Abstract
Although genetic factors contribute to almost half of all deafness cases, treatment options for genetic deafness are limited1–5. We developed a genome editing approach to target a dominantly inherited form of genetic deafness. Here we show that cationic lipid-mediated in vivo delivery of Cas9:guide RNA complexes can ameliorate hearing loss in a mouse model of human genetic deafness. We designed and validated in vitro and in primary fibroblasts genome editing agents that preferentially disrupt the dominant deafness-associated allele in the Tmc1 (transmembrane channel-like 1) Beethoven (Bth) mouse model, even though the mutant Bth allele differs from the wild-type allele at only a single base pair. Injection of Cas9:guide RNA:lipid complexes targeting the Bth allele into the cochlea of neonatal Bth/+ mice substantially reduced progressive hearing loss. We observed higher hair cell survival rates and lower auditory brainstem response (ABR) thresholds in injected ears compared with uninjected ears or ears injected with complexes that target an unrelated gene. Enhanced acoustic reflex responses were observed among injected compared to uninjected Bth/+ animals. These findings suggest protein:RNA complex delivery of target gene-disrupting agents in vivo as a potential strategy for the treatment of some autosomal dominant hearing loss diseases.
Keywords: Genome editing, CRISPR, Cas9, protein delivery, genetic hearing loss
While ~100 deafness-associated alleles have been identified, few treatments are available to slow or reverse genetic deafness4,5. Complementation of wild-type alleles, or silencing dominant negative mutant alleles, have shown promising results in animal models6, 7. Nonetheless, current approaches face potential challenges including immunogenicity, oncogenicity, and limitations of viral vectors8,9.
Cas9-based genome editing agents can mediate targeted gene disruption or repair10–13. For applications that seek a one-time, permanent modification of genomic DNA, we and others have shown that the delivery of non-replicable, transient Cas9:single guide RNA (sgRNA) ribonucleotide protein (RNPs) complexes in vivo offer improved DNA specificity and potentially greater safety and applicability14,15 compared with methods that introduce DNA expressing these agents. Approximately 20% of alleles associated with genetic deafness are dominantly inherited1. Since Cas9:sgRNA complexes can efficiently disrupt genes through end-joining processes, we sought to design Cas9:sgRNA complexes that selectively disrupt dominant alleles associated with hearing loss.
Many genes linked to genetic hearing loss affect the function of sensory hair cells, which transduce acoustic vibrations into electrical nerve signals. Tmc1 protein is an essential component of mechanotransduction channels in mammalian hair cells16. Mutations in TMC1 have been linked to recessive and dominant genetic deafness17. A dominant-negative missense mutation in TMC1 (p.M418K, c.T1253A) causes reduced single-channel current levels and calcium permeability16 and progressive post-lingual sensorineural hearing loss in humans18–20. The Bth/+ mouse model carries the orthologous missense mutation (p.M412K, c.T1235A) in the mouse Tmc1 gene and exhibits progressive auditory response threshold elevation and progressive hair cell loss beginning at one month21. Since the orthologous mutations in human and mouse both cause progressive, profound hearing loss, the Bth/+ mouse is a promising model for the development of treatment strategies21.
We began by developing a genome editing strategy that preferentially disrupts the mouse mutant Tmc1 (Bth) allele. To distinguish the mutant and wild-type alleles, we identified sgRNAs that target Tmc1 at sites that include the T1235A mutation and a nearby NGG protospacer-adjacent motif (PAM) sequence required by Streptococcus pyogenes Cas9. We identified three candidate sgRNAs (Tmc1-mut1, Tmc1-mut2, and Tmc1-mut3) that place the Bth mutation at position 11, 12, or 15, respectively, of the spacer, counting the PAM as positions 21-23 (Fig. 1a). Mismatches between the sgRNA and genomic DNA that are close to the PAM are poorly tolerated by Cas910, increasing the likelihood of selective editing of the Bth mutant allele. A fourth sgRNA, Tmc1-mut4, is a truncated version of Tmc1-mut3 designed to increase genome editing DNA specificity22. We evaluated the ability of these four sgRNAs when complexed with Cas9 to cleave either the wild-type Tmc1 or the Bth allele in vitro. All sgRNAs tested preferentially cleaved the Bth allele, with Tmc1-mut3 exhibiting the greatest selectivity (Extended Data Fig 1a and 1b).
Figure 1. Design of a genome-editing strategy to disrupt the Bth mutant allele.

(a) SpCas9 sgRNAs were designed to target the mutant Tmc1 Bth allele, in which T1235 is changed to A (red). The protospacer (blue arrows) of each Bth-targeting sgRNA contains a complementary T (red) that pairs with the T1235A mutation in the Bth allele, but that forms a mismatch with wild-type Tmc1 allele. (b) Lipid-mediated delivery of Cas9:sgRNA complexes into primary fibroblasts derived from wild-type or homozygous Bth/Bth mice. 100 nM of purified Cas9 protein and 100 nM of each sgRNA shown were delivered using Lipofectamine 2000. Indels were quantitated by HTS. Values and error bars represent the mean ± SD of three or more independent biological replicates.
We performed lipid-mediated delivery of Cas9:sgRNA RNP complexes into cultured primary fibroblasts derived from wild-type or homozygous Bth/Bth mice to evaluate the allele specificity of genomic DNA modification in mouse cells. We delivered Cas9 complexed with each of the four sgRNAs using Lipofectamine 2000 into both wild-type and Bth/Bth mutant fibroblasts. RNP delivery into these primary fibroblasts was 2- to 4-fold less efficient than with commonly used HEK293T cells (Extended Data Fig. 1c). The highest rate of targeted insertions and deletions (indels) in mutant Bth/Bth fibroblasts (10%) was observed with Cas9:Tmc1-mut3 RNPs, while lower indel frequencies (0.74%-4.1%) were observed using the other sgRNAs (Fig. 1b). In contrast, all tested sgRNAs edited the wild-type Tmc1 locus much less efficiently in wild-type fibroblasts (0.066%-1.6% indels) (Fig. 1b). Notably, Cas9:Tmc1-mut3 modified the mutant Bth allele 23-fold more efficiently than the wild-type allele (Fig. 1b and Extended Data Fig. 1d). We also prepared three corresponding wild-type Tmc1-targeting sgRNAs (Tmc1-wt1, Tmc1-wt2, and Tmc1-wt3) that lack the T1235A mutation. These sgRNAs edited wild-type fibroblasts on average 10-fold more efficiently than Bth/Bth fibroblasts (Extended Data Fig. 1e), confirming that the observed allele selectivities did not arise from the inability of the wild-type Tmc1 allele to be edited.
We tested 17 cationic lipids for their ability to deliver the Cas9:Tmc1-mut3 RNP into Bth/Bth fibroblasts. Several lipids supported substantial modification of the target locus, including RNAiMAX (7.7%), CRISPRMAX (8.9%), and Lipofectamine 2000 (12%) (Extended Data Fig. 2). In contrast, treatment of wild-type fibroblasts with Cas9:Tmc1-mut3 and the same suite of 17 lipids resulted in low (≤ 0.5%) indel rates (Extended Data Fig. 2a and 2b). These results collectively suggest that the target mutant Tmc1 locus can be preferentially disrupted by Cas9:guide RNA complexes.
Exposure of cells to Cas9:sgRNA agents typically results in the modification of both on-target and off-target loci10, 23. We used both the GUIDE-seq method23 and computational prediction24 to identify potential off-target loci that could be modified by Cas9:Tmc1-mut3 exposure. Ten off-target sites containing up to six mismatches in the protospacer region of Tmc1-mut3 sgRNA were identified by GUIDE-seq (Extended Data Fig. 3a). None of these off-target loci are known to be associated with hearing function (Extended Data Table 1a). We measured the indel frequency at each off-target site by HTS in Cas9:Tmc1-mut3 treated Bth/Bth primary mouse fibroblasts following plasmid DNA nucleofection or RNP delivery. Plasmid nucleofection resulted in 0.68%-8.1%indels observed at nine of the ten GUIDE-seq-identified off-target sites (Extended Data Fig. 3b and Table 1a). In contrast, following RNP delivery, modification of only one off-target site (off-T1, 1.2% indels) was detected (Extended Data Fig. 3b), consistent with our earlier findings that RNP delivery greatly reduces off-target editing compared with DNA delivery15. Among the computationally predicted off-target sites24, only the two (off-T1′ and off-T2′) that were also identified as off-targets by GUIDE-seq were observed to undergo modification (Extended Data Table 1b). Together, these results suggest that delivery of Cas9:Tmc1-mut3 RNP complexes into Bth/Bth cells lead to minimal off-target modification, and that phenotypes affecting hearing are unlikely to arise from off-target modification.
To evaluate the ability of the Cas9:Tmc1-mut3 sgRNA complex to target the Bth allele in hair cells in vivo, we complexed Cas9:Tmc1-mut3 sgRNA with Lipofectamine 2000 and injected the resulting mixture into the scala media of neonatal mice by cochleostomy. Neonatal cochlear hair cells express Tmc1 and Tmc2, both of which can enable sensory transduction. To isolate the effect of editing the Bth allele, we injected the Cas9:Tmc1-mut3 sgRNA:lipid complex into Tmc1 Bth/Δ;Tmc2 Δ/Δ mice16 to avoid transduction current contributions from Tmc2 and from wild-type Tmc1. We recorded sensory transduction currents from inner hair cells (IHCs) after injection with Cas9:Tmc1-mut3:lipid complex, or with a control targeting an unrelated gfp gene. We observed a significant decline in transduction current amplitudes in Tmc1 Bth/Δ;Tmc2 Δ/Δ mice following injection with Cas9:Tmc1-mut3:lipid complexes, consistent with disruption of the Bth allele in sensory hair cells in vivo, but not with Cas9:GFP sgRNA:lipid complexes (Fig. 2a and 2b).
Figure 2. Effects of Cas9:Tmc1-mut3 sgRNA:lipid injection on hair-cell function and hair-cell survival in mice.

(a) Representative transduction currents from inner hair cells (IHCs) of P0-P1 wild-type or Tmc1 Bth/Δ;Tmc2Δ/Δ mice that were uninjected, or injected with Cas9:Tmc1-mut3:lipid complex. (b) Maximal transduction current amplitudes for 135 IHCs from P1 wild-type C57B/L6 and Tmc1 Bth/Δ;Tmc2Δ/Δ mice. (i): uninjected wild-type C57B/L6 mice; (ii): wild-type C57B/L6 mice injected with Cas9:Tmc1-wt3:lipid; (iii): uninjected Tmc1 Bth/Δ;Tmc2Δ/Δ mice; (iv): Tmc1 Bth/Δ;Tmc2Δ/Δ mice injected with Cas9:GFP-targeting sgRNA:lipid; (v): Tmc1 Bth/Δ;Tmc2Δ/Δ mice injected with Cas9:Tmc1-mut3:lipid. Horizontal lines and error bars reflect mean ± SD. (c–e) Representative confocal microscopy images from (c) an uninjected Bth/+ cochlea; (d) the contralateral cochlea of the mouse in (c) injected with Cas9:Tmc1-mut3:lipid complex at P1; (e) an untreated wild-type C3H cochlea. Numbers in pink indicate approximate frequencies (in kHz) sensed by each region. Scale bars = 50 μm. (f, g) Quantification of IHC (f) and OHC (g) survival in Bth/+ mice relative to wild-type C3H mice 8 weeks after Cas9:Tmc1-mut3:lipid injection (blue) compared to uninjected (red) contralateral ears. Values and error bars reflect the means and SEMs of five biological replicates. Statistical tests in (b) are two-population T-tests, and in (f) and (g) are two-way ANOVA with Bonferroni correction: **p < 0.01, ***p < 0.001, and ****p < 0.0001.
In Tmc1 Bth/+;Tmc2 +/+ mice (abbreviated as Bth/+ mice below), IHCs followed by the outer hair cells (OHCs) undergo progressive death21. To determine the effect of Cas9:Tmc1-mut3 sgRNA on Bth/+ hair cell survival, we injected Cas9:Tmc1-mut3:lipid complex into the scala media of P1 mice and harvested injected and uninjected cochleae after 8 weeks. Uninjected ears exhibited significant loss of IHCs, and partial degeneration of OHCs (Fig. 2c, 2f, and 2g) compared with wild-type C3H ears (Fig. 2e). In injected ears, IHC and OHC survival were substantially enhanced in those regions (Fig. 2d, 2f, and 2g). Stereocilia bundles were observed on surviving IHCs in injected ears, but were absent in uninjected ears in the basal and middle turns (Extended Data Fig. 4a). These results suggest that Cas9:Tmc1-mut3:lipid injection in vivo promotes hair cell survival in Bth/+ mice. The strong differences between treated and untreated ears suggests that sporadic disruption of Bth may benefit not only edited hair cells, but also surrounding hair cells, consistent with previous findings25.
To study the effect of Cas9:Tmc1-mut3:lipid injection on cochlear function in Bth/+ mice, we measured auditory brainstem responses (ABRs) that measure the sound-evoked neural output of the cochlea, as well as distortion product otoacoustic emissions (DPOAEs) that measure the amplification provided by OHCs21. In uninjected ears we observed profound attenuation of cochlear neural responses, with ABR thresholds ranging from 70-90 dB at 4 weeks (Fig. 3a and Extended Data Fig. 4b). At 4 weeks, DPOAE threshold elevations in Bth/+ mice were smaller than ABR threshold elevations (Extended Data Fig. 5a), consistent with prior reports that IHCs are more severely affected than OHCs in Bth/+ mice. Four weeks after Cas9:Tmc1-mut3:lipid injection, treated Bth/+ ears showed substantially enhanced cochlear function, with lower ABR thresholds relative to uninjected ears at all frequencies below 45 kHz (Fig. 3a). Significant hearing preservation was detected from 8-23 kHz, with average ABR thresholds 15 dB lower for treated ears than untreated contralateral ears (Fig. 3a; Supplementary Table 1). DPOAE thresholds were slightly elevated in the injected ears, consistent with OHC damage, perhaps from the injection procedure (Extended Data Fig. 5). We also observed greater ABR Wave 1 amplitudes, and a more normal ABR waveform pattern, in injected ears compared to uninjected controls (Fig. 3b and 3c). Together, these results show that injection of neonatal Bth/+ mice with Cas9:Tmc1-mut3:lipid complexes reduces progressive hearing loss.
Figure 3. Cas9:Tmc1-mut3 sgRNA:lipid injections reduce hearing loss in Bth/+ mice.
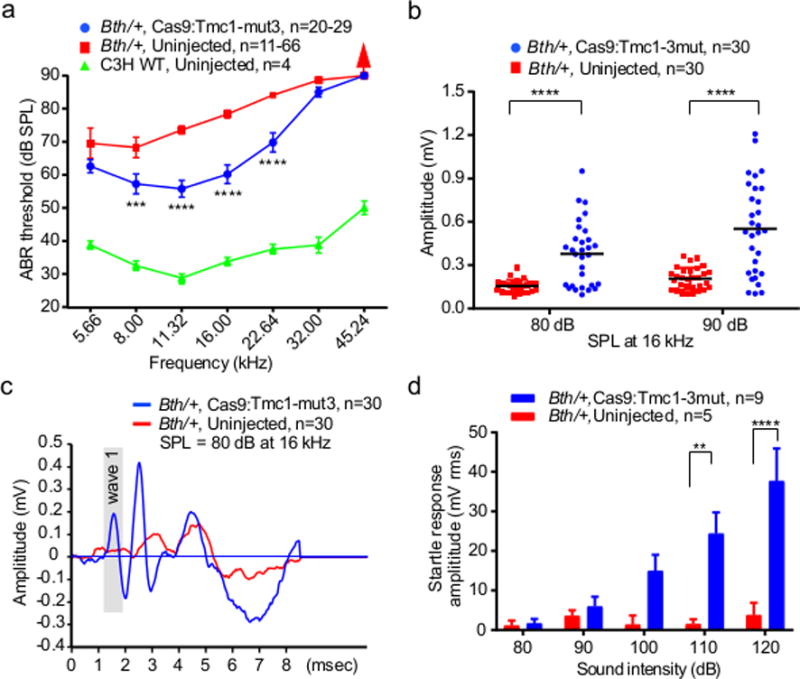
(a) ABR thresholds in Bth/+ ears injected with Cas9:Tmc1-mut3:lipid (blue), uninjected Bth/+ ears (red), and wild-type C3H ears (green) after 4 weeks. (b) Amplitudes of ABR Wave 1 at 16 kHz in Cas9:Tmc1-mut3:lipid-injected ears (blue) compared with uninjected ears (red) after 4 weeks. Horizontal bars are mean values. (c) Mean ABR waveforms in Cas9:Tmc1-mut3:lipid-injected ears (blue) and uninjected ears (red). (d) Startle responses in Cas9:Tmc1-mut3:lipid-injected mice (blue) and in uninjected mice (red) 8 weeks after treatment. Red arrows in (a) indicate no ABR response at the highest stimulus level tested (90 dB). See also Supplementary Table 2. Statistical tests were two-way ANOVA with Bonferroni correction for multiple comparisons: **p < 0.01, ***p < 0.001, and ****p < 0.0001. Values and error bars reflect mean ± SEM.
To test if hearing loss amelioration requires the mutant Bth allele-specific sgRNA, we injected Cas9:Tmc1-wt3:lipid complexes targeting the wild-type Tmc1 allele rather than the Bth mutant allele into P1-2 Bth/+ mice. After four weeks, ABR thresholds in the injected ears were similar to, or worse than, contralateral uninjected ears (Extended Data Fig. 6a; Supplementary Table 1), consistent with the inability of Cas9:Tmc1-wt3 to efficiently disrupt the Bth allele (Extended Data Fig. 1e), and possible disruption of wild-type Tmc1. Injection of Cas9:sgRNA:lipid complexes targeting an unrelated gene (gfp) did not significantly affect ABR thresholds at most tested frequencies in Bth/+ mice (Extended Data Fig. 6b). To test if preservation of cochlear function requires Cas9 nuclease activity, rather than transcriptional interference from Cas9 binding to Tmc1, we treated Bth/+ mice with catalytically inactive dCas910 complexed with Tmc1-mut1 and observed no evidence of hearing preservation (Extended Data Fig. 5d and 6c; Supplementary Table 1). To evaluate the effects of the treatment on normal mice, we injected Cas9:Tmc1-mut3:lipid into wild-type C3H mice, the genetic background of the Bth/+ mice. We observed similar or slightly elevated ABR thresholds in injected ears relative to uninjected ears 4 weeks after treatment (Extended Data Fig. 6d and 6e), suggesting that Cas9:Tmc1-mut3 does not modify wild-type Tmc1 efficiently enough to substantially affect hearing. Finally, injection of Cas9 and lipid without sgRNA did not improve ABR or DPOAE thresholds (Extended Data Fig. 6f and 6g). Collectively, these results establish that hearing preservation is dependent on sgRNA allele specificity, Cas9 DNA cleavage activity, and the presence of the Bth allele. We also characterized cochlear function of Bth/+ mice 8 weeks after treatment. Mean ABR thresholds following Cas9:Tmc1-mut3:lipid injection remained lower than uninjected controls from 5.7-23 kHz, although the average improvement decreased compared to 4 weeks post-treatment (Extended Data Fig. 4c and 4d), potentially due to continued progressive hearing loss in the non-edited hair cells.
As a behavioral measure of hearing rescue, we assessed acoustic startle responses 8 weeks after injection. In uninjected Bth/+ mice, no startle response was detected following stimuli at 120 dB. In contrast, significant startle responses were detected in Cas9:Tmc1-mut3:lipid-injected Bth/+ mice following stimulus at 110 and 120 dB (Fig. 3d and Extended Data Fig. 4e), demonstrating that hearing preservation upon treatment also preserves an acoustic behavioral reflex.
To evaluate the ability of each of the other Bth-targeting sgRNAs to mediate hearing rescue in vivo, we also injected Tmc1-mut1, Tmc1-mut2, and Tmc1-mut4 complexed with Cas9 into neonatal Bth/+ cochleae, and observed varying degrees of enhanced cochlear function (Extended Data Fig. 7). Thus, while Tmc1-mut3 resulted in the most robust hearing preservation, other sgRNAs targeting the mutant Bth allele also partially enhanced cochlear function.
To test if RNP delivery of editing agents in adult mouse inner ears supports genome editing in hair cells, we injected Cas9:GFP-targeting sgRNA:lipid complexes into the cochlea of 6-week-old Atoh1-GFP mice. Two weeks after injection, GFP fluorescence loss near the injection sites suggested target gene disruption at 25±2.1% efficiency (Extended Data Fig. 8), comparable to our previous observations of 20% GFP editing in neonatal hair cells15. These results suggest that this approach may be applicable to dominant genetic deafness that manifest with late-onset hearing loss.
To confirm that in vivo treatment of Bth/+ mice with Cas9:Tmc1-mut3 sgRNA disrupted the Bth allele, we sequenced DNA from cochlea tissue collected from injected Bth/+ and untreated Bth/+ mice. After injection at P1, tissues were harvested at P5 and separated into organ of Corti (containing hair cells), spiral ganglion, and spiral ligament samples (Extended Data Fig. 9a and 9b). We estimated the fraction of hair cells in dissected cochlear tissue to be only ~1.5% of the total cells used for DNA sequencing (Extended Data Fig. 9a and 9b). Nevertheless, we observed unambiguous indels at the Bth locus in cochlear tissue from treated mice (Fig. 4a). The organ of Corti samples contained average Tmc1 editing of 0.92% of total sequenced DNA, which corresponds to ~1.8% Bth allele disruption (Fig. 4a). We also isolated samples of much smaller numbers of cells (up to a few dozen, mostly hair cells) from treated mice. Decreasing the number of cells entering the genomic DNA amplification and sequencing process increased the observed editing percentage to as high as 10% Bth allele disruption, but also elevated background Tmc1 indel rates of untreated mice to an average of 0.82±0.57% and a maximum of 1.6%, likely reflecting increased noise from processing of minute quantities of genomic DNA. No indel frequencies above that of untreated controls at any of the above-identified off-target sites were observed in Cas9:Tmc1-mut3:lipid-treated tissues (Extended Data Fig. 9c). Together, these observations confirm in vivo editing of the Tmc1 locus from Cas9:Tmc1-mut3:lipid treatment with no detected editing at GUIDE-seq-identified off-target loci.
Figure 4. Genome modification at Tmc1 induced by lipid-mediated delivery of Cas9:Tmc1-mut3 RNP into Bth/+ mice.
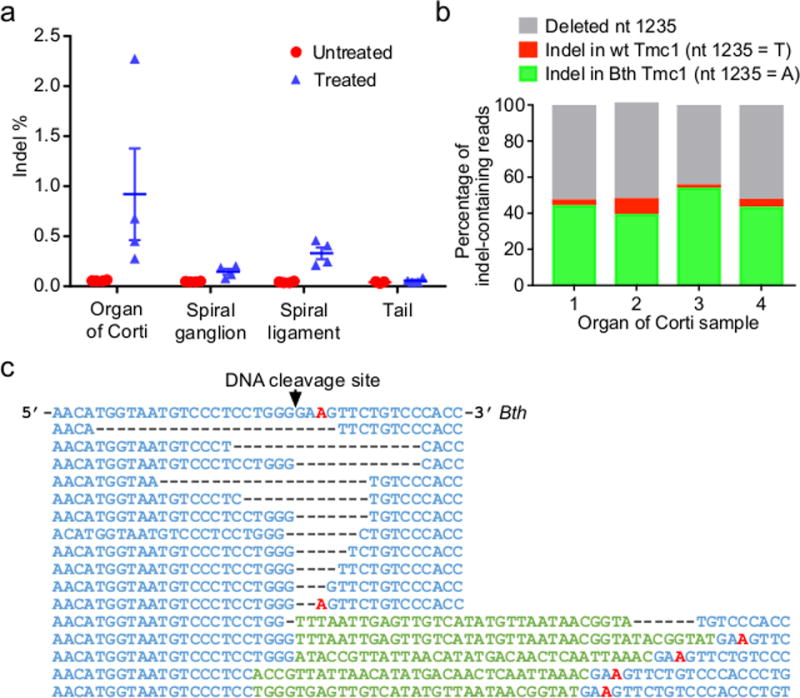
(a) Tmc1 indel frequencies from tissue samples 5 days after injection of Cas9:Tmc1-mut3:lipid (blue) or from uninjected mice (red). Horizontal lines and error bars reflect mean ± SEM. Note that Bth allele indel frequencies in these heterozygous mice are approximately double the observed indel frequencies. (b) Analysis of indel-containing Tmc1 sequencing reads from four injected organ of Corti samples in (a). (c) The most abundant 16 Tmc1 sequences, grouped by similarity, from organ of Corti samples in (b). The T1235A Bth mutation is red.
Importantly, an analysis of indel-containing Tmc1 sequencing reads from treated Bth/+ mice allowed us to directly assess the allele specificity of Cas9:Tmc1-mut3 in vivo. Of 11,694 sequencing reads containing indels from four treated organ of Corti samples, 6,118 (52%) contained an intact nucleotide at Tmc1 position 1235. Of these, 5,736 (94%) contained modification of the mutant Bth allele, while only 382 (6%) contained modification of the wild-type Tmc1 allele (Fig. 4b). Therefore, samples following treatment on average contained 15-fold higher modification of the Bth allele over the wild-type allele (Fig. 4b and 4c), demonstrating selective disruption of the Bth allele in Bth/+ mice, consistent with observed hearing phenotypes, even though the Bth and wild-type Tmc1 alleles differ only at a single base pair.
This work establishes cationic lipid-mediated Cas9:sgRNA complex delivery in vivo to achieve allele-specific gene disruption in a mouse model of a human genetic disease, resulting in amelioration of a disease phenotype. Our results suggest the potential of such an approach for the treatment of autosomal dominant hearing loss diseases related to hair cell dysfunction, and provide a complementary strategy to other approaches that use antisense oligos (ASO) or RNA interference6, 25. The genome editing strategy developed here may inform the future development of a DNA-free, virus-free, one-time treatment for certain genetic hearing loss disorders.
METHODS
Primary cell culture
Wild-type, Bth/+ and Bth/Bth fibroblasts were obtained from P5 pups. Mice were euthanized and cleaned with 70% ethanol. 1-2 cm2 underarm skin fragments were excised and immerged in cold HBSS (ThermoFisher). Subcutaneous fat was removed by forceps. Skin fragments were cut to ~1 mm2 pieces with a 25G 5/8″ Syringe (1180125058, Covidien). Tissues were digested with 0.5 mg/mL Liberase DL (Sigma 5401160001) at 37 °C for 1 hour with occasional pipetting up and down to break cell clumps. Warm culture medium (1:1 DMEM:F12 media (ThermoFisher) with 15% FBS (ThermoFisher) and 100 U/mL penicillin+streptomycin (ThermoFisher) was added to stop the enzyme digestion. The solution was filtered with a 70-μm cell strainer (Falcon) and centrifuged at 200 g for 5 min. The pellet was resuspended in culture medium and transferred to a 25-mL culture flask, then incubated at 37°C, 5% CO2, 3% O2. Fibroblasts were cultured for 2 ~ 3 days to reach ~90% confluence, then passaged in 100-mL flasks in DMEM plus GlutaMax (ThermoFisher) supplemented with 10% (v/v) fetal bovine serum (FBS) at 37 °C with 5% CO2.
Delivery of proteins complexed with cationic lipids in mouse fibroblasts
Cultured fibroblast cells were plated in 24-well format (500 μL well volume) in Dulbecco’s Modified Eagle’s Media plus GlutaMAX (DMEM, Life Technologies) with 10% FBS (no antibiotics) at a cell density sufficient to reach ~80% confluence at the time of usage. Purified sgRNA was incubated with Cas9 protein for 5 min before complexing with cationic lipid15, 26. Delivery of Cas9:sgRNA was performed by combining 100 nM RNP complex with 3 μL cationic lipid in 50 μL OPTIMEM media (Life Technologies) according to the manufacturer’s protocol for DNA plasmid transfection. The above mixture containing cationic lipid and RNP was then added to cells. All complexing steps were performed at room temperature. Cells were harvested and genomic DNA was extracted for sequencing ~96 h after treatment.
GUIDE-seq and data analysis
Mouse fibroblasts were transfected using 1,000 ng Cas9 plasmid (pCas9), 300 ng sgRNA plasmid (pTmc1-mut3 sgRNA), and 50 pmol GUIDE-seq double-stranded oligodeoxynucleotides (dsODN) using a LONZA 4D-Nucleofector. Transfection programs were optimized following manufacturer’s instructions (CA158 and CA189, P2 Primary Cell 4D-Nucleofector® X Kit). 400 ng of pmaxGFP Control Vector (LONZA) was added to the nucleofection solution to assess nucleofection efficiencies in primary cells. Media was replaced ~16 h after nucleofection and cells were harvested for genomic DNA extraction after ~96 h. For GUIDE-seq off-target DNA cleavage analysis, pCas9, pTmc1-mut3 sgRNA, pmaxGFP, and dsODN were nucleofected into Bth/+ heterozygous mouse primary fibroblasts. A sample nucleofected with dsODN only served as a negative control. ~400 ng of genomic DNA for each sample was sheared acoustically using a Covaris m220 sonicator to an average length of 500 bp in 130 μL TE buffer. Each sample was sequenced on an Illumina Miseq following previously described protocols23. Reads were consolidated first by their Illumina indexes and then by the 8-nt molecular index that defines a single pre-PCR template fragment. The consolidated reads were mapped to the mouse reference genome (GRCm38) using BWA-MEM. Off-target sites were identified by first mapping the start position of the amplified sequences using a 10-bp sliding window, then retrieving the reference sequence around the site. Given the size of some of the deletions, the number of base pairs used as the flanking sequence was increased to 100 bp. The retrieved sequences were aligned to the Cas9 target sequence using a Smith-Waterman local-alignment algorithm. The negative control sample treated with the dsODN but no Cas9 or sgRNA was used to assess background.
High-throughput DNA sequencing of genomic DNA samples
Treated cells or tissues were harvested after four days and genomic DNA was isolated using the Agencourt DNAdvance Genomic DNA Isolation Kit (Beckman Coulter) according to the manufacturer’s instructions. On-target and off-target genomic regions of interest were amplified by PCR with flanking HTS primer pairs listed in the Supplementary Sequences. PCR amplification was carried out with Phusion high-fidelity DNA polymerase (ThermoFisher) according to the manufacturer’s instructions using ~100 ng of genomic DNA as a template. PCR cycle numbers were chosen to ensure the reaction was stopped during the log-linear range of amplification. PCR products were purified using RapidTips (Diffinity Genomics). Purified DNA was amplified by PCR with primers containing sequencing adaptors. The products were purified by gel electrophoresis and quantified using the Quant-iT™ PicoGreen dsDNA Assay Kit (ThermoFisher) and KAPA Library Quantification Kit-Illumina (KAPA Biosystems). Samples were sequenced on an Illumina MiSeq as previously described27.
Sequencing reads were demultiplexed using MiSeq Reporter (Illumina), and individual FASTQ files were analyzed with a custom Matlab script (Supplementary Note). Each read was pairwise aligned to the appropriate reference sequence using the Smith-Waterman algorithm. Base calls with a Q-score below 31 were excluded from calculating editing frequencies. Sequencing reads were scanned for exact matches to two 10-bp sequences that flank both sides of a window in which indels might occur. If no exact matches were located, the read was excluded from analysis. If the length of this indel window exactly matched the reference sequence the read was classified as not containing an indel. If the indel window was one or more bases longer or shorter than the reference sequence, then the sequencing read was classified as an insertion or deletion, respectively.
General in vivo experiments
All in vivo experiments met NIH guidelines for the care and use of laboratory animals and were approved by the Massachusetts Eye & Ear Infirmary IACUC committee. Isogenic heterozygous Tmc1 Bth/+ mice maintained on a C3HeB/FeJ (C3H) background were obtained as a gift from Dr. Andrew Griffith21, and inbred with wild-type C3H mice obtained from Jackson Laboratory. Crossbred homozygous C3H-Tmc1 Bth/Bth were caged with C3H mice to generate heterozygous Tmc1 Bth/+ mice. All mice were genotyped by Transnetyx (Cordova, TN). For mechanotransduction experiments, two genotypes of Tmc mutant mice were used: Tmc1 Bth/Bth;Tmc2 Δ/Δ and Tmc1 Δ/Δ;Tmc2 Δ/Δ as previously reported16.
Microinjection into the inner ear of neonatal mice
A total of 106 Tmc1 Bth/+ or C3H mice (P0-2) of either sex were used for injections. The mice were randomly assigned to the different experimental groups. The final 25% of the experiments were performed in a double-blinded manner. At least 5 mice were injected in each group. All surgical procedures were done in a clean, dedicated space. Instruments were thoroughly cleaned with 70% ethanol and autoclaved prior to surgery. Fresh Cas9 and sgRNA were mixed before injection at a final concentration of 25 μM. 1 μL Lipofectamine 2000 was mixed with 1 μL Cas9:sgRNA RNP and incubated for 20 min at room temperature. Mice were anesthetized by hyperthermia on ice. Cochleostomy was performed by preauricular incision to expose the cochlear bulla. Anatomic landmarks included the stapedial artery and tympanic ring, which were identified before injection. Glass micropipettes (4878, WPI) were pulled with a micropipette puller (PP83, Narishige) to a final OD of ~10 μm. Needles held by a Nanoliter 2000 micromanipulator (WPI) were used to manually deliver the Cas9:sgRNA:lipid complexes into the scala media, which allows access to inner ear cells. The injection sites were base, middle, and apex-middle turn of the cochlea. The volume for each injection was 0.3 μL with a total volume of 0.9 μL per cochlea. The release rate was 69 nL/min, controlled by MICRO4 microinjection controller (WPI).
Microinjection into adult inner ear by canalostomy
Three 6-week-old Atoh1-GFP mice28 were injected with Cas9:GFP-targeting sgRNA:lipid complex, with the same concentration and volume for each component as used in injection into neonatal inner ear. Mice were anesthetized by intraperitoneal injection of combination of xylazine (10 mg/kg) and ketamine (100 mg/kg). The right post-auricular region was exposed by shaving and disinfected by 10% povidone iodine. For canalostomy, a 10-mm postauricular incision was made under the operating microscope, and the right pinna and the sternocleidomastoid muscle were extracted to expose the posterior semicircular canal (PSCC) that was located in the margin of temporal bone. We used a Bonn micro probe (Fine Science Tools, Foster City, CA) to drill a small hole on the PSCC, then left it open for a few minutes until no obvious perilymph leakage was observed. The tip of the polyimide tube (ID 0.0039 inches, OD 0.0049 inches, Microlumen) was inserted into the PSCC toward the ampulla. The hole was sealed with tissue adhesive (3M Vetbond, St. Paul, MN), and the lack of fluid leakage indicated the tightness of the sealing. The tubing was cut after injection, with approximately 5 mm of tubing left connected to the PSCC and sealed with tissue adhesive. The volume for each injection was 1 μL per cochlea. The release rate was 169 nL/min, controlled by MICRO4 microinjection controller (WPI). The skin was closed with 5-0 nylon suture (Ethicon Inc., Somerville, NJ). The total surgery time was approximately 20 min, including a 6-min injection period.
Acoustic testing
Auditory brainstem response (ABR) and distortion product otoacoustic emissions (DPOAE) recording were conducted as described previously29 at 32 °C in a soundproof chamber. Mice of either sex were anesthetized with xylazine (10 mg/kg, i.p.) and ketamine (100 mg/kg, i.p.). Acoustic stimuli were delivered through a custom acoustic assembly consisting of two miniature dynamic electrostatic earphones (CDMG15008-03A, CUI) to generate primary tones and a miniature microphone (FG-23329-PO7, Knowles) to record ear-canal sound pressure near the eardrum. Custom LabVIEW software controlling National Instruments 24-bit soundcards (6052E) generated all ABR/DPOAE stimuli and recorded all responses.
For ABR measurements, needle electrodes were inserted at the vertex and ventral edge of the pinna, with a ground reference near the tail. ABR potentials were evoked with 5 ms tone pips (0.5 ms rise–fall with a cos2 onset, delivered at 35/s). The response was amplified 10,000-fold, filtered (100 Hz–3 kHz passband), digitized, and averaged (1024 responses) at each SPL. The sound level was raised in 5 dB steps from 30 dB below threshold up to 90 dB SPL at frequencies from 5.66-45.24 kHz (in half-octave steps). Following visual inspection of stacked waveforms, “threshold” was defined as the lowest SPL level at which any wave could be detected. In general, thresholds were defined by three independent observers. Wave 1 amplitude was defined as the difference between the average of the 1-ms pre-stimulus baseline and the Wave 1 peak (P1), after additional high-pass filtering to remove low-frequency baseline shifts.
For DPOAE measurements, the cubic distortion product was measured in response to primaries f1 and f2. The primary tones were set so that the frequency ratio (f2/f1) was 1.2 and so that the f2 level was 10 dB below the f1 level. For each f2/f1 primary pair, primaries were swept in 5 dB steps from 20 dB SPL to 80 dB SPL (for f2). At each level, the amplitude of the DPOAE at 2f1-f2 was extracted from the averaged spectra, along with the noise floor. Threshold was computed by interpolation as the f2 level required to produce a DPOAE at 5 dB SPL.
Acoustic startle reflex
Mice were placed into a small, acoustically transparent cage resting atop a piezoelectric force plate in a sound attenuated booth. Acoustic stimuli and amplified force plate signals were encoded by a digital signal processor (Tucker-Davis Technologies, RX6) using LabView scripts (National Instruments). Mice were placed in silence for 2 min and 60 dB broadband white noise for 5 min to acclimate to the test environment before real measurements. Broadband white noise was presented at a background level of 60 dB SPL throughout the experiment and a 16-kHz tone was presented at randomized intervals from an overhead speaker (80 dB to 120 dB SPL, 20 msec duration with 0 msec onset and offset ramps). Ten repetitions were recorded for each of the intensities per test subject. Startle response amplitude was measured as the root mean square (RMS) voltage of the force plate signal shortly after sound presentation.
Immunohistochemistry and histology
Injected and non-injected cochleae were harvested after animals were sacrificed by CO2 inhalation. Temporal bones were fixed in 4% paraformaldehyde at 4°C overnight, then decalcified in 120 mM EDTA at least 1 week. The cochleae were dissected in pieces from the decalcified tissue for whole-mount immunofluorescence. Tissues were infiltrated with 0.3% Triton X-100 and blocked with 8% donkey serum for 1 h before applying the first antibody. 1:500 rabbit anti-MYO7A (#25-6790, Proteus BioSciences), 1:750 chicken anti-GFP (ab13970, Abcam) and 1:350 goat anti-SOX2 (sc-17320, Santa Cruz Biotechnology) were used at room temperature overnight. The second antibody was incubated for 1 h after three rinses with PBS rinses. All Alexafluor secondary antibodies were from Invitrogen: donkey anti-rabbit Alex488 (A21206) or Alex 594 (A21207), donkey anti goat Alex594 (A11058) or Alexa-488-phalloidin (A12379) and goat anti-chicken Alex488 (A-11039) were used as a 1:500 dilution. Specimens were mounted in ProLong Gold Antifade Mountant medium (P36930, Life Technologies). Confocal images were taken with a Leica TCS SP5 microscope using a 20× or 63× glycerin-immersion lens, with or without digital zoom. For IHC and OHC counting, we acquired z-stacks by maximum intensity projections of z-stacks for each segments by imageJ (NIH image), and composite images showing the whole cochlea were constructed in Adobe Photoshop CS3 to show the whole turn of cochlea. A frequency map was constructed for each case by measuring the spiral extent of all the dissected cochlear pieces and converting cochlear location to frequency using a plug-in of ImageJ (http://www.masseyeandear.org/research/ent/eaton-peabody/epl-histology-resources/imagej-plugin-for-cochlear-frequency-mapping-in-whole-mounts/). MYO7A positive IHCs and OHC were counted in the cochlear regions that respond to different sound frequencies, and any segments containing dissection-related damage were omitted from further analysis.
Hair cell transduction current recording
Tmc1 Bth/Bth;Tmc2 Δ/Δ and Tmc1 Δ/Δ;Tmc2 Δ/Δ littermates were injected with 0.9 μL Cas9:Tmc1-mut3 sgRNA:Lipofectamine 2000 or Cas9:GFP sgRNA: Wild-type C57B/L6 mice were injected with 0.9 μL Cas9:Tmc1-wt3 sgRNA:Lipofectamine 2000 Lipofectamine 2000 at P0-P1 via the cochleostomy approach. Cochleae were harvested at P5-P6 and cultured in MEM(1X) + GlutaMAX™-I medium with 1% FBS at 37 °C, 5% CO2 for up to 15 days. For recording, the organs of Corti were bathed in standard artificial perilymph containing 137 mM NaCl, 0.7 mM NaH2PO4, 5.8 mM KCl, 1.3 mM CaCl2, 0.9 mM MgCl2, 10 mM HEPES, and 5.6 mM D-glucose. Vitamins (1:50) and amino acids (1:100) were added to the solution from concentrates (Invitrogen, ThermoFisher Scientific), and NaOH was used to adjust the final pH to 7.4 (~310 mOsm/kg). Recording pipettes (2-4 MΩ) were pulled from R6 capillary glass (King Precision Glass) and filled with intracellular solution containing 135 mM CsCl, 5 mM HEPES, 5 mM EGTA, 2.5 mM MgCl2, 2.5 mM Na2-ATP, and 0.1 mM CaCl2; CsOH was used to adjust the final pH to 7.4 (~285 mOsm/kg). Whole-cell, tight-seal, voltage-clamp recordings were conducted at -84 mV at room temperature (22-24 °C) with an Axopatch 200B amplifier (Molecular Devices). Hair bundles were deflected with a stiff glass probe fabricated from capillary glass with a fire polisher (MF-200, World Precision Instruments) for creating a rounded probe tip of ~3-5 μm in diameter. Probes were mounted on a PICMA Chip piezo actuator (Physik Instrument, Karlsruhe, Germany) and driven by an LVPZT amplifier (E-500.00, Physik Instrumente, Karlsruhe, Germany). Sensory-transduction currents were recorded from uninjected and Cas9:sgRNA-treated hair cells. The data were filtered at 10 kHz with a low-pass Bessel filter and digitized at > 20 kHz with a 16-bit acquisition board (Digidata 1440A, Molecular Devices) and pClamp 10 software (Molecular Devices).
Inner ear tissue dissection for HTS
Tmc1 Bth/+ mice were injected with Cas9:sgRNA at P1 as described above. All dissection instruments were thoroughly cleaned with 70% ethanol and DRNAase Free (D6002, ARgos), then autoclaved prior to dissection. Mice were euthanized at P5. Temporal bones were removed and immersed in clean PBS pH 7.4 (10010001, ThermoFisher) individually. Different forceps were used for each ear. The organ of Corti, spiral ganglion, and spiral ligament from the injected and non-injected ear, and tail tissue were all harvested under microscope from each mouse.
Hair cells isolation for HTS
Tmc1 Bth/+ mice were injected with Cas9:Tmc1-mut3 sgRNA:Lipofectamine 2000 at P1 and euthanized at P5. Cochleae were dissected and immersed in 1 μM FM 1-43FX (PA1-915, ThermoFisher) dissolved in HBSS (ThermoFisher) for 10 sec at room temperature in the dark. Cochleae were rinsed three times with HBSS and placed in 100 μL Cell Recovery Solution (354253, Discovery Labware) for 10 min at 37 °C, then transferred to 100 μL TrypleE Express Enzyme (12604013, ThermoFisher). Sensory epithelium were extracted by forceps. After incubation for 10 min at 37 °C, the tissues were pipetted up and down 30 times. FM 1-43-positive cells were isolated by 1-μL pipette under a microscope (Axiovert 200M, Carl Zeiss), then subjected to whole genome amplification by MALBAC Single Cell WGA Kit (YK001A, Yikon Genomics).
Statistical analysis
Statistical analyses were performed by two-way ANOVA with Bonferroni corrections for multiple comparisons for ABRs, DPOAEs, and acoustic startle response; and by Student’s t-test for hair cell transduction currents using Prism 6.0 (GraphPad).
Data availability
High-throughput sequencing data is deposited in the NCBI Sequence Read Archive database under accession code SRP103108.
Code availability
Labview software for cochlear function testing is available here: http://www.masseyeandear.org/research/otolaryngology/investigators/laboratories/eaton-peabody-laboratories/epl-engineering-resources. Matlab scripts used to quantify the acoustic startle response are available on request. Indel identification scripts are in the Supplementary Information.
Extended Data
Extended Data Figure 1.

Allele-selective editing of wild-type or Bth mutant Tmc1 in cleavage assays in vitro and in lipid-mediated delivery into primary fibroblasts. (a) In vitro Cas9:sgRNA-mediated Tmc1 DNA cleavage. 100 nM of a 995-bp DNA fragment containing wild-type Tmc1 (lanes 1-5) or Bth mutant Tmc1 (lanes 6-10) was incubated with 300 nM of each of the four Cas9:sgRNAs shown for 15 min at 37 °C. Expected cleavage products are 774-778 bp and 217-221 bp. M = 100-bp ladder; the lower two heavy bands are 500 and 1,000 bp. (b) Quantification of DNA cleavage in (a) by densitometry using imageJ. (c) Transfection efficiency comparison of HEK293T cells and wild-type primary fibroblasts. 50 ng GFP plasmid, 10 nM Cas9:FitC-Tmc1-mut3 sgRNA RNP, or 10 nM Cas9:CrRNA-Tmc1-mut3:atto-550-TracrRNA RNP were delivered into HEK293T cells or wild-type primary fibroblasts using 3 μL Lipofectamine 2000. For samples with GFP plasmid, the fraction of GFP-positive cells was measured by flow cytometry 24 h after delivery. For samples with Cas9:FitC-Tmc1-mut3 RNP, or Cas9:CrRNA-Tmc1-mut3:atto-550-TracrRNA RNP, media was removed 6 h after delivery. The cells were trypsinized, washed three times with 500 μL PBS containing 20 U/mL heparin, and subjected to flow cytometry. (d) Wild-type or Bth mutant Tmc1 allele editing in primary fibroblasts derived from wild-type or Bth/Bth mice as a function of the dose of Cas9:Tmc1-mut3:lipid complex. 12.5, 25, 50, 100, 200, or 400 nM of Cas9:Tmc1-mut3 were delivered into the primary fibroblasts using Lipofectamine 2000 in DMEM-FBS. (e) Lipid-mediated delivery of Cas9:sgRNA complexes into primary fibroblasts derived from wild-type or Bth/Bth mice. 100 nM of purified Cas9 protein and each wild-type Tmc1-targeting sgRNA (Tmc1-wt1, Tmc1-wt2, or Tmc1-wt3) or Bth mutant-targeting sgRNA (Tmc1-mut1, Tmc1-mut2, or Tmc1-mut3) were delivered into wild-type fibroblasts (red) and Bth/Bth fibroblasts (blue) using Lipofectamine 2000 in DMEM-FBS. Primary fibroblast cells were harvested 96 h after treatment. Genomic DNA was extracted and indels were detected by HTS. Values and error bars reflect the mean ± standard deviation of three or more biological replicates.
Extended Data Figure 2.

Delivery of Cas9:Tmc1-mut3 sgRNA complexes into primary fibroblasts derived from wild-type or homozygous Bth/Bth mice using (a) seven commercially available lipids. LPF2000 = Lipofectamine 2000; RNAiMAX = Lipofectamine® RNAiMAX; LPF3000 = Lipofectamine 3000; CRISPRMAX = Lipofectamine CRISPRMAX; LTX = Lipofectamine LTX, or (b) ten biodegradable, bioreducible lipids. Lipid 1 = 75-O14B; Lipid 2 = 76-O14B; Lipid 3 = 80-O18B; Lipid 4 = 87-O16B; Lipid 5 = 113-O18B; Lipid 6 = 306-O12B; Lipid 7 = 306-O16B; Lipid 8 = 306-O18B; Lipid 9 = 400-O12B; Lipid 10 = 400-O16B. 100 nM purified Cas9:Tmc1-mut3 RNP was delivered using 3 μL of the cationic lipid shown in DMEM-FBS. Fibroblast cells were harvested 96 h after treatment, genomic DNA was extracted, and indels were detected by HTS. (c) Synthetic route and chemical structure of lipids. (d) Commercially available amine head groups used in lipid synthesis. Lipids were synthesized as previously described26. Values and error bars reflect the mean ± standard deviation of three or more biological replicates.
Extended Data Figure 3.

Off-target sites identified by GUIDE-seq after nucleofection of DNA plasmids encoding Cas9 and Tmc1-mut3 sgRNA into primary fibroblasts from Bth/+ mice. (a) 1,000 ng Cas9 plasmid, 300 ng Tmc1-mut3 sgRNA plasmid, 400 ng pmaxGFP plasmid, and 50 pmol double-stranded oligodeoxynucleotides (dsODN) were nucleofected into Bth/+ fibroblasts using a LONZA 4D-Nucleofector. Genomic DNA was extracted 96 h after nucleofection and subjected to GUIDE-seq as previously described25. Off-T1 to off-T10 are ten off-target sites detected by GUIDE-seq. Mismatches compared to the on-target site are shown and highlighted in color. The Bth allele targeted by sgRNA Tmc1-mut3 is shown in the top row. (b) Indel frequency at the Tmc1 locus and at each of the off-target loci in Cas9:Tmc1-mut3 treated Bth/Bth primary fibroblasts following plasmid DNA nucleofection or following RNP delivery. For RNP delivery, 100 nM Cas9:Tmc1-mut3 RNP was delivered to the Bth/Bth fibroblasts using 3 μL Lipofectamine 2000. Indels were detected by HTS at the Tmc1 on-target site and at each off-target site. Red: samples nucleofected with DNA plasmids encoding Cas9 and Tmc1-mut3 sgRNA; blue: samples treated with Cas9:Tmc1-mut3 RNPs; grey: control samples nucleofected with unrelated dsDNA only.
Extended Data Figure 4.

Cas9:Tmc1-mut3:lipid injection reduce hearing loss, improve acoustic startle response, and preserve stereocilia in Bth/+ mice. (a) Phalloidin labeling showed the preservation of stereocilia of IHCs in an ear 8 weeks after injection with Cas9:Tmc1-mut3 sgRNA at three frequency locations indicated, whereas the uninjected contralateral inner ear of the same mouse showed severe degeneration of stereocilia at locations corresponding to 16 and 32 kHz. The boxes indicate the stereocilia, which are shown at the bottom of each image at higher magnification. Scale bars: 10 μm. Similar results were observed in other injected ears that were immunolabeled (n = 5). (b) Representative ABR waveforms showing reduced threshold (red traces) at 16 kHz in a Cas9:Tmc1-mut3:lipid-injected Bth/+ ear (left) compared to an uninjected contralateral ear (right) after 4 weeks. (c) 8 weeks after Cas9:Tmc1-mut3 injection into Bth/+ ears (blue), mean ABR thresholds were significantly reduced at three frequencies. Uninjected Bth/+ ears (red) showed ABR thresholds > 85 dB at all frequencies after 8 weeks. ABR thresholds from wild-type C3H are shown in green. (d) ABR Wave 1 amplitudes following 90 dB SPL at 16 kHz were greater in injected Bth/+ ears than in uninjected ears 8 weeks after treatment. The horizontal bars represent mean values. (e) Startle responses at 16 kHz in individual Cas9:Tmc1-mut3 sgRNA-injected mice (blue) were significantly stronger (p < 0.001) than in uninjected mice (red) 8 weeks after treatment. Among the different frequencies assayed, the number of ears tested (n) varies within the range shown (see Supplementary Table 2). Statistical analyses of ABR thresholds, amplitudes, and startle responses were performed by two-way ANOVA with Bonferroni correction for multiple comparisons: *p < 0.05, **p < 0.01, and ****p < 0.0001. Values and error bars reflect mean ± SEM.
Extended Data Figure 5.

Effect of in vivo injection of Cas9:sgRNA:lipid complexes on DPOAE thresholds. DPOAE thresholds 4 weeks after injection were elevated compared with uninjected ears at three frequencies following treatment with Cas9:Tmc1-mut3 sgRNA (a), and were elevated at two frequencies following treatment with Cas9:Tmc1-wt3 sgRNA, (b) Cas9:GFP sgRNA (c), or dCas9:Tmc1-mut1 sgRNA (d). (e) 8 weeks after Cas9:Tmc1-mut3 sgRNA injection, DPOAE thresholds were elevated at three frequencies in the injected group. Mean DPOAE thresholds of untreated wildtype (WT) C3H mice at 4 weeks (a) or 8 weeks (e) weeks of age are also shown in purple. Statistical analysis of DPOAE thresholds was performed by two-way ANOVA with Bonferroni correction for multiple comparisons: **p < 0.01, ***p < 0.001, and ****p < 0.0001. Values and error bars reflect mean ± SEM. Among the different frequencies assayed, the number of ears tested (n) varies within the range shown (see Supplementary Table 2). The elevation of DPOAE thresholds despite enhanced hair cell survival (Fig. 2d and 2g) suggests that the surviving OHCs may not be fully functional. IHCs can respond to sound and excite auditory nerve fibers in the absence of OHC amplification, although at higher SPLs. Thus, an improvement in ABR thresholds and suprathreshold amplitudes can occur without concomitant DPOAE enhancement if the functional improvements are restricted to the surviving IHCs.
Extended Data Figure 6.
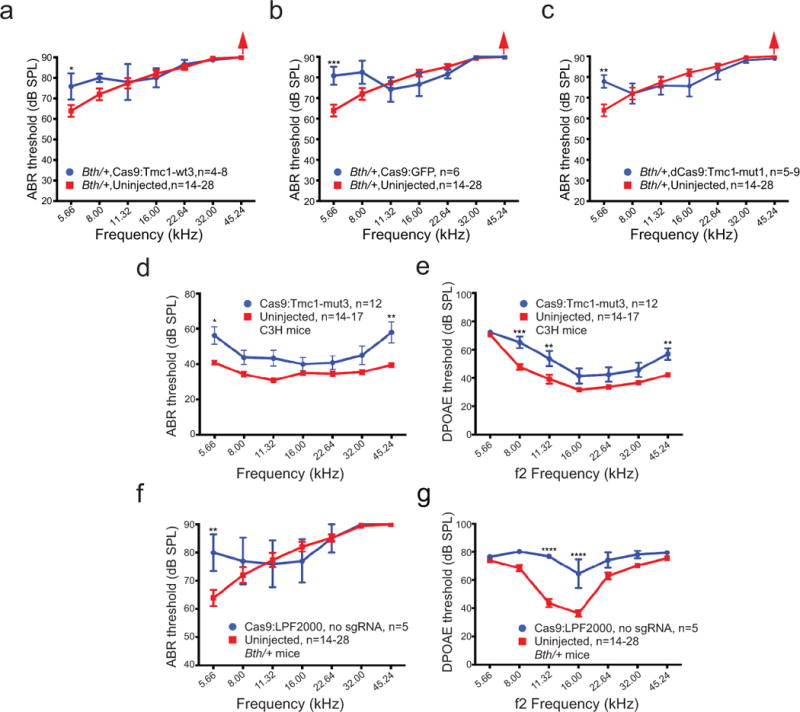
Hearing rescue is dependent on the Bth target specificity of the sgRNA, Cas9 nuclease activity, the presence of the Bth mutation, and the presence of the sgRNA. (a) In Bth/+ ears injected with Cas9:Tmc1-wt3:lipid, which targets the wild-type Tmc1 allele instead of the mutant Bth allele, ABR thresholds (blue) were comparable to or higher than those of uninjected controls (red) after 4 weeks. (b) Bth/+ ears injected with Cas9:GFP sgRNA:lipid (blue) did not show improved ABR thresholds 4 weeks after treatment. (c) Bth/+ ears injected with catalytically inactive dCas9:Tmc1-mut1:lipid did not show improved ABR thresholds 4 weeks after treatment. (d) ABR thresholds of wild-type C3H mice injected with Cas9:Tmc1-mut3:lipid showed similar patterns as the uninjected control inner ears at 4 weeks, except at 5.66 and 45.24 kHz where ABR thresholds were elevated. (e) Elevated DPOAE thresholds at three frequencies were observed after the treatment in (d). (f) Injection of Cas9:Lipofectamine 2000 (LPF2000) without sgRNA in Bth/+ mice did not improve ABR thresholds after 4 weeks. (g) Elevated DPOAE thresholds at 11 and 16 kHz were observed after the treatment in (f). Statistical analysis of ABR and DPOAE thresholds was performed by two-way ANOVA with Bonferroni correction for multiple comparisons: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. Values and error bars reflect mean ± SEM. Among the different frequencies assayed, the number of ears tested (n) varies within the range shown (see Supplementary Table 2).
Extended Data Figure 7.
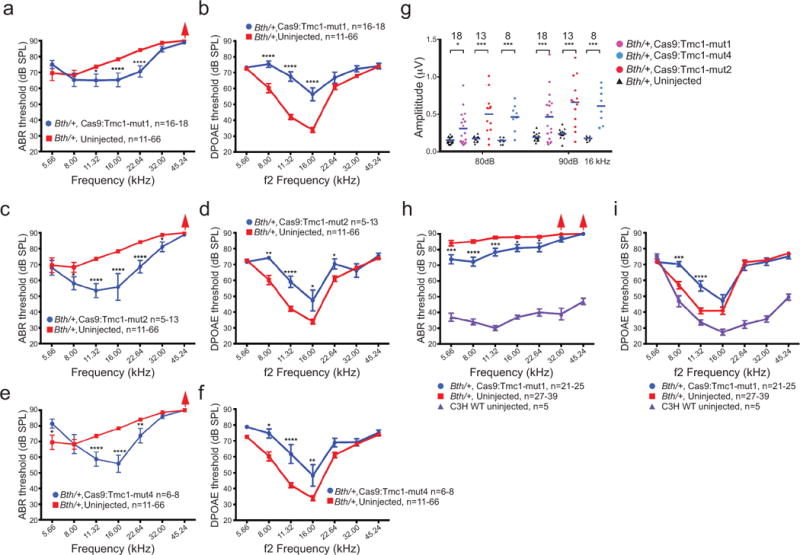
Hearing preservation following treatment with additional Tmc1-mut sgRNAs other than Tmc1-mut3. (a) Mean ABR thresholds were significantly reduced at three frequencies in ears injected with Cas9:Tmc1-mut1:lipid compared to uninjected Bth/+ ears after 4 weeks. (b) DPOAE thresholds were elevated in the same group of inner ears after Cas9:Tmc1-mut1 injection as in (a) after 4 weeks. (c) Mean ABR thresholds were significantly reduced at five frequencies in ears injected with Cas9:Tmc1-mut2:lipid compared to uninjected Bth/+ ears after 4 weeks. (d) DPOAE thresholds were elevated in the same group of inner ears after Cas9:Tmc1-mut2 injection as in (c) after 4 weeks. (e) Mean ABR thresholds were significantly reduced at three frequencies in ears injected with Cas9:Tmc1-mut4:lipid compared to uninjected Bth/+ ears after 4 weeks. (f) DPOAE thresholds were elevated in the same group of inner ears after Cas9:Tmc1-mut4:lipid injection as in (e) after 4 weeks. (g) Significantly stronger Wave 1 amplitudes were detected in ears injected with each of the Cas9:Tmc1-mut:lipid complexes shown at 16 kHz (80 and 90 dB SPL). (h) 8 weeks after Cas9:Tmc1-mut1:lipid injection into Bth/+ ears, mean ABR thresholds were significantly reduced at five frequencies compared to the uninjected Bth/+ ears, which showed ABR thresholds > 80 dB at all frequencies after 8 weeks. Mean ABR thresholds of untreated wildtype (WT) C3H mice of 8 weeks of age are shown in purple. Red arrows indicate no ABR response at the highest SPL level of 90 dB. (i) DPOAE thresholds were modestly elevated in the same group of inner ears after Cas9:Tmc1-mut1 injection as in (h) after 8 weeks. Mean DPOAE thresholds of untreated wildtype (WT) C3H mice of 8 weeks of age are shown in purple. Statistical analysis of ABR and DPOAE thresholds and Wave 1 amplitudes was performed by two-way ANOVA with Bonferroni correction for multiple comparisons: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. Values and error bars reflect mean ± SEM. Among the different frequencies assayed, the number of ears tested (n) varies within the range shown (see Supplementary Table 2).
Extended Data Figure 8.

RNP delivery of Cas9:sgRNA:lipid complexes results in genome editing in adult hair cells. 6-week-old adult Atoh1-GFP cochlea were injected with 1 μL 25 μM Cas9:GFP sgRNA:lipid complex by canalostomy, with the cochlea harvested two weeks after injection. (a) Genome editing was detected by the loss of GFP (green, with GFP absence noted using cyan shapes) in inner hair cells (IHCs) and outer hair cells (OHCs) (b) Hair cells were labeled with a hair cell marker MYO7A (red) in the apex turn of cochlea. (c, d) In uninjected contralateral Atoh1-GFP cochlea, all hair cells were GFP-positive. Scale bars = 10 μm. Similar results were observed in other injected ears that were immunolabeled (n = 3).
Extended Data Figure 9.

In vivo editing of the Tmc1 locus from Bth/+ ears injected with Cas9:Tmc1-mut3 sgRNA. A representation of the organ of Corti harvested at P5 for high-throughput DNA sequencing: (a) A confocal z-stack image showing the surface view of a dissected and labeled organ of Corti used for HTS. (b) A cross-sectional view of the organ of Corti (along the white line in (a)) showing the positions of hair cells (MYO7A), supporting cells (SOX2) and the cells from other cochlear regions that were used for quantification. LER: lesser epithelial ridge; GER: greater epithelial ridge; SE: sensory epithelium; Lib: The limbus region. DAPI-labeled nuclei are shown in blue. Quantification showed hair cells represented 1.45% ± 0.05% (mean ± SEM, n = 4) of all the cells in the dissected cochlea. Scale bars = 10 μm. (c) On-target and off-target in vivo editing of the Tmc1 locus in organ of Corti samples. No indels were observed at frequencies substantially above that of an untreated control sample at any of the ten off-target sites identified by GUIDE-seq (Off-T1 to Off-T10). Indels were detected by HTS at the Tmc1 on-target site and each off-target site from in vivo tissue samples dissected from the inner ear of neonatal mouse 5 days after Cas9:Tmc1-mut3 RNP injection (blue), or from untreated control samples (red).
Extended Data Table 1.
Off-target editing after nucleofection of DNA plasmids encoding Cas9 and Tmc1-mut3 sgRNA into primary fibroblasts derived from Bth/Bth mice. (a) Off-target sties identified by GUIDE-seq25. Mismatch positions are indicated counting the PAM as positions 21-23. Off-T3, off-T6, off-T7, off-T8 and off-T9 are located within predicted gene regions, while the rest are intergenic. 1000 ng Cas9 plasmid and 300 ng Tmc1-mut3 sgRNA plasmid were nucleofected into Bth/Bth fibroblasts using a LONZA 4D-Nucleofector and indels were detected by HTS at Tmc1 Bth on-target and each off-target site. Mismatches compared to the on-target sequence are shown in red and PAMs are in blue; (b) Off-target sites identified by computational prediction using the CRISPR Design Tool24. Among the top eight computationally predicted off-target sites, only two (off-T1′ and off-T2′ with two and three mismatches, respectively) were identified as bona fide off-targets in cells by GUIDE-seq. 1000 ng Cas9 plasmid and 300 ng Tmc1-mut3 sgRNA plasmid were nucleofected into Bth/Bth fibroblasts using a LONZA 4D-Nucleofector. Indels were detected by HTS at the Tmc1 Bth on-target site and at each predicted off-target site. Mismatches are shown in red and PAMs are in blue.
| a | |||||||
|---|---|---|---|---|---|---|---|
| 5′-Sequence-3′ | Mismatches (MMs) | NCBI accession | Predicted function | Location | Indels in Bth/Bth | ||
| Bth |
|
0MMs | N/A | chr9 | 31% | ||
| Off-T1 |
|
2MMs [4:13] | N/A | chr1 | 8.1% | ||
| Off-T2 |
|
3MMs [2:4:9] | N/A | chr16 | 4.4% | ||
| Off-T3 |
|
3MMs [1:3:7] | NC_000068.7 | CD82 antigen | chr2 | 2.6% | |
| Off-T4 |
|
3MMs [1:2:14] | N/A | chr12 | 3.9% | ||
| Off-T5 |
|
5MMs [1:3:4:6:16] | N/A | chr13 | 3.4% | ||
| Off-T6 |
|
2MMs [13:20] | NC_000067.6 | hemicentin-1 isoform | chr1 | 0.68% | |
| Off-T7 |
|
3MMs [2:5:18] | XM_006507026.3 | inositol 1,4,5-triphosphate receptor 2 | chr6 | 1.5% | |
| Off-T8 |
|
6MMs [3:4:5:7:8:13] | XR_389309.3 | unknown | chr5 | 1.2% | |
| Off-T9 |
|
3MMs [1:9:18] | NC_000070.6 | paralemmin A kinase anchor protein | chr4 | 1.4% | |
| Off-T10 |
|
3MMs [7:9:18] | N/A | chr10 | 0.048% | ||
| b | ||||||
|---|---|---|---|---|---|---|
| 5′-Sequence-3′ | Mismatches | NCBI accession | Location | Indels in Bth/Bth | ||
| Bth |
|
0MMs | chr9 | 31% | ||
| Off-T1′ |
|
2MMs [4:13] | chr1 | 8.1% | ||
| Off-T2′ |
|
3MMs [2:4:9] | chr16 | 4.4% | ||
| Off-T3′ |
|
4MMs [1:4:5:8] | NM_001312644.1 | chr12 | 0.037% | |
| Off-T4′ |
|
4MMs [3:4:8:10] | chr9 | 0.071% | ||
| Off-T5′ |
|
3MMs [2:7:20] | chr1 | 0.046% | ||
| Off-T6′ |
|
4MMs [1:2:3:7] | chr2 | 0.097% | ||
| Off-T7′ |
|
3MMs [2:4:15] | chr5 | 0.033% | ||
| Off-T8′ |
|
4MMs [1:4:5:11] | chr1 | 0.091% | ||
Supplementary Material
Acknowledgments
This work was supported by DARPA HR0011-17-2-0049 (to D.R.L.), and US NIH R01 EB022376 (to D.R.L.), R35 GM118062 (to D.R.L.), R01 DC006908 (to Z.Y.C), P30 DC05209 (to M.C.L.), R01 DC00138 (to M.C.L.), and R01 DC013521 (to J.R.H.). We are grateful for support from the David-Shulsky Foundation (to Z.Y.C.), a Frederick and Ines Yeatts Hair Cell Regeneration grant (to Y.T., V.L., M.H., and Y.S.), the Bertarelli Foundation (to J.R.H.), the Broad Institute (to D.R.L and Z.Y.C.), and HHMI (to D.R.L.). We thank Holly Rees, Shengdar Tsai, Michael Packer, and Dmitry Usanov, for assistance.
Footnotes
Author contributions
X.G. and Y.T. designed the research, performed genome editing (X.G.) and hearing biology (Y.T.) experiments, analyzed data, and wrote the manuscript. V.L., M.H., W.Y., B.P., Y.H., and H.W. designed experiments, performed hearing biology experiments and analyzed data. D.P., M.C.L., and W.K. designed hearing experiments and analyzed data. Y.S. and S.Y. supported hearing biology experiments. J.H. analyzed GUIDE-seq data. D.T. supported genome editing experiments. Y.L. and Q.X. designed and synthesized lipids. J.H., Z.C., and D.L. designed and supervised the research, and wrote the manuscript. All authors edited the manuscript.
Supplementary Information is available in the online version of the paper.
The authors declare competing financial interests: D.R.L. is a consultant and co-founder of Editas Medicine, Beam Therapeutics, and Pairwise Plants, companies that use genome editing. The co-authors have filed patent applications on aspects of this work. Correspondence and requests for materials should be addressed to D.R.L. and Z.Y.C.
References
- 1.Angeli S, Lin X, Liu XZ. Genetics of hearing and deafness. Anat Rec (Hoboken) 2012;295:1812–1829. doi: 10.1002/ar.22579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marazita ML, et al. Genetic epidemiological studies of early-onset deafness in the U.S. school-age population. Am J Med Genet. 1993;46:486–491. doi: 10.1002/ajmg.1320460504. [DOI] [PubMed] [Google Scholar]
- 3.Morton CC, Nance WE. Newborn hearing screening–a silent revolution. N Engl J Med. 2006;354:2151–2164. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- 4.Geleoc GS, Holt JR. Sound strategies for hearing restoration. Science. 2014;344:1241062. doi: 10.1126/science.1241062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muller U, Barr-Gillespie PG. New treatment options for hearing loss. Nat Rev Drug Discov. 2015;14:346–365. doi: 10.1038/nrd4533. [DOI] [PubMed] [Google Scholar]
- 6.Shibata SB, et al. RNA Interference Prevents Autosomal-Dominant Hearing Loss. Am J Hum Genet. 2016;98:1101–1113. doi: 10.1016/j.ajhg.2016.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan B, et al. Gene therapy restores auditory and vestibular function in a mouse model of Usher syndrome type 1c. Nat Biotechnol. 2017 doi: 10.1038/nbt.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun H, Huang A, Cao S. Current status and prospects of gene therapy for the inner ear. Hum Gene Ther. 2011;22:1311–1322. doi: 10.1089/hum.2010.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sacheli R, Delacroix L, Vandenackerveken P, Nguyen L, Malgrange B. Gene transfer in inner ear cells: a challenging race. Gene Ther. 2013;20:237–247. doi: 10.1038/gt.2012.51. [DOI] [PubMed] [Google Scholar]
- 10.Komor AC, Badran AH, Liu DR. CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell. 2017;168:20–36. doi: 10.1016/j.cell.2016.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Long C, et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Y, et al. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol. 2016;34:334–338. doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin H, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuris JA, et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol. 2015;33:73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan B, et al. TMC1 and TMC2 are components of the mechanotransduction channel in hair cells of the mammalian inner ear. Neuron. 2013;79:504–515. doi: 10.1016/j.neuron.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Camp GV, Smith R. Hereditary Hearing loss Homepage. 2017 http://hereditaryhearingloss.org.
- 18.Zhao Y, et al. A novel DFNA36 mutation in TMC1 orthologous to the Beethoven (Bth) mouse associated with autosomal dominant hearing loss in a Chinese family. PLoS One. 2014;9:e97064. doi: 10.1371/journal.pone.0097064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurima K, et al. Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nat Genet. 2002;30:277–284. doi: 10.1038/ng842. [DOI] [PubMed] [Google Scholar]
- 20.Kawashima Y, Kurima K, Pan B, Griffith AJ, Holt JR. Transmembrane channel-like (TMC) genes are required for auditory and vestibular mechanosensation. Pflugers Arch. 2015;467:85–94. doi: 10.1007/s00424-014-1582-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vreugde S, et al. Beethoven, a mouse model for dominant, progressive hearing loss DFNA36. Nat Genet. 2002;30:257–258. doi: 10.1038/ng848. [DOI] [PubMed] [Google Scholar]
- 22.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang F. CRISPR DESIGN. 2015 http://crispr.mit.edu.
- 25.Lenz JJ, Jodelka FM, et al. Rescue of hearing and vestibular function by antisense oligonucleotides in a mouse model of human deafness. Nat Med. 2013;19:345–50. doi: 10.1038/nm.3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang M, et al. Efficient Delivery of Genome-Editing Proteins using Bioreducible Lipid Nanoparticles. Proc Natl Acad Sci. 2015;113:2868–2873. doi: 10.1073/pnas.1520244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pattanayak V, et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lumpkin EA, et al. Math1-driven GFP expression in the developing nervous system of transgenic mice. Gene Expr Patterns. 2003;3:389–395. doi: 10.1016/s1567-133x(03)00089-9. [DOI] [PubMed] [Google Scholar]
- 29.Huang M, et al. Hair cell overexpression of Islet1 reduces age-related and noise-induced hearing loss. J Neurosci. 2013;33:15086–94. doi: 10.1523/JNEUROSCI.1489-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
High-throughput sequencing data is deposited in the NCBI Sequence Read Archive database under accession code SRP103108.


