Abstract
Parallel adaptive radiations have arisen following the colonization of islands by lizards and lakes by fishes. In these classic examples, parallel adaptive radiation is a response to the ecological opportunities afforded by the colonization of novel ecosystems and similar adaptive landscapes that favour the evolution of similar suites of ecomorphs, despite independent evolutionary histories. Here, we demonstrate that parallel adaptive radiations of cichlid fishes arose in South American rivers. Speciation-assembled communities of pike cichlids (Crenicichla) have independently diversified into similar suites of novel ecomorphs in the Uruguay and Paraná Rivers, including crevice feeders, periphyton grazers and molluscivores. There were bursts in phenotypic evolution associated with the colonization of each river and the subsequent expansion of morphospace following the evolution of the ecomorphs. These riverine clades demonstrate that characteristics emblematic of textbook parallel adaptive radiations of island- and lake-dwelling assemblages are feasible evolutionary outcomes even in labile ecosystems such as rivers.
Keywords: cichlid, convergence, Crenicichla, diversification, parallel evolution, species flock
1. Introduction
Adaptive radiation is an evolutionary response to ecological opportunity such that the rise of adaptations coincides with expansion of ecological diversity within a lineage [1–3]. These ecological opportunities may include access to novel resources, relaxed selection on ecologically important traits allowing for the exploration of novel phenotypes, or relaxed competition that allows for exploration of novel regions of the adaptive landscape [2]. Islands and lakes have served as the foundation for parallel adaptive radiations after their colonization by Anolis lizards and cichlid fishes, respectively [4,5]. Series of islands and lakes probably provide access to common resources, similar adaptive landscapes and thereby elicit the evolution of similar suites of ecomorphs despite independent evolutionary histories. The discrete nature of islands and lakes also constrains immigration and emigration such that in situ speciation results in monophyletic groups that are endemic to that ecosystem (i.e. species flocks).
Rapid speciation, phenotypic diversification and convergence are hallmarks of adaptive radiation [3,5]. Islands and lakes have provided unique examples of parallel adaptive radiation in which diversification on different islands/lakes has independently produced similar outcomes. For example, Anolis lizards have diversified into similar suites of habitat specialists on different islands of the Greater Antilles [4]. These island-specific clades exhibit phenotypic convergence upon inferred adaptive peaks and the onset of diversity dependence as ecological opportunities wane as niches fill towards capacity [6–8]. Likewise, lakes have provided similar opportunities for cichlid fishes, which have diversified in parallel in lakes throughout Africa and Middle America [5,9]. These lake-specific clades exhibit phenotypic convergence, diversification along common environmental gradients such as the benthic-to-pelagic habitat and the hard-shelled to soft-bodied prey axes, as well as diversity-dependent evolution [10–13]. In addition to these prominent examples, parallel adaptive radiations have also occurred in postglacial whitefish and stickleback [14,15], Hawaiian spiders and angiosperms [16–18], and island land snails [19]. Such parallel adaptive radiations provide a unique framework to study evolutionary determinism as well as idiosyncrasy while adaptive radiation unfolds independently among similar ecosystems.
Although a few riverine cichlid species flocks have been described [20,21], these flocks lack the dramatic accumulation of ecological roles and their associated adaptations that characterize their lake-dwelling counterparts and have not evolved in parallel (reviewed in [5,22]). These conflicting patterns of diversification may be because the adaptive landscape is very different in lakes and rivers. This disparity may be explained by a combination of factors (summarized from Seehausen [5]): (i) river communities are immigration-assembled such that co-occurring ecomorphs are often from disparate lineages that were united by unstable and shifting basin configurations (as opposed to speciation-assembled communities that arise in stable lakes); (ii) many ecomorphs observed in lakes may be implausible evolutionary results in rivers owing to niches that are uncommon or temporally unstable in rivers; and (iii) niches are partitioned to minute ecological scales in lakes where stable conditions favour the evolution of accommodating processes such as resource partitioning.
Despite the aforementioned paucity of in situ diversification and lack of cases of parallel evolution within rivers [5,22], one promising example has recently been proposed in the Paraná and Uruguay rivers of South America where the resident pike cichlid (Crenicichla) assemblages exhibit similar ecomorphs [23]. These communities are endemic and consist of species that co-occur throughout their drainage and exhibit dramatic ecomorphological divergence [23–26]. More recently, this putative example of parallel evolution was expanded following the discovery of additional species in the Paraná River that also have ecomorphological counterparts in the Uruguay River [27]. Furthermore, these two communities may each be monophyletic and distantly related to one another [23,24].
In this study, we test the hypothesis that the similar ecomorphs observed in the Paraná and Uruguay rivers arose via parallel phenotypic and trophic diversification. To test this hypothesis, we employed a series of statistical tests. Firstly, we establish the evolutionary independence of the two assemblages using a novel whole-genome single nucleotide polymorphism (SNP) (ddRADseq) phylogeny. Secondly, we evaluate if the similarities of these ecomorphs is because of selection towards similar adaptive peaks rather than similarity that may have arisen via a neutral model of evolution. Thirdly, we evaluate if these ecomorphs arose from similar ancestral states using ancestral state reconstruction of phenotypes and trophic guilds. Lastly, we test for bursts in the rates of diversification and morphological evolution throughout the evolutionary history of pike cichlids. We then discuss these riverine clades in the context of other prevalent examples of parallel adaptive radiations such as those of lizards on islands and fishes in lakes [5,8].
2. Material and methods
(a). Phylogeny construction
We sampled 64 Crenicichla (includung Teleocichla) species (approximately 63% of the valid species) that represent all major lineages (i.e. there is replication within all the recognized species groups [23]) and five additional South American cichlid taxa that comprised the outgroup (i.e. Retroculus sp., Satanoperca daemon, Apistogramma sp., Biotodoma wavrini and Geophagus sp.). We sampled all eight members of the Uruguay River species flock and all nine members of the Paraná River species flock. All tissues were stored in 95% ethanol. The ddRADseq library preparation and bioinformatic processing of the obtained tags were performed as described in Říčan et al. [28]. Homologous loci were aligned based on a reference mapping of reads onto the genome of Oreochromis niloticus GCA_000188235.1 (http://www.ensembl.org), and SNPs were called in Stacks v. 1.35 [29]. Only fixed (homozygotic) SNPs were extracted from loci with a minimum depth of five and present in a minimum of 70% of the samples. The resulting matrix included 25 128 variable sites. The tree was inferred from a concatenated SNP matrix in RAxML v. 8.2.4 [30] under a GTR + Γ model, which was estimated with jModelTest [31]. To account for potential ascertainment bias, we used the Stamatakis correction [32], which corrected for 326 937 unrepresented constant sites. We used 100 bootstrap replicates to evaluate branch support in RAxML. To further test the robustness of the phylogeny, particularly the monophyly of the putative species flocks which contain the species that may constitute parallel adaptive radiations, we constructed coalescent-based species trees with different methods and data matrices. The general tree topology and monophyly of both species flocks was supported by the RADseq markers (see the electronic supplementary material, figure S1) as well as ultraconserved elements [33]. The sequencing reads and datasets are deposited on the NCBI Sequence Repository Archive (SRA; BioProject ID: PRJNA420902) and Dryad Digital Repository (doi:10.5061/dryad.678rp), respectively.
To estimate divergence times and establish relative node ages within Crenicichla for diversification analyses, we used congruification [34] to impose secondary calibrations on nodes that were congruent with a time-calibrated phylogeny including cichlids by Friedman et al. [35]. Friedman et al. [35] used 14 fossil calibrations for the ages of 13 outgroup nodes distributed across Percomorpha and the root node using a relaxed-clock analysis in BEAST to infer their time-calibrated phylogeny, which included 156 percomoph species, including 91 cichlids. Specifically, three nodes were time-calibrated based on the maximum clade credibility age estimate from Friedman et al. [35] phylogeny: the most recent common ancestor (MRCA) of Crenicichla and Retroculus (29.2 Ma; 95% confidence interval (CI) 35.1–25.6 Ma), the MRCA of Crenicichla and Satanoperca (18.0 Ma; 95% CI 22.3–15.0 Ma), and the MRCA of Crenicichla and Geophagus (20.8 Ma; 95% CI 25.2–17.5 Ma). Divergence time estimation was then performed in treePL [36] using the input file generated by congruification. Following this procedure, the maximum likelihood phylogeny was pruned to include only the 57 species for which we have trait data (electronic supplementary material, figure S2).
(b). Morphological analyses
We quantified body and lower pharyngeal jaw (LPJ) shape of 220 formalin-fixed individuals representing 57 Crenicichla species (approx. 56% of the valid species) using landmark-based geometric morphometrics (electronic supplementary material, table S1). We sampled six members of the Uruguay River species flock and nine members of the Paraná River species flock. We used 15 homologous and eight sliding landmarks to describe the shape of the body in lateral view (electronic supplementary material, figure S3). We used three homologous and nine sliding landmarks to describe the shape of the LPJ in dorsal view (electronic supplementary material, figure S3). Sliding landmarks were evenly spaced between homologous landmarks. Only one side of each LPJ was landmarked owing to structure symmetry. All analyses were performed using the tps program suite. Images were consolidated and landmarked using tpsUtil [37] and tpsDIG2 [38], respectively. Procrustes fit and principal component (PC) scores were generated using tpsRelw [39]. Variation in scale, rotation and translation were removed from the analyses during the Procrustes fit. Additionally, we measured two variables with intuitive associations with the benthic-to-pelagic habitat axis and the soft-bodied to hard-shelled prey axis: mouth angle and LPJ mass, respectively. Mouth angle, based on the upper jaw (i.e. premaxilla), was measured directly from photographs in tpsDIG2 [38] using the measure function. Measurements were adjusted such that a perfectly horizontal (i.e. forming a parallel plane with the body) mouth corresponded with 90° (i.e. benthic-oriented mouths were less than 90° and superior-oriented mouths were greater than 90°). LPJ mass was measured on a digital scale to the nearest 0.00001 g. Magnitude was accounted for by calculating residuals from the regression with standard length. We calculated species means for PCs, angles and residuals for all subsequent analyses.
(c). Testing for convergence
To assess some key features of the adaptive landscape and quantify convergence, we used SURFACE analysis [40], which used stepwise Akaike information criterion to locate the number of regime shifts (k) on the phylogeny and then identify whether these shifts are towards convergent regimes (i.e. trait optima). This process involved iteratively adding regime shifts using a Hansen model, then iteratively removing shifts to identify convergent regimes (k′). The reduction in complexity (k − k′) corresponds to the number of regimes that are evolving towards common sets of traits (i.e. optima) and thus can be collapsed into a common regime (i.e. convergence; Δk). These evolutionary regimes were projected onto the phylogeny and morphospace to visualize the adaptive landscape. Convergence can result from Brownian motion (BM)-like processes [40,41]; therefore, we compared parameters estimated from observed trait data to those generated from data simulated from BM to assess if the observed degree of convergence is more than expected by chance. We generated 100 simulated datasets using the surfaceSimulate function [40].
We then estimated the evolutionary history of trophic guild diversification using maximum-likelihood (mk1 model) ancestral state reconstruction in Mesquite v. 3.02 [42] and stochastic character mapping with SIMMAP [43] implemented in Phytools [44]. The mk1 model assumes the probability of all trait changes are equal. Maximum-likelihood analyses find the ancestral states (i.e. internal nodes) that maximize the probability that the observed states (i.e. terminal nodes) would evolve under a stochastic model [45,46]. Stochastic character mapping [47,48] simulates precise histories of character evolution such that they depict the character states at nodes and along branches between nodes. Character changes along branches are predicted by the rates of character change [42]. We summarized the state frequencies at internal nodes from 1000 SIMMAP stochastic character histories. Species were pooled into general trophic guilds based on existing descriptions of Crenicichla trophic ecology: piscivore, invertivore, molluscivore, crevice-feeder and periphyton grazer (electronic supplementary material, table S2). The piscivore category includes only species that feed almost exclusively upon fishes. By contrast, the invertivore category includes species that feed primarily upon invertebrates but may also consume secondary fractions of fishes (i.e. generalist predators). To simultaneously evaluate the direction and magnitude of shape change and trophic guild evolution along branches of the phylogeny, we overlaid the phylogeny and trophic guild ancestral state reconstruction onto the PC scores (i.e. phylomorphospace; [49]) in Mesquite v. 3.02 [42]. In this procedure, internal (i.e. ancestral) node values are estimated using weighted squared-change parsimony [50,51].
(d). Testing for shifts in diversification rates
We estimated the distribution of discrete rate shifts in diversification and morphological evolution (mouth angle and size-relative LPJ mass) across the phylogeny and through time using BAMM 2.5.0 [52,53]. Priors for all BAMM runs were set using the setBAMMpriors command in BAMMtools [54], with the number of expected shifts set to 1.0. BAMM includes an implementation of an algorithm to account for non-random incomplete taxon sampling in the estimation of diversification rates by allowing for specification of clade-specific sampling fractions [53]. We specified sampling fractions for each species-group as the proportion of species in our phylogeny out of the total valid species in each species-group. Diversification (speciation–extinction) analyses were run for 25M generations, Markov chain Monte Carlo (MCMC) output was written every 20 000 generations and event data were written every 10 000 generations. Trait diversification analyses were run for 5 million generations, MCMC output was written every 2000 generations and event data were written every 5000 generations. BAMM output was further analysed in BAMMtools [54] to calculate and plot the 95% credible shift set. As recommended in the BAMM documentation, coda in R [55] was used to assess whether the MCMC chain included an adequate number of samples of the posterior distribution by determining if the effective sample size > 200 for the number of shifts and log likelihood. We assessed the number of rate shifts in the posterior distribution (with 10% burn-in excluded) relative to the prior distribution, which indicated that the posterior distribution was not sensitive to the prior as can occur in some situations [56] such as when there is a low probability of rate shifts [57,58].
To independently assess rates of morphological evolution through time, we used node-height tests. We calculated the absolute value of phylogenetically independent contrasts and regressed those against the height (i.e. time since root) of the node at which they were calculated. Contrasts are Brownian rate parameters [59]; therefore, a significant positive relationship between absolute rate contrasts and node height would indicate that rates of morphological evolution have increased through time [60].
3. Results
(a). Phenotypic convergence
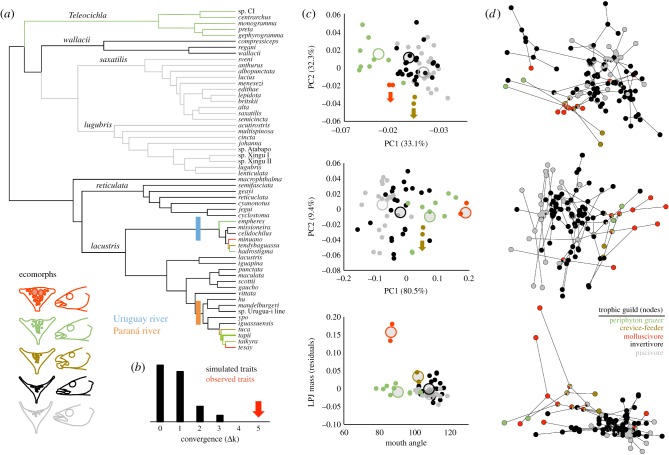
The Uruguay and Paraná River species flocks exhibited considerable overlap in body shape (figure 1a), LPJ shape (figure 1b), LPJ mass and mouth angle (figure 1c). Both species flocks also occupy a large volume of morphospace (figure 1). SURFACE identified non-convergent and convergent evolutionary regimes (figure 2a). Three convergent regimes were largely restricted to the Paraná and Uruguay River species flocks (figure 2a). The degree of phenotypic convergence (Δk = 5) was not sampled using trait data simulated under BM (Δk = 0–3; figure 2b), indicating highly non-random evolutionary processes generating similar ecomorphs in these species flocks.
Figure 1.
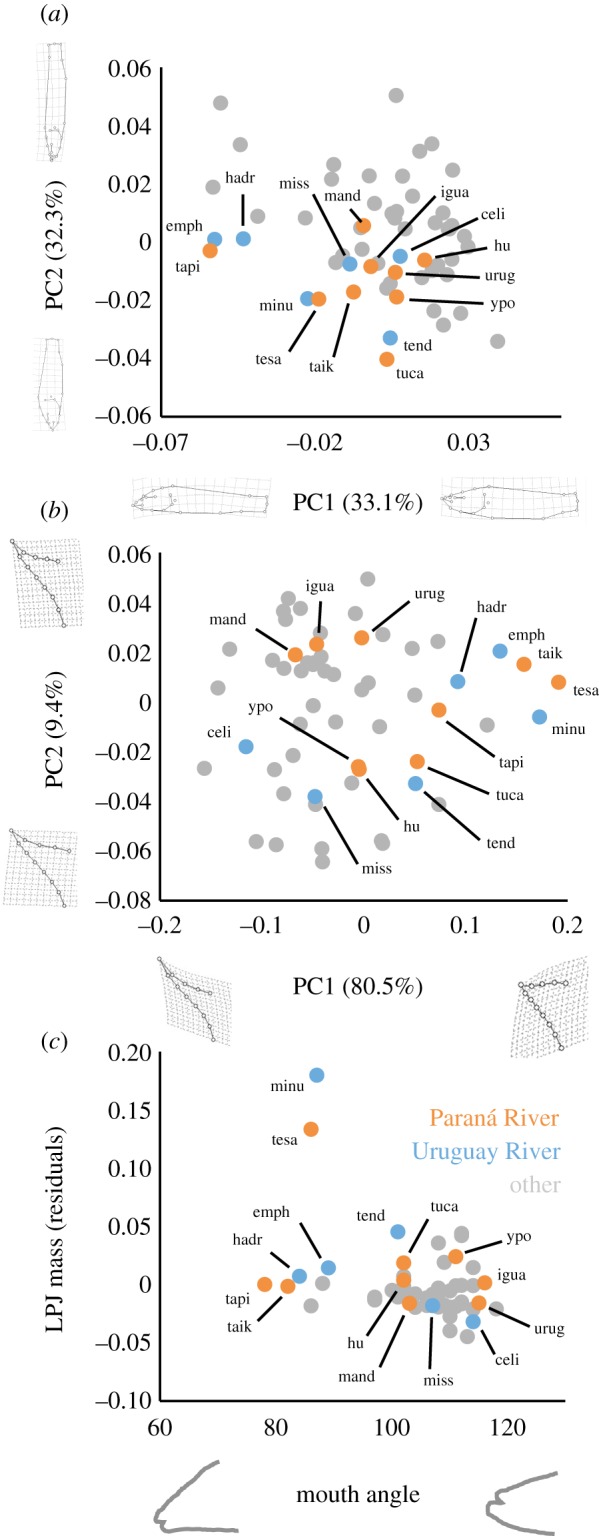
Major axes of body (a) and lower pharyngeal jaw (LPJ; b) shape variation, and (c) mouth angle and size-relative LPJ mass among 57 Crenicichla species. Wire frames depict shapes associated with the extremes of each axis. The Paraná and Uruguay River species flocks are highlighted in colour and designated with codes that correspond to the first letters of the species names shown in figure 2a.
Figure 2.
Morphological and trophic convergence between the Paraná (orange) and Uruguay River (blue) species flocks. (a) Non-convergent (greyscale branches) and convergent (like-coloured branches) evolutionary regimes across 57 Crenicichla species from SURFACE analysis. (b) The observed degree of convergence relative to an expected distribution under a Brownian motion model of evolution. (c) The distribution of inferred adaptive peaks (large circles) and species (small circles) across morphospace. (d) Phylomorphospace depicting the direction and magnitude of shape change (branches) and corresponding trophic guild evolution (nodes). The trophic guild colour codes apply only to (d). Arrows point towards optima that fell outside of the observed trait values.
These convergent regimes coincided, in part, with the independent evolution of periphyton grazing, crevice feeding and molluscivory within the Uruguay and Paraná radiations (electronic supplementary material, figure S4). The first convergent regime (green in figure 2) included C. tapii and C. taikyra (Paraná) as well as C. hadrostigma and C. empheres (Uruguay), which was characterized by an optimum defined by curved snouts, small mouths, robust LPJ and mostly benthic-oriented mouths (figure 2c). The second regime (brown in figure 2) included C. tuca (Paraná) and C. tendybaguassu (Uruguay) and was characterized by tapered bodies, terminal mouths and hypertrophied lips (figure 2c). The third regime (red in figure 2) included only C. tesay (Paraná) and C. minuano (Uruguay) and was characterized by an optimum defined by intermediate mouth size, hypertrophied LPJ and terminal mouths (figure 2c). The red and brown regimes) included optima that fell outside the observed trait space for body and/or LPJ shape. Outlier optima may indeed represent evolution towards distant optima; however, it may also represent rapid adaptation to a new optimum that results in the optimum being interpreted by the model as distant [61]. Given the rapid ecological diversification within these clades, it is likely that it represents rapid evolution to a new optimum. Several species within both species flocks are united within an ancestral non-convergent regime (black in figure 2). Outgroups generally fell into two non-convergent regimes. The C. lugubris and C. saxatilis were united with a non-convergent regime (grey in figure 2), as well as C. wallacii, C. reticulata and the (non-Paraná and Uruguay) C. lacustris group species were united within a non-convergent regime (black in figure 2). Lastly, the members of the Teleocichla group shared a regime with some members of the species flocks (green in figure 2).
(b). Trophic convergence
The ancestral trophic state of both species flocks was well resolved as invertivore based on our maximum-likelihood ancestral state reconstruction and stochastic character mapping (electronic supplementary material, figure S4). Within the Paraná River, this ancestral trophic state is retained in C. mandelburgeri, C. hu and C. ypo, whereas all other species exhibit a novel trophic state (figure 2d; electronic supplementary material, figure S4). Piscivory evolved independently in Crenicichla sp. Urugua-í line and C. iguassuensis; molluscivory evolved once, in C. tesay and C. taikyra; and periphyton grazing and crevice feeding evolved in C. tapii and C. tuca, respectively (figure 2d; electronic supplementary material, figure S4). Within the Uruguay River, the ancestral trophic state is retained only in C. empheres, whereas piscivory evolved once in C. celidochilus and C. missioneira, crevice feeding evolved in C. tendybaguassu, molluscivory evolved in C. minuano and periphyton grazing evolved in C. hadrostigma (figure 2d; electronic supplementary material, figure S4). Piscivory evolved in the Uruguay (C. celidochilus and C. missioneira) and Paraná (C. sp. Urugua-í line and C. iguassuensis) Rivers; however, piscivorous ecomorphs have not evolved in parallel because these species retain the ancestral body and LPJ morphologies indicating that ancestral morphologies may have been co-opted, in parallel, for novel trophic functions (figure 2a,d; electronic supplementary material, figure S4). A host of derived trophic states evolved independently in both rivers from similar ancestral states and in association with the evolution of novel trait combinations (figure 2d). Namely, these cases involved the evolution of molluscivory, crevice feeding, and periphyton grazing and the corresponding expansion of morphospace associated with novel trophic role-specific craniofacial and LPJ morphologies (figure 2d).
(c). Diversification rates
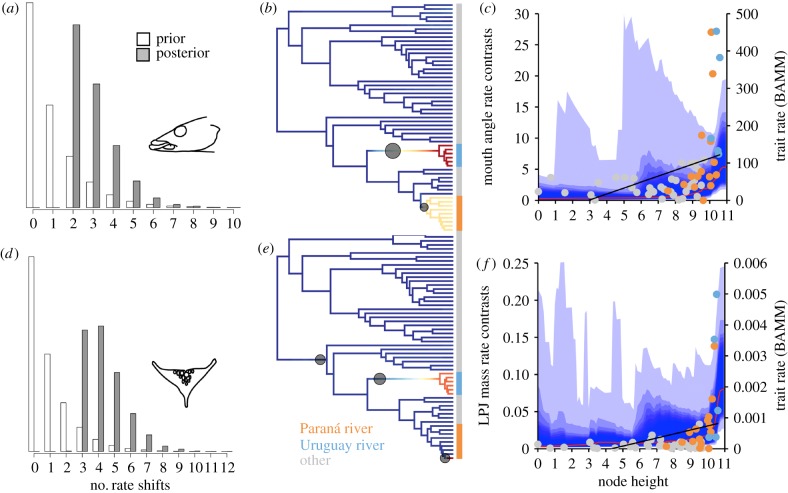
Rate shifts are not independent of one another such that there are many possible shift configurations. The most probable shift configuration for diversification rates includes no rate shifts (electronic supplementary material, figures S5 and S6). Less probable shift configurations include having one rate shift near the base of the clade containing both species flocks or having a shift associated with the origin of the Uruguay River species flock (electronic supplementary material, figures S5 and S6).
For the evolution of mouth angle, the most probable shift configuration includes two shifts near the base of each species flocks (figure 3a,b). Less probable shift configurations vary in the placement of the shift along the branch leading to the Uruguay River species flock and the placement of the shift near the base of the Paraná species flock, sometimes excluding some of the piscivores and invertivores (electronic supplementary material, figure S7). Rate through time plots revealed that evolutionary rates have accelerated through time (figure 3c). There was a significant positive relationship between absolute contrasts and the height of the nodes at which they were calculated (R2 = 0.161; F = 10.37; p = 0.002), indicating that rates of evolution have increased over time. Furthermore, node-height tests indicate that the rapid rate increase near present day is driven by the Parana and Uruguay species flocks (figure 3c).
Figure 3.
Rates of morphological evolution across the Crenicichla phylogeny. Prior and posterior distribution of rate shifts (a,d), most probable shift configuration from BAMM (b,e), rate through time (c,f), and node-height test (c,f) for mouth angle (a–c) and size-relative lower pharyngeal jaw (LPJ) mass (d–f). Less probable shift configurations are shown in the electronic supplementary material. Mean rate through time (red lines) is from BAMM analysis and the best fit lines (black lines) are fitted to the node-height tests.
For the evolution of size-relative LPJ mass, the most probable shift configuration included three shifts (figure 3d), including an early shift at the base of the clade containing both species flocks, and more recent shifts at the base of the Uruguay River species flock and within the Paraná River species flock (figure 3e; electronic supplementary material, figure S8). Less probable shift configurations varied in their inclusion of a shift near the base of the clade containing both species flocks, but generally had shifts at the base of or within both species flocks (electronic supplementary material, figure S8). Rate through time plots revealed that evolutionary rates have accelerated through time (figure 3f). There was a significant positive relationship between absolute contrasts and the height of the nodes at which they were calculated (R2 = 0.134; F = 8.35; p = 0.006), indicating that rates of evolution have increased over time. Furthermore, node-height tests indicate that the rapid rate increase near present day is driven by the Parana and Uruguay species flocks (figure 3f).
4. Discussion
Parallel diversification into similar suites of ecomorphs is a central theme of adaptive radiation within discrete ecosystems such as islands and lakes. Textbook examples of parallel adaptive radiations such as those of anole lizards and cichlid fishes provide predictable evolutionary outcomes such as non-random phenotypic evolution towards inferred adaptive peaks, the evolution of specializations and the onset of diversity dependence as niches fill towards capacity [5,7,10,62]. We found that pike cichlids exhibit parallel adaptive radiation in the Paraná and Uruguay Rivers (figure 4) that is consistent with those of island- and lake-dwelling clades and is uncharacteristic for river-dwelling fish assemblages.
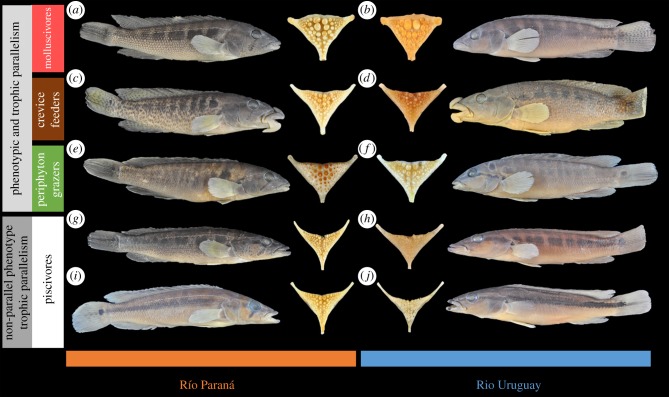
Figure 4.
Parallel and non-parallel evolution of ecomorphs in the Uruguay and Paraná River species flocks. Parallel evolution of molluscivores: Crenicichla tesay (a) and C. minuano (b), crevice feeders: C. tuca (c) and C. tendybaguassu (d), and periphyton grazers: C. tapii (e) and C. hadrostigma (f). Non-parallel morphological evolution, but trophic parallelism among piscivores: C. iguassuensis (g), C. missioneira (h), C. sp. Urugua-í line (i) and C. celidochilus (j).
Similar suites of ecomorphs have independently evolved within the Paraná and Uruguay Rivers from similar ancestral states (figure 2; electronic supplementary material, figure S4). The evolution of specialized trophic roles were strictly associated with the evolution of novel phenotypes and therefore contributed to a dramatic expansion of phenotypic diversity during these clades’ exploration of the adaptive landscape (figure 2d). Non-random phenotypic diversification towards inferred adaptive peaks associated with unique morphologies such as hypertrophied and atrophied pharyngeal jaws (i.e. molluscivores and piscivores, respectively), hypertrophied oral lips (i.e. crevice feeders) and benthic-oriented snouts (i.e. periphyton grazers) led to high degrees of convergence between these two clades. These trophic roles and their associated adaptations are rare among river-dwelling cichlids [22,63]; however, they are conspicuously associated with adaptive radiations of lake-dwelling clades. For example, African cichlids have diversified extensively within primary production-associated trophic roles (e.g. algae scrapers) in lakes Tanganyika and Malawi [22]. Rapid transitions to herbivory are otherwise rare among freshwater fishes [64]. Likewise, hypertrophied pharyngeal jaws associated with durophagy (i.e. eating hard-shelled prey) and hypertrophied lips associated with feeding from rocky crevices are also rare among Neotropical cichlids, but have often arose during adaptive radiation in African and Middle American lakes and are common sources of polymorphism between incipient species pairs in lakes and within some extremely polymorphic species (reviewed in [22]). These results indicate that while the ecomorphs are novel in terms of the recent evolutionary history of these river-dwelling clades, the specific ecological roles and associated phenotypic adaptations that arose are predictable when compared with lake-dwelling counterparts in Africa and Middle America.
The hallmark phylogenetic signature of adaptive radiation includes an initial burst in diversification rates following the colonization of a novel environment, followed by slowing after the onset of diversity dependence [65]. This period of slowing occurs in response to ecological constraints as niches fill towards capacity [60]. Pike cichlids probably colonized subtropical South America via stream capture between southern tributaries of the Amazon and the headwaters of the La Plata River basin [66–68]. We found some evidence of a burst in diversification rates at the base of the subtropical clade, but the most likely shift configuration contained zero shifts (electronic supplementary material, figures S5 and S6). The paucity of cichlid (and non-cichlid) lineages that occur in subtropical South America [69] may have resulted in competitive release from the comparatively diverse Amazonian lineages and communities, but such processes have not elicited dramatic bursts in species diversification as similar colonization events in the East African Great Lakes [70], perhaps owing to ecological constraints specific to riverine ecosystems. By contrast, bursts in morphological evolution were more closely associated with the Paraná and Uruguay River clades and the rise of the parallel ecomorphs (figure 3). Rates of phenotypic evolution increased through time, including dramatic increases near present day and are thus inconsistent with diversity dependence associated with niche filling [60]. These clades may be too young to exhibit such patterns of morphological evolution.
Diversification along the benthic-to-pelagic habitat axis has been prodigious among adaptive radiations of lake-dwelling cichlids [22,64], including within Lake Malawi [71], Lake Tanganyika [72], Cameroonian crater lakes [11], Ugandan crater lakes [13] and Nicaraguan crater lakes [9,73]. Most of the parallel ecomorphs that arose in the Paraná and Uruguay Rivers are strictly benthic such that they consume substrate-associated primary producers or invertebrates (i.e. periphyton grazers, crevice feeders and molluscivores); however, some species have specialized to exploit schooling fishes from the water column [26]. Vast depths and steep shoreline reefs that characterize many lakes provide dramatic ecological gradients that may have permitted the evolution of a variety of pelagic trophic roles including open-water piscivores as well as filter feeders [22]. Diversification along the benthic-to-pelagic habitat axis may be constrained within the Paraná and Uruguay River clades because the axis is more physically constrained in rivers and permits fewer evolutionary outcomes and less potential for niche packing [5]. Such environmental constraints may provide conditions that favour rapid morphological evolution but not in combination with rapid species diversification.
The Paraná and Uruguay River species flocks exhibit many characteristics generally attributed to the textbook examples of parallel adaptive radiation in island- and lake-dwelling organisms including that similar suites of ecomorphs evolved independently and in situ in the Uruguay and Paraná Rivers and that colonization-associated ecological opportunity elicited rapid phenotypic diversification. Normally, the adaptive landscape may be very different in lakes and rivers such that river assemblages are immigration-assembled rather than speciation-assembled and some regions of the adaptive landscape may be implausible evolutionary results in rivers owing to niches that are uncommon or temporally unstable in such environments [5]. Nevertheless, the Paraná and Uruguay Rivers have provided suitable circumstances for the rise of lake-like adaptive radiations. Namely, these rivers are mostly shallow, clear and have rocky substrate, which probably provided the opportunity for the exploitation of primary production-associated trophic roles (i.e. algae grazers) as is provided along the littoral zone of lakes. Second, these basins are home to few cichlid and non-cichlid lineages. Thus, after colonization of subtropical South American streams, pike cichlids may have experienced relaxed competition for mostly vacant niches. Lastly, pike cichlids colonized a novel adaptive zone among Neotropical cichlids associated with elongate bodies adapted for ram feeding [74], which may have predisposed the lineage to further trophic-based exploration of the adaptive landscape. This specific set of circumstances may have provided the opportunity for these clades to diversify in a pattern reminiscent of island-dwelling anoles [7] and lake-dwelling cichlids [5], such that similar suites of ecomorphs arose rapidly in parallel. Furthermore, the suites of parallel ecomorphs appear idiosyncratic within the immediate evolutionary histories of these clades, but are quite predictable evolutionary outcomes when viewed in the broader context of parallel adaptive radiations in other ecosystems such as lakes.
Supplementary Material
Acknowledgements
We are grateful to Luis Malabarba (UFRGS), Mark Sabaj (ANSP), Caleb McMahan (FMNH), Luiz Rocha (CAS) and Marcelo Loureiro for loans of specimens. Thanks are expressed to Davide Werneke for accessioning additional examined material at AUM. We thank Travis Ingram and Dan Rabosky for discussions about SURFACE and BAMM, respectively. For assistance with fieldwork in Uruguay, we thank Marcelo Loureiro, Alejandro Duarte, Wilson S. Serra and Daniel Hernandez. For assistance with fieldwork in Argentina, we thank Štěpánka Říčanová, Radka Piálková, Klára Dragová, Lukáš Drag and Jan Štefka. Feedback from three anonymous reviewers improved this manuscript. Access to computing and storage facilities owned by parties and projects contributing to the National Grid Infrastructure MetaCentrum provided under the programme ‘Projects of Large Infrastructure for Research, Development, and Innovations' (LM2010005) was greatly appreciated as well as the access to the CERIT-SC computing and storage facilities provided under the programme Center CERIT Scientific Cloud, part of the Operational Program Research and Development for Innovations, reg. no. CZ. 1.05/3.2.00/08.0144.
Ethics
Fishes were collected in accordance with Ministerio de Ecologia y Recursos Renovables, Misiones, Argentina (Resolucion 509/07 and 071), Administración de Parques Nacionales, Argentina (NEA 224, 328 and 35) and La Dirección Nacional de Recursos Acuáticos, Montevideo, Uruguay (resolucion 202/1383/2010 and 13/2014).
Data accessibility
The sequencing reads are deposited on the NCBI Sequence Repository Archive (SRA; BioProject ID: PRJNA420902). The concatenated tree file and morphological datasets are deposited on the Dryad Digital Repository (doi:10.5061/dryad.678rp) [75].
Authors' contributions
E.D.B. conceived and designed the study. E.D.B., L.P. and M.T. carried out laboratory work and analyses. E.D.B., L.P., O.R., J.C. and A.A. provided samples and specimens. E.D.B. drafted the manuscript. L.P., O.R., J.C., A.A., M.T. and J.W.A. provided critical revisions.
Competing interests
The authors declare no competing interests.
Funding
This research was partially funded by the Jim Smith Endowment Fund from the Ohio Cichlid Association to E.D.B., the US National Science Foundation grant DEB-1023403 to J.W.A. and the Czech Science Foundation GAČR 14-28518P grant to L.P.
References
- 1.Simpson GG. 1953. The major features of evolution. New York, NY: Columbia University Press. [Google Scholar]
- 2.Schluter D. 2000. The ecology of adaptive radiation. Oxford, UK: Oxford University Press. [Google Scholar]
- 3.Givnish TJ. 2015. Adaptive radiation versus ‘radiation' and ‘explosive diversification’: why conceptual distinctions are fundamental to understanding evolution. New Phytol. 207, 297–303. ( 10.1111/nph.13482) [DOI] [PubMed] [Google Scholar]
- 4.Losos J. 2009. Lizards in an evolutionary tree: ecology and adaptive radiation of anoles, vol. 10 Berkeley, CA: University of California Press. [Google Scholar]
- 5.Seehausen O. 2015. Process and pattern in cichlid radiations: inferences for understanding unusually high rates of evolutionary diversification. New Phytol. 207, 304–312. ( 10.1111/nph.13450) [DOI] [PubMed] [Google Scholar]
- 6.Harmon LJ, Kolbe JJ, Cheverud JM, Losos JB. 2005. Convergence and the multidimensional niche. Evolution 59, 409–421. ( 10.1111/j.0014-3820.2005.tb00999.x) [DOI] [PubMed] [Google Scholar]
- 7.Mahler DL, Revell LJ, Glor RE, Losos JB. 2010. Ecological opportunity and the rate of morphological evolution in the diversification of Greater Antillean anoles. Evolution 64, 2731–2745. ( 10.1111/j.1558-5646.2010.01026.x) [DOI] [PubMed] [Google Scholar]
- 8.Mahler DL, Ingram T, Revell LJ, Losos JB. 2013. Exceptional convergence on the macroevolutionary landscape in island lizard radiations. Science 341, 292–295. ( 10.1126/science.1232392) [DOI] [PubMed] [Google Scholar]
- 9.Elmer KR, Fan S, Kusche H, Spreitzer ML, Kautt AF, Franchini P, Meyer A. 2014. Parallel evolution of Nicaraguan crater lake cichlid fishes via non-parallel routes. Nat. Comm. 5, 5168 ( 10.1038/ncomms6168.) [DOI] [PubMed] [Google Scholar]
- 10.Kocher TD, Conroy JA, McKaye KR, Stauffer JR. 1993. Similar morphologies of cichlid fish in Lakes Tanganyika and Malawi are due to convergence. Mol. Phylogenet. Evol. 2, 158–165. ( 10.1006/mpev.1993.1016) [DOI] [PubMed] [Google Scholar]
- 11.Schliewen U, Rassmann K, Markmann M, Markert J, Kocher T, Tautz D. 2001. Genetic and ecological divergence of a monophyletic cichlid species pair under fully sympatric conditions in Lake Ejagham, Cameroon. Mol. Ecol. 10, 1471–1488. ( 10.1046/j.1365-294X.2001.01276.x) [DOI] [PubMed] [Google Scholar]
- 12.Wagner CE, Harmon LJ, Seehausen O. 2014. Cichlid species-area relationships are shaped by adaptive radiations that scale with area. Ecol. Lett. 17, 583–592. ( 10.1111/ele.12260) [DOI] [PubMed] [Google Scholar]
- 13.Machado-Schiaffino G, Kautt AF, Kusche H, Meyer A. 2015. Parallel evolution in Ugandan crater lakes: repeated evolution of limnetic body shapes in haplochromine cichlid fish. BMC Evol. Biol. 15, 1 ( 10.1186/s12862-015-0287-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rundle HD, Nagel L, Boughman JW, Schluter D. 2000. Natural selection and parallel speciation in sympatric sticklebacks. Science 287, 306–308. ( 10.1126/science.287.5451.306) [DOI] [PubMed] [Google Scholar]
- 15.Østbye K, et al. 2006. Parallel evolution of ecomorphological traits in the European whitefish Coregonus lavaretus (L.) species complex during postglacial times. Mol. Ecol. 15, 3983–4001. ( 10.1111/j.1365-294X.2006.03062.x) [DOI] [PubMed] [Google Scholar]
- 16.Blackledge TA, Gillespie RG. 2004. Convergent evolution of behavior in an adaptive radiation of Hawaiian web-building spiders. Proc. Natl Acad. Sci. USA 101, 16 228–16 233. ( 10.1073/pnas.0407395101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gillespie R. 2004. Community assembly through adaptive radiation in Hawaiian spiders. Science 303, 356–359. ( 10.1126/science.1091875) [DOI] [PubMed] [Google Scholar]
- 18.Givnish TJ, et al. 2009. Origin, adaptive radiation and diversification of the Hawaiian lobeliads (Asterales: Campanulaceae). Proc. R. Soc. B 276, 407–416. ( 10.1098/rspb.2008.1204) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson MS, Murray J, Clarke B. 2000. Parallel evolution in Marquesan partulid land snails. Biol. J. Linn. Soc. 69, 577–598. ( 10.1111/j.1095-8312.2000.tb01224.x) [DOI] [Google Scholar]
- 20.Sullivan JP, Lavoué S, Hopkins CD. 2002. Discovery and phylogenetic analysis of a riverine species flock of African electric fishes (Mormyroidae: Teleostei). Evolution 56, 597–616. ( 10.1111/j.0014-3820.2002.tb01370.x) [DOI] [PubMed] [Google Scholar]
- 21.Schwarzer J, Misof B, Ifuta SN, Schliewen UK. 2011. Time and origin of cichlid colonization of the lower Congo rapids. PLoS ONE 6, e22380 ( 10.1371/journal.pone.0022380) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burress ED. 2015. Cichlid fishes as models of ecological diversification: patterns, mechanisms, and consequences. Hydrobiologia 748, 7–27. ( 10.1007/s10750-014-1960-z) [DOI] [Google Scholar]
- 23.Piálek L, Říčan O, Casciotta J, Almirón A, Zrzavý J. 2012. Multilocus phylogeny of Crenicichla (Teleostei: Cichlidae), with biogeography of the C. lacustris group: species flocks as a model for sympatric speciation in rivers. Mol. Phylogenet. Evol. 62, 46–61. ( 10.1016/j.ympev.2011.09.006) [DOI] [PubMed] [Google Scholar]
- 24.de Lucena CAS, Kulander SO. 1992. The Crenicichla (Teleoctei: Cichlidae) species of the Uruguai River drainage in Brazil. Ichthyol. Explorat. Fresh. 3, 97–192. [Google Scholar]
- 25.Serra WS, Duarte A, Burress ED, Loureiro M. 2011. Perciformes, Cichlidae, Crenicichla tendybaguassu Lucena and Kullander, 1992: first record for Uruguay. Check List 7, 357–359. ( 10.15560/7.3.357) [DOI] [Google Scholar]
- 26.Burress ED, Duarte A, Serra WS, Loueiro M, Gangloff MM, Siefferman L. 2013. Functional diversification within a predatory species flock. PLoS ONE 8, e80929 ( 10.1371/journal.pone.0080929) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piálek L, Dragová K, Casciotta J, Almirón A, Říčan O. 2015. Description of two new species of Crenicichla (Teleostei: Cichlidae) from the lower Iguazú River with a taxonomic reappraisal of C. iguassuensis, C. tesay and C. yaha. Hist. Nat. 5, 5–27. [Google Scholar]
- 28.Říčan O, Piálek L, Dragova K, Novak J. 2016. Diversity and evolution of the Middle American cichlid fishes (Teleostei: Cichlidae) with revised classification. Vert. Zool. 66, 3–102. [Google Scholar]
- 29.Catchen JM, Amores A, Hohenlohe P, Cresko W, Postlethwait JH. 2011. Stacks: building and genotyping loci de novo from short-read sequences. G3 1, 171–182. ( 10.1534/g3.111.000240) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. ( 10.1093/bioinformatics/btu033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Posada D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256. ( 10.1093/molbev/msn083) [DOI] [PubMed] [Google Scholar]
- 32.Leaché AD, Banbury BL, Felsenstein J, Nieto-Montes de Oca A, Stamatakis A. 2015. Short tree, long tree, wrong tree: new acquisition bias corrections for inferring SNP phylogenies. Syst. Biol. 64, 1032–1047. ( 10.1093/sysbio/syv053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burress ED, Alda F, Duarte A, Loureiro M, Armbruster JW, Chakrabarty P. In press. Phylogenomics of pike cichlids (Cichlidae: Crenicichla): the rapid ecological speciation of an incipient species flock. J. Evol. Biol. ( 10.1111/jeb.13196) [DOI] [PubMed] [Google Scholar]
- 34.Eastman JM, Harmon LJ, Tank DC. 2013. Congruification: support for time scaling large phylogenetic trees. Methods Ecol. Evol 4, 688–691. ( 10.1111/2041-210X.12051) [DOI] [Google Scholar]
- 35.Friedman M, Keck BP, Dornburg A, Eytan RI, Martin CH, Hulsey CD, Wainwright PC, Near TJ. 2013. Molecular and fossil evidence place the origin of cichlid fishes long after Gondwanan rifting. Proc. R. Soc. B 280, 20131733 ( 10.1098/rspb.2013.1733) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith SA, O'Meara BC. 2012. treePL: divergence time estimation using penalized likelihood for large phylogenies. Bioinformatics 28, 2689–2690. ( 10.1093/bioinformatics/bts492) [DOI] [PubMed] [Google Scholar]
- 37.Rohlf FJ. 2004. Tpsutil, file utility program. version 1.58. Stony Brook, NY: Department of Ecology and Evolution, State University of New York at Stony Brook. [Google Scholar]
- 38.Rohlf FJ. 2006. Tpsdig, version 2.17. Stony Brook, NY: Department of Ecology and Evolution, State University of New York at Stony Brook. [Google Scholar]
- 39.Rohlf FJ. 2007. Tpsrelw version 1.54. Stony Brook, NY: Department of Ecology and Evolution, State University of New York at Stony Brook. [Google Scholar]
- 40.Ingram T, Mahler DL. 2013. SURFACE: detecting convergent evolution from comparative data by fitting Ornstein-Uhlenbeck models with stepwise Akaike information criterion. Methods Ecol. Evol. 4, 416–425. ( 10.1111/2041-210X.12034) [DOI] [Google Scholar]
- 41.Slayton CT. 2008. Is convergence surprising? An examination of the frequency of convergence in simulated datasets. J. Theor. Biol. 252, 1–14. ( 10.1016/j.jtbi.2008.01.008) [DOI] [PubMed] [Google Scholar]
- 42.Maddison WP, Maddison DR. 2015. Mesquite: a modular system for evolutionary analysis. Version 3.02. See http://mesquiteproject.org.
- 43.Bollback JP. 2006. Stochastic character mapping of discrete traits on phylogenies. BMC Bioinformatics 7, 88 ( 10.1186/1471-2105-7-88) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Revell LJ. 2012. Phytools: an R package for ohylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223. ( 10.1111/j.2041-210X.2011.00169.x) [DOI] [Google Scholar]
- 45.Schluter D, Price T, Mooers AØ, Ludwig D. 1997. Likelihood of ancestor states in adaptive radiation. Evolution 51, 1699–1711. ( 10.1111/j.1558-5646.1997.tb05095.x) [DOI] [PubMed] [Google Scholar]
- 46.Pagel M. 1999. The maximum likelihood approach to reconstructing ancestral character states of discrete characters on phylogenies. Syst. Biol. 48, 612–622. ( 10.1080/106351599260184) [DOI] [Google Scholar]
- 47.Nielsen R. 2002. Mapping mutations on phylogenies. Syst. Biol. 51, 729–739. ( 10.1080/10635150290102393) [DOI] [PubMed] [Google Scholar]
- 48.Huelsenbeck JP, Nielsen R, Bollback JP. 2003. Stochastic mapping of morphological characters. Syst. Biol. 52, 131–134. ( 10.1080/10635150390192780) [DOI] [PubMed] [Google Scholar]
- 49.Sidlauskas B. 2008. Continuous and arrested morphological diversification in sister clades of characiform fishes: a phylomorphospace approach. Evolution 62, 3135–3156. ( 10.1111/j.1558-5646.2008.00519.x) [DOI] [PubMed] [Google Scholar]
- 50.Maddison WP. 1991. Squared-change parsimony reconstructions of ancestral states for continuous-valued characters on a phylogenetic tree. Syst. Biol. 40, 304–314. ( 10.1093/sysbio/40.3.304) [DOI] [Google Scholar]
- 51.Revell LJ, Johnson MA, Schulte JA, Kolbe JJ, Losos JB. 2007. A phylogenetic test for adaptive convergence in rock-dwelling lizards. Evolution 61, 2898–2912. ( 10.1111/j.1558-5646.2007.00225.x) [DOI] [PubMed] [Google Scholar]
- 52.Rabosky DL. 2014. Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS ONE 9, e89543 ( 10.1371/journal.pone.0089543) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rabosky DL, Donnellan SC, Grundler M, Lovette IJ. 2014. Analysis and visualization of complex macroevolutionary dynamics: an example from Australian scincid lizards. Syst. Biol. 63, 610–627. ( 10.1093/sysbio/syu025) [DOI] [PubMed] [Google Scholar]
- 54.Rabosky DL, Grundler M, Anderson C, Shi JJ, Brown JW, Huang H, Larson JG. 2014. BAMMtools: an R package for the analysis of evolutionary dynamics on phylogenetic trees. Methods Ecol. Evol. 5, 701–707. ( 10.1111/2041-210X.12199) [DOI] [Google Scholar]
- 55.Plummer M, Best N, Cowles K, Vines K. 2006. CODA: Convergence diagnosis and output analysis for MCMC. R News 6, 7–11. [Google Scholar]
- 56.Moore BR, Höhna S, May MR, Rannala B, Huelsenbeck JP. 2016. Critically evaluating the theory and performance of Bayesian analysis of macroevolutionary mixtures. Proc. Natl Acad. Sci. USA 113, 9569–9574. ( 10.1073/pnas.1518659113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mitchell JS, Rabosky DL. 2016. Bayesian model selection with BAMM: effects of the model prior on the inferred number of diversification shifts. Methods Ecol. Evol. 8, 37–46. ( 10.1111/2041-210X.12626) [DOI] [Google Scholar]
- 58.Rabosky DL, Mitchell JS, Chang J. 2017. Is BAMM flawed? Theoretical and practical concerns in the analysis of multi-rate diversification models. Syst. Biol. 66, 477–498. ( 10.1093/sysbio/syx037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McPeek MA. 1995. Testing hypotheses about evolutionary change on single branches of a phylogeny using evolutionary contrasts. Am. Nat. 145, 686–703. ( 10.1086/285763) [DOI] [Google Scholar]
- 60.Freckleton RP, Harvey PH. 2006. Detecting non-Brownian trait evolution in adaptive radiations. PLoS. Biol. 4, e373 ( 10.1371/journal.pbio.0040373) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ingram T, Kai Y. 2014. The geography of morphological convergence in the radiations of Pacific Sebastes rockfishes. Am. Nat. 184, E115–E131. ( 10.1086/678053) [DOI] [PubMed] [Google Scholar]
- 62.Losos JB. 2011. Convergence, adaptation, and constraint. Evolution 65, 1827–1840. ( 10.1111/j.1558-5646.2011.01289.x) [DOI] [PubMed] [Google Scholar]
- 63.Burress ED. 2016. Ecological diversification associated with the pharyngeal jaw diversity of Neotropical cichlid fishes. J. Anim. Ecol. 85, 302–313. ( 10.1111/1365-2656.12457) [DOI] [PubMed] [Google Scholar]
- 64.Seehausen O, Wagner CE. 2014. Speciation in freshwater fishes. Ann. Rev. Ecol. Evol. Syst. 45, 621–651. ( 10.1146/annurev-ecolsys-120213-091818) [DOI] [Google Scholar]
- 65.Glor RE. 2010. Phylogenetic insights on adaptive radiation. Ann. Rev. Ecol. Evol. Syst. 41, 251–270. ( 10.1146/annurev.ecolsys.39.110707.173447) [DOI] [Google Scholar]
- 66.Albert JS, Carvalho TP. 2011. Neogene assembly of modern faunas. In Historical biogeography of neotropical freshwater fishes (eds Albert JS, Reis R), pp. 119–136. Berkeley, CA: University of California Press. [Google Scholar]
- 67.Brea M, Zucol A. 2011. The Paraná-Paraguay Basin: geology and paleoenvironments. In Historical biogeography of neotropical freshwater fishes (eds Albert JS, Reis R), pp. 69–88. Berkeley, CA: University of California Press. [Google Scholar]
- 68.Carvalho TP, Albert JS. 2011. The Amazon-Paraguay divide. In Historical biogeography of neotropical freshwater fishes (eds Albert J, Reis R), pp. 193–202. Berkeley, CA: University of California Press. [Google Scholar]
- 69.Albert JS, Bart HL Jr, Reis RE. 2011. Species richness and cladal diversity. In Historical biogeography of neotropical freshwater fishes (eds Albert JS, Reis RE), pp. 89–104. Berkeley, CA: University of California Press. [Google Scholar]
- 70.Burress ED, Tan M. 2017. Ecological opportunity alters the timing and shape of adaptive radiation. Evolution 17, 2650–2660. ( 10.1111/evo.13362) [DOI] [PubMed] [Google Scholar]
- 71.Hulsey CD, Roberts RJ, Loh YH, Rupp MF, Streelman JT. 2013. Lake Malawi cichlid evolution along a benthic/limnetic axis. Ecol. Evol. 3, 2262–2272. ( 10.1002/ece3.633) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cooper WJ, Parsons K, McIntyre A, Kern B, McGee-Moore A, Albertson RC. 2010. Bentho-pelagic divergence of cichlid feeding architecture was prodigious and consistent during multiple adaptive radiations within African rift-lakes. PLoS ONE 5, e9551 ( 10.1371/journal.pone.0009551) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kusche H, Recknagel H, Elmer KR, Meyer A. 2014. Crater lake cichlids individually specialize along the benthic–limnetic axis. Ecol. Evol. 4, 1127–1139. ( 10.1002/ece3.1015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.López-Fernández H, Arbour JH, Winemiller K, Honeycutt RL. 2013. Testing for ancient adaptive radiations in neotropical cichlid fishes. Evolution 67, 1321–1337. ( 10.1111/evo.12038) [DOI] [PubMed] [Google Scholar]
- 75.Burress ED, Piálek L, Casciotta JR, Almirón A, Tan M, Armbruster JW, Řĩcan O. Data from: Island- and lake-like parallel adaptive radiations replicated in rivers. Dryad Digital Repository ( 10.5061/dryad.678rp) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Burress ED, Piálek L, Casciotta JR, Almirón A, Tan M, Armbruster JW, Řĩcan O. Data from: Island- and lake-like parallel adaptive radiations replicated in rivers. Dryad Digital Repository ( 10.5061/dryad.678rp) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
The sequencing reads are deposited on the NCBI Sequence Repository Archive (SRA; BioProject ID: PRJNA420902). The concatenated tree file and morphological datasets are deposited on the Dryad Digital Repository (doi:10.5061/dryad.678rp) [75].