- 9.McQuaid CF, Sseruwagi P, Pariyo A, van den Bosch F. Cassava brown streak disease and the sustainability of a clean seed system. Plant Pathol. 2016;65:299–309. doi: 10.1111/ppa.12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anjanappa RB, et al. Characterization of Brown StreakVirus–Resistant Cassava. Mol. Plant-Microbe Interact. 2016;29:527–534. doi: 10.1094/MPMI-01-16-0027-R. [DOI] [PubMed] [Google Scholar]
- 11.Ogwok E, Patil BL, Alicai T, Fauquet CM. Transmission studies with Cassava brown streak Uganda virus (Potyviridae: Ipomovirus) and its interaction with abiotic and biotic factors in Nicotiana benthamiana. J. Virol. Methods. 2010;169:296–304. doi: 10.1016/j.jviromet.2010.07.030. [DOI] [PubMed] [Google Scholar]
- 12.Maruthi MN, Jeremiah CS, Mohammed IU, Legg JP. The role of the whitefly, Bemisia tabaci (Gennadius), and farmer practices in the spread of cassava brown streak ipomoviruses. J. Phytopathol. 2017;165:707–717. doi: 10.1111/jph.12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hillocks RJ, Raya M, Thresh JM. The association between root necrosis and above‐ground symptoms of brown streak virus infection of cassava in southern Tanzania. Int. J. Pest Manag. 1996;42:285–289. doi: 10.1080/09670879609372008. [DOI] [Google Scholar]
- 14.Ndyetabula IL, et al. Analysis of Interactions Between Cassava Brown Streak Disease Symptom Types Facilitates the Determination of Varietal Responses and Yield Losses. Plant Dis. 2016;100:1388–1396. doi: 10.1094/PDIS-11-15-1274-RE. [DOI] [PubMed] [Google Scholar]
- 15.Legg J, et al. A global alliance declaring war on cassava viruses in Africa. Food Secur. 2014;6:231–248. doi: 10.1007/s12571-014-0340-x. [DOI] [Google Scholar]
- 16.Hillocks RJ, Jennings DL. Cassava brown streak disease: a review of present knowledge and research needs. Int. J. Pest Manag. 2003;49:225–234. doi: 10.1080/0967087031000101061. [DOI] [Google Scholar]
- 17.Kaweesi T, et al. Field evaluation of selected cassava genotypes for cassava brown streak disease based on symptom expression and virus load. Virol. J. 2014;11:216. doi: 10.1186/s12985-014-0216-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kulembeka HP, et al. Diallel analysis of field resistance to brown streak disease in cassava (Manihot esculenta Crantz) landraces from Tanzania. Euphytica. 2012;187:277–288. doi: 10.1007/s10681-012-0730-0. [DOI] [Google Scholar]
- 19.Nzuki I, et al. QTL Mapping for Pest and Disease Resistance in Cassava and Coincidence with some Introgression Regions derived from M. glaziovii. Front. Plant Sci. 2017;8:1168. doi: 10.3389/fpls.2017.01168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masumba EA, et al. QTL associated with resistance to cassava brown streak and cassava mosaic diseases in a bi-parental cross of two Tanzanian farmer varieties, Namikonga and Albert. Theor. Appl. Genet. 2017;130:2069–2090. doi: 10.1007/s00122-017-2943-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ceballos H, Iglesias CA, Pérez JC, Dixon AGO. Cassava breeding: opportunities and challenges. Plant Mol. Biol. 2004;56:503–16. doi: 10.1007/s11103-004-5010-5. [DOI] [PubMed] [Google Scholar]
- 22.Ceballos H, Kawuki RS, Gracen VE, Yencho GC, Hershey CH. Conventional breeding, marker-assisted selection, genomic selection and inbreeding in clonally propagated crops: a case study for cassava. Theor. Appl. Genet. 2015;128:1647–1667. doi: 10.1007/s00122-015-2555-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolfe, M. D. et al. Prospects for genomic selection in cassava breeding. Plant Genome10, 10.3835/plantgenome2017.03.0015 (2017). [DOI] [PMC free article] [PubMed]
- 24.Maruthi, M. N., Bouvaine, S., Tufan, H. A., Mohammed, I. U. & Hillocks, R. J. Transcriptional response of virus-infected cassava and identification of putative sources of resistance for cassava brown streak disease. PLoS One9, 10.1371/journal.pone.0096642 (2014). [DOI] [PMC free article] [PubMed]
- 25.Amuge T, et al. A time series transcriptome analysis of cassava (Manihot esculenta Crantz) varieties challenged with Ugandan cassava brown streak virus. Sci. Rep. 2017;7:9747. doi: 10.1038/s41598-017-09617-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor NJ, et al. The VIRCA Project: virus resistant cassava for Africa. GM Crops Food. 2012;3:93–103. doi: 10.4161/gmcr.19144. [DOI] [PubMed] [Google Scholar]
- 27.Wagaba H, et al. Artificial microRNA-derived resistance to Cassava brown streak disease. J. Virol. Methods. 2016;231:38–43. doi: 10.1016/j.jviromet.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beyene G, et al. A Virus-Derived Stacked RNAi Construct Confers Robust Resistance to Cassava Brown Streak Disease. Front. Plant Sci. 2017;7:1–12. doi: 10.3389/fpls.2016.02052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolfe MD, et al. Genome-wide association and prediction reveals the genetic architecture of cassava mosaic disease resistance and prospects for rapid genetic improvement. Plant Genome. 2016;9:1–13. doi: 10.3835/plantgenome2015.11.0118. [DOI] [PubMed] [Google Scholar]
- 30.Esuma W, et al. Genome-wide association mapping of provitamin A carotenoid content in cassava. Euphytica. 2016;212:97–110. doi: 10.1007/s10681-016-1772-5. [DOI] [Google Scholar]
- 31.Rabbi, I. Y. et al. Genome-Wide Association Mapping of Correlated Traits in Cassava: Dry Matter and Total Carotenoid Content. Plant Genome10, 10.3835/plantgenome2016.09.0094 (2017). [DOI] [PMC free article] [PubMed]
- 32.Meuwissen THE, Hayes BJ, Goddard ME. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157:1819–1829. doi: 10.1093/genetics/157.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jannink J-LL, Lorenz AJ, Iwata H. Genomic selection in plant breeding: from theory to practice. Brief. Funct. Genomics. 2010;9:166–177. doi: 10.1093/bfgp/elq001. [DOI] [PubMed] [Google Scholar]
- 34.Lorenz, A. J. et al. Genomic Selection in Plant Breeding. Knowledge and Prospects. Advances in Agronomy110, (Elsevier Inc, 2011).
- 35.Ly D, et al. Relatedness and genotype x environment interaction affect prediction accuracies in genomic selection: A study in cassava. Crop Sci. 2013;53:1312–1325. doi: 10.2135/cropsci2012.11.0653. [DOI] [Google Scholar]
- 36.Elshire RJ, et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One. 2011;6:e19379. doi: 10.1371/journal.pone.0019379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jennings DL. Manihot melanobasis Müll. Arg.—a useful parent for cassava breeding. Euphytica. 1959;8:157–162. [Google Scholar]
- 38.Bredeson JV, et al. Sequencing wild and cultivated cassava and related species reveals extensive interspecific hybridization and genetic diversity. Nat. Biotechnol. 2016;34:562–570. doi: 10.1038/nbt.3535. [DOI] [PubMed] [Google Scholar]
- 39.Kawuki RSRS, et al. Eleven years of breeding efforts to combat cassava brown streak disease. Breed. Sci. 2016;66:560–571. doi: 10.1270/jsbbs.16005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anjanappa, R. B. et al. Molecular insights into cassava brown streak virus susceptibility and resistance by profiling of the early host response. Mol. Plant Pathol. 1–14 (2017). [DOI] [PMC free article] [PubMed]
- 41.Rwegasira GM, Rey ME. C. Response of Selected Cassava Varieties to the Incidence and Severity of Cassava Brown Streak Disease in Tanzania. J. Agric. Sci. 2012;4:237–245. [Google Scholar]
- 42.Lozano R, Hamblin MT, Prochnik S, Jannink J-L. Identification and distribution of the NBS-LRR gene family in the Cassava genome. BMC Genomics. 2015;16:1–14. doi: 10.1186/s12864-015-1554-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hillocks, R. J. & Thresh, J. M. Cassava: biology, production and utilization (CABI, 2002).
- 44.Legarra A, Christensen OF, Aguilar I, Misztal I. Single Step, a general approach for genomic selection. Livest. Sci. 2014;166:54–65. doi: 10.1016/j.livsci.2014.04.029. [DOI] [Google Scholar]
- 45.De Los Campos G, et al. Predicting quantitative traits with regression models for dense molecular markers and pedigree. Genetics. 2009;182:375–385. doi: 10.1534/genetics.109.101501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Habier D, Fernando RL, Kizilkaya K, Garrick DJ. Extension of the bayesian alphabet for genomic selection. BMC Bioinformatics. 2011;12:186. doi: 10.1186/1471-2105-12-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anthony P, et al. Stability of resistance to cassava brown streak disease in major agro-ecologies of Uganda. J. Plant Breed. Crop Sci. 2015;7:67–78. doi: 10.5897/JPBCS2013.0490. [DOI] [Google Scholar]
- 48.Federer WT, Nguyen N-K. Constructing Augmented Experiment Designs with Gendex. Biometrics Unit Tech. Reports. 2002;BU-1610-M:12. [Google Scholar]
- 49.Federer, W. T. & Crossa, J. Screening Experimental Designs for Quantitative Trait Loci, Association Mapping, Genotype-by EnvironmentInteraction, and Other Investigations. Front. Physiol. 3, 10.3389/fphys.2012.00156 (2012). [DOI] [PMC free article] [PubMed]
- 50.Hillocks RJ, Thresh JM. Cassava mosaic and cassava brown streak virus diseases inAfrica: a comparative guide to symptoms and aetiologies. Roots. 2000;7:1–8. [Google Scholar]
- 51.Mohammed, I. U., Abarshi, M. M., Muli, B., Hillocks, R. J. & Maruthi, M. N. The symptom and genetic diversity of cassava brown streak viruses infecting cassava in EastAfrica. Adv. Virol. 2012, 10.1155/2012/795697 (2012). [DOI] [PMC free article] [PubMed]
- 52.Garrick DJ, Taylor JF, Fernando RL. Deregressing estimated breeding values and weighting information for genomic regression analyses. Genet. Sel. Evol. 2009;41:55. doi: 10.1186/1297-9686-41-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bates D, et al. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 2017;67:1–113. [Google Scholar]
- 54.QIAGEN. DNeasy® Plant Handbook DNeasy Plant Mini Kit and tissues, or fungi Sample & Assay Technologies QIAGEN Sample and Assay Technologies. (2012).
- 55.Hamblin MT, Rabbi IY. The effects of restriction-enzyme choice on properties of genotyping-by-sequencing libraries: A study in Cassava (Manihot esculenta) Crop Science. 2014;54:2603–2608. doi: 10.2135/cropsci2014.02.0160. [DOI] [Google Scholar]
- 56.Glaubitz JC, et al. TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS One. 2014;9:e90346. doi: 10.1371/journal.pone.0090346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prochnik S, et al. The Cassava Genome: Current Progress, Future Directions. Trop. Plant Biol. 2012;5:88–94. doi: 10.1007/s12042-011-9088-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goodstein DM, et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:1–2. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Browning BL, Browning SR. Genotype Imputation with Millions of Reference Samples. Am. J. Hum. Genet. 2016;98:116–126. doi: 10.1016/j.ajhg.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Akdemir, D. & Okeke, U. G. EMMREML: Fitting Mixed Models with Known Covariance Structures. https://cran.r-project.org/package=EMMREML. R package version 3.1 (2015).
- 61.Endelman JB. Ridge Regression and Other Kernels for Genomic Selection with R Package rrBLUP. Plant Genome J. 2011;4:250. doi: 10.3835/plantgenome2011.08.0024. [DOI] [Google Scholar]
- 62.R Development Core Team. R: A Language and Environment for Statistical Computing. https://www.R-project.org/ (2017).
- 63.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Turner, S. D. qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. bioRxiv10.1101/005165 (2014).
- 65.Gota M, Gianola D. Kernel-based whole-genome prediction of complex traits: A review. Front. Genet. 2014;5:1–13. doi: 10.3389/fgene.2014.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Park T, Casella G. The Bayesian Lasso. J. Am. Stat. Assoc. 2008;103:681–686. doi: 10.1198/016214508000000337. [DOI] [Google Scholar]
- 67.Pérez P, De Los Campos G. Genome-wide regression and prediction with the BGLR statistical package. Genetics. 2014;198:483–495. doi: 10.1534/genetics.114.164442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Charmet G, Storlie E. Implementation of genome-wide selection in wheat. Russ. J. Genet. Appl. Res. 2012;2:298–303. doi: 10.1134/S207905971204003X. [DOI] [Google Scholar]
- 69.Strobl C, Malley J, Tutz G. An Introduction to Recursive Partitioning: Rationale, Application, and Characteristics of Classification and Regression Trees, Bagging, and Random Forests. Psychol. Methods. 2009;14:323–348. doi: 10.1037/a0016973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Breiman L. Random Forests. Mach. Learn. 2001;45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 71.Heslot N, Yang H-P, Sorrells ME, Jannink J-L. Genomic Selection in Plant Breeding: A Comparison of Models. Crop Sci. 2012;52:146–160. doi: 10.2135/cropsci2011.06.0297. [DOI] [Google Scholar]
- 72.Motsinger-Reif AA, Reif DM, Fanelli TJ, Ritchie MD. A comparison of analytical methods for genetic association studies. Genet. Epidemiol. 2008;32:767–778. doi: 10.1002/gepi.20345. [DOI] [PubMed] [Google Scholar]
- 73.Spindel J, et al. Genomic Selection and Association Mapping in Rice (Oryza sativa): Effect of Trait Genetic Architecture, Training Population Composition, Marker Number and Statistical Model on Accuracy of Rice Genomic Selection in Elite, Tropical Rice Breeding Lines. PLoS Genet. 2015;11:1–25. doi: 10.1371/journal.pgen.1004982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Charmet G, et al. Genome-wide prediction of three important traits in bread wheat. Mol. Breed. 2014;34:1843–1852. doi: 10.1007/s11032-014-0143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liaw a, Wiener M. Classification and Regression by random Forest. R news. 2002;2:18–22. [Google Scholar]
- 76.Bulik-Sullivan B, et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Purcell S, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rentería ME, Cortes A, Medland SE. Using PLINK for genome-wide association studies (GWAS) and data analysis. Methods Mol. Biol. 2013;1019:193–213. doi: 10.1007/978-1-62703-447-0_8. [DOI] [PubMed] [Google Scholar]
- 79.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The phenotypic and genotypic data generated and analyzed during this study are available in the Cassavabase repository, https://www.cassavabase.org/.