Abstract
Objective
Brown adipose tissue (BAT) thermogenesis depends on the mobilization and oxidation of fatty acids from intracellular lipid droplets (LD) within brown adipocytes (BAs); however, the identity and function of LD proteins that control BAT lipolysis remain incomplete. Proteomic analysis of mouse BAT subcellular fractions identified vacuolar protein sorting 13C (VPS13C) as a novel LD protein. The aim of this work was to investigate the role of VPS13C on BA LDs.
Methods
Biochemical fractionation and high resolution confocal and immuno-transmission electron microscopy (TEM) were used to determine the subcellular distribution of VPS13C in mouse BAT, white adipose tissue, and BA cell culture. Lentivirus-delivered shRNA was used to determine the role of VPS13C in regulating lipolysis and gene expression in cultured BA cells.
Results
We found that VPS13C is highly expressed in mouse BAT where it is targeted to multilocular LDs in a subspherical subdomain. In inguinal white adipocytes, VPS13C was mainly observed on small LDs and β3-adrenergic stimulation increased VPS13C in this depot. Silencing of VPS13C in cultured BAs decreased LD size and triglyceride content, increased basal free fatty acid release, augmented the expression of thermogenic genes, and enhanced the lipolytic potency and efficacy of isoproterenol. Mechanistically, we found that BA lipolysis required activation of adipose tissue triglyceride lipase (ATGL) and that loss of VPS13C greatly increased the association of ATGL to LDs.
Conclusions
VPS13C is present on BA LDs where is targeted to a distinct subdomain. VPS13C limits the access of ATGL to LD and loss of VPS13C elevates lipolysis and promotes oxidative gene expression.
Keywords: Brown adipose tissue, Thermogenesis, Free fatty acids, ATGL, Perilipin 1, Oxidative genes
Abbreviations: VPS13C, Vacuolar Protein Sorting 13C; BAT, brown adipose tissue; BAs, brown adipocytes; LDs, lipid droplets; ATGL, adipose triglyceride lipase; ABHD5, α-β hydrolase domain-containing protein 5; PLIN1, perilipin 1; GAPDH, Glyceraldehyde 3-phosphate dehydrogenase; HSL, hormone sensitive lipase; Cidea, cell death-inducing DNA fragmentation factor, alpha subunit-like effector A; Cox8b, Cytochrome c oxidase subunit 8B; Ucp1, uncoupling protein 1; Ppara, Peroxisome proliferator-activated receptor alpha; Ppargc1a, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha; Hadhb, hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein); beta subunit, Acadl; Long-chain specific acyl-CoA dehydrogenase, Ebf2; early B cell factor 2, Esrra; estrogen related receptor, alpha
Highlights
-
•
VPS13C is highly expressed on mouse brown adipose tissue lipid droplets.
-
•
VPS13C is present on lipid droplets in a unique subspheric pattern.
-
•
VPS13C silencing in BAs decreases lipid droplet size and triglyceride content.
-
•
VPS13C silencing in BAs increases lipolysis and oxidative gene expression.
-
•
VPS13C silencing in BAs enhances ATGL but not HSL targeting to LDs.
1. Introduction
Brown adipose tissue (BAT) plays a key role in nonshivering thermogenesis in rodents and certain newborn mammals, including humans, by mobilizing and oxidizing fatty acids. In adult humans, the prevalence of BAT correlates with improved glucose homeostasis and energy metabolism [1], [2], [3], [4], [5], suggesting that activation of BAT thermogenesis might be a therapeutic target for obesity and diabetes. A defining feature of brown adipocytes (BAs) is the presence of multiple small lipid droplets (LDs) that integrate triglyceride storage and mobilization [6]. LDs are often closely associated with other organelles including neighboring LDs, the endoplasmic reticulum (ER) and mitochondria [7], [8], and it is thought that these contacts allow efficient coupling of lipid synthesis, mobilization, and oxidation.
The LD proteome is unique among organelles and can vary across tissues. For example, perilipin 1 is an LD scaffold protein in both brown and white adipocytes that suppresses lipolysis in the basal state and facilitates lipolysis during beta-adrenergic receptor activation [9], [10], [11]. In contrast, perilipin 5 is present in oxidative tissues like BAT, heart, and soleus muscle, where it links LDs to mitochondria and likely facilitates fatty acid trafficking for oxidation [12], [13]. Thus, the LD proteome often reflects the specialized tissue function as it relates to fatty acid trafficking and LD interactions with other organelles.
Proteomic analysis of BAT subcellular fractions demonstrated that vacuolar protein sorting 13C (VPS13C) segregates with known LD proteins under basal and stimulated conditions, suggesting it is a bona fide LD protein. VPS13C is the mammalian ortholog of the yeast Vps13 and belongs to a family of genes involved in vacuolar function. Relatively little is known about mammalian VPS13C, but recent studies found that mutations in VPS13C cause early-onset Parkinson's disease in humans [14]. Interestingly, gene wide association studies have also linked VPS13C with diabetes, glucose levels and pro-insulin release [15], [16], [17]. In the study described below, we used a combination of mass spectrometry analysis, biochemical fractionation, confocal and transmission electron microscopy as well as metabolic assays to examine the role of VPS13C in BAs.
2. Materials and methods
2.1. Animals
All protocols involving animals were approved by the Institutional Animal Care and Use Committee of Wayne State University and followed the National Institutes of Health Guide for the care and use of Laboratory Animals (NIH Publications No. 8023, revised 1978). Male and female mice 129S1/SvImJ (129S1; stock number 002448), Tg(Ucp1-cre)1Evdr (stock number 024670), C57BL/6J (C57B/6; stock number 000664), and td-tomato (R26-LSL-tdTomato; stock number 007909) were purchased from Jackson Laboratory and bred in house. Mice were fed regular chow and kept at a 12 h light/dark cycle.
For β3-adrenergic receptor agonist studies, vehicle (phosphate-buffered saline, PBS) or CL 316,243 (Sigma–Aldrich) was infused at a dose of 0.75 nmol/h using osmotic minipumps (Durect-ALZET; Cupertino, CA) for 3 days. For mass spectrometry analysis, CL 316,243 was infused at the same dose for 1 day.
2.2. Cell culture
Unless specified, chemicals used were obtained from Sigma–Aldrich (St. Louis, MO). BAs were grown as pre-adipocytes in Dulbecco's Modified Eagles Medium (DMEM) (HyClone GE Lifesciences; Pittsburgh, PA) containing 10% fetal bovine serum (FBS) (Atlanta Biologicals; Flowery Branch, GA), penicillin/streptomycin (Hyclone, GE Lifesciences), 20 nM insulin and 1 nM 3,3′,5-triiodo-l-thyronine (T3; differentiation media) for 3 days until confluent [18], [19]. Adipocyte differentiation was induced by incubating cells in differentiation media containing 2 ug/mL dexamethasone, 0.5 mM isobutylmethylxanthine (IBMX) and 0.25 mM indomethacin for 1 day. The following day, medium was replaced with differentiation media (day 1) and after that media was changed every day (days 2–6). Unless specified, cells were harvested at day 4 after induction.
In experiments where free fatty acids were measured, cells were incubated in Krebs–Ringer Bicarbonate Buffer containing HEPES (H-KRBB) or Hanks Balanced Salt Solution (HBSS) (Gibco, Thermo Fisher Scientific) supplemented with 1% Bovine Serum Albumin (BSA) (Alkali Scientific Inc. Pompano Beach, FL). Cells were treated for 1 or 2 h and media was collected. Non-esterified fatty acids were measured using a fluorescence kit (WAKO Diagnostics; Richmond, VA) using the fluorogenic substrate amplex red (Cayman Chemical; Ann Arbor, MI) and glycerol was measured by colorimetry using free glycerol reagent (Sigma; St. Louis, MO). For pharmacological inhibition of ATGL, cells were pre-incubated with 50 μM atglistatin (N′-[4′-(dimethylamino)[1,1′-biphenyl]-3-yl]-N,N-dimethyl-urea; (Cayman Chemical; Ann Arbor, MI) or vehicle (dimethyl sulfoxide, DMSO) for 30 min before treatment with vehicle (PBS) or isoproterenol.
Triglyceride was extracted from BAs using a modified method from Barkley et al. [20]. Briefly, BAs were collected and homogenized in PBS. Neutral lipids were extracted three times with isooctane: ethyl-acetate (1:9) (Fisher Scientific). The organic layer containing neutral lipids was collected and dried for 2 h under N2. Triglycerides were resuspended in 5% NP-40 by sonication. Triglyceride content was measured using triglyceride determination kit (Sigma; St. Louis, MO). BA cells grown in parallel were used to measure protein content using bicinchoninic acid assay (BCA; Pierce, Thermo Fisher Scientific Grand Island, NY).
2.3. Lentivirus production
Lentiviruses were produced by transfecting 293T cells using VPS13C shRNA-containing vector (Sigma cat# TRCN0000246772) or control (Sigma cat# SHC216), packaging psPAX2 plasmid (from Didier Trono to Addgene plasmid# 12260) and envelope plasmid pMD2.G (from Didier Trono to Addgene plasmid# 12259) at a ratio 1:0.75:0.25 respectively. Media containing lentiviruses was collected 40 h and 64 h after transfection, combined, filtered and centrifuged at 35,000×g for 2 h. Lentiviruses were resuspended in OptiMEM (Gibco). BAs were infected with lentiviruses, in the presence of OptiMEM and Polybrene (Millipore). Thirty hours later, media was replaced for regular media (DMEM) containing 10% FBS.
2.4. Subcellular fractionation
Cells were washed with PBS (Gibco; Thermo Fisher Scientific Grand Island, NY) and scraped in 20% sucrose solution containing 20 mM 4-(2-hydroxyethyl)-1-piperazine ethane sulfonic acid (HEPES), 1 mM ethylenediaminetetraacetic acid (EDTA) and EDTA-free proteases inhibitor cocktail (Thermo Fisher Scientific). Cells were homogenized by 10 passages through a 26 1/2G needle. Homogenates were spun 5 min at 1000×g and the supernatant or post-nuclear fraction (PN) was recovered. The PN fraction was layered with 10 and 0% sucrose solution respectively (containing HEPES, EDTA and proteases inhibitors) and centrifuged at 100,000×g (Beckman TL-100 ultracentrifuge) for 30 min. Floating LDs were transferred to another tube and proteins were precipitated using acetone (Fisher Scientific). Precipitated proteins were resuspended in 2% sodium dodecyl sulfate (SDS) and heated at 70 °C for 10 min. The cytoplasmic fraction was recovered from the upper portion of 20% sucrose layer. Proteins were measured by bicinchoninic acid assay (BCA; Pierce, Thermo Fisher Scientific Grand Island, NY) in all the fractions.
In experiments where nuclei, mitochondria and membrane fractions were collected, PN fraction was spun at 10,000×g to pellet mitochondria. The resultant supernatant was centrifuged at 100,000×g for 30 min. LD and cytosol fractions were collected as described above and pellet was collected as membrane fraction. Nuclei, mitochondria and membrane pellets were then resuspended in Buffer A (20 mM HEPES, 100 mM KCl, 2 mM MgCl, pH = 7.4), passed through a 26G needle 10 times and spun again at their corresponding speed. Pellets were resuspended in 2% SDS and protein measured in each fraction by BCA assay.
2.5. Mass spectrometry
For mass spectrometry analysis BAT LD fractions from sham-operated or CL 316,243 treated mice were obtained as described above. LD fractions and total lysate were run on PAGE and gel was stained with Sypro Ruby. The whole lane was cut and submitted for mass spectrometry analysis to the Proteomics Core of Wayne State University.
2.5.1. In-gel digestion
Washed gel slices were reduced with Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), alkylated with iodoacetamide and digested with sequencing-grade trypsin (Promega) overnight at 37 °C. Peptides were eluted from the gel slices using 0.1% formic acid and acetonitrile and dried. Peptides were separated by reverse phase chromatography by HPLC using a C18 column and analyzed with an LTQ-XL mass spectrometer (Thermo). Abundant ions were fragmented by MS/MS using collision-induced dissociation (CID).
2.5.2. Data analysis
Peak lists from MS2 spectra were generated with Proteome Discoverer (ver 1.1; Thermo) and scored using Mascot (Matrix Science) and SEQUEST algorithms. A mouse protein sequence database was used (UniProt; downloaded on 2010-07-28 containing 16,358 entries). Parent and fragment tolerances were 3.5 and 0.7 kDa, respectively. A fixed modification of +57 (alkylation) on C, and variable modifications of +16 (oxidation) on M, and −17 (pyro cmC) and +42 (acetylation) on peptide N-termini were included in the searches. Data was imported in Scaffold (ver 3.0; Proteome Software) and subset database searches were performed with X!Tandem. Proteins were scored as positive identifications if they achieved a ≥99% protein probability, and with at least 2 unique peptide identifications with ≥80% probability.
The top 100 LD proteins in control samples were selected, spectral counts were normalized for total counts and converted to a log scale. Hierarchical clustering was performed using Heatmapper [21] using Pearson complete linkage.
2.6. VPS13C antibody production
Mouse VPS13C cDNA corresponding to peptide amino acids 3189 to 3708 (accession number NP_796158.2) was cloned and sent to Proteintech (Rosemont, IL) for peptide synthesis and antibody production. Affinity-purified rabbit polyclonal antibodies against VPS13C were used in Western blot and immunostaining experiments.
2.7. Protein isolation and Western blot analysis
Mice were euthanized by CO2 and cervical dislocation, tissue collected and immediately frozen in liquid Nitrogen. Frozen tissues were disrupted using a Polytron homogenizer in RIPA buffer (Teknova; Hollister, CA) containing EDTA-free proteases inhibitor cocktail (Pierce, Thermo Fisher Scientific). BAs from cell cultures were scraped and lysed in RIPA buffer with EDTA-free proteases inhibitor cocktail. In experiments for phospho-HSL, 1/100 Halt™ phosphatase inhibitor cocktail was added to the lysis buffer (Thermo Scientific). Protein levels were determined by bicinchoninic acid assay (Thermo Fisher Scientific, Grand Island, NY).
Equal amounts of proteins were heated at 70 °C for 10 min in the presence of 1M urea and 5.7% β-mercaptoethanol then loaded in wells of a 5% SDS-Tris polyacrylamide gel or 3–8% Tris-acetate polyacrylamide gradient gel (Invitrogen) for detection of VPS13C, or NuSep 4–20% Tris-HEPES gels (NuSep; New South Wales, Australia) for ATGL, HSL and perilipin 1. Samples were transferred to PVDF membranes overnight at 4 °C for VPS13C (transfer buffer containing 4% SDS) or 90 min for others. Membranes were blocked in blocking solution (5% non-fat milk in Tris buffer saline containing 0.05% Tween (TBS-T)) and antibodies were diluted in blocking solution and incubated for 2 h at room temperature for VPS13C or overnight 4 °C for other proteins. Antibodies used were: rabbit anti UCP1 1/10,000 (Alpha Diagnostic International; San Antonio, TX) rabbit anti-perilipin 1 1/2500 [24], rabbit anti-GAPDH 1/2000 (Santa Cruz; Dallas, TX; cat# sc25778), rabbit anti-prohibitin 1/5000 (Abcam; Cambridge, MA; cat# ab28172), goat anti-mouse ATGL 1/1000 (Everest Biotech; Upper Heyford Oxfordshire, UK; cat#EB8403); rabbit anti-mouse ATGL [22] 1/3000; rabbit anti-HSL 1/2000 (Cell Signaling; Danvers, MA; cat#4107S) [23], rabbit anti-phospho-HSL (S563) 1/2000 (Cell Signaling; Danvers, MA; cat#4139S) rabbit anti-α/β tubulin 1/5000 (Cell Signaling, Danvers, MA; cat# 2148S). Because of the large size difference between VPS13C, GAPDH and prohibitin, blots from Figure 1C correspond to two gels that were run in parallel. Membranes were incubated with horseradish peroxidase-labeled secondary antibodies (Jackson ImmunoResearch Laboratories; West Grove, PA) for 1 h at room temperature. Super signal west-dura extended duration substrate (Thermo Fisher) was used for detection of secondary antibodies. Blots were imaged using the Azure c600 imaging system (Azure Biosystems; Dublin, CA) and quantified using ImageJ [25].
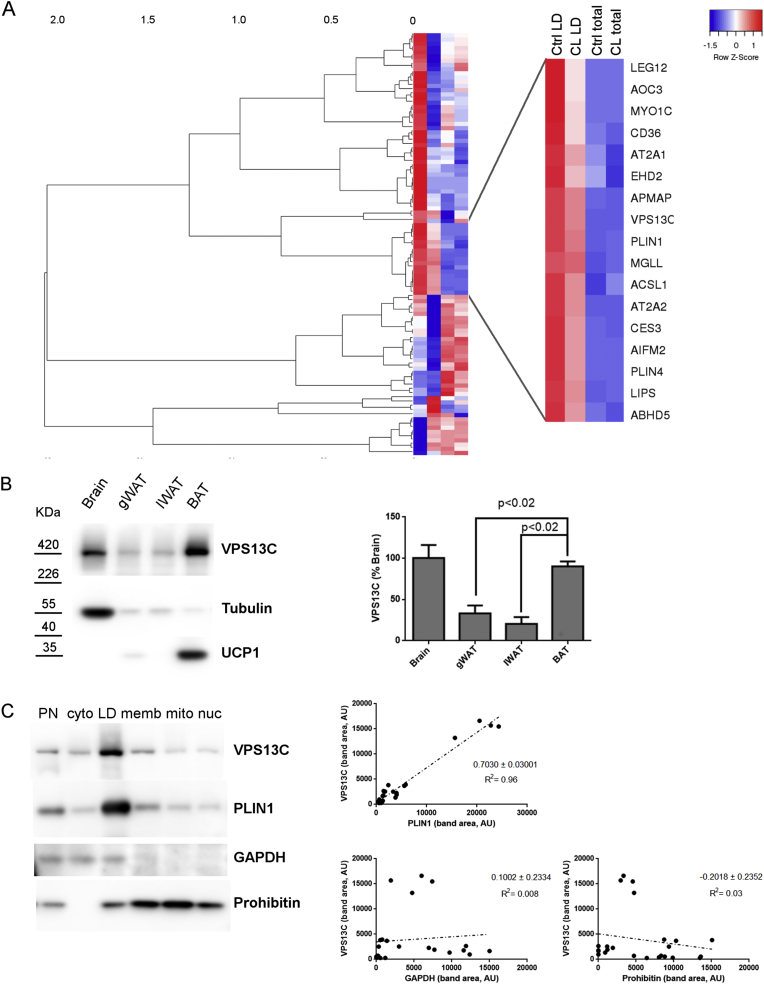
Figure 1.
VPS13C is highly expressed in mouse BAT and concentrates in LDs. A) Mass spectrometry results from subcellular BAT fractions were analyzed using hierarchical clustering analysis. BAT from mice treated with vehicle (ctrl) or 0.75 nmol/h CL 316,243 for 24 h (CL) were used to collect total lysates (total) and LD fractions. Colors in the heatmap represent row Z score calculated from normalized spectral counts that were converted to a log scale. Insert shows that the cluster where VPS13C is present includes well established LD proteins. B) VPS13C tissue distribution was assessed by Western blot, left panel: representative Western blot for VPS13C, tubulin and UCP1 (BAT marker); right panel: cumulative data (n = 4; *p < 0.05 vs BAT, ANOVA one way followed by Dunnett's multiple comparison test). C) VPS13C subcellular distribution, left panel: Western blot for VPS13C, perilipin 1 (PLIN1), GAPDH and prohibitin of BAT subcellular fractions; right panel: correlation between VPS13C and PLIN1, VPS13C and GAPDH and VPS13C and prohibitin expression levels (AU = arbitrary units).
2.8. Real-time quantitative PCR
RNA was extracted from cells using Qiagen (Valencia; CA) RNeasy mini kit and was reverse transcribed by using Superscript III (Invitrogen) and random or oligo dT primers. qPCR reaction was done using ABsolute Blue QPCR SYBR (Thermo Fisher Scientific) in Stratagene Mx3000P qPCR. Primers used were
| Gene Name | Forward Primer (5'->3′) | Reverse Primer (5'->3′) |
|---|---|---|
| Vps13c | GAAGCTAAAGTAAAAGCCCACGA | ACACATCAGAGGTGTTGACAATG |
| Ppara | CGGGTAACCTCGAAGTCTGA | CTAACCTTGGGCCACACCT |
| Ppargc1a | TTCACGGGTGTCAGCTTTGC | GGTCGCCCTTGTTCGTTCTG |
| Ebf2 | GCTGCGGGAACCGGAACGAGA | ACACGACCTGGAACCGCCTCA |
| Esrra | CTCAGCTCTCTACCCAAACGC | CCGCTTGGTGATCTCACACTC |
| Ucp1 | TGGCCTCTCCAGTGGATGTG | CGTGGTCTCCCAGCATAGAAG |
| Cidea | TGCTCTTCTGTATCGCCAGT | GCCGTGTTAAGGAATCTGCTG |
| Cox8b | TGCGAAGTTCACAGTGGTTC | CTCAGGGATGTGCAACTTCA |
| Hadhb | TTCGCGGACTCTAAGATTTCA | GCAGAGTGCAGTTGGGAAGA |
| Acadl | CGATTGCAAAAGCTTACGTG | AGAATCCGCATTAGCTGCAT |
| Ppia | GTGGTCTTTGGGAAGGTGAA | TTACAGGACATTGCGAGCAG |
2.9. Confocal microscopy
Mouse adipose tissues were fixed in 4% paraformaldehyde overnight and embedded in paraffin. Five-micrometer sections were obtained, deparaffinized and rehydrated using xylene, ethanol and ethanol/water. Antigen retrieval was carried out by three 5 min incubations at 95 °C in 10 mM citrate buffer. Tissue sections were blocked in 5% normal donkey or normal goat serum (NDS or NGS respectively) (Jackson ImmunoResearch Laboratories) in PBS for 1 h. Primary antibodies or their respective isotype-matched control antibodies were incubated at equivalent concentrations in blocking buffer overnight at 4 °C in a humidified chamber. Secondary antibodies were incubated in blocking buffer for 2 h at room temperature and protected from light. Antibodies used: rabbit VPS13C (custom made, Proteintech, Rosemont, IL) 1/250, goat anti-perilipin 1 (Everest Biotech; cat#EB 07728) 1/300. Secondary antibodies were obtained from Invitrogen and used at 1/1000 dilution, Alexa Fluor 488 goat anti-rabbit, Alexa Fluor 594 donkey anti-rabbit, Alexa Fluor 488 donkey anti-rabbit, Alexa Fluor 488 donkey anti-goat and Alexa Fluor 594 donkey anti goat (Invitrogen). Sections were incubated with 4′,6-diamidine-2′-phenylindole dihydrochloride (DAPI) 0.5 μg/mL for 15 min and mounted using Permafluor Paramount (Thermo Fisher Scientific).
BA cells were grown and differentiated on glass coverslips. Cells were washed with PBS and fixed with 4% paraformaldehyde at different time points during differentiation. In some experiments, cells were trypsinized and replated on glass coverslips one day after induction to avoid cell clumping and obtain clearer images. Cells were blocked with PBS containing 0.04% saponin and 5% NDS or NGS and incubated with rabbit anti-VPS13C, (1/250), rabbit anti-ATGL [22] 1/250, rabbit anti-perilipin 1 (1/250) or rabbit IgG at matching concentrations, then incubated with secondary antibodies and DAPI as above. Cells were post-fixed with 1% paraformaldehyde for 15 min and washed in PBS. Lipids were stained with LipidTOX deep red (Invitrogen; Thermo Fisher Scientific) 1/1000 for 30 min before imaging.
Fluorescence and brightfield microscopy were performed using an Olympus IX-81 microscope equipped with a spinning disc confocal unit and 40× (0.9NA) and 60× (1.2NA) water immersion objectives, using standard excitation and emission filters (Semrock; Rochester, New York) for visualizing DAPI, FITC (Alexa Fluor 488), Rhodamine (Alexa Fluor 594), and Cherry (LipidTOX) as described [9]. Raw data were acquired using IPLab and processed using ImageJ software [25] and image sets belonging to the same experiment were equally adjusted.
2.9.1. LD number and size
BA treated with control and VPS13C shRNA were trypsinized and re-plated on glass coverslips 2 days after induction. At day 4 after induction cells were fixed and stained with DAPI 0.5 μg/mL for 15 min and incubated with LipidTOX (1/2000) overnight. LDs and nuclei were imaged by confocal microscopy. Raw data was acquired and deconvolved using CellSens Software (Olympus; Waltham, MA). LD images were then thresholded and analyzed using ImageJ “analyze particles” plugin. Nuclei were manually counted. A total of 30 images per cell line from 3 independent experiments were analyzed (>3800 cells total). LD size distribution was calculated using GraphPad Prism software (Version 5.0; San Diego, CA).
2.10. Transmission electron microscopy (TEM)
2.10.1. Tissue preparation for embedding
Mice were euthanized by CO2 and cervical dislocation. BAT was removed and placed in H-KRBB with 4% BSA for dissection into 1–2 mm3 pieces and fixed in 4% paraformaldehyde and 0.05% glutaraldehyde in 0.12 M sodium phosphate buffer (sPBS) or 0.1 M Sorensen's buffer, pH 7.2 on ice at 4 °C for several hours. Tissues were either stored in phosphate buffer or 7% sucrose-containing Sorensen's buffer. Samples were dehydrated in a graded series of ethanol and embedded in LR White – medium hardness with en bloc staining by 70% ethanolic 2% uranyl acetate. After tissue infiltration, gelatin capsules were polymerized overnight at 70 °C. LR White ultrathin sections were cut at 70–90 nm on carbon-coated Formvar 200-mesh nickel grids by ultramicrotomy.
2.10.2. Immunogold TEM
Sections were blocked in sPBS containing 5% NGS, 5% BSA and 0.15% glycine for 1 h at 4 °C or in aqueous 100 mM glycine followed by incubation in 5% NGS and 1% egg albumin in sPBS-Tween 20 for 1.5 h at RT. Sections were incubated with rabbit anti-VPS13C (1:100) antibody or rabbit IgG (equivalent concentration) in sPBS containing 5% NGS, 5% BSA and 0.15% glycine overnight at 4 °C or in 5% NGS and 1% egg albumin in sPBS-Tween for 1 h at 37 °C and then overnight at 4 °C. Sections were incubated with either secondary goat anti-rabbit FluoroNanogold-Alexa Fluor 594 (1:25; Nanoprobes; Yaphank, NY, 1.4 nm) in 5% BSA and 0.15% glycine in sPBS, or (1:50) in 1% NGS in sPBS-Tween for 1.5 h at RT. Labeled sections were post-fixed in 3% glutaraldehyde and 1% tannic acid, pH 7.4, in 0.1 M sodium cacodylate buffer or in 2% glutaraldehyde in sPBS for 10 min at RT, the latter followed by an aqueous 100 mM glycine incubation for 15 min. Gold probes were enhanced with HQ silver (Nanoprobes) and gold toned with 0.05% aqueous gold chloride for 10 min at 4 °C, followed by 0.5% aqueous oxalic acid and 1% aqueous sodium thiosulfate treatment or not gold toned. Thereafter, grids were either post-fixed in 0.5% or 0.1% aqueous osmium tetroxide at 4 °C for 25 min. Finally, grids were post-stained with saturated 4 or 5% aqueous uranyl acetate and then post-stained with Reynolds lead citrate. Samples were imaged on a JEOL 2010 TEM at 200 kV or a Morgagni 368 TEM at 80 kV.
2.11. Statistical analysis
Statistical analyses were conducted using GraphPad Prism software (Version 5.0; San Diego, CA). Pairwise comparisons were analyzed using two-ways Student's t-test. When more than two groups were compared one or two-way analysis of variance (ANOVA) was used and when significant differences were detected (p < 0.05), individual comparisons were made using Dunnett's or Sidak's multiple comparison tests. All results are presented as mean ± S.E.M.
3. Results
3.1. VPS13C is highly expressed in mouse BAT and concentrates on LDs
To identify potentially novel LD proteins in BAT, we performed mass spectrometry analysis of BAT subcellular fractions in control mice and mice treated with the β3-adrenergic receptor agonist CL 316,243 for 24 h. Hierarchical cluster analysis of the top 100 proteins identified in LDs indicated that the distribution of VPS13C across different conditions and tissue fractions mirrored that of bona fide LD proteins like perilipin 1, perilipin 4, α-β hydrolase domain-containing protein 5 (ABHD5) and carboxylesterase 3 (Figure 1 A, Fig. S1 and Supplemental Tables 1 And 2), strongly suggesting that VPS13C is a BAT LD protein. To explore this possibility, we generated affinity purified polyclonal antibodies against VPS13C and examined its tissue distribution by immunoblotting. We found that VPS13C levels were the highest in the brain and BAT. Among adipose tissues, VPS13C levels were 3–4-fold higher in BAT compared to visceral (gonadal) and subcutaneous (inguinal) white adipose depots (Figure 1B).
To confirm VPS13C subcellular distribution we performed immunoblot analysis of fractionated mouse BAT. As expected from the proteomic analysis, VPS13C was highly enriched in LD fractions and its subcellular distribution correlated highly with perilipin 1 (r2 = 0.96, Figure 1C) whereas there was no correlation with the cytosolic marker GAPDH (r2 = 0.008) or the mitochondrial marker prohibitin (r2 = 0.03). Altogether, these data indicate that VPS13C is abundantly expressed in BAT and it concentrates on LDs.
Proteins binding LDs directly contain α-amphipathic helix or hydrophobic hairpin domains. Analysis of VPS13C protein sequence using Amphipaseek [26], [27] identified two regions around amino acids 2550 and 3535 with high amphipathic score that were predicted to be inserted in the membrane parallel to the membrane plane (in-plane), suggesting that VPS13C could possess an in-plane membrane anchor sequence (Supplemental Table 3). However, whether VPS13C attaches to the LD membrane is not known.
3.2. VPS13C is targeted to a unique LD subdomain
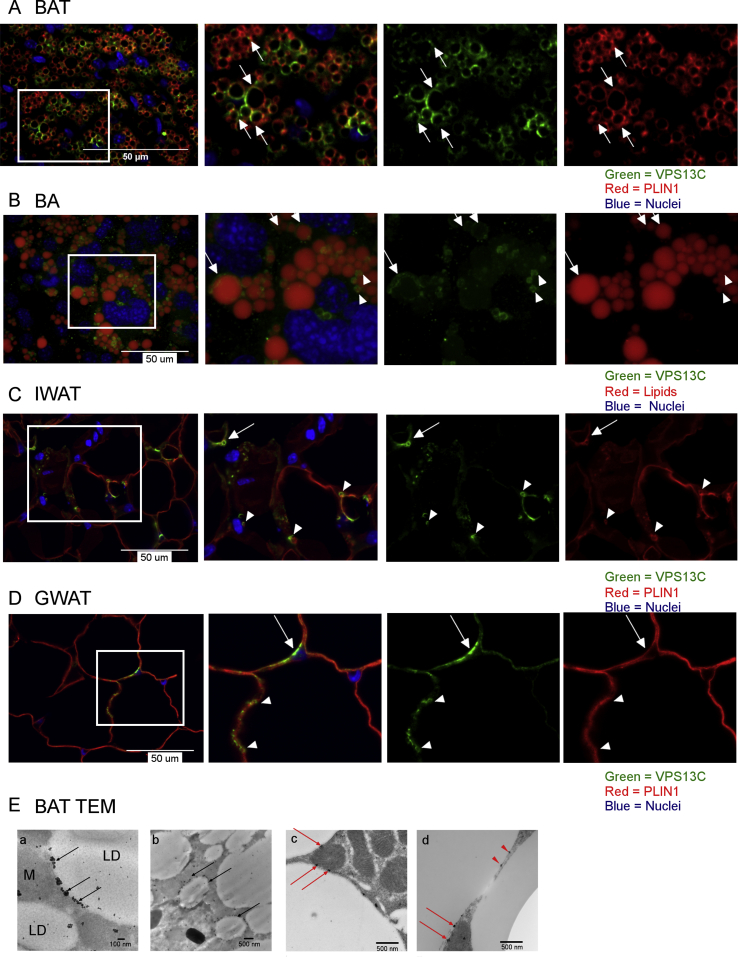
We next performed confocal microscopy of VPS13C in histological sections of mouse BAT. Double-label immunofluorescence imaging demonstrated that both VPS13C and perilipin 1 are targeted to LD surfaces (Figure 2A). However, whereas perilipin 1 appeared on virtually all LDs, the distribution of VPS13C was far more heterogeneous. High-resolution imaging clearly demonstrated that unlike perilipin 1, VPS13C did not surround large LDs uniformly, but rather localized in a unique subspherical pattern, while in smaller LDs VPS13C was more evenly distributed. Three-dimensional confocal imaging of cultured mouse BA cells further confirmed the heterogeneous subspherical targeting of VPS13C to larger LDs indicating that this is a cell-autonomous property (Figure 2B).
Figure 2.
VPS13C is targeted to a unique LD subdomain. Immunofluorescence images were obtained by confocal microscopy. Arrows show VPS13C subspherical distribution, arrowheads show VPS13C homogeneous distribution in small LDs. From left to right: composite image, magnified section and separate fluorescence channel images from magnified section. A) Mouse BAT paraffin sections immunostained for VPS13C and perilipin 1 (PLIN1). B) Mouse BAs from cell cultures were stained for VPS13C and lipids were stained with LipidTOX deep red (3D projection). C) Mouse inguinal white adipose tissue (IWAT) and D) Mouse gonadal white adipose tissue (GWAT) paraffin sections immunostained for VPS13C and perilipin 1 (PLIN1). E) Transmission electron micrographs of mouse BAT LR-White sections stained with VPS13C primary and anti-rabbit FluoroNanogold secondary antibodies; a- VPS13C concentrates in subspherical domain; b- VPS13C is homogeneously distributed in smaller LDs; c and d-sections enhanced with HQ Silver. M = mitochondria, LD = lipid droplets; black arrows show VPS13C on LDs; red arrows show VPS13C at LD-mitochondria interface; red arrowheads show VPS at LD–LD interface.
Although inguinal and gonadal adipose tissues expressed lower levels, VPS13C was also targeted to small LDs or restricted to a subspherical subdomain in unilocular mouse adipocytes (Figure 2C,D), supporting the notion that the targeting of VPS13C is conserved in different adipose depots.
Immunoelectron microscopy confirmed that VPS13C was present in some LDs and was strongly targeted to a sub-region of larger LDs, whereas the distribution on smaller LDs appeared more uniform (Figure 2E a-b). Interestingly, we found that VPS13C formed clusters and was often found at LD-mitochondria and LD–LD interfaces (Figure 2E).
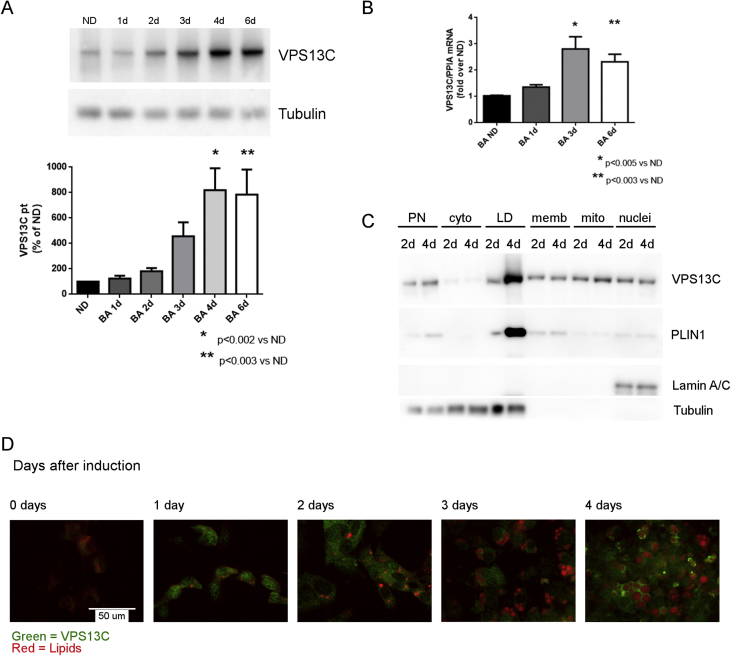
3.3. VPS13C levels increase during BA differentiation
Many adipocyte proteins are differentially expressed during differentiation, often reflecting the role of those proteins in adipocyte function. As shown in Figure 3A,B, VPS13C mRNA and protein were low in undifferentiated mouse pre-adipocytes and then rose sharply by 4 and 6 days following induction of differentiation. While VPS13C mRNA peaked on the 3rd day of differentiation, VPS13C protein levels continued to increase, peaking at 4 days after induction of differentiation. Cell fractionation and confocal microscopy analyses demonstrated that VPS13C selectively accumulates on LDs as BAs differentiate and that the upregulation of cellular VPS13C coincided with the appearance of intracellular LDs (Figure 3C,D).
Figure 3.
VPS13C levels increase during BA differentiation. Brown pre-adipocytes were grown in cell culture and harvested at 1–6 days after differentiation. A) protein levels (n = 4) and B) VPS13C mRNA (n = 3) were measured by Western blot and qt-PCR respectively (ND = non-differentiated). C) BAs Postnuclear (PN), cytosol (cyto), LD, membrane (memb), mitochondrial (mito) and nuclear (nuclei) fractions were collected at 2 and 4 days after differentiation. VPS13C was detected by Western Blot. D) Immunofluorescence staining of VPS13C in BAs at 0–4 days after induction of differentiation. VPS13C = green, lipids stained with LipidTOX deep red.
3.4. VPS13C increases in response to sustained adrenergic stimulation in inguinal white adipose tissue
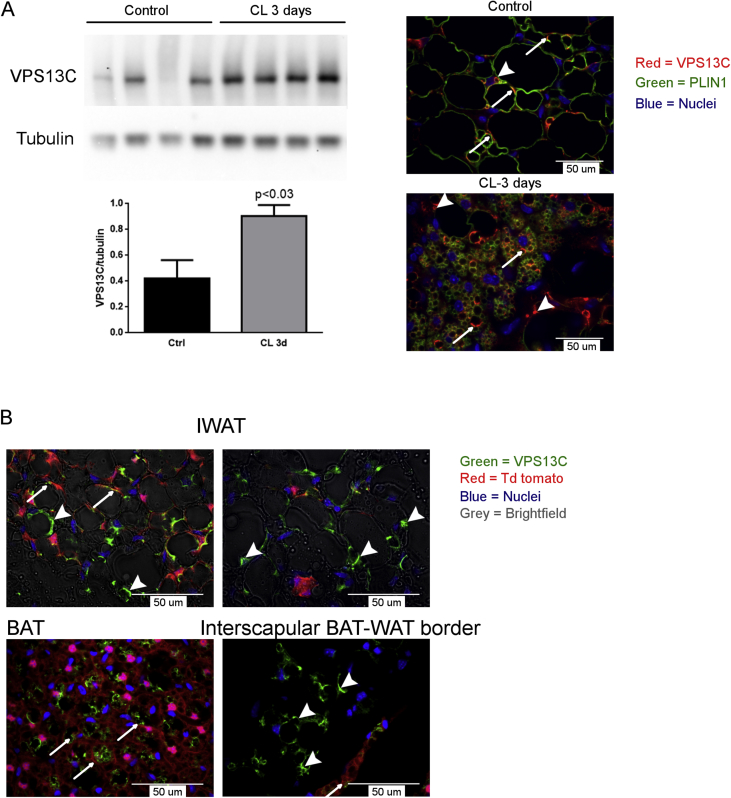
Sustained adrenergic activation of subcutaneous adipose tissue can induce the appearance of multilocular adipocytes, many of which express UCP1, which is the prototypic marker of thermogenic BA [28], [29], [30], [31]. Our results demonstrate that VPS13C is highly expressed in BAs, so it was of interest to examine in detail the expression of VPS13C in inguinal subcutaneous adipose tissue and its possible regulation by adrenergic stimulation. As shown in Figure 4 A, treatment with the β3-adrenergic receptor agonist CL 316,243 for 3 days more than doubled VPS13C levels in mouse inguinal white adipose tissue, and confocal microscopy demonstrated its targeting to numerous fragmented LDs within multilocular adipocytes.
Figure 4.
VPS13C increases in response to sustained adrenergic stimulation in inguinal white adipose tissue. A) Western blot and immunofluorescence analysis of total VPS13C levels in inguinal adipose tissue (IWAT) in control and mice infused with CL 316-243 (0.75 nmol/h) for 3 days (CL 3d). B) Inguinal white adipose tissue (IWAT), BAT, BAT-WAT intersection, sections from UCP-1-Cre td-tomato mice were stained for VPS13C. VPS13C is found in cells with (arrows) and without (arrowheads) a history of UCP-1 indicating that VPS13C is not restricted to brown adipocytes.
Because VPS13C levels are higher in BAs than in white adipocytes and because some adipocytes in inguinal adipose tissue exhibit a BA phenotype following adrenergic stimulation, we tested whether VPS13C was restricted to brown/beige adipocytes. Brown/beige adipocytes were identified by tracing cells with histories of UCP1 expression using the UCP1-Cre/td-tomato reporter system. Double label immunofluorescence experiments demonstrated that VPS13C expression was independent of UCP1 expression (Figure 4B). These results indicate that VPS13C expression is most closely associated with adipocytes having a multilocular, but not necessarily brown, phenotype.
3.5. VPS13C promotes lipid accumulation in BAs
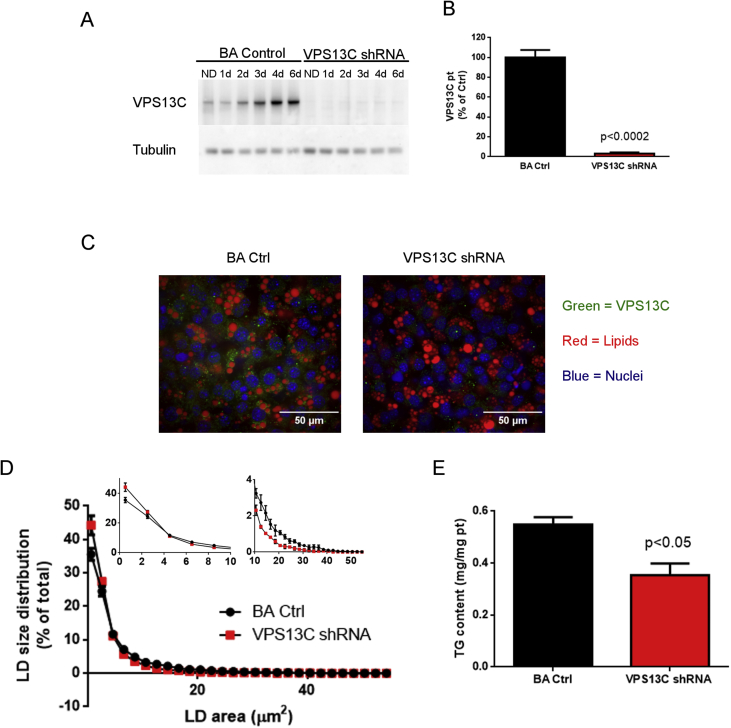
Lipolysis occurs on the surface of LDs and involves the interaction and trafficking of lipases, lipase activators and inhibitors, and resident LD proteins [11], [32]. To test whether VPS13C has a role on BA lipolysis we generated a mouse stable cell line knocked down for VPS13C using shRNA. As shown in Figure 5 (A, B, and C), VPS13C shRNA decreased VPS13C protein by 96%, demonstrating efficient silencing and confirming the specificity of our antibody. VPS13C knock down did not impair brown adipocyte differentiation, as indicated by similar levels of the transcription factors peroxisome proliferator-activated receptor gamma (Pparg) and CCAT/Enhancer Binding Protein Alpha (Cebpa) (Fig. S2). This contrasts with a recent report suggesting the involvement of VPS13C in the differentiation of 3T3-L1 cells [33]. Interestingly, VPS13C depletion affected LD size distribution, increasing the percentage of smaller LDs (<1.5 μm2) and reducing the proportion of bigger LDs (>10 μm2) (Figure 5D and Fig. S2C) without significantly affecting the total number of LDs (ctrl shRNA = 870 ± 68 vs VPS13C shRNA = 1066 ± 62 LDs/100 cells n = 3). Therefore, even when correcting by LD number, the average area occupied by LDs per cell was lower in cells knocked down for VPS13C than in control cells (ctrl shRNA = 45.3 ± 2.9 μm2/cell vs VPS13C shRNA = 36.3 ± 1.7μm2/cell, n = 3, p < 0.025). Consistent with imaging results, triglyceride content was significantly decreased in cells silenced for VPS13C (Figure 5E). Altogether these data suggest that lack of VPS13C reduces steady state LD triglyceride levels in BAs.
Figure 5.
VPS13C knock down decreases LD size and triglyceride content. Characterization of control and cells silenced for VPS13C: A) Representative Western blot for VPS13C at different differentiation stages (ND = non-differentiated, 1d-6d = 1–6 days after induction). B) Cumulative data at day 4 after differentiation (n = 3). C) Immunostaining for VPS13C at day 4 after differentiation, green: VPS13C, red: lipids stained with lipidTOX deep red. D) LD size distribution as percentage of total, quantified using confocal images in cells stained with LipidTOX and DAPI (n = 3). Insert shows magnified areas of the graph. E) Triglyceride (TG) content (n = 4).
3.6. VPS13C inhibits lipid hydrolysis in BAs
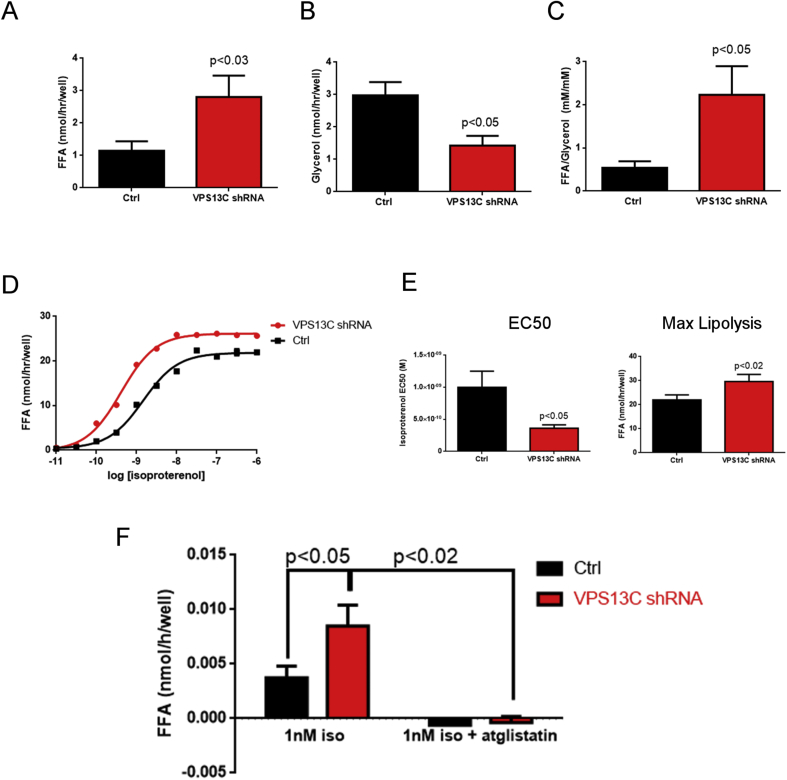
Given its targeting to LD and the reduced triglyceride content in cells lacking VPS13C we examined the effect of VPS13C silencing on BA lipolysis. As shown in Figure 6 A–C, VPS13C knock down increased basal free fatty acid release, as well as the relative release of free fatty acids to glycerol. We also found that knock down of VPS13C increased the sensitivity (as shown by lower EC50) and maximal activation of lipolysis induced by β-adrenergic receptor activation (Figure 6D,E).
Figure 6.
VPS13C knock down increases free fatty acid release in BA. A) Free fatty acid release in media (n = 5), B) Glycerol release in media (n = 4) C) Free fatty acid/glycerol ratio (n = 4). D) Free fatty acid release in response to isoproterenol. From left to right: representative experiment, sensitivity (EC50) and maximal response to isoproterenol (n = 4). E) Free fatty acid release in response to 1 nM isoproterenol in the presence and absence of the ATGL inhibitor atglistatin (n = 4).
3.7. VPS13C knock down increases targeting of ATGL to LDs
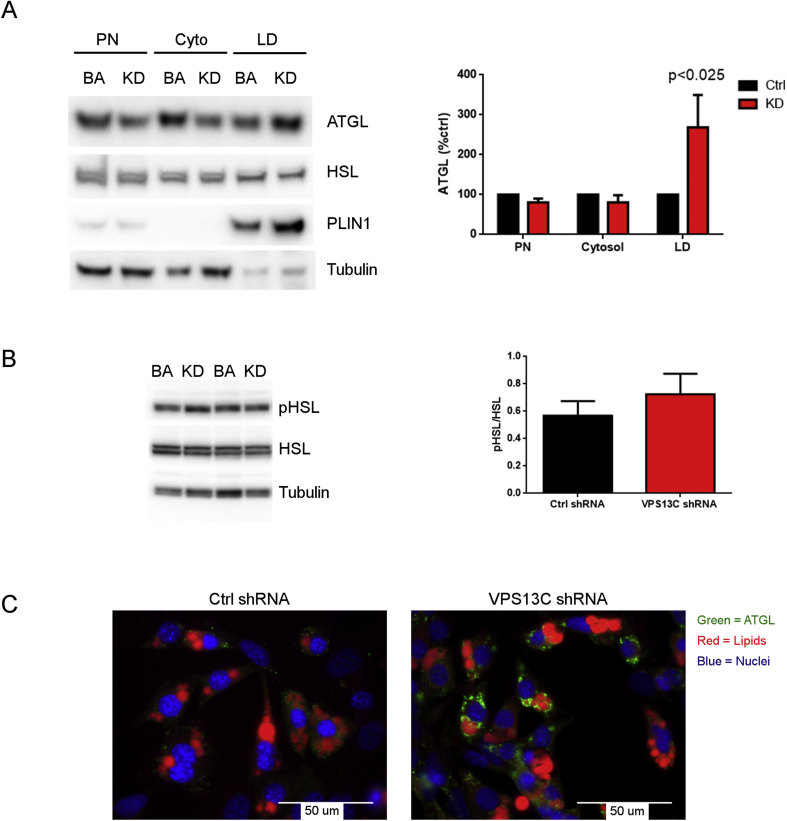
Together, ATGL and hormone sensitive lipase (HSL) account for >90% of adipocyte triglyceride lipase activity [34], [35], [36]. ATGL activity is directed specifically to triglyceride substrate, releasing free fatty acids, but not glycerol, and the greater release of free fatty acids relative to glycerol suggested that cells lacking VPS13C might exhibit greater ATGL activity. Accordingly, we found that atglistatin abolished isoproterenol-induced fatty acid release in both control and knock down cell lines (Figure 6E), indicating the central role of ATGL in mediating fatty acid release. Analysis by immunoblotting indicated that knock down of VPS13C did not alter total levels of ATGL or HSL. However, loss of VPS13C led to a more than 2-fold increase in the targeting of ATGL to LDs without affecting HSL phosphorylation or subcellular distribution (Figure 7A and 7B). Consistent with cell fractionation results, confocal microscopy demonstrated a striking increase in the targeting of ATGL to LDs in cells lacking VPS13C. (Figure 7C).
Figure 7.
VPS13C knock down increases targeting of ATGL to LDs. A) Western blot for ATGL in BA cell fractions. Left panel: representative Western blot; right panel: cumulative data (n = 4). KD = BA knocked down for VPS13C; PN = post-nuclear; cyto = cytoplasm; LD = lipid droplets. B) Western blot for phospho-HSL and HSL in total lysates from cells treated with control shRNA (ctrl) or shRNA against VPS13C (KD). Left panel: representative Western blot; right panel cumulative data (n = 4) C) Immunostaining for ATGL in control and VPS13C-silenced BAs.
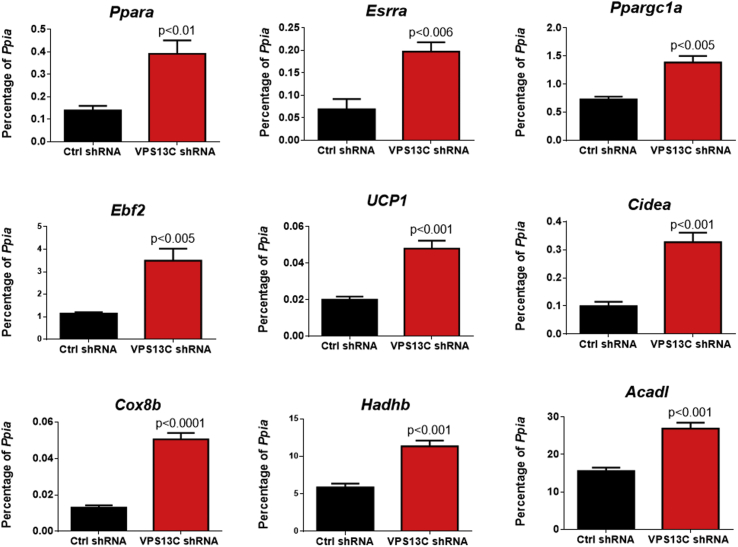
Lastly, we addressed the impact of VPS13C loss on metabolic gene expression in BA cells. Previous work has shown that lipolytic products promote the expression of genes involved in adipocyte oxidative metabolism and “browning,” in part through activation of nuclear receptors such as peroxisome proliferator-activated receptor alpha (Ppara) [19], [37], [38], [39]. We found that cells lacking VPS13C upregulated mRNA expression of the nuclear receptors Ppara and Esrra, as well as the coactivator protein Ppargc1a and the BA determination factor Ebf2. Downstream targets of these pathways, including Ucp1, Cidea, Cox8b, Hadhb, and Acadl were also upregulated (Figure 8). These data indicate that VPS13C suppresses lipolysis and oxidative gene expression, likely by reducing ATGL targeting to LDs.
Figure 8.
VPS13C knock down increases oxidative gene expression in BAs. Analysis of thermogenic gene expression by qRT-PCR in control and VPS13C knockdown cells 4 days after differentiation.
4. Discussion
VPS13C is a widely-expressed protein whose function in mammalian cells is poorly understood. Using immunochemical and mass spectrometry proteomic approaches we found that VPS13C is highly targeted to small LDs of both brown and white adipocytes.
While highly expressed in BAs, our results indicate that this expression pattern more closely reflects the structure of intracellular LDs in that multilocular white adipocytes also exhibit high levels of VPS13C that are independent of UCP1 expression.
Although VPS13C targets LDs, it is presently unclear whether this association represents a direct interaction with the LD surface or is indirectly mediated by close interactions with other LD proteins or even closely-associated organelles. Proteins targeted to LDs do not have typical transmembrane domains but rather amphipathic α-helices or hydrophobic hairpin domains [40]. Interestingly, analysis of VPS13C sequence using Amphiseek showed the presence of a couple amphipathic sections in-plane with the membrane, suggesting the possibility that VPS13C could have an in-membrane anchor domain and bind LDs directly as it has been hypothesized for yeast Vps13 and human VPS13A [41]. Alternatively, VPS13C could bind LDs by interacting with other LD proteins like galectin 12 [33], [42], which has also been observed in LD fractions [43].
Interestingly, the targeting of VPS13C to larger LDs (>1 μm diameter) was not uniform, as demonstrated by immunofluorescence and immunoelectron microscopy. In physical or optical sections VPS13C exhibited a unique subspherical distribution in tissue, whereas this structure appeared as a partial cap-like structure in three-dimensional confocal projections in BA cell culture. In addition, we observed that VPS13C was present in small vesicles or micro LDs decorating larger LDs in inguinal and gonadal white adipose tissue. The unique targeting to LD subdomain and the elevation of VPS13C expression following activation of lipolysis all suggest potential involvement in LD trafficking, fragmentation, or fusion.
VPS13C LD subspherical targeting contrasts with other known LD proteins like perilipin 1 and suggests that VPS13C might act as a tether between LDs and other organelles. Indeed, immunoelectron microscopy analysis showed that clusters of VPS13C were often observed on LD surfaces that apposed other organelles. The idea that VPS13C could mediate LD-organelle interaction is consistent with findings on the yeast VPS13C ortholog, in which Vps13 is present at membrane contact sites like vacuole-mitochondria patches (vCLAMP), nuclei-vacuole junctions (NVJ1) and endosome-mitochondrion contacts [44], [45]. Interestingly, some of the Vps13-containing contact sites act in parallel to the ER-mitochondria encounter structures (ERMES) [45] and in some cases gain of function Vps13 mutations phenotypically compensate for the loss of ERMES [44], [46]. Since orthologues of the yeast ERMES complex have not been described in metazoans, it is possible that Vps13-containing contact sites serve a similar function and that VPS13C could have a role in LD-organelle interaction. In addition, human VPS13C has been shown to interact with vesicle-associated membrane protein-associated protein A and B (VAPA and VAPB) [42], an ER protein known to mediate ER-organelle interactions and lipid transfer [47], [48], [49] supporting the hypothesis that VPS13C could mediate LD-ER contact. However, further analysis is needed to confirm the preferential localization of VPS13C at LD-organelle contact sites.
VPS13C protein levels increased in mouse BAs during differentiation, similar to what was found in 3T3-L1 cells [33], indicating that either VPS13C has a role in adipogenesis or that VPS13C function becomes more relevant as BAs differentiate. On one hand, VPS13C knock down in 3T3-L1 cells has been reported to inhibit adipocyte differentiation [33], suggesting that VPS13C could be necessary for BA differentiation as well. On the other hand, changes in cellular metabolism as cells progress from pre-adipocytes to fully differentiated BAs, such as higher free fatty acid mobilization and oxidation, may result in increased demand for VPS13C function and therefore augmented protein levels. Interestingly, the localization of VPS13C yeast ortholog, Vps13, varies with changes in both metabolism (induced by changes in carbon source) and developmental stage [44], [45], [50]. However, unlike in 3T3-L1 cells, we found no significant decreases between control and VPS13C knockdown BA cells in Peroxisome Proliferator-Activated Receptor Gamma (Pparg) and CCAT/Enhancer Binding Protein Alpha (Cebpa) mRNA levels as well as HSL protein levels. The discrepancies between brown adipocytes and 3T3-L1 cells may be due to differences in the cell type studied or growth conditions used, but is not likely due to inefficient VPS13C knock down since in our study VPS13C expression was reduced by more than 95%.
VPS13C knock down in mouse BAs decreased average LD size, increased basal and β-adrenergic-stimulated free fatty acid release and reduced triglyceride content. This increased fatty acid release was dependent on ATGL, as shown by pharmacological inhibition. Furthermore, knock down of VPS13C did not enhance HSL phosphorylation; however, it increased the targeting of ATGL, but not HSL, to LDs. It is possible that VPS13C and/or the LD subdomain it defines restricts the access of ATGL LD triglyceride. This is consistent with greater translocation of ATGL and its non-uniform targeting to the LD surface in cells lacking VPS13C. More speculatively, it is possible that loss of VPS13C disrupts fatty acid channeling to mitochondria and ER, resulting in greater fatty acid efflux and nuclear signaling. In this scenario, enhanced ATGL targeting to LDs could be a compensatory response. Regardless, our results indicate that VPS13C function is important for suppression of lipolysis and proper fatty acid trafficking.
In mammals, there are four Vps13 orthologues that are products of distinct genes. In humans, mutations in VPS13 A, B, and C result in neurodegenerative diseases, namely chorea-acanthocytosis [51], Cohen Syndrome [52], and autosomal recessive early-onset Parkinson's disease [14], respectively. Interestingly, gene-wide association studies have also linked single nucleotide polymorphisms near VPS13C/C2CD4A/C2CD4B to variations on fasting plasma glucose (lead SNP rs11071657) [15], glucose disposal (lead SNP rs17271305) [16] and glucose-stimulated insulin release (rs7172432 A allele) [17]. Altogether, these data suggest that VPS13C may have multiple functions depending on the tissue expressed and that the role of VPS13C on lipid homeostasis could impact glucose metabolism in mammals.
5. Conclusions
We have found that VPS13C is present on BA LDs where it occupies a unique LD subdomain. Deletion of VPS13C augments basal and β-adrenergic-induced lipolysis and this is likely due to increased ATGL trafficking to LDs. The targeting of VPS13C to a distinctive LD subdomain suggests a specialized role in the subcellular control of lipolysis and the trafficking of lipolytic products between LDs and other organelles.
Author contributions
V.D.R. participated in experimental design, analyzed the data, did all the experiments except TEM and sample acquisition for mass spectrometry and co-wrote the manuscript; V.A.K performed TEM experiments and contributed to the writing of the methods section; J.G.G. oversaw the project, participated in experimental design, prepared samples for mass spectrometry and co-wrote the manuscript; V.D.R., V.A.K. and J.G.G. reviewed and approved the final version.
Acknowledgments
This work was supported by grants from National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases (RO1 DK076629) to J.G. Granneman and from American Heart Association (15POST21860000) to V.D. Ramseyer.
The Wayne State University Proteomics Core, and this work are supported through the NIH Center Grant P30 ES 020957, the NIH Cancer Center Support Grant P30 CA 022453 and the NIH Shared Instrumentation Grant S10 OD 010700.
We thank members of CIMER for helpful discussions, Dr. Anelia Petkova for preparing tissue sections and Dr. Andrew F.X. Goldberg for providing advice on electron microscopy experiments.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.molmet.2017.10.014.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Cypess A.M., Lehman S., Williams G., Tal I., Rodman D., Goldfine A.B. Identification and importance of brown adipose tissue in adult humans. The New England Journal of Medicine. 2009;360(15):1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chondronikola M., Volpi E., Borsheim E., Porter C., Annamalai P., Enerback S. Brown adipose tissue improves whole-body glucose homeostasis and insulin sensitivity in humans. Diabetes. 2014;63(12):4089–4099. doi: 10.2337/db14-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Marken Lichtenbelt W.D., Vanhommerig J.W., Smulders N.M., Drossaerts J.M., Kemerink G.J., Bouvy N.D. Cold-activated brown adipose tissue in healthy men. The New England Journal of Medicine. 2009;360(15):1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- 4.Virtanen K.A., Lidell M.E., Orava J., Heglind M., Westergren R., Niemi T. Functional brown adipose tissue in healthy adults. The New England Journal of Medicine. 2009;360(15):1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- 5.Muzik O., Mangner T.J., Leonard W.R., Kumar A., Janisse J., Granneman J.G. 15O PET measurement of blood flow and oxygen consumption in cold-activated human brown fat. Journal of Nuclear Medicine. 2013;54(4):523–531. doi: 10.2967/jnumed.112.111336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farese R.V., Jr., Walther T.C. Lipid droplets finally get a little R-E-S-P-E-C-T. Cell. 2009;139(5):855–860. doi: 10.1016/j.cell.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodman J.M. The gregarious lipid droplet. Journal of Biological Chemistry. 2008;283(42):28005–28009. doi: 10.1074/jbc.R800042200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao Q., Goodman J.M. The lipid droplet-a well-connected organelle. Frontiers Cell and Developmental Biology. 2015;3:49. doi: 10.3389/fcell.2015.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Granneman J.G., Moore H.P., Krishnamoorthy R., Rathod M. Perilipin controls lipolysis by regulating the interactions of AB-hydrolase containing 5 (Abhd5) and adipose triglyceride lipase (Atgl) Journal of Biological Chemistry. 2009;284(50):34538–34544. doi: 10.1074/jbc.M109.068478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaudhry A., Granneman J.G. Differential regulation of functional responses by beta-adrenergic receptor subtypes in brown adipocytes. American Journal of Physiology. 1999;277(1 Pt 2):R147–R153. doi: 10.1152/ajpregu.1999.277.1.R147. [DOI] [PubMed] [Google Scholar]
- 11.Sztalryd C., Brasaemle D.L. The perilipin family of lipid droplet proteins: Gatekeepers of intracellular lipolysis. Biochimica et Biophysica Acta. 2017;1862(10 Pt B):1221–1232. doi: 10.1016/j.bbalip.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimmel A.R., Sztalryd C. Perilipin 5, a lipid droplet protein adapted to mitochondrial energy utilization. Current Opinion in Lipidology. 2014;25(2):110–117. doi: 10.1097/MOL.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Granneman J.G., Moore H.P., Mottillo E.P., Zhu Z. Functional interactions between Mldp (LSDP5) and Abhd5 in the control of intracellular lipid accumulation. Journal of Biological Chemistry. 2009;284(5):3049–3057. doi: 10.1074/jbc.M808251200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lesage S., Drouet V., Majounie E., Deramecourt V., Jacoupy M., Nicolas A. Loss of VPS13C function in autosomal-recessive parkinsonism causes mitochondrial dysfunction and increases PINK1/parkin-dependent mitophagy. American Journal of Human Genetics. 2016;98(3):500–513. doi: 10.1016/j.ajhg.2016.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dupuis J., Langenberg C., Prokopenko I., Saxena R., Soranzo N., Jackson A.U. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nature Genetics. 2010;42(2):105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saxena R., Hivert M.F., Langenberg C., Tanaka T., Pankow J.S., Vollenweider P. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nature Genetics. 2010;42(2):142–148. doi: 10.1038/ng.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grarup N., Overvad M., Sparso T., Witte D.R., Pisinger C., Jorgensen T. The diabetogenic VPS13C/C2CD4A/C2CD4B rs7172432 variant impairs glucose-stimulated insulin response in 5,722 non-diabetic Danish individuals. Diabetologia. 2011;54(4):789–794. doi: 10.1007/s00125-010-2031-2. [DOI] [PubMed] [Google Scholar]
- 18.Uldry M., Yang W., St-Pierre J., Lin J., Seale P., Spiegelman B.M. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metabolism. 2006;3(5):333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Mottillo E.P., Bloch A.E., Leff T., Granneman J.G. Lipolytic products activate peroxisome proliferator-activated receptor (PPAR) alpha and delta in brown adipocytes to match fatty acid oxidation with supply. Journal of Biological Chemistry. 2012;287(30):25038–25048. doi: 10.1074/jbc.M112.374041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutchins P.M., Barkley R.M., Murphy R.C. Separation of cellular nonpolar neutral lipids by normal-phase chromatography and analysis by electrospray ionization mass spectrometry. Journal of Lipid Research. 2008;49(4):804–813. doi: 10.1194/jlr.M700521-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Babicki S., Arndt D., Marcu A., Liang Y., Grant J.R., Maciejewski A. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Research. 2016;44(W1):W147–W153. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyoshi H., Perfield J.W., 2nd, Souza S.C., Shen W.J., Zhang H.H., Stancheva Z.S. Control of adipose triglyceride lipase action by serine 517 of perilipin A globally regulates protein kinase A-stimulated lipolysis in adipocytes. Journal of Biological Chemistry. 2007;282(2):996–1002. doi: 10.1074/jbc.M605770200. [DOI] [PubMed] [Google Scholar]
- 23.Sanders M.A., Madoux F., Mladenovic L., Zhang H., Ye X., Angrish M. Endogenous and synthetic ABHD5 ligands regulate ABHD5-perilipin interactions and lipolysis in fat and muscle. Cell Metabolisam. 2015;22(5):851–860. doi: 10.1016/j.cmet.2015.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore H.P., Silver R.B., Mottillo E.P., Bernlohr D.A., Granneman J.G. Perilipin targets a novel pool of lipid droplets for lipolytic attack by hormone-sensitive lipase. Journal of Biological Chemistry. 2005;280(52):43109–43120. doi: 10.1074/jbc.M506336200. [DOI] [PubMed] [Google Scholar]
- 25.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Combet C., Blanchet C., Geourjon C., Deleage G. NPS@: network protein sequence analysis. Trends in Biochemical Science. 2000;25(3):147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 27.Sapay N., Guermeur Y., Deleage G. Prediction of amphipathic in-plane membrane anchors in monotopic proteins using a SVM classifier. BMC Bioinformatics. 2006;7:255. doi: 10.1186/1471-2105-7-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cousin B., Cinti S., Morroni M., Raimbault S., Ricquier D., Penicaud L. Occurrence of brown adipocytes in rat white adipose tissue: molecular and morphological characterization. Journal of Cell Sciences. 1992;103(Pt 4):931–942. doi: 10.1242/jcs.103.4.931. [DOI] [PubMed] [Google Scholar]
- 29.Seale P., Conroe H.M., Estall J., Kajimura S., Frontini A., Ishibashi J. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. The Journal of Clinical Investigation. 2011;121(1):96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Contreras G.A., Lee Y.H., Mottillo E.P., Granneman J.G. Inducible brown adipocytes in subcutaneous inguinal white fat: the role of continuous sympathetic stimulation. American Journal of Physiology and Endocrinology Metabolisam. 2014;307(9):E793–E799. doi: 10.1152/ajpendo.00033.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee Y.H., Petkova A.P., Konkar A.A., Granneman J.G. Cellular origins of cold-induced brown adipocytes in adult mice. FASEB Journal. 2015;29(1):286–299. doi: 10.1096/fj.14-263038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Granneman J.G., Kimler V.A., Moore H.P. Cell Biology Symposium: imaging the organization and trafficking of lipolytic effectors in adipocytes. Journal of Animal Science. 2011;89(3):701–710. doi: 10.2527/jas.2010-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang R.Y., Xue H., Yu L., Velayos-Baeza A., Monaco A.P., Liu F.T. Identification of VPS13C as a galectin-12-binding protein that regulates Galectin-12 protein stability and adipogenesis. PLoS One. 2016;11(4):e0153534. doi: 10.1371/journal.pone.0153534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zimmermann R., Strauss J.G., Haemmerle G., Schoiswohl G., Birner-Gruenberger R., Riederer M. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306(5700):1383–1386. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
- 35.Schweiger M., Schreiber R., Haemmerle G., Lass A., Fledelius C., Jacobsen P. Adipose triglyceride lipase and hormone-sensitive lipase are the major enzymes in adipose tissue triacylglycerol catabolism. Journal of Biological Chemistry. 2006;281(52):40236–40241. doi: 10.1074/jbc.M608048200. [DOI] [PubMed] [Google Scholar]
- 36.Haemmerle G., Lass A., Zimmermann R., Gorkiewicz G., Meyer C., Rozman J. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science. 2006;312(5774):734–737. doi: 10.1126/science.1123965. [DOI] [PubMed] [Google Scholar]
- 37.Li P., Zhu Z., Lu Y., Granneman J.G. Metabolic and cellular plasticity in white adipose tissue II: role of peroxisome proliferator-activated receptor-alpha. American Journal of Physiology and Endocrinol Metabolisam. 2005;289(4):E617–E626. doi: 10.1152/ajpendo.00010.2005. [DOI] [PubMed] [Google Scholar]
- 38.Mottillo E.P., Balasubramanian P., Lee Y.H., Weng C., Kershaw E.E., Granneman J.G. Coupling of lipolysis and de novo lipogenesis in brown, beige, and white adipose tissues during chronic beta3-adrenergic receptor activation. Journal of Lipid Research. 2014;55(11):2276–2286. doi: 10.1194/jlr.M050005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mottillo E.P., Shen X.J., Granneman J.G. Role of hormone-sensitive lipase in beta-adrenergic remodeling of white adipose tissue. American Journal of Physiology and Endocrinol Metabolisam. 2007;293(5):E1188–E1197. doi: 10.1152/ajpendo.00051.2007. [DOI] [PubMed] [Google Scholar]
- 40.Thiele C., Spandl J. Cell biology of lipid droplets. Current Opinion in Cell Biology. 2008;20(4):378–385. doi: 10.1016/j.ceb.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 41.Drin G., Casella J.F., Gautier R., Boehmer T., Schwartz T.U., Antonny B. A general amphipathic alpha-helical motif for sensing membrane curvature. Nature Structural and Molecular Biology. 2007;14(2):138–146. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- 42.Huttlin E.L., Ting L., Bruckner R.J., Gebreab F., Gygi M.P., Szpyt J. The BioPlex network: a systematic Exploration of the human interactome. Cell. 2015;162(2):425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang R.Y., Yu L., Graham J.L., Hsu D.K., Lloyd K.C., Havel P.J. Ablation of a galectin preferentially expressed in adipocytes increases lipolysis, reduces adiposity, and improves insulin sensitivity in mice. Proceeding in National Academy of Science U S A. 2011;108(46):18696–18701. doi: 10.1073/pnas.1109065108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lang A.B., John Peter A.T., Walter P., Kornmann B. ER-mitochondrial junctions can be bypassed by dominant mutations in the endosomal protein Vps13. Journal of Cell Biology. 2015;210(6):883–890. doi: 10.1083/jcb.201502105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park J.S., Thorsness M.K., Policastro R., McGoldrick L.L., Hollingsworth N.M., Thorsness P.E. Yeast Vps13 promotes mitochondrial function and is localized at membrane contact sites. Molecular Biology Cell. 2016;27(15):2435–2449. doi: 10.1091/mbc.E16-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xue Y., Schmollinger S., Attar N., Campos O., Vogelauer M., Carey M.F. Endoplasmic reticulum-mitochondria junction is required for iron homeostasis. Journal of Biological Chemistry. 2017 doi: 10.1074/jbc.M117.784249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hua R., Cheng D., Coyaud E., Freeman S., Di Pietro E., Wang Y., Kim P.K. VAPs and ACBD5 tether peroxisomes to the ER for peroxisome maintenance and lipid homeostasis. Journal of Cell Biology. 2017;216(2):367–377. doi: 10.1083/jcb.201608128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Vos K.J., Morotz G.M., Stoica R., Tudor E.L., Lau K.F., Ackerley S. VAPB interacts with the mitochondrial protein PTPIP51 to regulate calcium homeostasis. Human Molecular Genetics. 2012;21(6):1299–1311. doi: 10.1093/hmg/ddr559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peretti D., Dahan N., Shimoni E., Hirschberg K., Lev S. Coordinated lipid transfer between the endoplasmic reticulum and the Golgi complex requires the VAP proteins and is essential for Golgi-mediated transport. Molecular Biology Cell. 2008;19(9):3871–3884. doi: 10.1091/mbc.E08-05-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park J.S., Neiman A.M. VPS13 regulates membrane morphogenesis during sporulation in Saccharomyces cerevisiae. Journal of Cell Science. 2012;125(Pt 12):3004–3011. doi: 10.1242/jcs.105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Velayos Baeza A., Dobson-Stone C., Rampoldi L., Bader B., Walker R.H., Danek A. Chorea-acanthocytosis. In: Pagon E.-i.-c. Roberta A., Adam Margaret P., Ardinger Holly H., editors. GeneReviews® [Internet] University of Washington; Seattle: 2014. [Google Scholar]
- 52.Wang H., Falk M.J., Wensel C., Traboulsi E.I. Cohen Syndrome. In: Adam E.-i.-c. Margaret P., Ardinger Holly H., Pagon Roberta A., editors. GeneReviews® [Internet] University of Washington; Seattle: 2016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.