Abstract
Understanding how disease risk varies over time and across heterogeneous populations is critical for managing disease outbreaks, but this information is rarely known for wildlife diseases. Here, we demonstrate that variation in host and pathogen factors drive the direction, duration and intensity of a coral disease outbreak. We collected longitudinal health data for 200 coral colonies, and found that disease risk increased with host size and severity of diseased neighbours, and disease spread was highest among individuals between 5 and 20 m apart. Disease risk increased by 2% with every 10 cm increase in host size. Healthy colonies with severely diseased neighbours (greater than 75% affected tissue) were 1.6 times more likely to develop disease signs compared with colonies with moderately diseased neighbours (25–75% affected tissue). Force of infection ranged from 7 to 20 disease cases per 1000 colonies (mean = 15 cases per 1000 colonies). The effective reproductive ratio, or average number of secondary infections per infectious individual, ranged from 0.16 to 1.22. Probability of transmission depended strongly on proximity to diseased neighbours, which demonstrates that marine disease spread can be highly constrained within patch reefs.
Keywords: outbreak, transmission, force of infection, coral disease, effective reproductive ratio
1. Introduction
Rates and processes of disease transmission in the ocean are almost completely unknown. Theoretically, the openness and connectivity within marine populations may facilitate disease spread, while limited adult dispersal (long-distance dispersal typically occurs in planktonic larvae) may buffer marine populations from wide-scale disease transmission [1]. Disease transmission rates are affected by variation in host–pathogen contact, host traits and habitat. However, few studies have addressed how variation in size and spatial structure of host populations influence disease spread in the ocean. Here, we test hypotheses about how host heterogeneity and spatial heterogeneity affect transmission rates during a coral disease outbreak.
Variation in host susceptibility to disease is influenced by a variety of factors such as body size, immune capacity and genetics. Large body size has been associated with disease risk for many marine organisms [2,3]. The size–disease risk relationship may be related to surface area exposed to pathogens or host senescence [4]. Immune capacity, which can be related to host senescence, influences the host's ability to resist and respond to infection, and can vary seasonally with nutritional status, and during periods of reproduction and environmental stress [5]. Genetic diversity can also lead to within-population variation in disease resistance and has been shown to affect disease spread and severity in a variety of marine organisms (e.g. sea lions [6]; brown algae [7]). Clonal marine invertebrates such as corals, sponges and bryozoans may be particularly vulnerable to large epidemics because genetic homogeneity enables the build-up of virulent pathogens, and sessile organisms are physically limited in their ability to ward off infectious agents [1].
Habitat configuration can also influence disease spread by increasing geographic connectivity or creating physical barriers preventing pathogen and/or host movement. On land, streams can act as semipermeable barriers to the movement of mammals affected by rabies [8] but can also serve as corridors for the spread of waterborne pathogens such as the Port Orford cedar tree pathogen, Phytophthora lateralis [9]. It is plausible, therefore, that discontinuities in mosaic marine habitats such as coral reefs, seagrass meadows and oyster reefs similarly affect disease spread in the ocean. In coral reefs, several studies have shown associations between colony configuration and clustering of disease cases [10–13], while other studies have found no evidence of disease clustering [4]; these opposing results probably reflect differences in modes of transmission for different diseases.
In this study, we investigated how host, pathogen and environmental factors influence marine disease transmission through time by visually assessing disease spread using a naturally occurring coral disease outbreak as a case study. In winter 2015, an outbreak spread rapidly through Montipora capitata, a dominant Hawaiian reef-building coral species [14] in central Kāne‘ohe Bay, O‘ahu, Hawai‘i (electronic supplementary material, figure S1). Histology revealed a previously undescribed tissue loss disease characterized by microscopic lesions with dissociation of gastrodermal cells and mucus hypertrophy; however, no pathogen was identified [15]. For this study, we refer to this newly discovered disease with no known pathogen as Montipora capitata disease unknown (MCX). We investigated how MCX risk varied through time and with host characteristics and spatial proximity to other diseased colonies.
2. Methods
(a). Field surveys
Immediately after the MCX outbreak was discovered [15], we identified 200 visually healthy branching M. capitata coral colonies and monitored their health state over the course of the outbreak. The 200 focal colonies were distributed among three patch reefs adjacent to the epicentre of the outbreak in Kāne‘ohe Bay (electronic supplementary material, figure S1). We recorded geographic coordinates and measured host, pathogen and environmental factors for each focal colony between 5 and 7 February 2015. We recorded health state for each focal colony daily by assessing the presence and severity of macroscopic disease lesions (visibly actively affected tissue) for three weeks (8 February to 2 March 2015) with weekly follow-up surveys for two weeks (3–16 March 2015). We categorized coral health state as healthy (no visible actively affected tissue), low, moderate or high disease severity (less than 25%, 25–75% or greater than 75% visibly actively affected tissue, respectively), or removed (previously diseased colonies with no remaining visibly actively affected tissue). No colonies experienced re-infection during the study.
To investigate the role of host factors on disease risk, we measured host size, sediment cover and M. capitata cover (table 1). We measured host size as the longest horizontal axis diameter for each focal colony. We visually estimated sediment cover, which can accumulate from re-suspended sediment and fish excrement (potential disease vectors [16]), as the percentage of host surface tissue covered in sediment. We also quantified the percentage of M. capitata cover in 2 × 1 m belt transects surrounding each focal colony.
Table 1.
Measurements of host, pathogen and environmental factors. Pathogen factors were measured 4 days prior to (initial) and 1 to 3 days after (final) the monitoring period (equations in electronic supplementary material). For lesion severity, low, moderate and high correspond to less than 25%, 25–75% and greater than 75% of the colony affected by disease, respectively.
| factor | metric description | range |
|---|---|---|
| host | host size (maximum diameter) | 12–135 cm |
| host | sediment cover | 0–33% of tissue covered |
| host | host cover | 4–67% cover of M. capitata colonies in 2 m2 |
| environment | benthic cover (coral, sponge, and algae) | 12–100% cover in 2 m2 |
| environment | diversity: species richness | 2–7 |
| environment | diversity: Simpson's diversity index | 0.12–0.72 |
| environment | depth | 0.5–7 m |
| environment | reef position | flat, crest, slope |
| pathogen | distance to diseased neighbours | initial: 0.00–8.23 m |
| final: 0.16–7.90 m | ||
| pathogen | lesion severity | initial: low–high |
| final: moderate–high | ||
| pathogen | size-weighted lesion severity | initial: 4–90 cm2 |
| final: 9–150 cm2 | ||
| pathogen | distance- and size-weighted lesion severity | initial: 0.05–2643 |
| final: 0.21–256 |
To investigate the role of local environment on disease risk, we measured depth, reef position and benthic cover (table 1). We measured water depth, which can indicate solar radiation (both necessary for coral's symbiotic photosynthetic algae to translocate energy and harmful to coral in excessive sunlight conditions), as the distance between the coral surface and ocean surface. We categorized reef position as reef flat, crest or slope. We measured percentage cover of coral (identified to species), algae and sponge in 2 × 1 m belt transects surrounding each focal colony, and calculated two metrics of benthic diversity from the belt transect data: species richness, which indicates the number of unique species, and Simpson's diversity index, which accounts for the number of unique species and their relative abundances. While we did not evaluate climate factors in this study, it is valuable to note that temperature-induced coral bleaching occurred in Kāne‘ohe Bay in 2014 and 2015, and heavy rainfall preceded the MCX outbreak.
To investigate the role of pathogen load on disease risk, we identified the three nearest neighbours exhibiting disease signs, and measured their size, lesion severity and distance to the focal colony. We calculated four proxies of pathogen load for each focal colony based on measurements for the three nearest diseased neighbours (equations in electronic supplementary material). Briefly, the four proxies were (i) distance to diseased neighbour (metres); (ii) lesion severity (per cent cover of actively affected tissue), where low = 0–25%, moderate = 25–75%, and high = 75–100%; (iii) size-weighted lesion severity, calculated as the product of lesion severity and colony diameter; and (iv) distance- and size-weighted lesion severity, calculated as size-weighted lesion severity divided by distance2. We measured and calculated these metrics for each focal colony at the beginning (4 days prior) and end (1–3 days after) of the monitoring period.
(b). Associating host, pathogen and environment factors with disease risk
To assess variation in disease risk as a function of host, pathogen and environment factors, we used a spatio-temporal point process regression model [17]. Disease risk is the likelihood an individual contracts a disease. In the regression model, disease risk is described by the conditional intensity term, or instantaneous rate of infection, 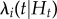 (equation (2.1)). Disease risk is characterized in the model through a multiplicative endemic component and an additive epidemic component
(equation (2.1)). Disease risk is characterized in the model through a multiplicative endemic component and an additive epidemic component
| 2.1 |
where the conditional intensity, 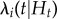 , reflects the probability that at time (t), individual (i) transitions from susceptible to infected, given Ht, the event history for individual (i). The event history describes whether a transition from susceptible to infected or infected to removed occurred up to, but not including, time (t) (equation (2.1)). γi(t) is the right-continuous at-risk indicator for individual (i), equalling 1 if the individual is at-risk of infection at time (t) and 0 otherwise. hi(t) is the endemic model component and
, reflects the probability that at time (t), individual (i) transitions from susceptible to infected, given Ht, the event history for individual (i). The event history describes whether a transition from susceptible to infected or infected to removed occurred up to, but not including, time (t) (equation (2.1)). γi(t) is the right-continuous at-risk indicator for individual (i), equalling 1 if the individual is at-risk of infection at time (t) and 0 otherwise. hi(t) is the endemic model component and  is the epidemic model component.
is the epidemic model component.
Endemic risk, hi(t), is the risk of infection from external sources. The endemic model component is similar to a Cox proportional hazards model with a time-dependent baseline risk, exp(h0(t)), with external covariates zi(t):
| 2.2 |
where T is the duration between event transitions and β is a vector of coefficients for covariates zi. In our model, we included host size, depth, reef position, sediment cover, M. capitata cover, benthic diversity measures and pathogen load proxies in the vector of static endemic covariates (zi) (table 1).
The epidemic model component, 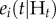 , characterizes individual-to-individual disease transmission and depends entirely on internal event history Ht. The epidemic component assumes disease risk for individual (i) is a distance- and time-weighted sum over the infected individuals I, just before time (t),
, characterizes individual-to-individual disease transmission and depends entirely on internal event history Ht. The epidemic component assumes disease risk for individual (i) is a distance- and time-weighted sum over the infected individuals I, just before time (t),
| 2.3 |
where g1(u) reflects isotopic spatial clustering between positions of individuals i and j (si−sj), and, g2(v) reflects time-dependent infectivity. In our model, the parametric function g1(u) was a Euclidean distance, and we used this function to calculate the number of infected neighbours within different Euclidean distances at each time point, to determine the distance where disease transmission was greatest. g2(v) is measured in time units where individual j transitions to an infected state at time 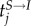 . Equation (2.4) reflects the functions for g1(u) and g2(v):
. Equation (2.4) reflects the functions for g1(u) and g2(v):
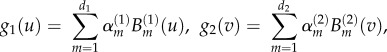 |
2.4 |
where 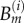 are basis functions (truncated power splines) and
are basis functions (truncated power splines) and 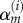 are the respective coefficients.
are the respective coefficients.
We conducted this analysis using the surveillance package v. 1.13.1 in R statistical software v. 3.3.1.
(c). Calculating force of infection and Reff
Force of infection (FOI) is the per capita rate at which susceptible individuals acquire a disease and is a key parameter for quantifying how quickly and extensively a disease can spread through a population [18,19]. We calculated the mean and standard error of the mean of FOI at each time point (t) from individual FOI trajectories, which were calculated in the point process regression model. Incidence, which can be measured more directly, is equal to the product of FOI and the susceptible proportion of the population. The effective reproductive ratio, Reff, is the average number of secondary infections in a population (including both susceptible and non-susceptible hosts) and is calculated as
| 2.5 |
where S is the number of susceptible colonies at time (t) and γ is the recovery rate, calculated as the average duration a colony exhibited disease signs.
3. Results
(a). Field surveys
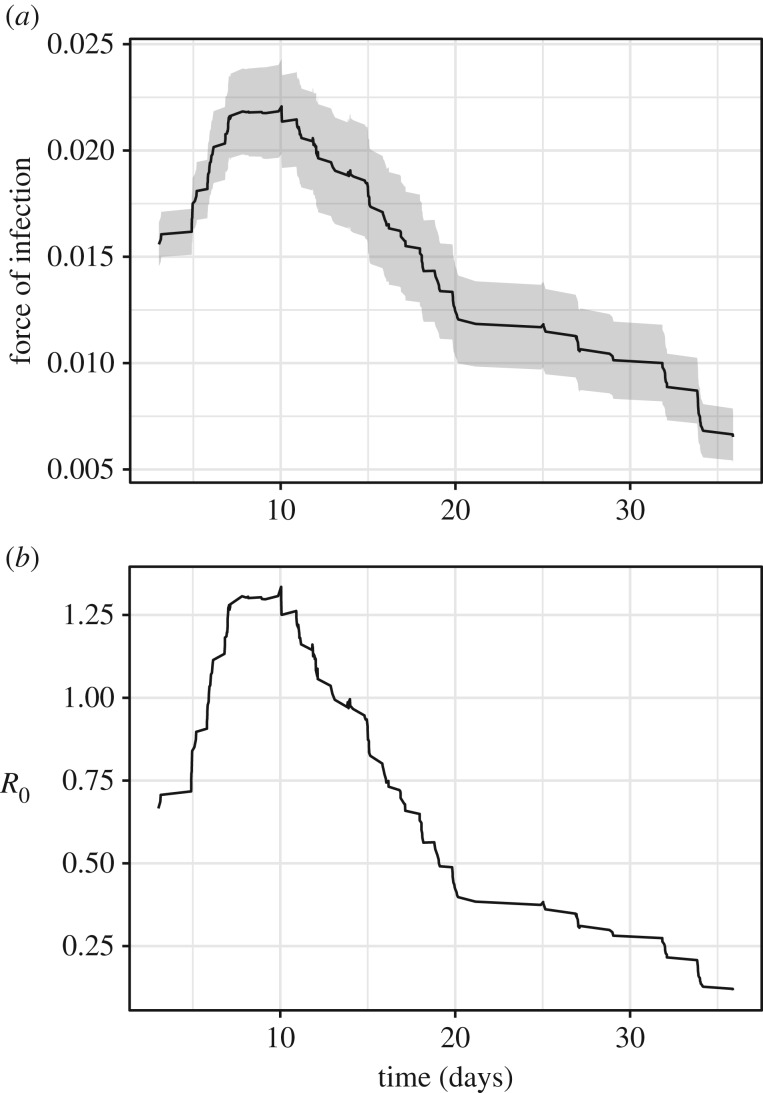
During the five-week study period, 106 of the 200 (53%) focal colonies developed disease signs. Of the diseased colonies, 83 colonies exhibited low disease severity, 22 colonies exhibited moderate disease severity and 1 colony exhibited high disease severity (electronic supplementary material, figure S1). Over 70% of diseased colonies contracted MCX within the first two weeks of the study period, with the outbreak peaking on day 11 with a total of 65 colonies (32.5%) exhibiting disease signs by that day (figure 1).
Figure 1.
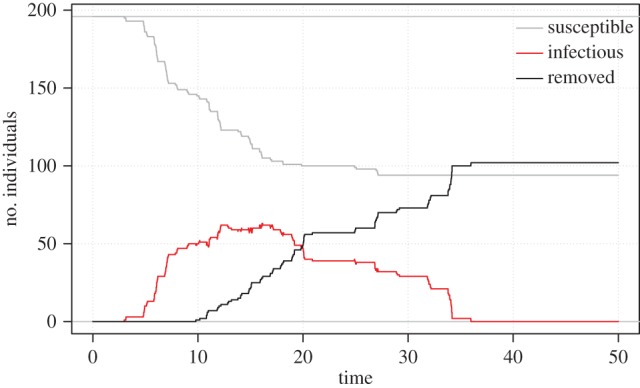
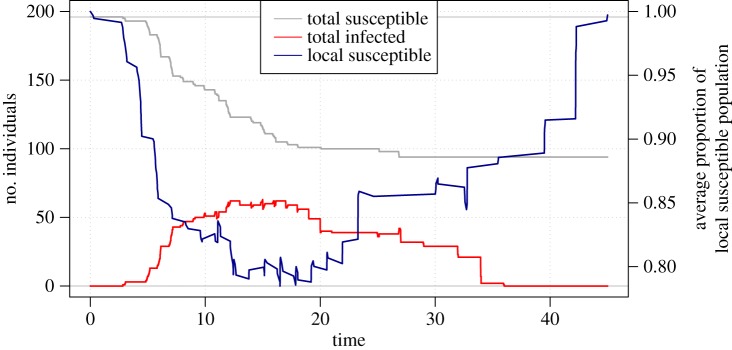
Susceptible infected removed plot. Outbreak trajectories of susceptible colonies (grey line), infected colonies (red line) and removed colonies (black line). Removed refers to colonies that became diseased during the monitoring period and then the disease lesion stopped progressing.
(b). Associating host and pathogen factors with disease risk
There were no differences in disease risk among reefs, so we pooled colonies from all reefs for our analysis. We estimated an endemic background rate of infection in a population with no current MCX cases as 0.000007 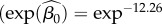 from electronic supplementary material, table S1).
from electronic supplementary material, table S1).
Of the host and environmental factors that we measured (table 1; electronic supplementary material, figures S2–S4), disease risk only varied with host size. Disease risk increased by 2% with every 10 cm increase in host diameter (exp0.00254×10; electronic supplementary material, table S1). The other host factors (sediment cover and M. capitata cover) and environmental factors (depth, reef position, coral cover, benthic diversity) that we analysed did not affect disease risk. Of the four pathogen load proxies that we measured at the beginning and end of the study period (table 1; electronic supplementary material, figures S2–S4), we found that disease risk was positively related to one factor: initial lesion severity. Colonies with moderate lesion severity neighbours at the outset of the study were 1.13 times more likely to develop disease signs compared with colonies with low lesion severity neighbours at the outset of the study (calculated as e0.118/e0 from coefficients in electronic supplementary material, table S1). Colonies with high lesion severity neighbours at the outset of the study were 1.6 times more likely to develop disease signs compared with colonies with moderate lesion severity neighbours at the outset of the study (calculated as e0.593/e0.118 from coefficients in electronic supplementary material, table S1). Other pathogen load proxies included in the analysis (distance to neighbour, size-weighted lesion severity and distance- and size-weighted lesion severity) did not perform as well.
Probability of disease transmission was highest among colonies located between 5 and 20 m apart. The number of diseased individuals in our sample population and their distances to susceptible focal colonies continuously changed over the course of the outbreak (figure 1), enabling us to determine the optimal distance for disease transmission to occur. By comparing seven models that accounted for the number of diseased individuals within concentric circles surrounding each focal colony at distances ranging from 0 to 50 m (while holding host size and initial lesion severity constant), we determined that MCX transmission was highest between 5 and 20 m, with optimal transmission at 15 m (indicated by the model with the lowest AIC value in electronic supplementary material, table S2).
(c). Calculating force of infection, recovery rate, and Reff
FOI and the effective reproductive ratio (Reff) changed through time, while recovery rate remained constant. FOI ranged from 7 to 20 disease cases per 1000 coral colonies with a mean of 15 disease cases per 1000 colonies (figure 2a). Mean recovery rate (analogous to the infectious period) was 7.15 days (s.e.m. ± 1.3 days) and was independent of all predictor variables that we measured in this study. Reff ranged from 0.16 to 1.22 with a mean of 0.71 (figure 2b). Reff exceeded 1, the theoretical outbreak threshold, between days 6 and 11 of the study period.
Figure 2.
Epidemiological parameters through time. (a) Average daily FOI where shaded area represents ± s.e. of the mean. (b) Average daily effective reproductive ratio (Reff).
4. Discussion
Disease risk varied strongly with colony size, with proximity to and lesion severity of diseased neighbours, and through time, demonstrating heterogeneity in disease risk for sessile organisms. FOI showed the expected exponential increase in disease cases followed by a decline as susceptible colonies were exhausted, indicating that evaluating an average FOI (rather than tracking FOI through time) could lead to underestimates of population-level transmission.
The positive relationship between disease risk and proximity to and severity of diseased neighbours indicates that MCX is probably an infectious disease that spreads locally and has density-dependent transmission. The optimal range for disease transmission to occur (5–20 m) suggests that MCX spreads via local water movement or by a vector with a small home range. The spatial dependence of disease transmission may help explain why MCX briefly reached epidemic levels before rapidly diminishing. In acute outbreaks, Reff decreases when the number of susceptible individuals is depleted below some threshold, forcing the outbreak to die out. In this study, the total number of susceptible colonies was never exhausted (figure 1); however, the local pool of susceptible colonies (between 5 and 20 m) was partially depleted by the peak of the outbreak (figure 3). Other processes that could have contributed to these patterns include a saturation point for infectious particles in the water column, a change in virulence over time, and/or a change in environmental conditions that led to lower susceptibility or higher disease resistance. For example, the El Niño-like conditions followed by a heavy rainfall event in Kāne‘ohe Bay could have increased host susceptibility prior to the MCX outbreak.
Figure 3.
Susceptible infected plot. Outbreak trajectories of total susceptible colonies (grey line), locally susceptible colonies (located between 5 and 20 m of each focal colony; blue line) and infected colonies (red line).
Like diseases in many other marine taxa, MCX risk increased with host size [3,20,21]. A positive relationship between disease risk and host size may arise because larger colonies have greater surface area exposed to pathogens in the water column and filter more seawater compared with smaller colonies, or because of colony senescence [4,10,22]. If the surface area hypothesis were true, we would expect to see lesions on different areas of the colony, at a rate proportional to surface area. Since we never observed multiple lesions on a single colony, our research does not support this hypothesis. By contrast, we often observed lesions at the basal portion of colonies, lending support to the water filtration hypothesis. Host senescence is another plausible hypothesis for the observed patterns; however, because M. capitata fragments easily, it is difficult to infer age from size.
While the MCX outbreak affected over half the sample population, most colonies survived, which could have positive and negative implications for reef recovery. In this study, most of the affected colonies experienced less than 25% partial mortality. Partial mortality is a survival mechanism for coral, allowing the rest of the colony to facilitate growth and persistence (rather than putting energy into wound repair and recolonization) following stressful events [23]. However, partial mortality may impede reef recovery by increasing coral vulnerability to predation, environmental stress and opportunistic pathogens [24], and by reducing growth rates and reproductive output [25]. Our results also complement a simulation study of white plague spreading through a heterogeneous reefscape, and could lead to similar changes in coral cover and/or composition through time [26]. For instance, MCX could alter M. capitata size-frequency distributions by disproportionately affecting large colonies, which could have cascading effects throughout the ecosystem. On the other hand, Montipora capitata has uniformly low dispersal in Hawaii, with over 90% self-recruitment within sites [18], which could aid local reef recovery.
Although marine populations are often characterized as ‘open’ systems, host structure, habitat patches and hydrodynamics may create barriers restricting pathogen spread. Thus, even though some notable marine disease outbreaks have caused widespread mass mortalities (e.g. abalone [27]; seastars [4]), it is unclear whether these patterns are typical. Our study indicates that marine disease spread can be highly localized, and, coral may be a good model system to study disease dynamics for sessile and/or clonal marine organisms.
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank the University of Hawai‘i Diving Safety Program and many field assistants for logistical and field support. Maps were created in ArcMap 10.4.1. We thank two anonymous reviewers for valuable feedback on our manuscript.
Ethics
Fieldwork was conducted in an ethical manner in accordance with the Guidelines for the Use of Animals in Research. No animals were touched or harmed in this study.
Data accessibility
Primary data and R code available in supplementary files.
Authors' contributions
J.M.C. and M.J.D. participated in the study design and collection of field data. All authors participated in data analysis and interpretation, wrote the manuscript and gave final approval for publication.
Competing interests
We have no competing interests.
Funding
We would like to thank the PADI Foundation and Charles H. and Margaret B. Edmondson Research Fund for financial support for this project.
References
- 1.Mccallum HI, Kuris A, Harvell CD, Lafferty KD, Smith GW, Porter J. 2004. Does terrestrial epidemiology apply to marine systems? Trends Ecol. Evol. 19, 585–591. ( 10.1016/j.tree.2004.08.009) [DOI] [Google Scholar]
- 2.Bruno JB, Ellner SP, Vu I, Kim K, Harvell CD. 2011. Impacts of aspergillosis on sea fan coral demography: modeling a moving target. Ecol. Monogr. 81, 123–139. ( 10.1890/09-1178.1) [DOI] [Google Scholar]
- 3.Eisenlord ME, et al. 2016. Ochre star mortality during the 2014 wasting disease epizootic: role of population size structure and temperature. Phil. Trans. R. Soc. B 371, 20150212 ( 10.1098/rstb.2015.0212) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Irikawa A, Casareto BE, Suzuki Y, Agostini S, Hidaka M van Woesik R. 2011. Growth anomalies on Acropora cytherea corals. Mar. Poll. Bull. 62, 1702–1707. ( 10.1016/j.marpolbul.2011.05.033) [DOI] [PubMed] [Google Scholar]
- 5.Ellis RP, Parry H, Spicer JI, Hutchinson TH, Pipe RK, Widdicombe S. 2011. Immunological function in marine invertebrates: responses to environmental perturbation. Fish Shellfish Immunol. 30, 1209–1222. ( 10.1016/j.fsi.2011.03.017) [DOI] [PubMed] [Google Scholar]
- 6.Acevedo-Whitehouse K, Gulland F, Greig D, Amos W. 2003. Inbreeding: disease susceptibility in California sea lions. Nature 422, 35 ( 10.1038/422035a) [DOI] [PubMed] [Google Scholar]
- 7.Gachon CMM, Strittmatter M, Müller DG, Kleinteich J, Küpper FC. 2009. Detection of differential host susceptibility to the marine oomycete pathogen Eurychasma Dicksonii by real-time PCR: not all algae are equal. Appl. Environ. Microbiol. 75, 322–328. ( 10.1128/AEM.01885-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith DL, Lucey B, Waller LA, Childs JE, Real LA. 2002. Predicting the spatial dynamics of rabies epidemics on heterogeneous landscapes. Proc. Natl Acad. Sci. USA 99, 3668–3672. ( 10.1073/pnas.042400799) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kauffman MJ, Jules ES. 2006. Heterogeneity shapes invasion: host size and environment influence susceptibility to a nonnative pathogen. Ecol. Appl. 16, 166–175. ( 10.1890/05-0211) [DOI] [PubMed] [Google Scholar]
- 10.Jolles AE, Sullivan P, Alker AP, Harvell CD. 2002. Disease transmission of aspergillosis in sea fans: inferring process from spatial pattern. Ecology 83, 2373–2378. ( 10.1890/0012-9658(2002)083%5B2373:DTOAIS%5D2.0.CO;2) [DOI] [Google Scholar]
- 11.Zvuloni A, Artzy-Randrup Y, Stone L, Kramarsky-Winter E, Barkan R, Loya Y. 2009. Spatio-temporal transmission patterns of black-band disease in a coral community. PLoS ONE 4, e4993 ( 10.1371/journal.pone.0004993) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zvuloni A, Artzy-Randrup Y, Katriel G, Loya Y, Stone L. 2015. Modeling the impact of white-plague coral disease in climate change scenarios. PLoS Computat. Biol. 11, e1004151 ( 10.1371/journal.pcbi.1004151) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lentz JA, Blackburn JK, Curtis AJ. 2011. Evaluating patterns of a white-band disease (WBD) outbreak in acropora palmata using spatial analysis: a comparison of transect and colony clustering. PLoS ONE 6, e21830 ( 10.1371/journal.pone.0021830) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Concepcion GT, Baums IB, Toonen RJ. 2014. Regional population structure of montipora capitata across the Hawaiian Archipelago. Bull. Mar. Sci. 90, 257–275. ( 10.5343/bms.2012.1109) [DOI] [Google Scholar]
- 15.Work T. 2015. Diagnostic case report. Honolulu, HI: US Geological Survey Biological Resources Division, National Wildlife Health Center. [Google Scholar]
- 16.Sheridan C, Baele JM, Kushmaro A, Frejaville Y, Eeckhaut I. 2014. Terrestrial runoff influences white syndrome prevalence in SW Madagascar. Mar. Environ. Res. 101, 44–51. ( 10.1016/j.marenvres.2014.08.003) [DOI] [PubMed] [Google Scholar]
- 17.Höhle M. 2009. Additive-multiplicative regression models for spatio-temporal epidemics. Biom. J. 51, 961–978. ( 10.1002/bimj.200900050) [DOI] [PubMed] [Google Scholar]
- 18.Hens N, Aerts M, Faes C, Shkedy Z, Lejeune O, Van Damme P, Beutels P. 2010. Seventy-five years of estimating the force of infection from current status data. Epidemiol. Infect. 138, 802–812. ( 10.1017/S0950268809990781) [DOI] [PubMed] [Google Scholar]
- 19.Muench H. 1934. Derivation of rates from summation data by the catalytic curve. J. Am. Stat. Assoc. 29, 25–38. ( 10.1080/01621459.1934.10502684) [DOI] [Google Scholar]
- 20.Dube D, Kim K, Alker AP, Harvell CD. 2002. Size structure and geographic variation in chemical resistance of sea fan corals gorgonia ventalina to a fungal pathogen. Mar. Ecol. Prog. Ser. 231, 139–150. ( 10.3354/meps231139) [DOI] [Google Scholar]
- 21.Groner ML. et al. 2014. Host demography influences the prevalence and severity of eelgrass wasting disease. Dis. Aquat. Organ. 108, 165–175. ( 10.3354/dao02709) [DOI] [PubMed] [Google Scholar]
- 22.Muller E, Van Woesik R. 2014. Genetic susceptibiity, colony size, and water temperature drive white-pox disease on the coral Acropora palmata. PLoS ONE 9, e110759 ( 10.1371/journal.pone.0110759) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lasker HR, Coffroth MA. 1999. Responses of clonal reef taxa to environmental change. Amer. Zool. 39, 92–103. ( 10.1093/icb/39.1.92) [DOI] [Google Scholar]
- 24.Guzner B, Novplansky A, Shalit O, Chadwick NE. 2010. Indirect impacts of recreational scuba diving: patterns of growth and predation in branching stony corals. Bull. Mar. Sci. 86, 727–742. [Google Scholar]
- 25.Lirman D. 2000. Fragmentation in the branching coral Acropora palmata (Lamarck): growth, survivorship, and reproduction of colonies and fragments. J. Exp. Mar. Biol. Ecol. 251, 41–57. ( 10.1016/S0022-0981(00)00205-7) [DOI] [PubMed] [Google Scholar]
- 26.Brandt ME, McManus JW. 2009. Dynamics and impact of the coral disease white plague: insights from a simulation model. Dis. Aquat. Organ. 87, 117–133. ( 10.3354/dao02137) [DOI] [PubMed] [Google Scholar]
- 27.Lafferty KD, Kuris AM. 1993. Mass mortality of abalone Haliotis cracherodii on the California channel islands: tests of epidemiological hypotheses. MEPS 96, 239 ( 10.3354/meps096239) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Primary data and R code available in supplementary files.