The cause of diarrheal disease is usually determined by screening for several microorganisms by various methods, and sole detection is used to assign the agent as the cause of disease. However, it has become increasingly clear that many infections are caused by coinfections with several pathogens and that the dose of the infecting pathogen is important. We quantified the absolute numbers of enterotoxigenic E. coli (ETEC) and Vibrio cholerae directly in diarrheal fluid. We noted several events where both pathogens were found but also a large dose dependency. In three samples, we found ETEC as the only pathogen sought for. These isolates belonged to globally distributed ETEC clones and were the dominating species in stool with active toxin expression. This suggests that certain superior virulent ETEC lineages are able to outcompete the gut microbiota and be the sole cause of disease and hence need to be specifically monitored.
KEYWORDS: ETEC, Vibrio cholerae, diarrhea, enterotoxin, quantification
ABSTRACT
The bacterial pathogens enterotoxigenic Escherichia coli (ETEC) and Vibrio cholerae are major causes of diarrhea. ETEC causes diarrhea by production of the heat-labile toxin (LT) and heat-stable toxins (STh and STp), while V. cholerae produces cholera toxin (CT). In this study, we determined the occurrence and bacterial doses of the two pathogens and their respective toxin expression levels directly in liquid diarrheal stools of patients in Dhaka, Bangladesh. By quantitative culture and real-time quantitative PCR (qPCR) detection of the toxin genes, the two pathogens were found to coexist in several of the patients, at concentrations between 102 and 108 bacterial gene copies per ml. Even in culture-negative samples, gene copy numbers of 102 to 104 of either ETEC or V. cholerae toxin genes were detected by qPCR. RNA was extracted directly from stool, and gene expression levels, quantified by reverse transcriptase qPCR (RT-qPCR), of the genes encoding CT, LT, STh, and STp showed expression of toxin genes. Toxin enzyme-linked immunosorbent assay (ELISA) confirmed active toxin secretion directly in the liquid diarrhea. Analysis of ETEC isolates by multiplex PCR, dot blot analysis, and genome sequencing suggested that there are genetic ETEC profiles that are more commonly found as dominating single pathogens and others that are coinfectants with lower bacterial loads. The ETEC genomes, including assembled genomes of dominating ETEC isolates expressing LT/STh/CS5/CS6 and LT/CS7, are provided. In addition, this study highlights an emerging important ETEC strain expressing LT/STp and the novel colonization factor CS27b. These findings have implications for investigations of pathogenesis as well as for vaccine development.
IMPORTANCE The cause of diarrheal disease is usually determined by screening for several microorganisms by various methods, and sole detection is used to assign the agent as the cause of disease. However, it has become increasingly clear that many infections are caused by coinfections with several pathogens and that the dose of the infecting pathogen is important. We quantified the absolute numbers of enterotoxigenic E. coli (ETEC) and Vibrio cholerae directly in diarrheal fluid. We noted several events where both pathogens were found but also a large dose dependency. In three samples, we found ETEC as the only pathogen sought for. These isolates belonged to globally distributed ETEC clones and were the dominating species in stool with active toxin expression. This suggests that certain superior virulent ETEC lineages are able to outcompete the gut microbiota and be the sole cause of disease and hence need to be specifically monitored.
INTRODUCTION
Diarrhea is the second leading cause of mortality in children younger than 5 years worldwide (1). Two of the major causes of severe diarrhea in low-resource countries, and for travelers to these countries, are the bacterial pathogens Vibrio cholerae (O1), which causes epidemic cholera, and enterotoxigenic Escherichia coli (ETEC) (2–4). These Gram-negative bacteria both infect the small intestine where they, by means of colonization factors (CFs), i.e., specific proteinaceous polymers, attach to the surface of the endothelial cells. An important CF for V. cholerae is the toxin-coregulated pilus (TCP) (5–7). For ETEC, more than 25 various CFs have to date been identified, of which the globally most prevalent ones are the CFA/I and the coli surface antigens 1 to 6 and 21 (CS1 to CS6 and CS21, respectively) (8–10). In some geographical areas, other coli surface antigens are also frequent, for example, CS7, CS14, and CS17 in Bangladesh (11). Both V. cholerae and ETEC are also defined by their toxin secretions. V. cholerae secretes cholera toxin (CT), whereas ETEC secretes heat-labile toxin (LT) and heat-stable toxin (ST). ST from ETEC strains pathogenic to humans consists of two variants, STh and STp (12, 13). The ETEC LT and the V. cholerae CT are in fact very similar in structure as well as function, and they cross-react immunologically (3, 14). These two toxins both bind to GM1 receptors on intestinal epithelial cells and trigger increased levels of intracellular cyclic AMP (cAMP), activation of protein kinase A (PKA), and subsequent activation of the ion channel cystic fibrosis transmembrane conductance regulator (CFTR). These steps of activation result in massive outflow of water and electrolytes from the epithelial cells with severe diarrhea as a result (3). The STs of ETEC cause diarrhea in a similar way, also deregulating the CFTR receptor, but by binding to guanylate cyclase receptors instead of GM1 receptors. For ETEC, individual isolates typically coexpress several CFs and/or toxins, such as CS5/CS6 with LT/STh and CS7 with LT. The genes encoding colonization factors and toxins are often located on the same plasmids (15, 16). Whole-genome sequencing has revealed that some ETEC strains carrying specific combinations of toxins and CFs on a conserved genomic background have spread globally, leading to the development of ETEC clades expressing certain CF-toxin combinations that are stable over time (10).
V. cholerae and ETEC are similar at symptom level, and in multipathogen diarrheas, ETEC is often underestimated or even missed (2). Coinfections with two or more diarrhea-causing pathogens are common. For example, it has been shown that for European travelers to the tropics, coinfections with various pathogens, including ETEC, enteroaggregative E. coli (EAEC), Shigella, and norovirus, were found in 61% of the diarrhea-suffering patients (17). With the development of new, molecular biology-based, diagnostic techniques, the probability of detecting all diarrhea-causing agents with high sensitivity increases (2). With these new techniques, underestimation of the role of ETEC in gastroenteritis outbreaks has been highlighted (18) and disease clinically diagnosed as cholera has been rediagnosed as ETEC after virulence determination using PCR (19). The incidence of coinfections by V. cholerae with ETEC is thus more common than previously thought. As an example, for patients attending the hospital ward at the International Centre for Diarrhoeal Diseases Research in Bangladesh (icddr,b), 2 to 16% of the patients were found positive for both V. cholerae and ETEC (11, 20). These findings prompted us to study the relationship between ETEC and V. cholerae in cases of severe diarrheal coinfections further. In particular, we aimed to study the CF/toxin profiles of the ETEC strains, both in single infection and in coinfection. Using qualitative culture, real-time quantitative PCR (qPCR) of the toxin profiles, and full-genome sequencing, directly in the diarrheal stool sample as well as in cultured isolated bacterial strains, new information about ETEC colonization factor/toxin profiles in coinfections with other E. coli strains and/or with V. cholerae could be retrieved. These pieces of information might be of importance for studies of ETEC virulence and pathology as well for the development of vaccines against ETEC.
RESULTS
Coinfections with ETEC, V. cholerae, and other enterobacteria are frequent in watery diarrhea.
During the diarrheal peak period in March to April 2006, 35 surveillance stool samples were randomly collected from children and adults seeking care for diarrheal disease at the hospital ward at icddr,b in Dhaka, Bangladesh. Seven collected samples did not meet the inclusion criteria and were therefore not analyzed. A total of 28 samples from children (n = 11, ages 2 to 17 years, median age of 12 years) and adults (n = 17, ages 18 to 54 years, median age of 35 years) were included in the study. The routine standard analyses for detection and surveillance of diarrheal pathogens at the icddr,b are culture analysis on MacConkey agar plates followed by multiplex PCR analysis on 3 to 10 pooled colonies for pathogenic E. coli and culture on selective taurocholate-tellurite-gelatin agar (TTGA) plates for detection of V. cholerae. Stool samples are by routine also tested for Salmonella spp. and Shigella spp. on selective plates. By use of these methods, 18 MacConkey agar plates were found positive for growth of E. coli-like bacteria, either as only E. coli (4 of the 18 plates) or as mixtures of E. coli with other bacteria, i.e., Klebsiella and other non-lactose fermenters, and/or V. cholerae, as presented in Table 1. Of the original 28 samples, 17 were positive for V. cholerae, which corresponds to 60%. All V. cholerae-positive samples were also found positive for other enterobacteria. None of the samples contained Salmonella or Shigella. Furthermore, the 18 samples positive for E. coli bacteria were tested for the presence of ETEC by multiplex PCR, showing that ETEC was present in 8 of the samples, corresponding to 29% of all the collected samples. Coinfections with ETEC and V. cholerae were found in 4 samples, corresponding to 14% of all the collected samples (Table 1). No significant correlation with age groups was found for either single infections or coinfections. Of the seven samples for which only E. coli colonies were detected on the MacConkey plates, four (samples 4, 5, 16, and 18) showed no growth of V. cholerae. These samples tested positive for ETEC toxins by use of multiplex PCR, suggesting that ETEC was the major pathogen.
TABLE 1 .
Diarrheal stool samples and presence of E. coli, ETEC, and V. cholerae by culture analysis
| Stool sample/ collection no. |
Date collected (mo-day) |
Diarrhea type | MacConkey culturea |
V. cholerae cultureb |
ETEC toxin + CFc |
ETEC % of total E. colid |
|---|---|---|---|---|---|---|
| 1/001 | 3-20 | Rice water | E. coli mix | Inaba | STh CF− | ND |
| 2/002 | 3-20 | Rice water | E. coli mix | Inaba | ||
| 3/005 | 3-21 | Rice water | E. coli mix | |||
| 4/007 | 3-21 | Brown watery | E. coli pure | LT CS7 | 100 | |
| 5/008 | 3-22 | Brown watery | E. coli pure | STh/LT CS5/CS6 | 100 | |
| 6/009 | 3-22 | Yellow watery | E. coli pure | Ogawa | ||
| 7/013 | 3-27 | Yellow | E. coli pure | Ogawa | ||
| 8/014 | 3-27 | Rice water | E. coli mix | |||
| 9/018 | 3-30 | Yellow water | E. coli mix | Ogawa | STp CF− | 60 |
| 10/020 | 3-30 | Yellow water | E. coli mix | Inaba | ||
| 11/023 | 4-2 | Yellow water | E. coli pure | Ogawa | ||
| 12/024 | 4-3 | Yellow water | E. coli mix | Ogawa | ||
| 13/025 | 4-3 | Yellow water | E. coli mix | |||
| 14/EN 94 | 4-3 | Yellow water | E. coli mix | Ogawa | ||
| 15/026 | 4-4 | Rice water | E. coli mix | Ogawa | LT CF− | 0 |
| 16/027 | 4-4 | Rice water | E. coli pure | STp/LT CF− | 66 mixe | |
| 17/030 | 4-5 | Rice water | E. coli mix | Ogawa | LT CF− | 4 |
| 18/033 | 4-5 | Brown rice water | E. coli pure | STp/LT CF− | 96 |
Culture on MacConkey agar plates; mix, other nonfermenting and fermenting colonies detected indicating the presence of Klebsiella and other species; E. coli pure, only E. coli-like colonies detected.
Presence of V. cholerae detected by growth on TTGA plates; the serotypes Inaba and Ogawa were detected by agglutination tests.
Toxin and colonization factor profiles were determined on isolated colonies tested by multiplex PCR for toxin genes and CF dot blot analysis (48).
Percent ETEC per total E. coli bacteria was determined by toxin multiplex PCR performed on 50 individual E. coli colonies from MacConkey plates. (The number of positive colonies was divided by 50 tested E. coli colonies for each sample.) ND, not determined.
Sample 027 was a mix of several ETEC strains; of the 50 tested, 33 were ETEC, 4 were only LT postive, 2 were only STh positive, 20 were STp and LT positive, 1 was LT and STh positive, and 6 were positive for STh, STp, and LT.
Determination of the ratio of ETEC CFU to total E. coli CFU in the diarrhea samples.
ETEC was detected in the samples where only E. coli colonies, and no other lactose-fermenting bacteria, grew on MacConkey agar. To determine the ETEC frequency in these samples, the numbers of ETEC CFU per total E. coli CFU were examined. The total numbers of E. coli in the samples were determined by quantitative culturing using serial dilutions. Isolated colonies, 50 in total, were picked randomly from the dilution plates and analyzed by toxin multiplex PCR. As presented in Table 1, the ETEC percentage of total number of E. coli-like CFU was then calculated by dividing the ETEC-positive colonies by 50, i.e., the total number of analyzed colonies. For two of the samples, sample 4 and sample 5, 50 out of 50 colonies were determined as positive for either LT (sample 4) or both LT and STh (sample 5), suggesting that they were pure ETEC. For sample 18, 96% of the colonies were determined as ETEC expressing LT and STp. For sample 9, 60% of the E. coli strains were STp positive, while in sample 17, only 4% were LT positive. The results from the individual colonies largely agreed with initial toxin profiles determined for the diarrheal samples, with the exception of sample 16. This sample was originally scored as an E. coli LT/STp-only infection, but in the analysis of the 50 colonies, it was found to contain multiple ETEC toxin profiles (Table 1). In addition, the fraction of ETEC in E. coli in the sample was 66%, indicating a mixed infection with several ETEC strains and other E. coli strains. Regarding sample 1, this sample was not tested, and in sample 15, the original ETEC isolate could not be found among the tested 50 colonies. In total, six representative isolates, E2264 to E2269, were collected from the remaining six ETEC-positive samples and stored in freeze medium. The colonization factor profiles of these strains were then tested using dot blot and PCR analysis, showing the presence of CS7 in sample 4 and CS5/CS6 in sample 5. For the other ETEC strains, the CF profiles could not be determined (Table 1).
qPCR quantification of ETEC and V. cholerae toxin genes.
Since variations in ETEC frequency occur in diarrheal stool, we next sought to determine the absolute numbers of ETEC and V. cholerae per milliliter of watery stool. The DNA copy numbers of ETEC and V. cholerae were determined in the 18 E. coli-positive stool samples by use of qPCR analysis and primers specific for estA1 STp, estA2 to estA4 STh, eltB LT, and ctxB CT together with standard curves of known copy numbers. Gene loci for all four toxin genes were detected in a majority of the samples, and the amounts varied between 0 copies and 2 × 108 copies per ml, as presented in Table 2. Higher numbers of toxin gene copies were found in samples that tested positive for ETEC toxins than in samples that were negative for ETEC in the previous culture analyses. Samples 4, 5, 16, and 18, which were all positive for LT ETEC, contained between 1 × 107 and 6 × 107 LT gene copies per ml. Furthermore, sample 4 and sample 5 contained, in comparison with the others, very low copy numbers (0 and 150 copies per ml stool, respectively) of the V. cholerae CT gene. Hence, these samples likely represent true ETEC-only diarrheas. The two other LT ETEC-positive samples, sample 15 and sample 17, contained few or no LT ETEC bacteria per total amount of E. coli in the culture analysis, and the levels of ETEC LT gene copies detected by qPCR were correspondingly 3 orders of magnitude lower (2 × 104 to 3 × 104) than in the other ETEC-positive samples. The copy number of eltB was additionally found at levels between 103 and 105 copies per ml in the samples that tested negative for ETEC in culture.
TABLE 2 .
Absolute gene copy numbers for ctxB (CT), eltB (LT), estA2 to estA4 (STh), and estA1 (STp) calculated by qPCR on DNA extracted directly from 1 ml of liquid diarrheaa
| Sample (presence of ETEC and/or V. cholerae toxins) |
Copy no./ml |
CFU/ml |
||||
|---|---|---|---|---|---|---|
| ctxB CT | eltB (LT) |
estA2 to estA4 (STh) |
estA1 (STp) | Total E. coli | ETEC (% of total E. coli) |
|
| 1/001 (STh/CT) | 7.79 × 106 | 8.6 × 104 | 2.97 × 106 | 0 | ND | ND |
| 2/002 (CT) | 3.88 × 107 | 5.9 × 104 | 8.5 × 103 | 0 | ND | |
| 3/005 (–) | 1.29 × 104 | 1.97 × 105 | 1.31 × 104 | 0 | 3.0 × 105 | |
| 4/007 (LT) | 0 | 3.75 × 107 | 3.6 × 103 | 0 | 6.7 × 108 | 6.7 × 108 (100) |
| 5/008 (LT + STh) | 1.51 × 102 | 1.09 × 107 | 2.12 × 107 | 0 | 2.7 × 107 | 2.7 × 107 (100) |
| 6/009 (CT) | 1.1 × 104 | 5.29 × 104 | 4.4 × 103 | 0 | ND | |
| 7/013 (CT) | 8.91 × 105 | 1.75 × 103 | 4.0 × 102 | 0 | 5.5 × 107 | |
| 8/014 (–) | 7.6 × 101 | 1.52 × 104 | 3.1 × 103 | 0 | 7.8 × 106 | |
| 9/018 (STp/CT) | 1.55 × 106 | 1.10 × 104 | 4.65 × 103 | 4.3 × 103 | 1.0 × 107 | 6.0 × 106 (60) |
| 10/ 020 (few CT) | 3.74 × 102 | 5.95 × 103 | 1.07 × 104 | 9.0 × 102 | 2.5 × 107 | |
| 11/023 (CT) | 4.59 × 104 | 1.14 × 104 | 3.8 × 103 | 4.85 × 103 | 8.5 × 107 | |
| 12/024 (CT) | 6.68 × 105 | 1.5 × 104 | 3.45 × 103 | 8.2 × 103 | 8.0 × 105 | |
| 13/025 (–) | 3.42 × 102 | 6.0 × 103 | 1.3 × 103 | 3.35 × 103 | 8.0 × 106 | |
| 14/EN 94 (CT) | 5.46 × 104 | 4.4 × 103 | 3.0 × 103 | 3.95 × 103 | 2.0 × 105 | |
| 15/026 (LT/CT) | 1.74 × 108 | 2.78 × 104 | 8.55 × 103 | 2.04 × 104 | 2.5 × 107 | 0 |
| 16/027 (LT + STh + STp) | 3.62 × 102 | 4.25 × 107 | 7.15 × 107 | 2.5 × 104 | 7.0 × 106 | 4.62 × 106 (66) |
| 17/030 (LT/CT) | 1.2 × 105 | 1.78 × 104 | 2.05 × 103 | 5.55 × 103 | 9.6 × 104 | 3.84 × 103 (4) |
| 18/033 (LT + STp) | 5.23 × 105 | 6.25 × 107 | 9.5 × 102 | 1.83 × 106 | 6.3 × 107 | 6.05 × 107 (96) |
Copy numbers corresponding to samples with culture-positive V. cholerae and/or ETEC and corresponding toxin profiles are indicated in bold. Quantitative culture of total amount of E. coli in the liquid diarrhea samples was used to estimate total number of E. coli bacteria in samples. The percentage of ETEC CFU determined on multiplex PCR analysis of 50 individual colonies was used to calculate the assumed numbers of ETEC CFU per milliliter of diarrhea. ND, not determined.
The gene encoding heat-stable toxin STp was detected in the STp-positive sample 9 (9 × 103 copies) and sample 16 (2 × 104) and in high numbers in sample 18 (2 × 108), as presented in Table 2. Sample 18 was by quantitative culture and multiplex PCR determined to contain 6.3 × 107 E. coli bacteria per ml diarrheal fluid, and 96% of the colonies were LT and STp positive in culture analysis. Hence, the qPCR results and quantitative culture corroborate a dissemination concentration of 107 to 108 gene equivalents per ml diarrheal stool in this patient. A similar concentration was also found for sample 5, which contained 100% ETEC and a total E. coli count of 2.7 × 107 CFU per ml. The LT and STh gene copy numbers of this sample were 1 × 107 (eltB) and 2 × 107 (estA3 and estA4), respectively. The gene copy numbers for STh were for sample 16 estimated to be 7 × 107 and for sample 1 estimated to be 3 × 107, whereas all samples that were negative for STh in culture analyses showed levels of between 102 and 104 copies of estA2 to estA4 per ml (Table 2).
The gene counts for V. cholerae ctxB in the analyzed samples varied from none to almost 2 × 108 gene copies per ml (Table 2). Low (≤200 copies) or absent levels were found in sample 4 and sample 5, both detected as 100% ETEC. Low levels (≤400 copies) were also found in samples 8, 10, 13, and 16. For sample 10, only around 25 colonies in total of V. cholerae were detected on culture plates, which corroborates the low counts detected with qPCR. In the other samples with low copy numbers of ctxB, samples 8, 13, and 16, no growth of V. cholerae was consequently detected in culture analysis. The samples that scored positive for V. cholerae in culture showed gene copy numbers of between 104 and 108 per ml. In addition, sample 18, which was detected as V. cholerae negative by culture, also contained 5 × 105 ctxB copies per ml. Taken together, these results suggest that low levels of ETEC and V. cholerae are continuously present in a majority of diarrheal stools from patients and that these levels are difficult to detect by routine culture analyses.
Toxin production and secretion determined directly in diarrheal liquid stool samples by GM1-ELISA.
The diarrheal stool samples were further analyzed for the presence of the translated ST, LT, and CT by use of GM1 enzyme-linked immunosorbent assay (GM1-ELISA). Seven of the eight stool samples that had been scored as positive for ETEC were tested using GM1-ELISA and inhibition GM1-ELISA (Table 3). In the three tested samples that were culture positive for V. cholerae (samples 1, 9, and 15), CT was detected by LT-39, an antibody that detects both LT and CT, as well as CT-Wi monoclonal antibody (MAb), which is more specific for CT. Cholera toxin was also detected in sample 18, which was estimated to contain approximately 5 × 105 V. cholerae bacteria per ml by qPCR analysis but was V. cholerae negative in culture. The toxin ELISA results thus confirmed the qPCR results for this sample. Two of the samples, sample 4 and sample 5, were detected as virtually pure ETEC samples, with undetectable levels of V. cholerae toxin in qPCR. For sample 4, traces of LT were detected in the pellet fraction using the LT-specific MAb LT-80, but the amount was close to the lower detection limit of the assay. In sample 5, however, positive results were obtained using MAbs LT-80 and LT-39 but not MAb CT-Wi, suggesting that the toxin found indeed was LT and not CT. In addition, this sample was positive only in the bacterial supernatant fraction and not in the pellet, indicating that LT was actively secreted during infection. ST was detected in the supernatants of sample 1, sample 9, and sample 16, for which STh or STp had already been detected by toxin multiplex PCR. Sample 5, additionally, showed trace amounts of ST. Furthermore, ST was detected in sample 15, in both pellet and supernatant, whereas the same toxin was not detected by culture analysis followed by multiplex PCR, and only approximately 104 copies of estA2 to estA4 were detected by real-time PCR analysis (Table 2). In contrast, no ST was detected in sample 18, for which high levels of STp had been detected by both culture/multiplex PCR and real-time PCR analysis. The analyses show that toxins are present in diarrheal stool and distributed to the environment. In addition, both ETEC and V. cholerae evidently actively secrete LT and CT during acute infection and dissemination.
TABLE 3 .
ETEC and cholera toxins detected in ETEC-positive diarrhea samples from patientsa
| Stool sample |
Identified ETEC strain |
Cholera toxin |
ETEC toxin |
Amt of toxin (ng/ml) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| LT-39 |
LT-80 |
CT-Wi |
ST-1 |
||||||||
| Pellet | Supernatant | Pellet | Supernatant | Pellet | Supernatant | Pellet | Supernatant | ||||
| 1 | NR | CT | STh | 15.8 | 61.3 | ||||||
| 4 | E2264 | LT | Trace | Trace | |||||||
| 5 | E2265 | STh/LT | 25.2 | 24 | Trace | ||||||
| 9 | E2266 | CT | STp | 4.8 | 4.8 | 2.2 | 3.4 | 53 | |||
| 15 | NR | CT | LT | 24.2 | 24.2 | 22 | 40.3 | 26 | 15 | ||
| 16 | E2267 | STp/LT | 20.3 | ||||||||
| 17 | E2268 | CT | LT | ND | ND | ND | ND | ND | ND | ND | ND |
| 18 | E2269 | STp/LT | 4.9 | 24.2 | 4.9 | 16.8 | |||||
Production and secretion of the toxins were quantified by GM1-ELISA. The LT-39 MAb detects both CT and LT, while the other MAbs are specific for ST (both STh and STp), LT, and CT, respectively. The pellet fraction represents toxins associated with the bacterial fraction, and the supernatant represents the amount of secreted toxin in the sample. NR, not recovered; ND, not determined.
Gene expression of the ETEC and V. cholerae toxin genes.
Toxin levels in stool might not indicate active transcription and translation. To investigate whether active toxin transcription occurs in ETEC and V. cholerae in diarrheal stool, the expression of the toxin genes in the two pathogens was measured by extracting RNA from the bacterial pellet from liquid diarrheal samples and reverse transcription of the RNA to cDNA. The relative expression of the ctxB (CTB subunit) gene, the ETEC toxin genes estA2 to estA4 and estA1 (STh and STp, respectively), and eltB (LTB subunit) was determined in a fixed concentration of total RNA converted to cDNA (15 ng/PCR mixture). The mRNA expression of the CT-, LT-, and ST-encoding genes per 15 ng total cDNA was found to largely correspond to the toxin profiles, as seen in Table 4. Higher gene expression levels of the ctxB mRNA were found in samples 1, 9, and 12 and particularly in sample 15 compared to the other samples. These samples also had correspondingly high levels (2 × 105 to 2 × 108) of ctxB DNA copies (Table 2). The highest mRNA levels for eltB and the ST-encoding mRNAs were found in samples 1, 4, 5, 16, and 18, all confirmed to have large amounts of ETEC (Tables 1 and 2). For sample 15, no gene expression of ETEC mRNA toxin genes could be determined, whereas sample 8, for which no pathogen was found in culture, showed expression of eltB and estA2 to estA4. These results, together with the DNA data, suggest that an LT and STh-positive ETEC infection was missed in culture analysis in this patient. The results show that toxin gene expression levels generally correlate with numbers of the respective pathogen in the stool. Calculations of expressed gene copy numbers (mRNA) divided by genomic copy numbers (DNA), i.e., gene expression per genome equivalent, showed that toxin gene expression levels in ETEC and V. cholerae are similar.
TABLE 4 .
Gene copy numbers determined by RT-qPCRc
| Stool sample |
ETEC strain |
Cholera toxin |
ETEC toxin |
Copy no./15 ng of cDNA for gene: |
|||
|---|---|---|---|---|---|---|---|
| ctxB | eltB |
estA2 to estA4 |
estA1 | ||||
| 1/001 | xa | CT | STh | 118 | 5.6 | 92 | |
| 3/005 | 55 | ||||||
| 4/007 | E2264 | LT | 2,705 | ||||
| 5/008 | E2265 | STh/LT | 823 | 80,223 | |||
| 8/014 | 24 | 151 | |||||
| 9/018 | E2266 | CT | STp | 44 | |||
| 12/024 | CT | 167 | 0.2 | ||||
| 15/026 | xa | CT | LT | 1,880 | |||
| 16/027 | E2267 | STp/LTb | 227 | 3,969 | |||
| 17/030 | E2268 | CT | LT | 0.07 | 0.07 | 1.3 | |
| 18/033 | E2269 | STp/LT | 3.4 | 4,020 | 25,887 | ||
Isolate not recovered.
This sample was a mixture of several different ETEC isolates.
Correlation between toxin profile determined by multiplex PCR and detection of corresponding gene is indicated in bold.
Whole-genome sequencing of ETEC isolates.
The results presented above suggest that ETEC might be underestimated in cases of cholera-like liquid diarrhea, as well as that certain ETEC clones such as LT CS7 (sample 4) and LT STh CS5 plus CS6 (sample 5) can manifest as monocultures of a single clonal infection. In order to investigate the genetics of the collected ETEC strains and to be able to correlate the collected ETEC with worldwide ETEC infections, whole-genome sequence (WGS) analysis was performed. For isolates E2264 (sample 4) and E2265 (sample 5) PacBio sequencing was performed to gain even more complete information about the genetic details. These two strains were of specific interest since they were detected in ETEC-only infections and since they belong to ETEC lineages that have persisted over time and have spread globally (10). The four other recovered isolates, E2266 (sample 9), E2267 (sample 16), E2268 (sample 17), and E2269 (sample 18), were sequenced using Illumina MiSeq sequencing. The details of the sequencing are provided in Tables S1 to S4 in the supplemental material. The genomes of the six isolates were annotated and analyzed using CGE in silico multilocus sequence typing (MLST), PlasmidFinder, and plasmid MLST (pMLST) as well as ResFinder VirulenceFinder and ARG-annot. In addition, pathogenic E. coli virulence genes were identified using BLAST analysis.
Assembly and annotation statistics of the PacBio sequencing of E2264 and E2265. Download TABLE S1, DOCX file, 0.02 MB (16.8KB, docx) .
Copyright © 2018 Begum et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Assembly and annotation statistics of the Illumina sequencing of E2266 to E2269. Download TABLE S2, DOCX file, 0.02 MB (16.5KB, docx) .
Copyright © 2018 Begum et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ETEC virulence factors identified in the ETEC strains. Download TABLE S3, DOCX file, 0.02 MB (17.1KB, docx) .
Copyright © 2018 Begum et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Antibiotic resistance genes found in the ETEC strains. Download TABLE S4, DOCX file, 0.02 MB (17KB, docx) .
Copyright © 2018 Begum et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In silico MLST confirmed that E2265 (sample 5) belongs to the major ETEC lineage 5 (ST-443) and that E2264 belongs to the novel ST-5305, a sublineage within ETEC lineage 3 (10). The STs identified for E2266 (sample 9), E2267 (sample 16), E2268 (sample 17), and E2269 (sample 18) were ST-226, ST-100, ST-5474, and ST-4493, respectively. A search in the Achtman MLST database (https://enterobase.warwick.ac.uk/warwick_mlst_legacy) revealed that ST-226 seemingly is associated with enteroaggregative E. coli (EAEC) as well as with CS26-positive ETEC (21). ST-100 (E2267, sample 16) has previously been found in porcine ETEC and also recently in human ETEC isolates (22). ST-5474 and ST-4493 have, in contrast, not previously been associated with ETEC. ST-4493 has been found in environmental E. coli isolates, whereas ST-5474 was not recorded in the Achtman database.
The genomes were furthermore searched for CF and other virulence genes, as well as for antibiotic resistance. The E2264 (sample 4) isolate was determined as LT/CS7 ETEC and also showed predicted antibiotic resistance to sulfonamide (sul2), beta-lactams (ampC and blaTEM-1b), tetracycline (tetB), trimethoprim (dfrA8), and streptomycin (strAB). The E2269 (sample 18) isolate was shown to harbor the LT, STp, and CS27b virulence genes. The last is a member of the CS18/CS18-like family of emerging new putative ETEC colonization factors (21, 22). E2269, additionally, harbored putative resistance genes for sulfonamide (sul2), beta-lactams (ampC and blaTEM-1b), tetracycline (tetA-like), trimethoprim (dfrA1), erythromycin [mph(A)-like], and streptomycin (strAB and aadA1). For isolate E2265, which was detected as an LT/STh/CS5/CS6 ETEC, the CF profile could be verified, and for isolate E2268, a CS13/CS23-like colonization factor could be detected. For both E2265 and E2268, no predicted antibiotic resistance genes were found using ResFinder, while ampC was detected by ARG-annot, indicating chromosomally encoded ampicillin resistance. E2266, originally scored as an STp ETEC, was predicted to be resistant to beta-lactams (ampC and blaTEM-1b), trimethoprim (dfrA1), and erythromycin [mph(A)-like]. The WGS data analysis could not, however, confirm any virulence genes for this isolate. Moreover, E2267, scored as LT STh, was predicted to be resistant to beta-lactams (ampC), streptomycin (aadA1), and trimethoprim (dfrA1), and the colonization factor was determined as CS14. Furthermore, all strains, except E2268, contained the plasmid segregation protein ParM, which has been identified as ETEC specific (23).
PacBio analysis of E2264 and E2265 revealed large virulence plasmids and additional plasmids with transfer systems and antibiotic resistance.
Since two stool samples, sample 4 and sample 5, contained 100% ETEC of two commonly isolated lineages of ETEC, PacBio sequencing was employed to further analyze these isolates. The PacBio analysis revealed that ETEC 2264 (sample 4), in addition to the chromosome, contained 3 plasmids. The largest plasmid (E2264_p112045) had a size of 112,045 bp and contained 128 putative open reading frames (ORFs). Among these, the ORFs containing the tra genes traD and traI, which may have helicase activity, as well as the genes encoding the heat-labile enterotoxin A and B subunits for production of LT (orf66 and orf67) and the CS7 operon subunit precursor (orf75 to orf81), were found. The distance between LT and CS7 was 3,986 bp, and the sequence contained two conserved, hypothetical proteins and four transposase genes. The six CS7 operon genes found were, in 5′ to 3′ direction, positioned in the order D, F, E, C, B, and A. The second largest plasmid (E2264_p77345) of ETEC 2264 was 77,345 bp long and contained 93 ORFs. This plasmid contained a high number of tra genes and antibiotic resistance genes. The tra genes, which are involved in conjugative transfer of plasmids between bacteria, encountered were traA, traD, traE, traG, traH, traK, traL, traM, traN, traP, traQ, traT, traU, traV, traX, and traY. Among these genes are the genes encoding the pilus subunit (traA), genes for regulation of traA, and genes for pilus assembly as well as genes for nicking and unwinding of DNA (24–26). In addition, six ORFs involved in antibiotic resistance were found: ones for resistance to tetracycline (orf63), trimethoprim (orf81), beta-lactams (orf83), and sulfonamide (orf87) and finally strA (orf88) and strB (orf89), coding for streptomycin resistance. The third plasmid (E2264_p45777) of ETEC 2264 was 45,777 bp long and contained 45 ORFs. This plasmid contained the gene encoding the secreted autotransporter serine protease EatA (ETEC autotransporter A) (orf25) (27). Sequence comparison of EatA using BLAST (NCBI) demonstrated high homology with a vast number of sequences of E. coli origin (97 to 99% homology with 7 samples, ≥71% homology with 84 samples) and also with a few sequences from Shigella.
ETEC 2265 contained two plasmids in addition to the chromosome. The largest plasmid, E2265_p142359, was 142,359 bp long and harbored 189 ORFs, including genes encoding important virulence factors like eatA (orf66), the csfA to -F operon encoding CS5 (orf79 to orf84), the cssABCD operon encoding CS6 (orf96 to orf99), and the estA3 and estA4 gene encoding STh (orf107). The plasmid also contained an aatPABCD operon encoding a membrane transporter initially described in enteroaggregative E. coli (EAEC) and a cexA-like gene located directly upstream of aatPABC (28). Plasmid E2265_p142359 also contained several tra genes: traM, traJ, traA, traL traE, traY, traK, traB, and traP. The second plasmid, E2265_p88757, was 88,757 bp long and contained 101 ORFs, including the eltAB operon (orf81 and orf82), as well as a high number of tra genes: traM, traA, traL, traE, traK, traB, traV, traC, trbL, traW, traU, trbC, traN, traF, trbB, traH, traG, traS, traT, traD, traI, and traX.
Scoary analysis of the genomes did not reveal any unique single-pathogen infection profiles for the ETEC.
The results indicate that three of the diarrheal samples collected in this study, E2264, E2265, and E2269, might be true ETEC infections without other coinfecting pathogens. In order to investigate whether these isolates differed from the three isolates with ETEC found as mixed infections with other E. coli strains and/or with V. cholerae (E2266, E2267, and E2268), the pan-, core, and accessory genomes were determined for the six sequenced isolates (Fig. 1). The pan-genome of the six isolates comprised 7,537 genes; of these, 3,502 genes were common to all isolates and constituted the core genome. Scoary analysis to determine if genes were significantly associated with the three ETEC-only infections did not reveal any significant results. The only differences found were larger genomes of E2264, E2265, and E2269 than of isolates E2266, E2267, and E2268 (see Table S1 in the supplemental material).
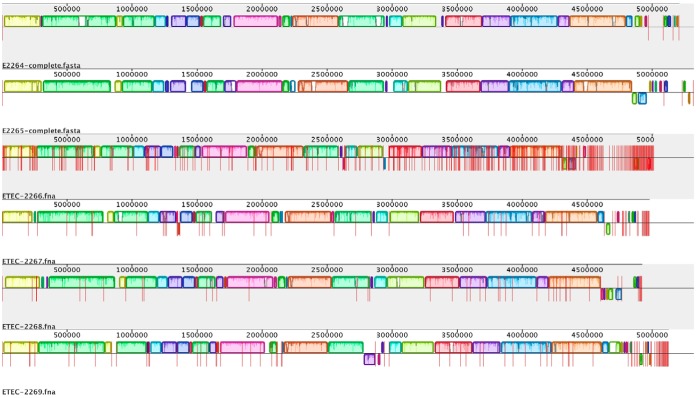
FIG 1 .
Mauve analysis of the six ETEC isolates E2264, E2265, E2266, E2267, E2268, and E2269. ETEC isolates E2264 and E2265 were PacBio sequenced and contain one chromosome and three and two plasmids, respectively, that are separated by lines in the figures. The other four ETEC isolates were sequenced by Illumina sequencing with individual contigs separated by lines in the figure. Genome analysis revealed conserved chromosomal organization, as represented by colors in the figure. The total pan-genome contained 7,537 genes, and the core genome consisted of 3,502 genes.
DISCUSSION
In this study, the prevalence and virulence factor profiles of ETEC and V. cholerae in watery stool from patients with cholera-like diarrhea were investigated by use of genetic sequencing and molecular analysis as well as traditional culture methods. In total, 28 samples were randomly collected at the hospital ward at icddr,b in Dhaka, Bangladesh. Of these, 17 tested positive for V. cholerae and 8 tested positive for ETEC (Table 1). Quantitative culture of bacterial load on MacConkey plates and analysis of ETEC toxin profile by PCR found that some samples were dominated by ETEC while others were mixed with other bacteria. Three of the ETEC samples, samples 4, 5, and 18, were detected as pure or almost pure ETEC infections, while samples 1, 9, 15, 16, and 17 were found as coinfectants with V. cholerae and/or with various undetermined E. coli and Enterobacteriaceae strains. Coinfections with various pathogens are quite common among patients with diarrhea (29, 30). For example, Paschke et al. have previously reported a prevalence of coinfections in as many as 60.5% of travelers to tropical and subtropical parts of the world who suffer from diarrhea as well as in 12.5% of the travelers to the same areas with no disease symptoms (17). The coinfection rate of the two pathogens investigated in this work was found to be 14%, which is higher than the 2% previously reported by Chowdhury et al. (11) but similar to the results from the work of Begum et al. (20) in the same geographical area.
Since both V. cholerae and ETEC cause massive diarrheas with a daily loss of several liters of fluid during the acute phase, we wanted to investigate the concentration of the bacteria in the stool samples. By use of quantitative real-time PCR, we could determine the gene copy number of the genes encoding the disease-causing V. cholerae and ETEC toxins, to up to 108 copies per ml (Table 2). These numbers were corroborated by the results of the quantitative culture and other studies reporting between 107 and 108 CFU of diarrheal pathogens per milliliter or gram of stool (31, 32) and up to 108 to 109 pathogen gene copies per gram of stool (30). Toxin gene copy numbers in ETEC have been determined to be between 1 and 16 copies per cell (32, 33), indicating that quantification of gene copies might overestimate bacterial load up to 10-fold. Regardless, shedding of 107 to 108 bacteria per ml of stool in patients, who can lose several liters of fluid per day, is probably one of the main factors contributing to the large epidemic outbursts of diarrhea caused by these pathogens.
One of the findings of this work was that the use of highly sensitive molecular methods detects low levels of ETEC and V. cholerae in a majority of the stool samples negative in culture (Table 2). These low levels are probably not causing symptoms and may constitute a background pathogenic microbiota in individuals living in areas of endemicity. They could also mean that the rates of coinfections are significantly higher than previously reported. Recent studies have, however, highlighted the importance of pathogen load for disease manifestation (30, 34, 35). High levels of ETEC in stool were linked to clinical manifestation of diarrhea in ETEC-challenged volunteers (35). In the same study, it was found that E. coli 16S (i.e., ETEC) dominated the microbiota in volunteers who developed diarrheal symptoms but was low in asymptomatic volunteers. Even though severe diarrheas caused by ETEC and by V. cholerae are very similarly manifested, V. cholerae is frequently identified as the disease-causing pathogen. The development of quantitative molecular diagnostics, including real-time PCR and other PCR methods for identification of the pathogens, may aid in finding the major disease-causing pathogen(s) in an infection (30, 36). With the use of these techniques, ETEC has been shown to be frequently underestimated (32). It might also turn out that a high proportion of the cholera cases are, in fact, mixed infections, since we detected E. coli or other species in all V. cholerae-positive stool samples.
Although the heat-labile CT and LT of V. cholerae and ETEC have similar actions, cholera is generally regarded to cause a more severe disease outcome. This has been explained by differences in the amount of toxin that the bacteria secrete. V. cholerae is generally considered to secrete most of its produced CT, while ETEC retains >50% of the LT intracellularly, either in the periplasm or associated with the membrane lipopolysaccharides (LPS) under laboratory conditions (37, 38). ST is also considered to cause a more severe disease outcome than LT (39, 40), and infection with LT ETEC has subsequently often been detected in asymptomatic patients (17). The reason for this difference is unknown but might be addressed by the difference in secretion. Thus, in order to study the toxin secretion in more detail, the 8 samples that by culture analysis were identified to contain ETEC were further analyzed for both ETEC and V. cholerae toxin production and secretion (Table 3). Using quantitative ELISA and monoclonal antibodies specific for the different toxins, it was seen that for V. cholerae, CT was detected both in the bacterial fraction and in the supernatant, indicating that approximately one-third of the toxin produced was actually retained in the bacteria (Table 3). For ETEC, similar levels of LT were detected both in the bacterial cells and in the supernatant, suggesting that LT is secreted to a relatively large extent. ST was mainly found as secreted toxin, but for one of the samples, ST was localized to the bacterial cells. This implies that the concept that V. cholerae always secretes CT and that ST is always secreted by ETEC, while most LT is retained in the periplasm of ETEC, does not hold true during infection. Our results indicate that disseminating ETEC and V. cholerae actively both transcribe and secrete toxins during shedding in diarrheal stool, which further supports the epidemic nature of these pathogens. Since ETEC toxins were also found secreted in the liquid diarrhea in levels that were equal to V. cholerae and since the toxin gene expression levels and pathogen loads were similar for the two pathogens, the explanation for the more severe nature of V. cholerae infections is still elusive. However, since these results are from only a few stool samples, further studies are needed.
Quantitative culture of bacterial load and analysis of ETEC toxin profile by PCR in this study found that some samples were dominated by ETEC while others were mixed with other bacteria. Six ETEC isolates were recovered and used for further analysis. Half of the ETEC isolates, E2264, E2265, and E2269, from samples 4, 5, and 18, respectively, were detected as pure or almost pure ETEC infections. The other isolates, E2266, E2267, and E2268, recovered from samples 9, 16, and 17, respectively, were found as coinfectants with V. cholerae and with various undetermined E. coli and Enterobacteriaceae strains (Table 1). All the 6 isolated strains were whole genome sequenced, using Illumina or Pacific Bioscience (PacBio) sequencing, in order to further analyze the genomic content.
Moreover, the toxin and colonization factor profiles of the isolated ETEC strains were investigated. For E2264 (sample 4) and E2265 (sample 5), the genetic analysis could confirm the CF profiles detected by dot blot analysis of CS7 and CS5/CS6, as well as toxin profiles of LT and LT/STh, respectively (Table 1). These toxin/CF profiles are common and widespread over the world (10). The E2264 isolate belongs to a subtype of ETEC lineage 3 expressing LT and CS7, which is frequently isolated globally (10, 20, 41). The E2265 isolate belonged to ETEC lineage 5, a clonal group that expresses the toxins LT and STh and colonization factors CS5 and CS6. Recent reports indicate that ETEC expressing LT/STh/CS5/CS6 is the most common ETEC pathotype isolated in Dhaka (20). Both these strains were detected as pure ETEC infections in this study, suggesting that these toxin/CF profiles are beneficial for single infections. A study performed in Dhaka in 2011, using whole-genome sequencing of several isolates recovered from individual patients in a setup similar to this study, identified one patient for whom all analyzed isolates constituted a clonal expansion of L5 (LT/STh CS5/CS6) isolates (42), which further confirms that L5 isolates are able to outcompete the normal microbiota and cause serious infections. The other four strains were detected as CF negative in the initial dot blot analysis; however, sequence analysis revealed CF profiles for three of them: E2267 (sample 16) was detected as CS14 positive, E2269 (sample 18) was detected positive for CS27b, and E2268 (sample 17) was shown to harbor genes for a CF profile similar to CS13/CS23. No CF could be detected for E2266 (sample 9), suggesting that this strain either lacks CF or expresses a kind of colonization factor that has not yet been discovered or that the plasmid(s) was lost prior to sequencing. Nonetheless, this STp/CF combination rendered a strain that was the major infecting pathogen of the E. coli species still beneficial for coinfections. The strain E2267 showed an STp/LT/CS14 profile, whereas E2268 was shown positive for LT in combination with new CFs that are very similar, but not identical, to CS13/CS23. Both ETEC strains were found in coinfections. The E2269 strain also showed a new toxin-CF combination. This novel type of ETEC, expressing LT, STp, and CS27b, is worth keeping an eye on. It was detected in an almost pure ETEC infection and is definitely potent as a single-infection pathogen. The LT/STp/CS27b ETEC of MLST ST-4493 was not described during the time of isolation of the strains in this paper. The novel CF CS27b was first described by Nada and coworkers in 2011 (21). Recent publications have indicated that ETEC strains previously noted as CF negative might express a novel group of CS18- and CS20-like CFs, to which CS27b belongs (21, 22). These new CFs are not detectable using traditional dot blot techniques and might be missed using the present PCR methods in use; therefore, more information about the various ETEC toxin-CF combinations is needed. The matter is, however, complex, and no consensus regarding the CF/toxin profiles and disease outcome has been determined to date (9).
Next, we sought to determine why certain ETEC toxin-CF combinations manifest as single infections. Scoary analysis of the genomes, however, did not reveal any unique gene profiles that could explain the single-pathogen infections seen for samples 4, 5, and 18. Analysis of antibiotic resistance genes was performed to determine if single infections are more resistant. Two of the single-pathogen infectants, E2264 (sample 4) and E2269 (sample 18), showed multiple genes for antibiotic resistance. In contrast, using ResFinder, no acquired resistance genes were found for E2265 (sample 5), while ARG-annot confirmed chromosome-bound ampicillin resistance by ampC (see Table S4 in the supplemental material). This isolate has been described previously (43, 44). Regarding the ETEC strains found as coinfecting pathogens, E2266 (sample 9) and E2267 (sample 16) harbored four and three antibiotic resistance genes each, respectively. Also in this group, one of the three strains, this time E2268 (sample 17), harbored no presumed plasmid-borne resistance genes except ampC. For E2268, the lack of acquired resistance genes coincides with the lack of the ETEC-specific ParM gene (23), which was detected in all the other strains. This strain might thus be somewhat less potent, considering that only 4% of the E. coli isolates of this sample were determined as ETEC, or it might be highly specialized in occurring in coinfections. The high frequency of resistance genes detected in some isolates might not be surprising considering that a test of the drinking water in Dhaka some years ago revealed that 36% of the E. coli isolates were multiresistant, of which 26% were positive for extended-spectrum beta-lactamases (45). The presence of antibiotic resistance might thus not be important for diarrheal virulence.
Nevertheless, in this work we have shown, although with a limited number of samples, that there might be specific CF/toxin profiles associated with either single ETEC infections or multipathogen infections. Two of the isolates, E2264 (sample 4) and E2265 (sample 5), were found to belong to globally successful ETEC lineages that have been described previously (10). Hence, although the isolates described in this work were collected a decade ago, they are indeed still relevant and frequently detected pathogens. Here, we used second- (Illumina) and third-generation (PacBio) sequencing technologies, the latest developed in the last 5 years, which allowed us to perform de novo assemblies and characterize ETEC plasmids. The genome data will be useful for deeper studies of pathogen genomics.
Recent studies on global diarrhea in the GEMS and MAL-ED studies have identified STh-expressing ETEC to be a major contributor to diarrhea (40). The identification in this study of potent single infections by LT/CS7 and LT/STp/CS27b ETEC does indicate that focus on STh-expressing ETEC might be an oversimplification. Indeed, Del Canto et al. recently described clonal CS27b ETEC isolated from Chile, Pakistan, India, and Bangladesh (22), indicating that LT/STp/CS27b is an emerging virulent ETEC type. Given this, we propose that preventive efforts and vaccine strategies against ETEC should be focused on globally spread ETEC lineages.
MATERIALS AND METHODS
Ethics statement.
The collected samples were part of the icddr,b 2% surveillance system routine, approved by the Research Review Committee (RRC) and Ethical Review Committee (ERC) of icddr,b, Dhaka, Bangladesh, as described previously (10, 20). Exclusion criteria in this study were the presence of agents other than Enterobacteriaceae and Vibrio or if the patient reported having had antibiotic treatment prior to hospitalization. Informed oral consent was obtained from adult patients, or from caregivers or guardians of children, for collection of stool specimens, according to the hospital policy. The ERC has approved verbal consent and voluntary participation, and subjects may refuse participation without compromise of patient care. All patients were treated for their clinical conditions, e.g., dehydration, after sample collection. Consenting individuals were assured about the nondisclosure of name or identity of the participants. The ETEC strains collected and analyzed in this study were deposited at the ETEC culture collection of the University of Gothenburg and in the group of Å. Sjöling. Permission to use the ETEC strain collection was granted by the Regional Ethical Board of Gothenburg, Sweden (Ethics Committee reference 088-10).
Bacterial growth and detection of ETEC and V. cholerae.
Watery diarrheal stool samples were collected from the hospital ward at the International Centre for Diarrhoeal Diseases Research in Bangladesh (icddr,b) during the diarrheal peak season in March to April 2006 and brought to the adjacent laboratory for culture of bacteria. The clinical criteria for admission were moderate to severe watery diarrhea requiring hospitalization.
The collected liquid diarrheal samples were serially diluted in phosphate-buffered saline (PBS) and cultured on MacConkey agar plates for determination of CFU of E. coli per milliliter. E. coli was distinguished from other lactose-fermenting bacteria, including Klebsiella, Citrobacter, and Enterobacter, by ocular investigation. Only the E. coli-like colonies were picked for subsequent analyses. The same serial diluted samples were cultured in parallel on selective taurocholate-tellurite-gelatin agar (TTGA) plates to determine growth of V. cholerae (46). The V. cholerae serotype, Inaba or Ogawa, was identified by an agglutination test (47). To verify the presence of ETEC among the samples containing E. coli-like bacteria, ETEC toxin multiplex PCR was performed as described previously (48). In short, 6 to 10 colonies from the MacConkey agar plates were pooled, boiled in 500 µl MilliQ water, and tested for the presence of the genes encoding the ETEC toxins LT, STh, and STp. Individual colonies were collected from the ETEC-positive plates, and the toxin profile was retested and confirmed by multiplex PCR. One representative isolate was saved in freeze medium at −80°C. Furthermore, the colonization factor profiles of these representative isolates were determined by multiplex PCR and by dot blot analysis, as previously described (48, 49).
Determination of the proportion of ETEC to total number of E. coli bacteria in stool.
To determine the percentage of ETEC per total number of E. coli-like colonies in the diarrheal stool samples, 50 E. coli-like colonies were randomly collected from the original ETEC-positive MacConkey agar plates. The colonies were individually boiled for 10 min in MilliQ water and tested by toxin multiplex PCR (48). The percentage of ETEC per total E. coli bacteria in each sample was calculated by dividing the number of toxin-positive colonies by 50 (total number of analyzed E. coli colonies).
Collection of bacterial pellet and supernatant from diarrheal liquid for DNA and RNA analysis.
From the original watery diarrheal samples, DNA and RNA were extracted for molecular quantification of DNA as well as for gene expression analyses of both ETEC and V. cholerae. The diarrheal samples were first centrifuged at 1,000 × g for 5 min to separate mucus and solid particles from the bacterium-containing supernatant. From the remaining supernatant, three separate samples of 1 ml each were collected and additionally centrifuged for 5 min at 16,000 × g followed by subsequent separation of bacterial pellet and supernatant. The first centrifuged sample was saved for subsequent determination of the amounts of toxins secreted by the bacteria or associated with the bacterial cells by GM1-ELISA (described below). For this sample, the pellet was dissolved in PBS and sonicated before it was frozen at −70°C, and the corresponding supernatant was frozen separately at −70°C. These samples were stored at maximum for 1 day at −70°C, after which they were analyzed. For the second and third 1-ml samples from the same stool sample, the bacterial pellets were collected and immediately frozen at −70°C for subsequent DNA and RNA extraction, respectively.
Detection of ETEC and cholera toxins in diarrhea samples by GM1-ELISA.
In order to detect the toxins produced by ETEC and V. cholerae in the diarrheal samples, the pellet and supernatant samples were analyzed by GM1-ELISA for detection of CT and LT and by inhibition GM1-ELISA for detection of ST. The procedures have been described previously (48, 50). With the use of in-house monoclonal antibodies specific for ST (ST-1), LT (LT-80), and CT (CT-Wi), as well as an antibody that recognizes both LT and CT (LT-39), the total amount of ST, LT, or CT could be determined in the supernatant (secreted toxin) and in the pellet fraction (toxin associated with the bacterial cell membrane or cytoplasm). All antibodies were produced at the department of Microbiology and Immunology, Sahlgrenska Academy, University of Gothenburg. Threefold dilution series and reference toxin standards of known concentrations were used to determine the amount of each toxin present in both pellet and supernatant of the samples.
DNA and RNA extraction from diarrhea samples.
The second and third 1-ml samples from the diarrhea sample preparation were used for DNA and RNA extraction and subsequent qPCR analyses of gene copy numbers and gene expression. Bacterial DNA extractions from the frozen pellets were performed with the QIAamp stool DNA kit (Qiagen, Hilden, Germany) as described by the manufacturer and in previous studies (33, 51). The extracted DNA was kept at −20°C until analysis. RNA extraction was performed with the RNeasy kit (Qiagen, Hilden, Germany), and a DNase protocol (Qiagen) was included to remove genomic DNA as described previously (52). Extracted RNA was analyzed on an agarose gel to determine integrity, and the concentration was measured at 260 nm using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). cDNA was prepared from 200 ng RNA from each sample using the QuantiTect cDNA kit (Qiagen) with an additional DNase step included in the protocol. The cDNA was stored at −20°C until further analysis.
qPCR quantification of ETEC and V. cholerae in diarrhea samples.
Quantitative PCR (qPCR) was performed to determine the total amount of ETEC bacteria in the DNA samples using primers specific for the eltB (LT), estA1 (STp), and estA2 to estA4 (STh) genes (33) and ctxB (CT) (52). For ETEC, a standard curve for each gene was generated by PCR amplification using the respective real-time PCR primer and a toxin-positive ETEC strain as the DNA template. The PCR products were purified using the QIAquick PCR purification kit (Qiagen), and the concentrations were determined on a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). The PCR product copy number was determined as described previously using Avogadro’s number and the molecular weight of the PCR product (53). Tenfold serial dilutions between 5 × 108 and 5 copies/µliter were prepared and stored at −20°C until further use. To determine the total number of V. cholerae bacteria in the diarrheal sample, real-time PCR was performed using the same conditions as for ETEC. A standard curve was generated by manually counting the bacteria of the N16961 El Tor V. cholerae reference strain using a Neubauer improved counting chamber (Hausser Scientific, VWR International) at a magnification of ×40. Counted bacteria were diluted in 10-fold serial dilutions to the same concentrations as described above and used as a standard curve. Real-time PCRs were run in duplicates in 96-well plates (Applied Biosystems) with a total volume of 20 μl in each reaction mixture. The PCR mix contained 10 μl SYBR green real-time PCR master mix (Life Technologies), 10 pmol of each primer, 6 μl water, and 2 μl DNA. Negative controls and a standard curve were included in each PCR run. The numbers of bacteria per 1 ml liquid diarrheal sample were calculated by using the settings for absolute quantification in the ABI 7500 real-time PCR instrument and by assuming that one gene copy equals one bacterium.
Determination of toxin gene expression per bacterium in stool samples.
The same primers and qPCR conditions were used in reverse transcriptase qPCR (RT-qPCR) for gene expression analysis. As the template, the cDNA was used, and each sample was analyzed in duplicate. A no-RT reaction mixture containing only RNA for each sample was run in duplicate to confirm that the detected expression was not due to genomic DNA amplification. The real-time PCR was run on an ABI 7500 using SYBR green and standard amplification conditions, as described above, in a reaction volume of 20 µl. The gene copy number per bacterial genome was calculated by dividing the numbers of gene transcripts per milliliter of sample with the gene copy number per milliliter of sample as described previously (52).
Whole-genome Illumina sequencing.
In order to investigate the genetics of the collected ETEC strains, and to be able to correlate the collected ETEC with worldwide ETEC infections, whole-genome sequence (WGS) analysis was performed. For E2264 (sample 4) and E2265 (sample 5), PacBio sequencing was performed to gain even more complete information about the genetic details since these strains were of specific interest due to their being found in ETEC-only infections (described below). The four other ETEC strains were sequenced using Illumina MiSeq sequencing. The ETEC strains, stored at −80°C, were plated on LB agar plates and incubated for 24 h at 37°C. One large bacterial colony from each plate was selected and washed in 300 µl MilliQ water, after which the bacterial DNA was extracted using the DNeasy Blood & Tissue kit from Qiagen according to the manufacturer’s instructions. The DNA concentration was measured using a Qubit 2.0 fluorometer (Invitrogen). Sequencing libraries were prepared using the TruSeq Nano kit (Illumina, San Diego, CA) with a mean fragment length of 900 bp. Libraries were sequenced on the MiSeq platform v.3 chemistry, 2 by 300 bp, generating a coverage of >100× for all strains.
PacBio sequencing.
DNA for Pacific Biosciences (PacBio) sequencing was prepared from ETEC isolates grown in LB medium to an optical density at 600 nm (OD600) of 0.3. DNA was extracted by the Qiagen Genomic-tip 500/G kit according to the manufacturer’s instructions (Qiagen, Hilden, Germany). For each sample, one DNA aliquot was sheared into 10-kbp fragments using a Genemachines HydroShear instrument (Digilab, Marlborough, MA, USA) and a second aliquot was sheared into 2-kb fragments using a Covaris instrument (Covaris, Woburn, MA). SMRTbell templates were constructed according to the manufacturer’s instructions (Pacific Biosciences, Menlo Park, CA, USA). Each library was sequenced on 1 SMRT cell on a Pacific Biosciences RSII sequencer according to the manufacturer’s instructions with 4-h movie time.
Assembly and annotation.
The reads from the 10-kb PacBio sequencing library were assembled using HGAP3 from SMRTportal v2.3 (Pacific Biosciences, Menlo Park, CA, USA) with default settings. The 2-kb PacBio libraries were assembled using Falcon (Pacific Biosciences, Menlo Park, CA, USA) with settings allowing high coverage for plasmid assembly.
Illumina raw reads were trimmed and filtered using TrimGalore! (54), applying the quality cutoff Q30 and keeping only reads longer than 30 bp. Filtered reads were de novo assembled using SPAdes v 3.10.1 (55), and resulting assembly files were filtered for very-low-coverage contigs and contigs shorter than 500 bp before they were ordered to the E2265 complete PacBio sequence using the Mauve order contigs tool (56).
The resulting draft and complete genomes were annotated with the prokka annotation pipeline v. 1.1.12b (57) using the E24377A (CP000800.1) ETEC proteome as primary annotation source. Summary statistics from the sequencing, assembly, and annotation were collected using MultiQC v1.0 (58) and are shown in Table S1 (PacBio) and Table S2 (Illumina) in the supplemental material.
Functional and comparative genomic analysis.
To perform initial functional analysis, we used the CGE pipeline v 1.1 (59), which performs resistance gene prediction using ResFinder, in silico MLST, plasmid prediction, and pMLST. Resistance gene prediction was also performed by ARG-annot (http://en.mediterranee-infection.com/article.php?laref=283%26titre=arg-annot).
Whole-genome alignment was performed and visualized using progressiveMauve v. 2015/2/25 (60). Pan-genome analysis was performed using Roary v. 3.6.2 (61), using a blastp identity cutoff of 85%. Comparison between the ETEC-only infection genomes and mixed-infection genomes was performed using the Scoary tool v. 1.6.10 (62).
Statistical analyses.
Statistical analyses were performed using GraphPad Prism version 7.0. P values of <0.05 were considered significant.
Accession number(s).
The sequences of E2266 to E2269 have been deposited at DDBJ/ENA/GenBank under the accession numbers NQYN00000000 (E2266), NQYM00000000 (E2267), NQYL00000000 (E2268), and NQYK00000000 (E2269). The versions described in this paper are versions NQYN01000000, NQYM01000000, NQYL01000000, and NQYK01000000, respectively. The assembled chromosome and three plasmids of E2264 have been deposited in GenBank under accession numbers CP023349, CP023350, CP023351, and CP023352, respectively. The assembled chromosome and two plasmids of E2265 have been deposited in GenBank under accession numbers CP023346, CP023347, and CP023348, respectively.
ACKNOWLEDGMENTS
The PacBio sequencing and assembly were performed by the National Genomics Infrastructure, NGI, Science for Life Laboratory, Uppsala, Sweden. We thank Christian Tellgren-Roth at the NGI Science for Life Laboratory, Uppsala, for bioinformatic support. The Illumina sequencing was performed at the National Genomics Infrastructure, NGI, Science for Life Laboratory, Stockholm, Sweden. All bioinformatic analyses were performed on resources provided by SNIC through the Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX). Professor Ann-Mari Svennerholm at University of Gothenburg, Sweden, is greatly appreciated for support during the initial stages of this work.
All authors declare no conflict of interest.
This study was supported by the joint Formas Sida/SAREC Foundation for Sustainable Research in Developing Countries 213-2005-294, the Swedish Research Council grants 521-2011-3435 and 348-2014-2639 to Å.S., and the Swedish Foundation for Strategic Research (SSF) grant no. SB-2012-0072. The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Walker CLF, Aryee MJ, Boschi-Pinto C, Black RE. 2012. Estimating diarrhea mortality among young children in low and middle income countries. PLoS One 7:e29151. doi: 10.1371/journal.pone.0029151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu J, Kabir F, Manneh J, Lertsethtakarn P, Begum S, Gratz J, Becker SM, Operario DJ, Taniuchi M, Janaki L, Platts-Mills JA, Haverstick DM, Kabir M, Sobuz SU, Nakjarung K, Sakpaisal P, Silapong S, Bodhidatta L, Qureshi S, Kalam A, Saidi Q, Swai N, Mujaga B, Maro A, Kwambana B, Dione M, Antonio M, Kibiki G, Mason CJ, Haque R, Iqbal N, Zaidi AKM, Houpt ER. 2014. Development and assessment of molecular diagnostic tests for 15 enteropathogens causing childhood diarrhoea: a multicentre study. Lancet Infect Dis 14:716–724. doi: 10.1016/S1473-3099(14)70808-4. [DOI] [PubMed] [Google Scholar]
- 3.Sánchez J, Holmgren J. 2005. Virulence factors, pathogenesis and vaccine protection in cholera and ETEC diarrhea. Curr Opin Immunol 17:388–398. doi: 10.1016/j.coi.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 4.Begue RE, Castellares G, Hayashi KE, Ruiz R, Meza R, English CK, Gotuzzo E, Sanchez JL, Oberst R. 1994. Diarrheal disease in Peru after the introduction of cholera. Am J Trop Med Hyg 51:585–589. doi: 10.4269/ajtmh.1994.51.585. [DOI] [PubMed] [Google Scholar]
- 5.Krebs SJ, Taylor RK. 2011. Protection and attachment of Vibrio cholerae mediated by the toxin-coregulated pilus in the infant mouse model. J Bacteriol 193:5260–5270. doi: 10.1128/JB.00378-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, Lim MS, Li S, Brock M, Pique ME, Woods VL Jr, Craig L. 2008. Vibrio cholerae toxin-coregulated pilus structure analyzed by hydrogen-deuterium exchange mass spectrometry. Structure 16:137–148. doi: 10.1016/j.str.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Millet YA, Alvarez D, Ringgaard S, von Andrian UH, Davis BM, Waldor MK. 2014. Insights into Vibrio cholerae intestinal colonization from monitoring fluorescently labeled bacteria. PLoS Pathog 10:e1004405. doi: 10.1371/journal.ppat.1004405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaastra W, Svennerholm AM. 1996. Colonization factors of human enterotoxigenic Escherichia coli (ETEC). Trends Microbiol 4:444–452. doi: 10.1016/0966-842X(96)10068-8. [DOI] [PubMed] [Google Scholar]
- 9.Isidean SD, Riddle MS, Savarino SJ, Porter CK. 2011. A systematic review of ETEC epidemiology focusing on colonization factor and toxin expression. Vaccine 29:6167–6178. doi: 10.1016/j.vaccine.2011.06.084. [DOI] [PubMed] [Google Scholar]
- 10.von Mentzer A, Connor TR, Wieler LH, Semmler T, Iguchi A, Thomson NR, Rasko DA, Joffre E, Corander J, Pickard D, Wiklund G, Svennerholm AM, Sjöling Å, Dougan G. 2014. Identification of enterotoxigenic Escherichia coli (ETEC) clades with long-term global distribution. Nat Genet 46:1321–1326. doi: 10.1038/ng.3145. [DOI] [PubMed] [Google Scholar]
- 11.Chowdhury F, Rahman MA, Begum YA, Khan AI, Faruque AS, Saha NC, Baby NI, Malek MA, Kumar AR, Svennerholm AM, Pietroni M, Cravioto A, Qadri F. 2011. Impact of rapid urbanization on the rates of infection by Vibrio cholerae O1 and enterotoxigenic Escherichia coli in Dhaka, Bangladesh. PLOS Negl Trop Dis 5:e999. doi: 10.1371/journal.pntd.0000999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bölin I, Wiklund G, Qadri F, Torres O, Bourgeois AL, Savarino S, Svennerholm AM. 2006. Enterotoxigenic Escherichia coli with STh and STp genotypes is associated with diarrhea both in children in areas of endemicity and in travelers. J Clin Microbiol 44:3872–3877. doi: 10.1128/JCM.00790-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joffré E, von Mentzer A, Svennerholm AM, Sjöling Å. 2016. Identification of new heat-stable (STa) enterotoxin allele variants produced by human enterotoxigenic Escherichia coli (ETEC). Int J Med Microbiol 306:586–594. doi: 10.1016/j.ijmm.2016.05.016. [DOI] [PubMed] [Google Scholar]
- 14.Clements JD, Finkelstein RA. 1978. Immunological cross-reactivity between a heat-labile enterotoxin(s) of Escherichia coli and subunits of Vibrio cholerae enterotoxin. Infect Immun 21:1036–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wajima T, Sabui S, Kano S, Ramamurthy T, Chatterjee NS, Hamabata T. 2013. Entire sequence of the colonization factor coli surface antigen 6-encoding plasmid pCss165 from an enterotoxigenic Escherichia coli clinical isolate. Plasmid 70:343–352. doi: 10.1016/j.plasmid.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 16.Tobias J, Von Mentzer A, Loayza Frykberg P, Aslett M, Page AJ, Sjöling Å, Svennerholm AM. 2016. Stability of the encoding plasmids and surface expression of CS6 differs in enterotoxigenic Escherichia coli (ETEC) encoding different heat-stable (ST) enterotoxins (STh and STp). PLoS One 11:e0152899. doi: 10.1371/journal.pone.0152899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paschke C, Apelt N, Fleischmann E, Perona P, Walentiny C, Löscher T, Herbinger KH. 2011. Controlled study on enteropathogens in travellers returning from the tropics with and without diarrhoea. Clin Microbiol Infect 17:1194–1200. doi: 10.1111/j.1469-0691.2010.03414.x. [DOI] [PubMed] [Google Scholar]
- 18.Montero D, Vidal M, Pardo M, Torres A, Kruger E, Farfán M, O’Ryan M, Luo Q, Fleckenstein J, Del Canto F, Vidal R. 2017. Characterization of enterotoxigenic Escherichia coli strains isolated from the massive multi-pathogen gastroenteritis outbreak in the Antofagasta region following the Chilean earthquake, 2010. Infect Genet Evol 52:26–29. doi: 10.1016/j.meegid.2017.04.021. [DOI] [PubMed] [Google Scholar]
- 19.Vicente AC, Teixeira LF, Iniguez-Rojas L, Luna MG, Silva L, Andrade JR, Guth BE. 2005. Outbreaks of cholera-like diarrhoea caused by enterotoxigenic Escherichia coli in the Brazilian Amazon rainforest. Trans R Soc Trop Med Hyg 99:669–674. doi: 10.1016/j.trstmh.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Begum YA, Baby NI, Faruque AS, Jahan N, Cravioto A, Svennerholm AM, Qadri F. 2014. Shift in phenotypic characteristics of enterotoxigenic Escherichia coli (ETEC) isolated from diarrheal patients in Bangladesh. PLoS Negl Trop Dis 8:e3031. doi: 10.1371/journal.pntd.0003031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nada RA, Shaheen HI, Khalil SB, Mansour A, El-Sayed N, Touni I, Weiner M, Armstrong AW, Klena JD. 2011. Discovery and phylogenetic analysis of novel members of class b enterotoxigenic Escherichia coli adhesive fimbriae. J Clin Microbiol 49:1403–1410. doi: 10.1128/JCM.02006-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Del Canto F, O’Ryan M, Pardo M, Torres A, Gutiérrez D, Cádiz L, Valdés R, Mansilla A, Martínez R, Hernández D, Caro B, Levine MM, Rasko DA, Hill CM, Pop M, Stine OC, Vidal R. 2016. Chaperone-usher pili loci of colonization factor-negative human enterotoxigenic Escherichia coli. Front Cell Infect Microbiol 6:200. doi: 10.3389/fcimb.2016.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sahl JW, Sistrunk JR, Baby NI, Begum Y, Luo Q, Sheikh A, Qadri F, Fleckenstein JM, Rasko DA. 2017. Insights into enterotoxigenic Escherichia coli diversity in Bangladesh utilizing genomic epidemiology. Sci Rep 7:3402. doi: 10.1038/s41598-017-03631-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frost LS, Ippen-Ihler K, Skurray RA. 1994. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol Rev 58:162–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anthony KG, Klimke WA, Manchak J, Frost LS. 1999. Comparison of proteins involved in pilus synthesis and mating pair stabilization from the related plasmids F and R100-1: insights into the mechanism of conjugation. J Bacteriol 181:5149–5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ochi S, Shimizu T, Ohtani K, Ichinose Y, Arimitsu H, Tsukamoto K, Kato M, Tsuji T. 2009. Nucleotide sequence analysis of the enterotoxigenic Escherichia coli Ent plasmid. DNA Res 16:299–309. doi: 10.1093/dnares/dsp015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel SK, Dotson J, Allen KP, Fleckenstein JM. 2004. Identification and molecular characterization of EatA, an autotransporter protein of enterotoxigenic Escherichia coli. Infect Immun 72:1786–1794. doi: 10.1128/IAI.72.3.1786-1794.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pilonieta MC, Bodero MD, Munson GP. 2007. CfaD-dependent expression of a novel extracytoplasmic protein from enterotoxigenic Escherichia coli. J Bacteriol 189:5060–5067. doi: 10.1128/JB.00131-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang ZD, DuPont HL. 2017. Etiology of travellers’ diarrhea. J Travel Med 24(Suppl 1):S13–S16. doi: 10.1093/jtm/tax003. [DOI] [PubMed] [Google Scholar]
- 30.Liu J, Platts-Mills JA, Juma J, Kabir F, Nkeze J, Okoi C, Operario DJ, Uddin J, Ahmed S, Alonso PL, Antonio M, Becker SM, Blackwelder WC, Breiman RF, Faruque AS, Fields B, Gratz J, Haque R, Hossain A, Hossain MJ, Jarju S, Qamar F, Iqbal NT, Kwambana B, Mandomando I, McMurry TL, Ochieng C, Ochieng JB, Ochieng M, Onyango C, Panchalingam S, Kalam A, Aziz F, Qureshi S, Ramamurthy T, Roberts JH, Saha D, Sow SO, Stroup SE, Sur D, Tamboura B, Taniuchi M, Tennant SM, Toema D, Wu Y, Zaidi A, Nataro JP, Kotloff KL, Levine MM, Houpt ER. 2016. Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: a reanalysis of the GEMS case-control study. Lancet 388:1291–1301. doi: 10.1016/S0140-6736(16)31529-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harro C, Chakraborty S, Feller A, DeNearing B, Cage A, Ram M, Lundgren A, Svennerholm AM, Bourgeois AL, Walker RI, Sack DA. 2011. Refinement of a human challenge model for evaluation of enterotoxigenic Escherichia coli vaccines. Clin Vaccine Immunol 18:1719–1727. doi: 10.1128/CVI.05194-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Youmans BP, Ajami NJ, Jiang ZD, Petrosino JF, DuPont HL, Highlander SK. 2014. Development and accuracy of quantitative real-time polymerase chain reaction assays for detection and quantification of enterotoxigenic Escherichia coli (ETEC) heat labile and heat stable toxin genes in travelers’ diarrhea samples. Am J Trop Med Hyg 90:124–132. doi: 10.4269/ajtmh.13-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lothigius A, Janzon A, Begum Y, Sjöling A, Qadri F, Svennerholm AM, Bölin I. 2008. Enterotoxigenic Escherichia coli is detectable in water samples from an endemic area by real-time PCR. J Appl Microbiol 104:1128–1136. doi: 10.1111/j.1365-2672.2007.03628.x. [DOI] [PubMed] [Google Scholar]
- 34.Platts-Mills JA, Gratz J, Mduma E, Svensen E, Amour C, Liu J, Maro A, Saidi Q, Swai N, Kumburu H, McCormick BJ, Kibiki G, Houpt ER. 2014. Association between stool enteropathogen quantity and disease in Tanzanian children using TaqMan array cards: a nested case-control study. Am J Trop Med Hyg 90:133–138. doi: 10.4269/ajtmh.13-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pop M, Paulson JN, Chakraborty S, Astrovskaya I, Lindsay BR, Li S, Bravo HC, Harro C, Parkhill J, Walker AW, Walker RI, Sack DA, Stine OC. 2016. Individual-specific changes in the human gut microbiota after challenge with enterotoxigenic Escherichia coli and subsequent ciprofloxacin treatment. BMC Genomics 17:440. doi: 10.1186/s12864-016-2777-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, Silapong S, Jeanwattanalert P, Lertsehtakarn P, Bodhidatta L, Swierczewski B, Mason C, McVeigh AL, Savarino SJ, Nshama R, Mduma E, Maro A, Zhang J, Gratz J, Houpt ER. 2017. Multiplex real time PCR panels to identify fourteen colonization factors of enterotoxigenic Escherichia coli (ETEC). PLoS One 12:e0176882. doi: 10.1371/journal.pone.0176882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mudrak B, Kuehn MJ. 2010. Heat-labile enterotoxin: beyond G(M1) binding. Toxins 2:1445–1470. doi: 10.3390/toxins2061445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonzales L, Sanchez S, Zambrana S, Iñiguez V, Wiklund G, Svennerholm AM, Sjöling A. 2013. Molecular characterization of enterotoxigenic Escherichia coli isolates recovered from children with diarrhea during a 4-year period (2007 to 2010) in Bolivia. J Clin Microbiol 51:1219–1225. doi: 10.1128/JCM.02971-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qadri F, Das SK, Faruque AS, Fuchs GJ, Albert MJ, Sack RB, Svennerholm AM. 2000. Prevalence of toxin types and colonization factors in enterotoxigenic Escherichia coli isolated during a 2-year period from diarrheal patients in Bangladesh. J Clin Microbiol 38:27–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kotloff KL. 2017. The burden and etiology of diarrheal illness in developing countries. Pediatr Clin North Am 64:799–814. doi: 10.1016/j.pcl.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 41.Mansour A, Shaheen HI, Amine M, Hassan K, Sanders JW, Riddle MS, Armstrong AW, Svennerholm AM, Sebeny PJ, Klena JD, Young SY, Frenck RW Jr. 2014. Pathogenicity and phenotypic characterization of enterotoxigenic Escherichia coli isolates from a birth cohort of children in rural Egypt. J Clin Microbiol 52:587–591. doi: 10.1128/JCM.01639-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sahl JW, Sistrunk JR, Fraser CM, Hine E, Baby N, Begum Y, Luo Q, Sheikh A, Qadri F, Fleckenstein JM, Rasko DA. 2015. Examination of the enterotoxigenic Escherichia coli population structure during human infection. mBio 6:e00501-15. doi: 10.1128/mBio.00501-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu F, Yang X, Wang Z, Nicklasson M, Qadri F, Yi Y, Zhu Y, Lv N, Li J, Zhang R, Guo H, Zhu B, Sjöling Å, Hu Y. 2015. Draft genomes of four enterotoxigenic Escherichia coli (ETEC) clinical isolates from China and Bangladesh. Gut Pathog 7:10. doi: 10.1186/s13099-015-0059-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nicklasson M, Sjöling Å, von Mentzer A, Qadri F, Svennerholm AM. 2012. Expression of colonization factor CS5 of enterotoxigenic Escherichia coli (ETEC) is enhanced in vivo and by the bile component Na glycocholate hydrate. PLoS One 7:e35827. doi: 10.1371/journal.pone.0035827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Talukdar PK, Rahman M, Rahman M, Nabi A, Islam Z, Hoque MM, Endtz HP, Islam MA. 2013. Antimicrobial resistance, virulence factors and genetic diversity of Escherichia coli isolates from household water supply in Dhaka, Bangladesh. PLoS One 8:e61090. doi: 10.1371/journal.pone.0061090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Brien M, Colwell R. 1985. Modified taurocholate-tellurite-gelatin agar for improved differentiation of Vibrio species. J Clin Microbiol 22:1011–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rahman M, Sack DA, Mahmood S, Hossain A. 1987. Rapid diagnosis of cholera by coagglutination test using 4-H fecal enrichment cultures. J Clin Microbiol 25:2204–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sjöling A, Wiklund G, Savarino SJ, Cohen DI, Svennerholm AM. 2007. Comparative analyses of phenotypic and genotypic methods for detection of enterotoxigenic Escherichia coli toxins and colonization factors. J Clin Microbiol 45:3295–3301. doi: 10.1128/JCM.00471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodas C, Iniguez V, Qadri F, Wiklund G, Svennerholm AM, Sjöling A. 2009. Development of multiplex PCR assays for detection of enterotoxigenic Escherichia coli colonization factors and toxins. J Clin Microbiol 47:1218–1220. doi: 10.1128/JCM.00316-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Svennerholm AM, Wikström M, Lindblad M, Holmgren J. 1986. Monoclonal antibodies against Escherichia coli heat-stable toxin (STa) and their use in a diagnostic ST ganglioside GM1-enzyme-linked immunosorbent assay. J Clin Microbiol 24:585–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Janzon A, Sjöling A, Lothigius A, Ahmed D, Qadri F, Svennerholm AM. 2009. Failure to detect Helicobacter pylori DNA in drinking and environmental water in Dhaka, Bangladesh, using highly sensitive real-time PCR assays. Appl Environ Microbiol 75:3039–3044. doi: 10.1128/AEM.02779-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Janzon A, Bhuiyan T, Lundgren A, Qadri F, Svennerholm AM, Sjöling A. 2009. Presence of high numbers of transcriptionally active Helicobacter pylori in vomitus from Bangladeshi patients suffering from acute gastroenteritis. Helicobacter 14:237–247. doi: 10.1111/j.1523-5378.2009.00692.x. [DOI] [PubMed] [Google Scholar]
- 53.Sjöling A, Qadri F, Nicklasson M, Begum YA, Wiklund G, Svennerholm AM. 2006. In vivo expression of the heat stable (estA) and heat labile (eltB) toxin genes of enterotoxigenic Escherichia coli (ETEC). Microbes Infect 8:2797–2802. doi: 10.1016/j.micinf.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 54.Krueger F. 2014. TrimGalore! version 0.3.7. Bioinformatics Group, Babraham Institute, Cambridge, United Kingdom: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/. [Google Scholar]
- 55.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rissman AI, Mau B, Biehl BS, Darling AE, Glasner JD, Perna NT. 2009. Reordering contigs of draft genomes using the Mauve aligner. Bioinformatics 25:2071–2073. doi: 10.1093/bioinformatics/btp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 58.Ewels P, Magnusson M, Lundin S, Käller M. 2016. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thomsen MC, Ahrenfeldt J, Cisneros JL, Jurtz V, Larsen MV, Hasman H, Aarestrup FM, Lund O. 2016. A bacterial analysis platform: an integrated system for analysing bacterial whole genome sequencing data for clinical diagnostics and surveillance. PLoS One 11:e0157718. doi: 10.1371/journal.pone.0157718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brynildsrud O, Bohlin J, Scheffer L, Eldholm V. 2016. Erratum to: Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol 17:262. doi: 10.1186/s13059-016-1132-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Assembly and annotation statistics of the PacBio sequencing of E2264 and E2265. Download TABLE S1, DOCX file, 0.02 MB (16.8KB, docx) .
Copyright © 2018 Begum et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Assembly and annotation statistics of the Illumina sequencing of E2266 to E2269. Download TABLE S2, DOCX file, 0.02 MB (16.5KB, docx) .
Copyright © 2018 Begum et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ETEC virulence factors identified in the ETEC strains. Download TABLE S3, DOCX file, 0.02 MB (17.1KB, docx) .
Copyright © 2018 Begum et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Antibiotic resistance genes found in the ETEC strains. Download TABLE S4, DOCX file, 0.02 MB (17KB, docx) .
Copyright © 2018 Begum et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.