Abstract
Secondary plastids derived from green algae occur in chlorarachniophytes, photosynthetic euglenophytes, and the dinoflagellate genus Lepidodinium. Recent advances in understanding the origin of these plastids have been made, but analyses suffer from relatively sparse taxon sampling within the green algal groups to which they are related. In this study we aim to derive new insights into the identity of the plastid donors, and when in geological time the independent endosymbiosis events occurred. We use newly sequenced green algal chloroplast genomes from carefully chosen lineages potentially related to chlorarachniophyte and Lepidodinium plastids, combined with recently published chloroplast genomes, to present taxon-rich phylogenetic analyses to further pinpoint plastid origins. We integrate phylogenies with fossil information and relaxed molecular clock analyses. Our results indicate that the chlorarachniophyte plastid may originate from a precusor of siphonous green algae or a closely related lineage, whereas the Lepidodinium plastid originated from a pedinophyte. The euglenophyte plastid putatively originated from a lineage of prasinophytes within the order Pyramimonadales. Our molecular clock analyses narrow in on the likely timing of the secondary endosymbiosis events, suggesting that the event leading to Lepidodinium likely occurred more recently than those leading to the chlorarachniophyte and photosynthetic euglenophyte lineages.
Introduction
The spread of plastids by secondary endosymbiosis – the uptake of an alga containing a primary plastid by a heterotrophic eukaryotic host – has driven the evolution of many photosynthetic lineages of global ecological and economic importance. Haptophytes, diatoms and most photosynthetic dinoflagellates, for example, contain red-algal derived plastids originating from secondary (or subsequent higher) endosymbiotic events1–3, and together these organisms constitute major primary producers in marine environments. Other lineages contain a secondary plastid originating from a green algal ancestor4–7. Due in part to their ubiquity and diversity8,9, lineages with secondary red plastids have undergone intense scrutiny, whereas organisms with secondary green plastids are less well-studied.
Currently, three lineages are known to contain secondary plastids derived from green algae: euglenophytes, chlorarachniophytes and the dinoflagellate genus Lepidodinium10. Photosynthetic euglenophytes occur in both freshwater and marine environments11, and these taxa and their secondarily heterotrophic descendants comprise the class Euglenophyceae, forming a clade within the broader Euglenida sensu Adl et al.12. Chlorarachniophytes, a small group of exclusively marine unicellular algae, are often amoeboid in form and belong to the supergroup Rhizaria. Together with cryptophytes they are unique among lineages with secondary plastids in that they retain a vestigial nucleus from the algal endosymbiont, termed a nucleomorph13,14. Dinoflagellates are a large group of flagellated marine and freshwater protists belonging to the supergroup Alveolata. Approximately half are photosynthetic, harbouring a secondary plastid derived from a red algal ancestor, and these plastids contain the pigment peridinin15. Some dinoflagellate lineages, however, have subsequently acquired a plastid from a different source through additional secondary or tertiary endosymbioses (the latter involving uptake of an alga already containing a secondary plastid)16. Yet other species contain temporary but functional kleptoplastids (stolen plastids) acquired from algal prey17,18. The genus Lepidodinium contains a secondary plastid derived from a green algal ancestor. Two species are currently recognised, L. viride and L. chlorophorum19. Given that the secondary peridinin-type plastid is present in the last common dinoflagellate ancestor, the green Lepidodinium plastid is postulated to have originated from an additional secondary event via so-called ‘serial secondary endosymbiosis’16,20.
While there is ongoing controversy about exactly how many secondary (and perhaps higher) endosymbiosis events have led to the numerous lineages containing a secondary red plastid21, the situation for secondary green plastids is more clear-cut, and it is now apparent that euglenophytes, chlorarachniophytes and Lepidodinium acquired their plastids in three independent evolutionary events. Moreover, phylogenies of plastid genes recover the three lineages branching with different, relatively unrelated groups of green algae, indicating a distinct plastid origin in each case4,5,22. For Lepidodinium, recent phylogenomic analyses of the plastid genome (plDNA) recovered a well-supported sister relationship with the pedinophyte Pedinomonas minor4, whereas euglenophyte plastids are sister to prasinophytes from the order Pyramimonadales22. The ancestor of the chlorarachniophyte plastid remains uncertain23; although recent plDNA phylogenies recovered a well-supported relationship between chlorarachniophyte plastids and the green-algal order Bryopsidales, the closest relative of this clade within the green algal phylogeny was not resolved5. Overall, analyses of the origin of secondary green plastids suffer from relatively sparse taxon sampling within the green algal groups to which they are related. In recent years, complete plastid genomes (plDNA) from diverse green algae have been sequenced, encouraging us to revisit the question.
In this study we use an expanded dataset of publically available sequences, together with strategically selected newly sequenced plastid genomes, to re-examine the origins of secondary green plastids using a phylogenomics approach. In doing so, we aim to pinpoint the closest extant relatives of the ancestral green algae that were involved in the three distinct endosymbiosis events, and to infer the timing of these events using relaxed molecular clock methods.
Materials and Methods
Culturing, DNA extraction, sequencing, plDNA assembly, and gene annotation
The pedinophyte strain YPF-701 (NIES Microbial Culture Collection strain NIES-2566) was cultured in Guillard’s F/2 media at 20 °C on a 14-h light/10-h dark cycle. Avrainvillea mazei HV02664 (representing the early-branching family Dichotomosiphonaceae of the Bryopsidales) and Neomeris sp. HV02668 (representing the Dasycladaceae, an early-branching Dasycladales family) were collected from Tavernier (FL, USA) and Islamorada (FL, USA), respectively, and preserved in silica-gel. For collection details of Ostreobium sp. HV05042 see24. For pedinophyte YPF-701 cells were harvested by centrifugation (10 min, 3,000 g). Total genomic DNA was extracted using a modified CTAB protocol25, in which the CTAB extraction buffer was added directly to the cell pellet. Due to technology availability and variation of DNA yields, two sequencing strategies were used. For Avrainvillea mazei HV02664, a TruSeq Nano LT library (~350 bp inserts) was prepared for sequencing of 2 × 100 bp paired-end reads using the Illumina HiSeq 2000 platform. For the other two strains, libraries (~500 bp inserts) were prepared using a Kapa Biosystems kit for sequencing of 2 × 150 bp paired-end reads using the Illumina NextSeq platform. All libraries were sent for sequencing at Novogene (Hong Kong).
Sequence reads were assembled using SPAdes 3.8.126 using the –careful option. Contigs matching to pedinophyte or Ulvophycean chloroplast genome reference sequences were imported into Geneious 9.1.3 (http://www.geneious.com), where completeness and circularity were manually evaluated. Final contigs were annotated following Verbruggen and Costa27 and Marcelino et al.28. In brief, annotations were obtained from MFannot, DOGMA and ARAGORN and imported into Geneious. All annotations were vetted manually and a master annotation track was created from them.
Phylogenetic analyses
Maximum Likelihood phylogenies
All 151 chloroplast genomes for green algae, photosynthetic euglenophytes, Lepidodinium chlorophorum and chlorarachniophytes used in this study are shown in Table S7. For each protein-coding gene, protein sequences were aligned using MAFFT 7.21529, after which the aligned amino acid residues were reverse translated into the corresponding coding nucleotide sequences (in fixed codon positions) using TranslatorX30. Genes that were present in >50% of total taxa (64 genes) were included in subsequent analyses. For each alignment, poorly aligned regions were removed via an automated algorithm using the Gblocks software31 version 0.91b with options −t = c − b5 = h. Single-gene alignments were concatenated to produce a multigene supermatrix (Dataset A, 34,452 nucleotides) using Geneious (Biomatters) (see Supplementary Table S7 for missing data percentages), and an amino-acid translation of the nucleotide alignment was generated. The nucleotide alignment was partitioned by gene and codon position and Partition Finder32 was used to determine the best-fit partitioning scheme. Partition Finder was run multiple times, once for each of the following independent models: GTR, HKY, JC69, and K80. The amino-acid alignment was partitioned by gene, and Partition Finder was used to assign one of the following models to each partition: LG, WAG, MTREV, JTT, CPREV, DAYHOFF, BLOSUM62. For nucleotide analyses, individual maximum likelihood (ML) trees were estimated for each model/partitioning scheme, using the concatenated dataset with RAxML v8.2.633 and 500 non-parametric bootstrap replicates. RAxML amino-acid analyses were also performed with 500 non-parametric bootstrap replicates. For both nucleotide and amino-acid analyses a gamma model of rate heterogeneity with four categories was used. For amino-acid analyses, empirical amino-acid frequencies were applied to partitions where recommended by Partition Finder.
For site-stripping analyses, per-site substitution rates were calculated for our Dataset A alignments using HyPhy34, and the fastest evolving sites were removed using SiteStripper v.1.01 (http://www.phycoweb.net/software/SiteStripper/index.html, last accessed November 24, 2016), leaving a percentage length of the original alignment (95%, 90%, 85%, 80%, 75%, 70%, 65%, 60%, 55%, 50%). ML trees were estimated with RAxML as above under a GTR model, retaining partitions, with 100 non-parametric bootstrap replicates.
In addition to analyses with Dataset A, we also performed ML analyses on a second dataset including some very recently released green-algal plDNAs as well as sequences recently generated in our laboratory (Dataset B, see Supplementary Table S9 for taxa list), to evaluate whether the new sequences affected the branching positions of secondary plastid lineages and hence our overall conclusions. The Dataset B nucleotide supermatrix (69 genes, 35,739 nucleotides) was generated as per above, and an amino-acid translation was produced. Trees were generated using RAxML (LG model for amino-acid analysis; GTR model with partitioning by codon position for nucleotide analysis), with 100 rapid bootstrap replicates. A tree was also estimated from the amino-acid alignment using IQ-TREE35 with the site-heterogeneous LG + G4 + C60 + F + PMSF model36, using the topology estimated by the RAxML amino-acid analysis as a guide tree to compute PMSF profiles.
Bayesian phylogenies
Bayesian analyses of both nucleotide and amino-acid alignments (Dataset A) were performed using Phylobayes 4.1b37 with the CATGTR model. Two independent Markov chain Monte Carlo runs were performed for each analysis for a total of 10,000 cycles. A consensus topology was calculated using the bpcomp program; the first 2,000 cycles from each chain were discarded as burn-in, trees from both chains were sampled every ten cycles, and a majority-rule posterior consensus tree was generated. For both nucleotide and amino-acid analyses the largest discrepancy observed across all bipartitions (maxdiff) was 1, indicating that at least one of the runs was stuck in a local maximum in each case.
Relaxed molecular clock analyses
To select genes for the molecular clock analyses we followed the methods of Castresana (2000) using the SortaDate package, using the ML tree topology recovered in the amino-acid analyses (Dataset A) and amino-acid translations of single-gene alignments. A dataset was compiled from the top-ranked 11 genes (tufA, rps7, rps4, rpl14, rpl5, rpl2, psbC, psbB, petA, atpI, atpA). In addition, we included atpB and rbcL to allow testing of fossil calibrations from the Chlorophyceaen family Hydrodictyaceae. See Supplementary Information File S1 for full methods and details of node calibrations and chronogram analyses.
Data availability
Plastid sequence data is deposited in Genbank under accession KY347917 (pedinophyte strain YPF-701), KY509314 (Ostreobium sp. HV05042), KY495826-KY495874 (Neomeris sp.), and KY509313 (Avrainvillea mazei). All other data generated or analysed during this study are either publically available or included in this published article (and its Supplementary Information files).
Results and Discussion
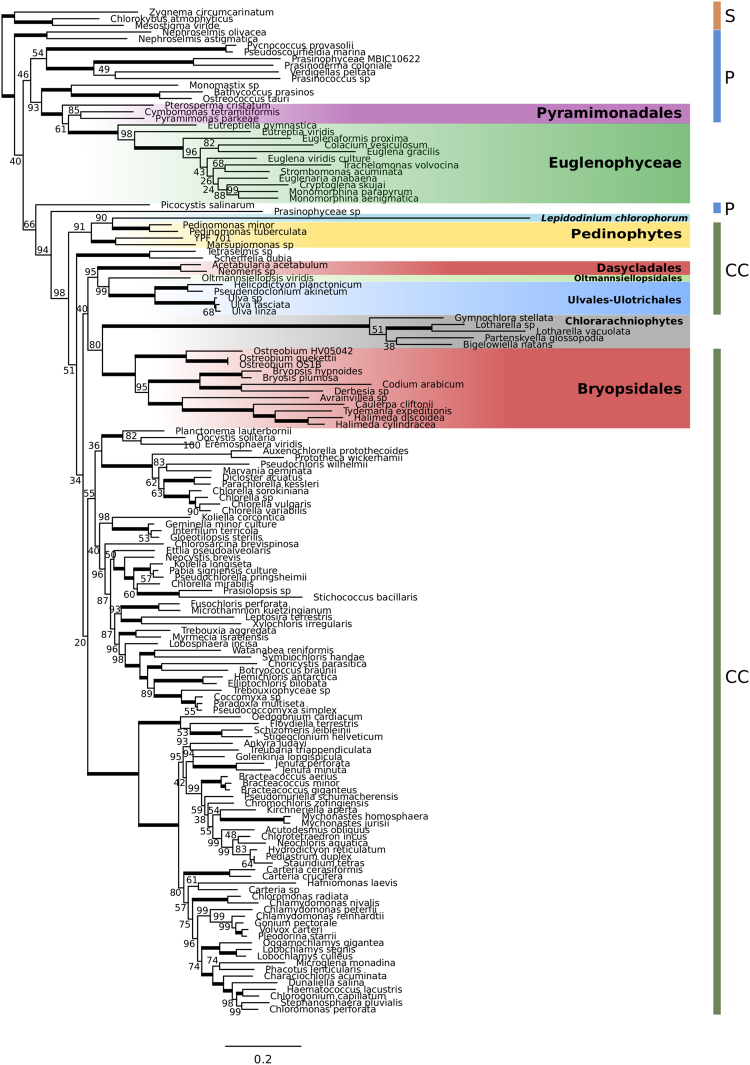
To increase the density of taxon sampling in lineages potentially related to secondary endosymbionts, we sequenced plDNAs from lineages in the Ulvophyceae (Ostreobium sp. HV05042, complete; Avrainvillea mazei, near complete; Neomeris sp., fragmented) and Pedinophyceae (strain YPF-701, complete), and combined them with publically available complete or near-complete plastid genome data (see Supporting Information Table S7). Accelerated rates of evolution have been observed in secondary green plDNAs, reflected in long branches in phylogenetic analyses4,38. This could potentially result in long-branch attraction (LBA), a phylogenetic artefact in which long branches cluster together even if they are not closely related39. Increased sampling of putatively related lineages minimises long branches in the clades which contain them, reducing potential LBA between secondary plastids and their closest green-algal relatives. Using our dataset of 151 taxa and 64 genes (Dataset A; see methods), we carried out Maximum likelihood (ML) and Bayesian analyses using both nucleotide and amino-acid alignments. Our phylogenies support the three independent gains of secondary green plastids (Fig. 1; see below for discussion of additional analyses, Supplementary Figs S1–S7). The topology of the backbone green-algal tree largely corresponds to previously published studies, with a series of early-branching prasinophyte lineages, a monophyletic “core Chlorophyta” and little support for the monophyly of traditionally recognized Ulvophyceae and Trebouxiophyceae40.
Figure 1.
RAxML phylogenetic analysis inferred from an amino acid alignment of 64 plastid genes from streptophytes, green algae, photosynthetic euglenophytes, the “green” dinoflagellate Lepidodinium chlorophorum, and chlorarachniophytes. The dataset was partitioned by gene according to the best-fit model assigned by PartitionFinder. Thick branches have full ML bootstrap support. Coloured vertical bars to the right of the phylogeny are labelled: S, Streptophytes; P, prasinophytes; CC, core Chlorophyta. Branch lengths are proportional to the number of substitution per site.
Euglenophytes
Previous plastid phylogenomic studies have recovered a sister relationship between the Euglenophyceae and the prasinophyte order Pyramimonadales7,22,41,42. These studies, together with lines of evidence such as plDNA gene-linkage conservation22 and individual gene similarity43 between Euglenophyceae and pyramimonadalean taxa, suggest that the secondary endosymbiosis event leading to the Euglenophyceae involved a pyramimonadalean alga.
In this study, we have collated publically available data to include the most comprehensive taxon sampling of chloroplast genomes in both the Euglenophyceae (12 taxa) and Pyramimonadales (three taxa) to date. Consistent with previous investigations, ML analyses and Bayesian phylogenies using both amino-acid and nucleotide data recover the Euglenophyceae branching most closely to the prasinophyte order Pyramimonadales (Fig. 1, Supplementary Figs S1, S6, S7). As observed in an earlier plDNA phylogenomic study7, Eutreptiella branches as the earliest lineage within the Euglenophyceae, followed by Eutreptia. To test the robustness of the Pyramimonadales + Euglenophyceae relationship (and other relationships described below) using nucleotide data, we also performed ML analyses using different substitution models in addition to the best-fit (GTR) model (Supplementary Figs S2–S4). A Pyramimonadales + Euglenophyceae clade is recovered with strong support in these phylogenies. However, in most cases the pyramimonadalean lineage closest to the Euglenophyceae clade is not well-resolved. In all ML analyses, Cymbomonas and Pyramimonas group together with good to full support, whereas the branching position of Pterosperma differs (Fig. 1, Supplementary Figs S1–S5). Generally, ML bootstrap support for the branching order of Pterosperma is low, with the exception of nucleotide analyses using either a JC69 or K80 substitution model: both phylogenies recovered Pterosperma branching as sister to Cymbomonas and Pyramimonas (89% and 90% bootstrap support [BS], respectively), and together this clade formed a sister group to the euglenophytes (95% and 98% BS, respectively). In Bayesian analyses either Cymbomonas or Pyramimonas branches most closely to the Euglenophyceae (nucleotide vs amino-acid alignments, respectively), but relationships between pyramimonadalean taxa are not well-resolved (Supplementary Figures S6 and S7). This lack of resolution might be due to the limited data available for Pterosperma; complete plDNA sequence is lacking, and only 10 genes were included in our analyses. To evaluate whether systematic biases caused by fast-evolving sites could also be contributing to low support, we calculated site-specific substitution rates for our concatenated nucleotide data set and progressively removed the fastest-evolving sites (see methods). As shown in Supplementary Fig. S8 and Table S8, the ML topology described above is recovered in the 95, 90, 85, 80, 75, 70, 65, and 60% datasets, with support for the branching position of Pterosperma ranging from 45–63% BS. Overall, the exact pyramimonadalean lineage (or closely related lineage) that was the source of the euglenophyte plastid remains an open question.
Although members of the Euglenophyceae are found in both marine and freshwater environments, it has been suggested that the secondary endosymbiosis event leading to this lineage occurred in a marine habitat6,41. This hypothesis is consistent with the marine habitat of early-branching lineages within the extant photosynthetic euglenophytes, Eutreptiella and Eutreptia7, as well as Rapaza44. The fact that Pyramimonas comprises marine species has also been presented as corroborating evidence for a marine origin41. However, phylogenetic analyses of the origin of the euglenophyte plastid have so far only included marine pyramimonadalean taxa, whereas many species occur in freshwater environments, including members of the genus Pyramimonas. It might still be the case, then, that the euglenophyte plastid is more closely related to freshwater pyramimonadalean lineages. Moreover, the closest known non-photosynthetic relative of the Euglenophyceae, the euglenid lineage Peranema45, largely comprises freshwater species, suggesting that the host cell involved in secondary endosymbiosis could have been of freshwater origin. Together, these facts leave open the possibility of a freshwater origin of the Euglenophyceae.
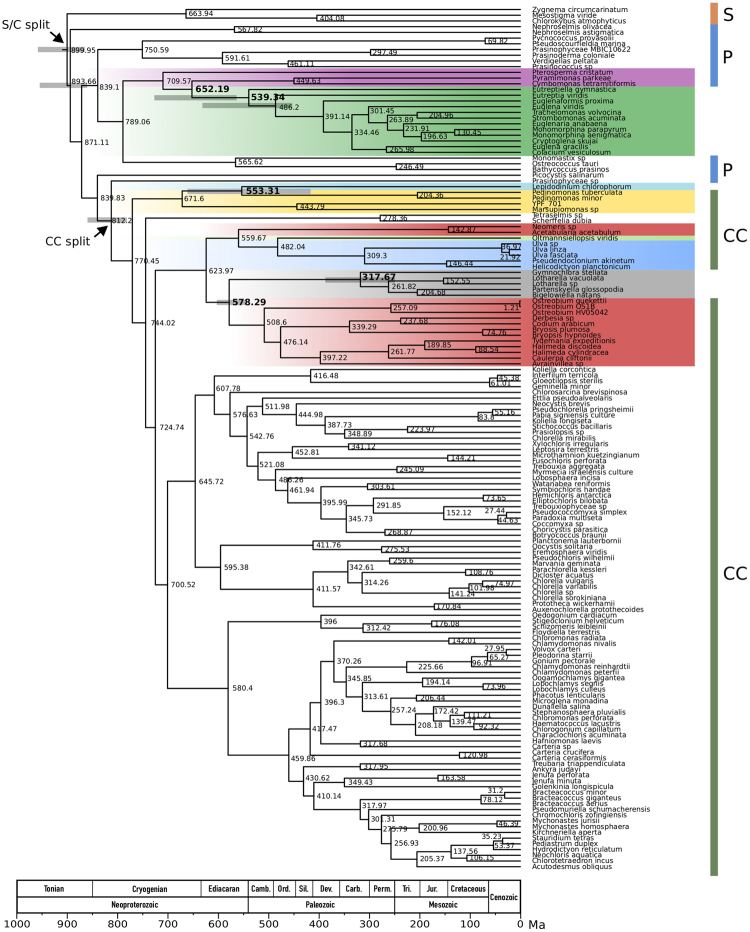
Our relaxed molecular clock analyses indicate that the secondary endosymbiosis event leading to the Euglenophyceae occurred between ~652 and 539 million years ago (Ma) (Fig. 2). The 95% confidence intervals (CI) for these dates overlap (~563–728 and ~453–631 Ma, respectively), and together encompass the late Proterozoic to early Paleozoic. The two timeframe estimates correspond to the split between the Pyramimonadales and the Euglenophyceae plastid lineage, and the start of the radiation of Euglenophyceae, respectively. Thus, the divergence of the euglenophyte plastid lineage may have followed closely on the heels of the initial diversification of marine Pyramimonadales, as recorded by fossil phycomata46, biomarker molecular fossils47 and molecular clock estimates48. However, the uncertain branching position of Pterosperma in our phylogenetic analyses means that this date estimate should be interpreted with caution. While we cannot determine at which point in time along the branch leading to Euglenophyceae the secondary endosymbiosis event took place, fossil pellicles indicate that photosynthetic euglenophytes existed at least 450 million years ago49,50. Phylogenies of nuclear SSU rDNA recover the photosynthetic marine euglenid Rapaza viridis branching as the nearest sister lineage to the remaining photosynthetic euglenophytes44. Plastid sequence data from Rapaza would therefore be informative in estimating the date of Euglenophyceae radiation, and in narrowing the potential time-frame for the secondary endosymbiosis event. Overall, the time-frame suggested by our analyses is consistent with molecular dating of euglenophyte host cells using nuclear data; Parfrey et al.51 recovered plastid-containing euglenophytes diverging from a non-photosynthetic bacteriotrophic euglenophyte ~760 Ma, implying that secondary plastid endosymbiosis in the euglenophyte lineage occurred no earlier than this date. Due to our broad time-frame estimate, it is difficult to speculate on global environmental conditions under which secondary endosymbiosis could have taken place; photosynthetic euglenophytes may have originated within freshwater or marine refugia during one of the Cryogenian ice ages or in cold, oligotrophic waters between ice ages.
Figure 2.
Chronogram of streptophytes, green algae, photosynthetic euglenophytes, the “green” dinoflagellate Lepidodinium chlorophorum, and chlorarachniophytes. Node ages were inferred using Bayesian inference assuming a relaxed molecular clock and a set of node age constraints derived from the fossil record as well as node age estimates from previous studies (see Supporting Information File 1). Values at nodes indicate average node ages and grey bars at selected nodes represent 95% confidence intervals. Coloured vertical bars to the right of the phylogeny are labelled: S, Streptophyta; P, prasinophytes; CC, core Chlorophyta. “S/C split” = divergence of the Streptophyta and Chlorophyta. “CC split” = divergence of the prasinophytes and core Chlorophyta. Timeline abbreviations: Camb., Cambrian; Ord., Ordovician; Sil., Silurian; Dev., Devonian; Carb., Carboniferous; Perm., Permian; Tri., Triassic; Jur., Jurassic.
Dinoflagellates: Lepidodinium
In the case of the ‘serial secondary’ green dinoflagellate plastid, earlier plastid multi-gene analyses42 indicated that the Lepidodinium plastid is most closely related to members of the core chlorophytes, with more recent whole-plDNA phylogenomics indicating a sister relationship between Lepidodinium and the pedinophyte Pedinomonas minor4. Pedinophytes are an early-diverging lineage of the core Chlorophyta (CC in Fig. 1)40. A caveat of the whole-plDNA study4 was that only a single pedinophyte species was included, and so it could not be determined whether the Lepidodinium plastid is descended from a pedinophyte or an unknown alga closely related to pedinophytes.
Using additional pedinophyte sequences, our ML analysis of amino-acid data recovers Lepidodinium branching within a strongly supported monophyletic pedinophyte clade (91% BS, Fig. 1). Although the branch leading to Lepidodinium is very long and potentially leads to issues such as long-branch attraction, this does not appear to be the case here. Our dataset includes four pedinophyte species, representing the broadest taxon sampling to date of this group in phylogenetic analyses of the Lepidodinium plastid. Consequently, with the exception of Lepidodinium, branches within the pedinophyte clade are of average length compared to other green-algal groups in the phylogeny. Further, a strongly supported Lepidodinium + pedinophyte clade is recovered in nucleotide ML phylogenies using the best-fit model (GTR), as well as Bayesian analyses using a site-heterogeneous CATGTR model, which is more robust against LBA artefacts52 (Supplementary Figs S1, S6 and S7). In ML analyses of nucleotide data using the much simpler JC69 and K80 models Lepidodinium branches as sister to chlorarachniophytes (Supplementary Figs S3–S4), but this is likely due to a poor fit of the model to the data, and/or potentially long-branch attraction given the long branch leading to the chlorarachniophyte clade. Overall we are confident that, consistent with previous investigations, the Lepidodinium plastid is indeed most closely related to a pedinophyte lineage. In our analyses Lepidodinium branches together with a clade consisting of two Pedinomonas species, with Marsupiomonas sp. and the newly sequenced strain YPF-701 forming a sister group to the Lepidodinium/Pedinomonas clade. Hence, our investigation confirms that the Lepidodinium plastid did indeed originate from a pedinophyte lineage, rather than a related lineage.
Our molecular clock analysis estimates ~553 Ma for the divergence of the Lepidodinium plastid lineage from its pedinophyte sisters, with a 95% CI of ~416–661 Ma (Fig. 2). However, both nuclear SSU and LSU rRNA phylogenies indicate that the Lepidodinium dinoflagellate host was a member of the dinoflagellate genus Gymnodinium19,53,54. The Gymnodiniaceae is nested phylogenetically within a core dinoflagellate clade that began to radiate, according to fossils and molecular clocks, only about 220 million years ago55–57. Thus, the incorporation of a plastid into the Lepidodinium lineage likely postdates the plastid-lineage divergence time estimated from living taxa by more than 300 million years, possibly occurring as part of the broader radiation of phytoplankton in Mesozoic oceans8. As data for only a single Lepidodinium species is available we cannot date the radiation of the Lepidodinium lineage, and so we lack an estimate for the upper (i.e. most recent) boundary for the timeframe during which endosymbiosis likely occurred. Overall, our results suggest that the Lepidodinium plastid lineage is likely to have had an extensive history as a free-living pedinophyte after diverging from other known lineages. It is possible that free-living members are now extinct; alternatively, further sampling of putative pedinophyte diversity might uncover extant members more closely related to the Lepidodinium plastid.
Chlorarachniophytes
Of the three known lineages with secondary green plastids, the ancestor of the chlorarachniophyte plastid has been the most difficult to pin down. The most taxon-rich analysis to date (55 plDNA genes) included five chlorarachniophyte species and recovered a monophyletic chlorarachniophyte clade branching as sister to the order Bryopsidales (three taxa included) with moderate to strong support5. The Bryopsidales, together with the order Dasycladales, comprise the siphonous green algae, whose members consist of a giant single filamentous or branched multinucleate cell58,59. The recovered tree topology suggested that the ancestor of the chlorarachniophyte plastid might have belonged to Bryopsidales or a closely related group, but could not distinguish between these scenarios due to limited taxon sampling of putative closely related taxa; the Dasycladales were not represented, and the closest relative of the chlorarachniophyte/Bryopsidalean clade was not recovered with statistical support5.
In our study we include data from eleven Bryopsidalean taxa from eight families, including several newly sequenced early-branching lineages, providing dense sampling within groups that are closely related to the chlorarachniophyte plastid and allowing us to test the hypothesis that chlorarachniophyte plastids may derive from a lineage within Bryopsidales. Importantly, we also include new data from Neomeris sp. and published data for Acetabularia acetabulum, members of the order Dasycladales. In some published analyses the Dasycladales group is sister to the Bryopsidales with strong support40,58,60, and the inclusion of this lineage in our analyses potentially allows a more-refined resolution of the branching position of the chlorarachniophytes within the green algae.
Our analyses recover chlorarachniophytes branching as sister to the Bryopsidales in all maximum likelihood phylogenies (Fig. 1, Supplementary Figs S1–S5), with bootstrap support for this clade ranging from strong with more sophisticated models (93%, partitioned nucleotide dataset, GTR model) to non-significant using very simple models of sequence evolution (42%, partitioned nucleotide dataset, HKY model). Analyses with our amino-acid dataset recovered the clade with good support (80%, Fig. 1). The consistent recovery of a chlorarachniophyte + Bryopsidales group using different models and data matrices suggests that the relationship is genuine, despite potential long-branch attraction artefacts caused by the extended branch leading to the chlorarachniophyte clade. In Bayesian analyses using a CATGTR model (Supplementary Figs S6–S7), chlorarachniophytes branch within the core Chlorophyta with strong support.
In contrast to the consistent relationship between the chlorarachniophytes and the Bryopsidales, the branching location of the siphonous Dasycladales varied in different analyses. In the best-fit model analysis of nucleotide data (GTR, partitioned), the Dasycladales branched as a sister group to a chlorarachniophyte + Bryopsidales clade with good support (85% BS, Fig. S1), lending support to the hypothesis that the ancestor of the chlorarachniophyte plastid was a lineage of siphonous green algae, rather than a closely related group. The same relationship was recovered using a partitioned HKY model, albeit with no statistical support (17% BS, Supplementary Fig. S2). Analyses with either a partitioned JC69 or K80 model also recovered a Dasycladales + chlorarachniophyte + Bryopsidales clade (66% and 85% BS, respectively), except that Lepidodinium branched inside the clade as sister to the chlorarachniophytes (Supplementary Figs S3–S4). Analyses of our amino-acid dataset, on the other hand, recovered Dasycladales grouping as sister to an Oltmannsiellopsidales + Ulvales-Ulotrichales clade with strong support (95% BS; Fig. 1). This relationship was also recovered with strong support in Bayesian analyses of both nucleotide and amino-acid data (Bayesian Posterior Probability of 1; Supplementary Figs S6 and S7). Notably, a weak association between Dasycladales and an Oltmannsiellopsidales + Ulvales–Ulotrichales clade has been reported previously61, although other analyses recover a sister relationship between the Dasycladales and Trentepohliales40,62. To test whether the branching positions of the Dasycladales and Bryopsidales were affected by inclusion of chlorarachniophyte sequences, we performed a partitioned ML analysis of both nucleotide and amino-acid data with all secondary plastid sequences removed (Supplementary Figs S9 and S10). In both cases the same respective topologies were recovered for these groups: a Dasycladales + Bryopsidales clade received 97% BS in the nucleotide analysis, and a Dasycladales + Oltmannsiellopsidales + Ulvales-Ulotrichales clade received 98% BS in the amino-acid analysis, suggesting that the secondary plastid sequences are not affecting these phylogenetic relationships. Finally, to examine whether inclusion of some very recently-released ulvophycean plDNAs63 affected the topologies recovered with our datasets, we performed maximum likelihood analyses with more early-branching Ulvophyceae lineages (Dataset B, Supplementary Table S9; see highlighted taxa). The position of chlorachniophyte plastids in these phylogenies was entirely consistent with analyses above (Supplementary Figs S11 and S12). Further, analysis of the amino-acid alignment from Dataset B using a more sophisticated site-heterogeneous LG + G4 + C60 + F + PMSF model36, which estimates site-specific amino-acid profiles based on C60 empirical frequency profiles64, also recovered a chlorarachniophyte + Bryopsidales clade (73% BS), with a Dasycladales + Oltmannsiellopsidales + Ignatiales + Ulvales-Ulotrichales clade receiving 99% BS (Supplementary Fig. S13).
To investigate possible reasons behind these two well-resolved but conflicting topologies we carried out a series of analyses, as described in Supplementary Information File S1. Briefly, we calculated site-wise support for the conflicting topologies from our alignments, and showed that the strong support for a Dasycladales + chlorarachniophyte + Bryopsidales clade in our nucleotide matrix largely originated from third-codon positions. Typically, the rate of nucleotide substitution is higher at third codon positions65,66, leading to substitutional saturation and erosion of accurate phylogenetic signal. Therefore, removal of third codon positions for protein-coding genes is a commonly employed strategy. Given this reasoning and the overall balance of evidence from our analyses (Supplementary Information File S1, Supplementary Fig. S5), we suggest that the Dasycladales + Oltmannsiellopsidales + Ulvales-Ulotrichales clade represents the most likely species relationship hypothesis based on plastid genome data, and that this clade might be sister to the chlorarachniophyte + Bryopsidales group (Fig. 1, Supplementary Fig. S5). This topology is broadly consistent with that recovered by Suzuki et al.5, although in that study the chlorodendrophyceaen Tetraselmis branched inside the Ulvophycean + chlorarachniophyte clade as sister to the Ulvales-Ulotrichales group, and the sister relationship between the Ulvales-Ulotrichales and the Chlorarachniophyte + Bryopsidales groups was not recovered with statistical support.
Overall, our phylogenetic results leave open several possibilities for the origin of chlorarachniophyte plastids. If the relationship recovered in Fig. 1 is correct, they could be derived from a single-celled ulvophycean lineage, and a siphonous physiology could have evolved independently in the lineages leading to the Dasycladales and Bryopsidales following this secondary endosymbiosis event. Alternatively, the Dasycladales + Bryopsidales grouping observed in our nucleotide analyses could represent the correct species relationship. Notably, phylogenies derived from mostly nuclear data60 also recover a strongly supported Dasycladales + Bryopsidales clade. If this relationship is indeed correct, it implies that the ancestor of the chlorarachniophyte plastid was a lineage of siphonous green algae.
Phylogenetic analyses suggest that the host cell that gave rise to the chlorarachniophytes was a Cercozoan amoeba67,68, likely similar to the small marine predator Minorisa minuta69. Cercozoans are an extremely diverse phylum of protists within the supergroup Rhizaria, and include gliding zooflagellates, filose amoebae and plasmodiophorid plant parasites68. It is difficult to envisage how secondary endosymbiosis involving a cercozoan host cell and a green seaweed could take place. Bryopsidales are siphonous macroalgae that are often branched and have multinucleate cells with many plastids. However, two observations suggest a possible mechanism. Firstly, amoeboid life stages of the chlorarachniophyte Cryptochlora perforans have been found to be chemotactically attracted to damaged algal filaments of the Bryopsidalean alga Boodleopsis pusilla, and subsequently perforate and penetrate such filaments and eat part of their contents70. Secondly, a unique and extraordinary wound response has been observed in bryopsidalean seaweeds: in cells of Bryopsis plumosa, protoplasm is extruded from injured cells, cell organelles aggregate, protoplasts are formed with a new cell membrane, and new cell walls are produced71. One can therefore envisage a cercozoan-like host cell feeding on siphonous algal protoplasts, seemingly providing an excellent opportunity for secondary endosymbiosis to take place. Alternatively, endosymbiosis in this lineage could have involved a cercozoan host capturing a microscopic precursor species to siphonous green algae (cf. Cocquyt et al.60). If the chlorachniophyte plastid is indeed derived from a lineage of siphonous greens, it is probable that the endosymbiotic event took place in a near-shore benthic marine environment, as all extant siphonous green algae are marine (with one derived exception), as are the closest known non-photosynthetic extant relatives of chlorarachniophytes69.
Relaxed molecular clock analyses suggest that the green algal lineage leading to the chlorarachniophyte plastid diverged from the Bryopsidales ~578 Ma (95% CI ~546–603), indicating that secondary endosymbiosis occurred after this time (Fig. 2). The date of radiation of extant sampled chlorarachniophytes (which must have occurred after plastid acquisition) was estimated at ~318 Ma (95% CI ~250–388). Thus, our analyses suggest that endosymbiosis occurred at some point between 578–318 Ma. The older boundary of this timeframe is consistent with the late Proterozoic and early Paleozoic radiation of the siphonous green algae with which chlorarachniophyte plastids may be associated, as recorded by the skeletal remains and occasional organic compressions of the former lineages58,72. Moreover, it is consistent with molecular clock analyses estimated from nuclear genes, which recover the chlorarachniophyte Bigelowiella diverging from other cercozoans around 1,000 Ma51. While additional sampling of putative later-branching cercozoans that are closely related to the chlorarachniophyte host lineage could bring this time estimate forward, overall this window (1,000–578 Ma) is consistent with suggestions of a putative cryptic endosymbiosis of a red algal plastid-containing eukaryote in the common ancestor of extant chlorarachniophytes, prior to acquisition of the green-algal plastid73.
Conclusions
Here we present advances in pinpointing the origin of secondary green plastids in the three lineages known to contain them. We improve the density of taxon sampling in green algal lineages closely related to secondary plastids, using new plastid genomes presented in this study along with new publically available data, allowing a more precise understanding of these relationships. We show that the Lepidodinium plastid originated from a pedinophyte rather than a closely related lineage. It will be interesting to see whether future discoveries of pedinophyte diversity can identify extant free-living members more closely related to the Lepidodinium plastid. While the euglenophyte plastid appears likely to have originated from a prasinophyte within the order Pyramimonadales, we cannot rule out an unknown closely related lineage. For chlorarachniophytes, our analyses suggest that the secondary plastid originated from a precursor of siphonous green algae or a closely related unknown lineage.
Nearly all known ulvophyte lineages were sampled here, and the main un-sampled lineage has highly deviant chloroplast genomes74, so further clarification does not seem likely unless as-yet-unknown lineages are discovered. Moreover, overall placement of the chlorarachniophyte plastid within the Ulvophyceae and the core Chlorophyta more broadly is hampered by weak signal in chloroplast genome datasets, which leads to poorly resolved or incongruent relationships between the main clades of core Chlorophyta66. Phylogenetic studies using nuclear endosymbiont-derived genes may further resolve the origin of the chlorarachniophyte plastid, but very little nuclear data is currently available. Finally, we integrate our phylogenies with fossil information to present estimates of timeframes during which secondary plastid endosymbiosis is likely to have occurred for each of the three secondary lineages, using a relaxed molecular clock model to better account for the changes in molecular evolutionary rates during secondary endosymbiosis. On balance, our results suggest that the secondary endosymbiosis event leading to Lepidodinium occurred more recently than endosymbiosis events leading to the euglenophyte and chlorarachniophyte lineages.
Electronic supplementary material
Acknowledgements
This work was supported by the Australian Research Council (FT110100585, DP150100705), the University of Melbourne (ECR and SPRINT grants to HV) and the NASA Astrobiology Institute (AHK). We thank the Victorian Life Sciences Computation Initiative (VLSCI) for providing access to computational resources (project UOM0007). The authors declare no conflict of interest. M. Chiela M. Cremen and Vanessa R. Marcelino are thanked for their contributions towards data generation.
Author Contributions
C.J. generated data. C.J. and H.V. designed and performed the research, carried out data analysis, collection, and interpretation, and wrote the manuscript. A.K. contributed towards data generation, interpreted analyses, and contributed to writing of the manuscript. C.X.C. contributed to writing of the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-18805-w.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Stiller JW, et al. The evolution of photosynthesis in chromist algae through serial endosymbioses. Nat. Commun. 2014;5:5764. doi: 10.1038/ncomms6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ševčíková T, et al. Updating algal evolutionary relationships through plastid genome sequencing: did alveolate plastids emerge through endosymbiosis of an ochrophyte? Sci. Rep. 2015;5:10134. doi: 10.1038/srep10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dorrell RG, et al. Chimeric origins of ochrophytes and haptophytes revealed through an ancient plastid proteome. Elife. 2017;6:1–45. doi: 10.7554/eLife.23717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamikawa R, et al. Plastid genome-based phylogeny pinpointed the origin of the green-colored plastid in the dinoflagellate Lepidodinium chlorophorum. Genome Biol. Evol. 2015;7:1133–1140. doi: 10.1093/gbe/evv060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki S, Hirakawa Y, Kofuji R, Sugita M. Plastid genome sequences of Gymnochlora stellata, Lotharella vacuolata, and Partenskyella glossopodia reveal remarkable structural conservation among chlorarachniophyte species. J. Plant Res. 2016;129:581–590. doi: 10.1007/s10265-016-0804-5. [DOI] [PubMed] [Google Scholar]
- 6.Marin B. Origin and Fate of Chloroplasts in the Euglenoida. 2004;155:13–14. doi: 10.1078/1434461000159. [DOI] [PubMed] [Google Scholar]
- 7.Bennett MS, Wiegert KE, Triemer RE. Characterization of Euglenaformis gen. nov. and the chloroplast genome of Euglenaformis [Euglena] proxima (Euglenophyta) Phycologia. 2014;53:66–73. doi: 10.2216/13-198.1. [DOI] [Google Scholar]
- 8.Falkowski PG, et al. The Evolution of Modern Eukaryotic Phytoplankton. Science. 2004;305:354–360. doi: 10.1126/science.1095964. [DOI] [PubMed] [Google Scholar]
- 9.de Vargas, C. et al. Eukaryotic plankton diversity in the sunlit ocean. Science348 (2015). [DOI] [PubMed]
- 10.Keeling PJ. The endosymbiotic origin, diversification and fate of plastids. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2010;365:729–48. doi: 10.1098/rstb.2009.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marin B, Palm A, Klingberg M, Melkonian M. Phylogeny and taxonomic revision of plastid-containing euglenophytes based on SSU rDNA sequence comparisons and synapomorphic signatures in the SSU rRNA secondary structure. Protist. 2003;154:99–145. doi: 10.1078/143446103764928521. [DOI] [PubMed] [Google Scholar]
- 12.Adl SM, et al. The revised classification of eukaryotes. J. Eukaryot. Microbiol. 2012;59:429–493. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki S, Shirato S, Hirakawa Y, Ishida KI. Nucleomorph genome sequences of two chlorarachniophytes, Amorphochlora amoebiformis and Lotharella vacuolata. Genome Biol. Evol. 2015;7:1533–1545. doi: 10.1093/gbe/evv096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishida, K., Yabuki, A. & Ota, S. The chlorarachniophytes: evolution and classification. Systematics 171–182 (2007).
- 15.Hackett JD, Anderson DM, Erdner DL, Bhattacharya D. Dinoflagellates: A remarkable evolutionary experiment. American Journal of Botany. 2004;91:1523–1534. doi: 10.3732/ajb.91.10.1523. [DOI] [PubMed] [Google Scholar]
- 16.Dorrell, R. & Howe, C. Integration of plastids with their hosts: Lessons learned from dinoflagellates. Proc. Natl. Acad. Sci.USA 112, 10247–10254 (2015). [DOI] [PMC free article] [PubMed]
- 17.Nishitani G, et al. Multiple plastids collected by the dinoflagellate Dinophysis mitra through kleptoplastidy. Appl. Environ. Microbiol. 2012;78:813–821. doi: 10.1128/AEM.06544-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wisecaver JH, Hackett JD. Transcriptome analysis reveals nuclear-encoded proteins for the maintenance of temporary plastids in the dinoflagellate Dinophysis acuminata. BMC Genomics. 2010;11:366. doi: 10.1186/1471-2164-11-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen G, Botes L, De Salas M. Ultrastructure and large subunit rDNA sequences of Lepidodinium viride reveal a close relationship to Lepidodinium chlorophorum comb. nov. (=Gymnodinium chlorophorum) Phycol. Res. 2007;55:25–41. doi: 10.1111/j.1440-1835.2006.00442.x. [DOI] [Google Scholar]
- 20.Watanabe MM, Suda S, Inouye I, Sawaguchi T, Chihara M. Lepidodinium viride gen. et sp. nov. (Gymnodiales, Dinophyta), a green dinoflagellate with a chlorophyll a- and b-containing endosymbiont. J. Phycol. 1990;26:741–751. doi: 10.1111/j.0022-3646.1990.00741.x. [DOI] [Google Scholar]
- 21.Archibald JM. Endosymbiosis and eukaryotic cell evolution. Curr. Biol. 2015;25:R911–R921. doi: 10.1016/j.cub.2015.07.055. [DOI] [PubMed] [Google Scholar]
- 22.Turmel M, Gagnon MC, O’Kelly CJ, Otis C, Lemieux C. The chloroplast genomes of the green algae Pyramimonas, Monomastix, and Pycnococcus shed new light on the evolutionary history of prasinophytes and the origin of the secondary chloroplasts of euglenids. Mol. Biol. Evol. 2009;26:631–648. doi: 10.1093/molbev/msn285. [DOI] [PubMed] [Google Scholar]
- 23.Rogers MB, Gilson PR, Su V, McFadden GI, Keeling PJ. The complete chloroplast genome of the chlorarachniophyte Bigelowiella natans: Evidence for independent origins of chlorarachniophyte and euglenid secondary endosymbionts REF 22. Mol. Biol. Evol. 2007;24:54–62. doi: 10.1093/molbev/msl129. [DOI] [PubMed] [Google Scholar]
- 24.Verbruggen, H., Marcelino, V. R., Guiry, M. D., Cremen, M. C. M. & Jackson, C. J. Phylogenetic position of the coral symbiont Ostreobium (ulvophyceae) inferred from chloroplast genome data, 10.1111/jpy.12540 (2017). [DOI] [PubMed]
- 25.Cremen MCM, Huisman JM, Marcelino VR, Verbruggen H. Taxonomic revision of Halimeda (Bryopsidales, Chlorophyta) in south-Western Australia. Aust. Syst. Bot. 2016;29:41–54. doi: 10.1071/SB15043. [DOI] [Google Scholar]
- 26.Bankevich A, et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verbruggen H, Costa JF. The plastid genome of the red alga. Laurencia. J. Phycol. 2015;51:586–589. doi: 10.1111/jpy.12297. [DOI] [PubMed] [Google Scholar]
- 28.Marcelino, V. R., Cremen, M. C. M., Jackson, C. J. & Larkum, A. W. D. Evolutionary dynamics of chloroplast genomes in low light: a case study of the endolithic green alga Ostreobium quekettii. 1–28, 10.1093/gbe/evw206 (2016). [DOI] [PMC free article] [PubMed]
- 29.Katoh K, Toh H. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics. 2010;26:1899–1900. doi: 10.1093/bioinformatics/btq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abascal F, Zardoya R, Telford MJ. TranslatorX: Multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010;38:7–13. doi: 10.1093/nar/gkq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castresana J. Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 32.Lanfear R, Calcott B, Ho SYW, Guindon S. Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 2012;29:1695–701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- 33.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pond SLK, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–9. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H-C, Minh BQ, Susko E, Roger AJ. Modeling Site Heterogeneity with Posterior Mean Site Frequency Profiles Accelerates Accurate Phylogenomic Estimation. Syst. Biol. 2017;0:1–19. doi: 10.1093/sysbio/syx068. [DOI] [PubMed] [Google Scholar]
- 37.Lartillot N, Lepage T, Blanquart S. PhyloBayes 3: A Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25:2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- 38.Tanifuji G, et al. Nucleomorph and plastid genome sequences of the chlorarachniophyte Lotharella oceanica: convergent reductive evolution and frequent recombination in nucleomorph-bearing algae. BMC Genomics. 2014;15:374. doi: 10.1186/1471-2164-15-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bergsten, J. A review of long-branch attraction. Cladistics 21, 163–193 (2005). [DOI] [PubMed]
- 40.Fucikova K, et al. New phylogenetic hypotheses for the core Chlorophyta based on chloroplast sequence data. Front. Ecol. Evol. 2014;2:1–12. [Google Scholar]
- 41.Hrdá, Š., Fousek, J., Szabová, J., Hampl, V. & Vlček, Č. The plastid genome of Eutreptiella provides a window into the process of secondary endosymbiosis of plastid in euglenids. PLoS One7 (2012). [DOI] [PMC free article] [PubMed]
- 42.Matsumoto T, et al. Green-colored Plastids in the Dinoflagellate Genus Lepidodinium are of Core Chlorophyte Origin. Protist. 2011;162:268–276. doi: 10.1016/j.protis.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 43.Wiegert KE, Bennett MS, Triemer RE. Evolution of the Chloroplast Genome in Photosynthetic Euglenoids: A Comparison of Eutreptia viridis and Euglena gracilis (Euglenophyta) Protist. 2012;163:832–843. doi: 10.1016/j.protis.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 44.Yamaguchi A, Yubuki N, Leander BS. Morphostasis in a novel eukaryote illuminates the evolutionary transition from phagotrophy to phototrophy: description of Rapaza viridis n. gen. et sp. (Euglenozoa, Euglenida) BMC Evol. Biol. 2012;12:29. doi: 10.1186/1471-2148-12-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leander BS, Triemer RE, Farmer MA. Character evolution in heterotrophic euglenids. Eurpean J. Protistol. 2001;37:337–356. [Google Scholar]
- 46.Moczydłowska M. The early Cambrian phytoplankton radiation: acritarch evidence from the Lükati Formation, Estonia. Palynology. 2011;35:103–145. doi: 10.1080/01916122.2011.552563. [DOI] [Google Scholar]
- 47.Brocks, J. J. et al. The rise of algae in Cryogenian oceans and the emergence of animals. Nature548, 578–581 (2017). [DOI] [PubMed]
- 48.Sanchez-Baracaldo, P., Raven, J., Pisani, D. & Knoll, A. H. Early photosynthetic eukaryotes inhabited terrestrial and low salinity habitats. Proc. Natl. Acad. Sci. USA114, E7737–7745 (2017). [DOI] [PMC free article] [PubMed]
- 49.Gray J, Boucot AJ. Is Moyeria a euglenoid? Lethaia. 1989;22:447–456. doi: 10.1111/j.1502-3931.1989.tb01449.x. [DOI] [Google Scholar]
- 50.Leander BS, Witek RP, Farmer MA. Trends in the Evolution of the Euglenid Pellicle. Evolution (N. Y). 2001;55:2215–2235. doi: 10.1111/j.0014-3820.2001.tb00737.x. [DOI] [PubMed] [Google Scholar]
- 51.Parfrey LW, Lahr DJG, Knoll AH, Katz LA. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc. Natl. Acad. Sci. USA. 2011;108:13624–9. doi: 10.1073/pnas.1110633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lartillot N, Brinkmann H, Philippe H. Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol. Biol. 2007;7:S4. doi: 10.1186/1471-2148-7-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takishita K, et al. Origins of plastids and glyceraldehyde-3-phosphate dehydrogenase genes in the green-colored dinoflagellate Lepidodinium chlorophorum. Gene. 2008;410:26–36. doi: 10.1016/j.gene.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 54.Murray S, Flø Jørgensen M, Ho SYW, Patterson DJ, Jermiin LS. Improving the analysis of dinoflagellate phylogeny based on rDNA. Protist. 2005;156:269–286. doi: 10.1016/j.protis.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 55.Janouškovec, J. et al. Major transitions in dinoflagellate evolution unveiled by phylotranscriptomics. Proc. Natl. Acad. Sci. USA114, E171–E180 (2017). [DOI] [PMC free article] [PubMed]
- 56.Dorrell RG, et al. Progressive and biased divergent evolution underpins the origin and diversification of peridinin dinoflagellate plastids. Mol. Biol. Evol. 2016;34:1–47. doi: 10.1093/molbev/msw235. [DOI] [PubMed] [Google Scholar]
- 57.Fensome RA, MacRae RA, Moldowan JM, Taylor MFJ, Williams GL. The early Mesozoic radiation of dinoflagellates. 1996;22:329–338. [Google Scholar]
- 58.Verbruggen H, et al. A multi-locus time-calibrated phylogeny of the siphonous green algae. Mol. Phylogenet. Evol. 2009;50:642–653. doi: 10.1016/j.ympev.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 59.Sun L, et al. Chloroplast Phylogenomic Inference of Green Algae Relationships. Sci. Rep. 2016;6:20528. doi: 10.1038/srep20528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cocquyt E, Verbruggen H, Leliaert F, De Clerck O. Evolution and cytological diversification of the green seaweeds (Ulvophyceae) Mol. Biol. Evol. 2010;27:2052–2061. doi: 10.1093/molbev/msq091. [DOI] [PubMed] [Google Scholar]
- 61.Leliaert F, Lopez-Bautista JM. The chloroplast genomes of Bryopsis plumosa and Tydemania expeditiones (Bryopsidales, Chlorophyta): compact genomes and genes of bacterial origin. BMC Genomics. 2015;16:204. doi: 10.1186/s12864-015-1418-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Melton JT, Leliaert F, Tronholm A, Lopez-Bautista JM. The complete chloroplast and mitochondrial genomes of the green macroalga Ulva sp. UNA00071828 (Ulvophyceae, Chlorophyta) PLoS One. 2015;10:1–21. doi: 10.1371/journal.pone.0121020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Turmel M, Otis C, Lemieux C. Divergent copies of the large inverted repeat in the chloroplast genomes of ulvophycean green algae. Sci. Rep. 2017;7:994. doi: 10.1038/s41598-017-01144-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Quang LS, Gascuel O, Lartillot N. Empirical profile mixture models for phylogenetic reconstruction. Bioinformatics. 2008;24:2317–2323. doi: 10.1093/bioinformatics/btn445. [DOI] [PubMed] [Google Scholar]
- 65.Bofkin L, Goldman N. Variation in evolutionary processes at different codon positions. Mol. Biol. Evol. 2007;24:513–521. doi: 10.1093/molbev/msl178. [DOI] [PubMed] [Google Scholar]
- 66.Zhong. Evolution of the Chlorophyta: Insights from chloroplast phylogenomic analyses. 3, 10.1002/mrd.22357 (2017).
- 67.Keeling PJ. Foraminifera and Cercozoa are related in actin phylogeny: two orphans find a home? Mol. Biol. Evol. 2001;18:1551–1557. doi: 10.1093/oxfordjournals.molbev.a003941. [DOI] [PubMed] [Google Scholar]
- 68.Howe AT, et al. Novel Cultured Protists Identify Deep-branching Environmental DNA Clades of Cercozoa: New Genera Tremula, Micrometopion, Minimassisteria, Nudifila, Peregrinia. Protist. 2011;162:332–372. doi: 10.1016/j.protis.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 69.del Campo J, Not F, Forn I, Sieracki ME, Massana R. Taming the smallest predators of the oceans. ISME J. 2012;7:351–358. doi: 10.1038/ismej.2012.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Calderon-Saenz E, Schnetter R. Morphology, biology, and systematics of Cryptochlora perforans (Chlorarachniophyta), a phagotrophic marine alga. Plant Syst. Evol. 1989;163:165–176. doi: 10.1007/BF00936512. [DOI] [Google Scholar]
- 71.Kim GH, Klotchkova TA, Kang YM. Life without a cell membrane: regeneration of protoplasts from disintegrated cells of the marine green alga Bryopsis plumosa. J. Cell Sci. 2001;114:2009–2014. doi: 10.1242/jcs.114.11.2009. [DOI] [PubMed] [Google Scholar]
- 72.LoDuca ST, Behringer ER. Functional morphology and evolution of early Paleozoic dasycladalean algae (Chlorophyta) Paleobiology. 2009;35:63–76. doi: 10.1666/07075.1. [DOI] [Google Scholar]
- 73.Yang, Y., Matsuzaki, M., Takahashi, F., Qu, L. & Nozaki, H. Phylogenomic analysis of ‘red’ genes from two divergent species of the ‘green’ secondary phototrophs, the chlorarachniophytes, suggests multiple horizontal gene transfers from the red lineage before the divergence of extant chlorarachniophytes. PLoS One9 (2014). [DOI] [PMC free article] [PubMed]
- 74.Del Cortona, A. et al. The plastid genome in cladophorales green algae is encoded by hairpin plasmids. Curr. Biol.27, 3771–3782 (2017). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Plastid sequence data is deposited in Genbank under accession KY347917 (pedinophyte strain YPF-701), KY509314 (Ostreobium sp. HV05042), KY495826-KY495874 (Neomeris sp.), and KY509313 (Avrainvillea mazei). All other data generated or analysed during this study are either publically available or included in this published article (and its Supplementary Information files).