Abstract
Objective
Nicotinamide phosphoribosyl transferase (NAMPT) is the rate-limiting enzyme in the salvage pathway that produces nicotinamide adenine dinucleotide (NAD+), an essential co-substrate regulating a myriad of signaling pathways. We produced a mouse that overexpressed NAMPT in skeletal muscle (NamptTg) and hypothesized that NamptTg mice would have increased oxidative capacity, endurance performance, and mitochondrial gene expression, and would be rescued from metabolic abnormalities that developed with high fat diet (HFD) feeding.
Methods
Insulin sensitivity (hyperinsulinemic-euglycemic clamp) was assessed in NamptTg and WT mice fed very high fat diet (VHFD, 60% by kcal) or chow diet (CD). The aerobic capacity (VO2max) and endurance performance of NamptTg and WT mice before and after 7 weeks of voluntary exercise training (running wheel in home cage) or sedentary conditions (no running wheel) were measured. Skeletal muscle mitochondrial gene expression was also measured in exercised and sedentary mice and in mice fed HFD (45% by kcal) or low fat diet (LFD, 10% by kcal).
Results
NAMPT enzyme activity in skeletal muscle was 7-fold higher in NamptTg mice versus WT mice. There was a concomitant 1.6-fold elevation of skeletal muscle NAD+. NamptTg mice fed VHFD were partially protected against body weight gain, but not against insulin resistance. Notably, voluntary exercise training elicited a 3-fold higher exercise endurance in NamptTg versus WT mice. Mitochondrial gene expression was higher in NamptTg mice compared to WT mice, especially when fed HFD. Mitochondrial gene expression was higher in exercised NamptTg mice than in sedentary WT mice.
Conclusions
Our studies have unveiled a fascinating interaction between elevated NAMPT activity in skeletal muscle and voluntary exercise that was manifest as a striking improvement in exercise endurance.
Keywords: Nicotinamide adenine dinucleotide, Nicotinamide phosphoribosyl transferase, High fat feeding, Mitochondrial gene expression, Insulin sensitivity, Exercise
Abbreviations: CD, chow diet; HFD, 45% high fat diet; LFD, 10% low fat diet; Mck, muscle creatine kinase; NAD+, nicotinamide adenine dinucleotide; NAM, nicotinamide; NAMPT, nicotinamide phosphoribosyl transferase; NMN, nicotinamide mononucleotide; NMNAT, NMN adenylyl transferase; PARPs, poly ADP-ribose polymerases; PGC-1α, PPAR-γ coactivator-1 alpha; PPAR-γ, peroxisome proliferator-activated receptor gamma; PPi, pyrophosphate; Rd, insulin stimulated glucose disposal; qRT-PCR, quantitative reverse transcriptase polymerase chain reaction; RER, respiratory exchange ratio; SIRT1, sirtuin-1; VCO2, carbon dioxide production; VHFD, 60% very high fat diet; VO2, oxygen consumption
Highlights
-
•
Skeletal muscle NAMPT overexpression increases NAD+ via elevated NAMPT activity.
-
•
Elevated NAMPT partially protects against very-high-fat-diet-induced weight gain.
-
•
Elevated NAMPT amplifies exercise-induced improvements in exercise endurance.
-
•
Fascinating interaction between elevated NAMPT activity in muscle and exercise.
1. Introduction
Nicotinamide adenine dinucleotide (NAD+) is an essential co-substrate for several enzyme classes such as sirtuins and poly ADP-ribose polymerases (PARPs) that regulate a myriad of signaling pathways governing metabolism, healthy aging, and lifespan extension [1]. NAD+ can be generated de novo from dietary tryptophan or NAD+ precursors. However, the NAD+ salvage pathway is the dominant pathway for NAD+ biosynthesis in mammals [2], [3], [4]. In this pathway, nicotinamide phosphoribosyl transferase (NAMPT), a homo-dimeric type II phosphoribosyl transferase [5], [6], catalyzes the reversible condensation of nicotinamide (NAM) and 5′-phosphoribosyl-1-pyrophosphate (PRPP) to yield nicotinamide mononucleotide (NMN) and pyrophosphate (PPi) [7], [8]. NMN is subsequently converted to NAD+ in the presence of ATP by the enzyme NMN adenylyl transferase (NMNAT) [7].
We previously showed that NAMPT protein content in skeletal muscle was higher in athletes compared to sedentary individuals, and positively correlated with mitochondrial function, insulin sensitivity, and oxidative capacity in humans [9]. Interestingly, the level of NAMPT protein in skeletal muscle was increased in sedentary individuals who were exercise trained for 3 weeks [9]. Exercise defends against environmental insults, such as high fat diet (HFD) feeding, that cause metabolic complications. While the molecular underpinnings of the beneficial effects of exercise are far from fully understood, emerging data has pointed to NAD+ elevation as a key driver. For example, exercise increases the activity of the NAD+-dependent deacetylase sirtuin-1 (SIRT1) [10], [11] through elevation of NAD+ [12], [13]. In turn, SIRT1 controls the activity of peroxisome proliferator-activated receptor gamma (PPAR-γ) coactivator-1 alpha (PGC-1α) [14], a master regulator of mitochondrial biogenesis and function [10], [15].
To further probe the putative salutary effects of increased NAMPT activity, we generated a mouse transgenic line that overexpressed NAMPT in skeletal muscle (NamptTg). We hypothesized that NAMPT overexpression in skeletal muscle would rescue mice from metabolic abnormalities that developed with HFD feeding and would increase oxidative capacity, endurance performance, and mitochondrial gene expression.
2. Materials and methods
2.1. Mice
Mice overexpressing the NAMPT transgene (C57BL/6J-Tg(Mck-NAMPT)Pbef2Srs) under the control of the muscle creatine kinase (Mck) promotor were generated at the Pennington Biomedical Research Center (PBRC) Transgenic Core by pronuclear injection using C57BL/6J embryos following standard techniques [16]. 1256 bp of the Mck promoter was amplified from p1256MCKCAT (a gift from Dr. M. W. Hulver) and inserted into pCMV-sport to make pCS-Mck1256. 2.6 kb of NAMPT cDNA was excised from pCMV-SPORT6-NAMPT and then inserted into pCS-Mck1256 between Mck and pA to yield pMck-NAMPT (Figure S1). To identify transgenic founder mice, DNA was isolated from tail biopsies at 21 days of age for PCR genotyping. Wild type (WT) C57BL/6J littermates were used as controls.
2.2. Animal studies
We performed three separate animal studies. Study 1 was conducted at the PBRC in Baton Rouge, LA, USA. Male C57BL/6J-Tg(Mck-NAMPT)Pbef2Srs (shortened to NamptTg for the remainder of the text) and wild type (WT) C57BL/6J mice were fed low fat diet (LFD) (10% by kcal) (D12450B, Research Diets, New Brunswick, NJ) or high fat diet (HFD) (45% by kcal) (D12451, Research Diets, New Brunswick, NJ) for 30 weeks from weaning. Body weight and body composition was assessed every 2 weeks at the same time of day. Studies 2 and 3 were conducted at the Sanford Burnham Prebys Medical Discovery Institute in Orlando, FL, USA. In study 2, male NamptTg and WT C57BL/6J mice were fed standard chow diet (CD) (2016, Harlan Teklad, Indianapolis, IN) or very high fat diet (VHFD) (60% by kcal) (D12492, Research Diets, New Brunswick, NJ) for 16 weeks from weaning. Body weight and body composition were assessed every week at the same time of day. Indirect calorimetry was performed and food intake, water intake, and spontaneous physical activity were assessed after 14 weeks on the diet. Hyperinsulinemic-euglycemic clamps were performed after 16 weeks on the diet. In study 3, male NamptTg and WT C57BL/6J mice were fed CD (2016, Harlan Teklad, Indianapolis, IN) for 11 weeks from weaning. Body weight and body composition were assessed every week at the same time of day. Half of the mice were given access to running wheels (Mini Run Around 4½″, Super Pet, Elk Grove Village, IL) equipped with odometers (F12 Bike Computer, Easton-Bell Sports, Rantoul, IL) on week 4 of the study. Time spent running and distance completed were recorded every 24 h for the first 4 weeks that the mice had access to the wheels. Aerobic capacity (VO2max) and exercise endurance tests were performed prior to giving mice access to the wheels and following 6 (VO2max) and 7 (exercise endurance) weeks of voluntary exercise training, respectively. Mice were given one week between tests to recover. Wheels were removed from the cages 24 h prior to VO2max tests and endurance tests. On week 11, mice were fasted for 5 h and then euthanized via CO2 followed by cervical dislocation, and tissues were collected and weighed. Wheels were removed from cages 24 h prior to euthanasia.
All mice were individually caged and maintained at 22–24 °C with light from 7:00am to 7:00pm. Lights were equipped with a dimmer such that a gradual increase/decrease in light occurred 30 min prior to lights being fully on/off. All animal studies and procedures were approved by the appropriate Institutional Animal Care and Use Committees.
2.3. Quantitative reverse transcriptase-PCR
In study 1, tissues were snap-frozen in liquid nitrogen immediately following dissection. RNA was extracted via column purification using the Qiagen RNeasy Fibrous Mini Kit (Qiagen, Valencia, CA). RNA quantity was determined using an ND-1000 Nanodrop Spectrophotometer (Thermo Fisher Scientific). The concentration of NAMPT mRNA was determined by qRT-PCR using Taqman primers and fluorescent probes as the detection system on an ABI 7900HT (Applied Biosystems, Foster City, CA) using the following parameters: one cycle of 48 °C for 30 min, then 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. NAMPT expression data was normalized to the housekeeping gene peptidylprolyl isomerase B (PPIB). Primers and probes were designed using Primer Express version 2.1 (Applied Biosystems). Sequences of primer/probe sets are shown in Table S1. For all other genes in study 1, cDNA was synthesized with the iSCRIP cDNA synthesis kit (Bio-Rad, Hercules, CA) using 200 ng of RNA. qRT-PCR reactions were performed using 1 μM of primers and LightCycler® 480 SYBR Green Master (Roche, Branford, CT) on a LightCycler® 480 detection system (Roche). Calculations were performed by a comparative method (2-ΔΔCT) using 18S RNA as an internal control. Primers were designed using the Integrated DNA Technologies (IDT) software, and the primer sequences can be found in Table S2. In study 3, total RNA was isolated as previously described [17]. Briefly, RNA was isolated from 50 to 100 mg of skeletal muscle tissues (red quadriceps, white quadriceps) with Qiazol reagent (Invitrogen, Carlsbad, CA). The quantity and purity of RNA was determined using a ND-1000 Nanodrop spectrophotometer (Thermo Fisher Scientific). Primer-probe sets were pre-designed Single Tube Taqman® Gene expression assays. qRT-PCR reactions were performed using Taqman Fast Virus 1-step reaction mix Standard protocol (Life Technologies, Grand Island, NY). Data were normalized by dividing the target gene by the geometric mean of the internal control genes (RPLP0, GAPDH).
2.4. Western blots
Tissues were collected immediately following euthanasia and flash frozen in liquid nitrogen. Homogenates were prepared by Polytron homogenization in RIPA buffer containing protease inhibitor and phosphatase inhibitor cocktails (Sigma, St. Louis, MO). Protein content was quantified by bicinchoninic acid (BCA) assay (Thermo Fisher Scientific). 15–25 μg of protein was run on a 10% SDS-PAGE gel (Bio-Rad) and transferred to a PVDF membrane (Millipore, Billerica, MA). Membranes were incubated overnight with antibodies to NAMPT (A300-372; Bethyl, Montgomery, TX), complex III (MS304, MitoSciences, Eugene, OR), and α-tubulin (ab7291, Abcam) and then probed with IRDye 680 goat anti-mouse IgG or IRDye 800CW goat anti-rabbit IgG (926-32220 and 92632211, respectively; LI-COR, Lincoln, NE). Bands were visualized using an Odyssey Digital Infrared Imaging System (LI-COR) and quantified using Odyssey Application Software version 3.0 (LI-COR).
2.5. Tissue skeletal muscle NAMPT enzyme activity
NAMPT enzyme activity in fractionated extracts from mixed gastrocnemius, mixed quadriceps, heart, and liver of CD fed mice was measured using a NMN production assay [18]. For this assay, the partially-purified NAMPT was incubated with NAM (10 μM), PRPP (50 μM), and ATP (2 mM) in TMT (50 mM Tris-HCl, 10 mM MgCl2, 1 mM Tris(hydroxy-propyl)-phosphine (THP), pH 7.5) buffer. An aliquot (37.5 μL) of the NMN-containing sample was sequentially mixed with 15 μL of 20% acetophenone (in DMSO) and 15 μL of 2M KOH. The mixture was placed on ice for approximately 10 min. Next, 67.5 μL of 100% formic acid was added to each sample, vortexed, and then incubated at 37 °C for 20 min. Samples (100 μL) were transferred to a 96-well opaque bottom plate and fluorescence (Ex/Em = 382/445 nm) was measured using a SpectraMax M5 plate reader (Molecular Devices, Sunnyvale, CA). NAM, NMN, ATP, PRPP, and acetophenone were purchased from Sigma Aldrich, St. Louis, MO.) THP was purchased from Santa Cruz Biotechnology, Santa Cruz, CA.
2.6. Ultra-performance liquid chromatography and tandem mass spectrometry
Mouse tissues were flash frozen in liquid nitrogen after dissection. Frozen tissues were lyophilized overnight and powdered using a Precellys Evolution tissue homogenizer (Bertin Corp., Rockville, MD). Approximately 5 mg of powdered tissue were homogenized in 0.5 M perchloric acid in the Precellys homogenizer. Then, a 100 μL aliquot of homogenate were mixed with 100 μL of 1 M ammonium formate along with isotopically-labeled nucleotide internal standards. This mixture was vortexed and centrifuged at 14,000 × g for 10 min at 10 °C. Then, the supernatant was filtered through a 3 kDa filter plate prior to quantitation of NMN, NAD+ and NAM by LC/MS/MS.
2.7. Body composition
Conscious mice were immobilized in ventilated tubes and placed in a Bruker Bio-Analyzer Minispec NMR machine (Bruker Optics, Billerica, MA) for determination of fat mass, fat free mass, and fluid.
2.8. Hyperinsulinemic-euglycemic clamp
Clamps were performed in conscious, unrestrained mice as previously described [19], [20], [21], [22]. Catheters were surgically implanted in the left common carotid artery and right jugular vein for sampling and infusions, respectively. Following a 5 day recovery, mice were fasted for 5 h. A primed-continuous infusion (2.5 μCi prime, 0.05 μCi/min continuous) of [3-3H]glucose (Perkin Elmer, Waltham, MA) was begun at t = −90 min prior to the beginning of the insulin infusion. Basal glucose and glucose specific activity were determined from blood samples obtained at t = −15 and −5 min prior to the insulin infusion. Basal plasma insulin levels were determined from blood samples obtained at t = −5 min prior to the insulin infusion. The clamp began at t = 0 min (end of the 5 h fast) with a continuous infusion of insulin (Humulin R, Eli Lilly) at a rate of 2.5 mU/kg/min. The [3-3H]glucose infusion was increased to 0.15 μCi/min for the remainder of the experiment. Euglycemia (∼150 mg/dL) was maintained by measuring blood glucose every 10 min starting at t = 0 min and infusing 50% dextrose as necessary. Mice received saline washed erythrocytes from donors beginning at t = 0 min and continuously throughout the clamp at a rate of 5.5 mL/min to prevent a fall of >5% hematocrit. Whole body glucose disappearance (Rd) was determined using Steele non-steady-state equations [23], [24]. At t = 155 min, mice were anesthetized with sodium pentobarbital.
2.9. Energy balance analysis
Oxygen consumption (VO2), carbon dioxide production (VCO2), food intake, water intake, and ambulatory activity were measured using a Comprehensive Lab Animal Monitoring System (CLAMS; Columbus Instruments, Columbus, OH). Respiratory exchange ratio (RER) was calculated as VCO2/VO2. Food intake was measured using a precision scale. Water intake was measured using a volumetric drinking dispenser. Ambulatory activity was estimated by the number of infrared beam breaks along the X-axis of the metabolic cage.
2.10. VO2max and exercise endurance test
VO2max tests were conducted as previously described [21]. Mice were acclimated to the treadmill 2 days prior to the stress test with a 10 min run at 10 m/min. On the day of the experiment, mice were placed in an enclosed, single-lane treadmill connected to the CLAMS and allowed to acclimate for 30 min. VO2 and VCO2 measurements were continuously made every min. Resting VO2 was calculated as the average of the 5 min before the beginning of the stress test. Mice began running at 10 m/min, 0% grade. The speed was increased by 3 m/min every 4 min until exhaustion. Mice were encouraged to run by an electric grid at the back of the treadmill (1.5 mA, 200 msec pulses, 4 Hz). Mice were defined as exhausted when they spent more than 5 continuous secs on the electric grid. VO2max was achieved when VO2 no longer increased despite an increase in treadmill speed. VO2max was expressed as the change in VO2 from resting (ΔVO2max). For exercise endurance tests, mice were run for 10 min at 10 m/min, 0% grade; then speed was increased to 20 m/min, 0% grade until exhausted, with exhaustion defined as before.
2.11. Fiber typing
Fiber typing was performed as previously described [25], with modifications. Previously frozen gastrocnemius muscle was embedded in Tissue-Tek Optimal Cutting Temperature (OCT) compound, and frozen in liquid nitrogen. 10 um serial cross-sections were taken on a Leica CM 1900 UV cryostat (Leica Biosystems, Richmond, IL), mounted on Super Frost Plus slides (Fisher Scientific, Hampton, NH) and blocked with 10% goat serum. Two primary antibody cocktails were applied to serial sections. The first contained anti-BF-F3 (Developmental Studies Hybridoma Bank [DSHB], Iowa City, IA) and anti-NOQ75d (ab11083, Abcam, Cambridge, MA), whereas the second contained anti-SC-71 (DSHB) and anti-6H1 (DSHB). The secondary antibody cocktail contained Alexa Fluor 568 goat anti-mouse IgG (A11004, Invitrogen, Grand Island, NY) and Alexa Fluor 647 goat anti-mouse IgM (A21238, Invitrogen). Sections were then incubated with 50 mg/ml lectin from Bandeiraea simplicifolia conjugated to FITC (L2895, Sigma–Aldrich, St. Louis, MO) in the dark. Cover slips were mounted with Prolong Gold antifade reagent. Images were taken with a Carl Zeiss Axio Observer.Z1 microscope using a 10X objective, AxioCam MRm camera and AxioVison 4.8 software. Individual images were taken across an entire cross-section and stitched together with the AxioVison software. Fiber types were counted manually using ImageJ software. Cells from three complete cross sections of gastrocnemius muscle were counted in two sequential slides per mouse, one for each primary antibody cocktail.
2.12. Skeletal muscle and liver glycogen content
15 mg of frozen skeletal muscle (gastrocnemius) and liver tissue were incubated at 100 °C (dry heat/oven) in 0.5 mL of 2N HCl for 2 h, then neutralized with 1.5 mL of 0.67N NaOH. Following neutralization, muscle samples were shaken until dissolved. 20 μL of the dissolved muscle samples and a glucose standard (0.473 mM) were then added to borosilicate tubes containing 1 mL of the reagent cocktail (50 mM Tris base, 25 mM HCl, 1 mM MgCl2, 0.5 mM Dithiothreitol, 0.3 mM ATP, 0.05 NADP, 1 U/mL hexokinase + glucose-6-phosphodehydrogenase). Samples were then incubated at room temperature for 5–10 min 200 μL from each reaction mixture were transferred to a 96-well black plate, and fluorescence was detected using a Biotek plate reader (Excitation 360 nM, Emission 460 nM). Glycogen content was calculated by the following equation: (ΔFsample/ΔFstandard) × (mM concentration standard × mL standard volume) × muscle dilution/mg of tissue × 1000 = μmoles glucosyl units/grams tissue
2.13. Statistics
Data are expressed as mean ± SEM. One-way ANOVA was used to detect significant differences between groups. A repeated measures two-way ANOVA with group as between and time as within factor was used to detect significant differences when repeated measures were performed. An unpaired Student's t-test was used to detect differences when only 2 groups were considered. Statistical significance was set at p < 0.050. Data were analyzed using JMP version 12 (SAS institute, Cary, NC) and GraphPad Prism v6.07 (GraphPad Software, Inc. La Jolla, CA).
3. Results
3.1. Basic characterization of the NamptTg mouse model
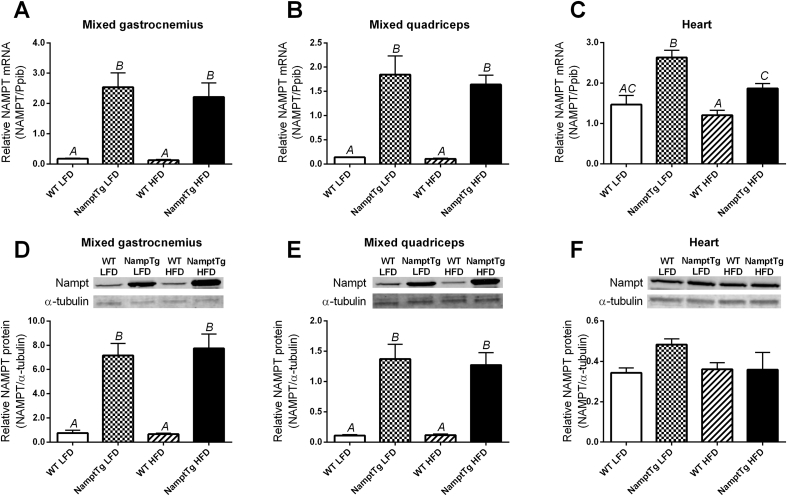
Expression of NAMPT mRNA was ∼15-fold higher in mixed gastrocnemius and mixed quadriceps from NamptTg versus WT mice (p < 0.010, Figure 1A–B). NAMPT protein levels in skeletal muscle were ∼10-fold higher in NamptTg versus WT mice (p < 0.001, Figure 1D–E). Expression of NAMPT mRNA was ∼1.6-fold higher in heart tissue from NamptTg mice versus WT mice (p < 0.050, Figure 1C), likely due to leaky Mck-directed expression in cardiac tissue [26]. Nevertheless, higher NAMPT mRNA expression did not result in increased NAMPT protein content in heart tissue (p = 0.125, Figure 1F). HFD feeding did not impact NAMPT mRNA expression or NAMPT protein content (p > 0.050, Figure 1A–F). NAMPT protein content was unchanged in liver and in epididymal white adipose tissue from NamptTg versus WT mice (data not shown).
Figure 1.
NAMPT overexpression in skeletal muscle increased NAMPT mRNA and protein content in skeletal muscle. Quantitative reverse transcriptase-PCR (qRT-PCR) analyses of NAMPT mRNA expression in (A) mixed gastrocnemius, (B) mixed quadriceps, and (C) heart; and western blot (WB) analyses of NAMPT protein content in (D) mixed gastrocnemius, (E) mixed quadriceps, and (F) heart. WT LFD = Wild Type Low Fat Diet; NamptTg LFD = NamptTg Low Fat Diet; WT HFD = Wild Type High Fat Diet; NamptTg HFD = NamptTg High Fat Diet. Letters indicate significant differences between groups (p < 0.050). Data are mean ± SEM.
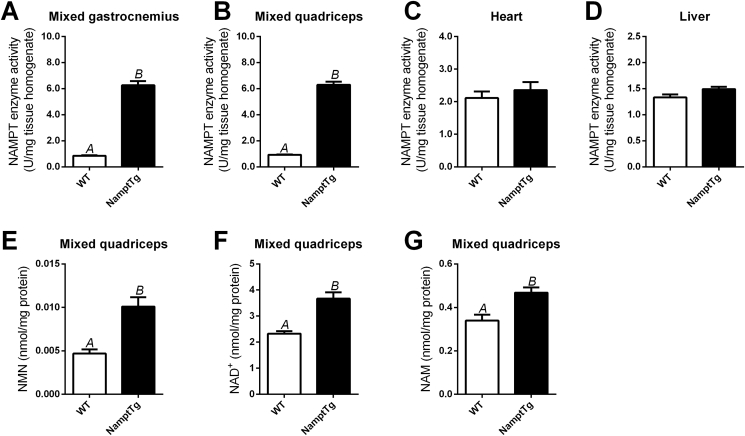
The levels of NAMPT enzyme activity in mixed gastrocnemius and mixed quadriceps from NamptTg mice were ∼7-fold higher than NAMPT enzyme activities in the corresponding muscles from WT mice (p < 0.001, Figure 2A–B). NAMPT enzyme activity in heart and liver were unchanged by NAMPT overexpression (p > 0.050, Figure 2C–D). The addition of the highly potent and specific NAMPT inhibitor FK-866 completely suppressed NMN production, confirming that NAMPT was solely responsible for the observed activity (data not shown).
Figure 2.
NAMPT overexpression in skeletal muscle increased NAMPT enzyme activity, NMN and NAD+in skeletal muscle. NAMPT enzyme activity in (A) mixed gastrocnemius, (B) mixed quadriceps, (C) heart, and (D) liver; and ultra-performance liquid chromatography and tandem mass spectrometry (UPLC-MS/MS) analyses of (E) nicotinamide mononucleotide (NMN), (F) nicotinamide adenine dinucleotide (NAD+), and (G) nicotinamide (NAM) in mixed quadriceps of chow fed WT and NamptTg mice. Letters indicate significant differences between groups (p < 0.050). Data are mean ± SEM.
NMN, the product of NAMPT activity, was ∼2-fold higher in skeletal muscle from NamptTg versus WT mice (p = 0.004, Figure 2E). The NAD+ levels in skeletal muscle were ∼1.6-fold higher in NamptTg versus WT mice (p = 0.002, Figure 2F). Surprisingly, the levels of skeletal muscle NAM, a substrate for NAMPT, were ∼1.4-fold higher in NamptTg versus WT mice (p = 0.010, Figure 2G).
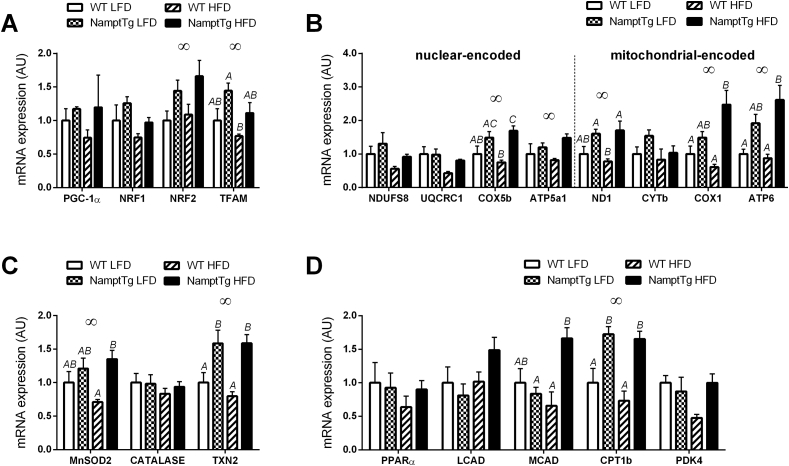
3.2. NAMPT overexpression in skeletal muscle increased mitochondrial gene expression in skeletal muscle
Skeletal muscle gene expression of several genes related to mitochondrial biogenesis (NRF2, TFAM) (Figure 3A), oxidative phosphorylation (COX5b, ATP5a1, ND1, COX1, ATP6) (Figure 3B), oxidative stress modulation (MnSOD2, TXN2) (Figure 3C), and fuel selection (CPT1b) (Figure 3D) was significantly higher in NamptTg versus WT mice. When fed LFD, WT mice displayed significantly lower gene expression of TXN2 and CPT1b than NamptTg mice fed LFD (Figure 3C–D). When fed HFD, WT mice displayed significantly lower gene expression of COX5b, ND1, COX1, ATP6, MnSOD2, TXN2, MCAD, and CPT1b than NamptTg mice fed HFD (Figure 3A–D). NamptTg and WT mice exhibited similar increases in body weight and fat mass percentage when fed HFD or LFD (data not shown).
Figure 3.
NAMPT overexpression in skeletal muscle increased mitochondrial gene expression in skeletal muscle. Gene expression of genes related to (A) mitochondrial biogenesis, (B) oxidative phosphorylation, (C) oxidative stress modulation, and (D) substrate selection. WT LFD = Wild Type Low Fat Diet; NamptTg LFD = NamptTg Low Fat Diet; WT HFD = Wild Type High Fat Diet; NamptTg HFD = NamptTg High Fat Diet. Letters indicate significant differences between groups (p < 0.050). ∞ p < 0.050 all WT versus all NamptTg. Data are normalized to WT LFD, and are mean ± SEM.
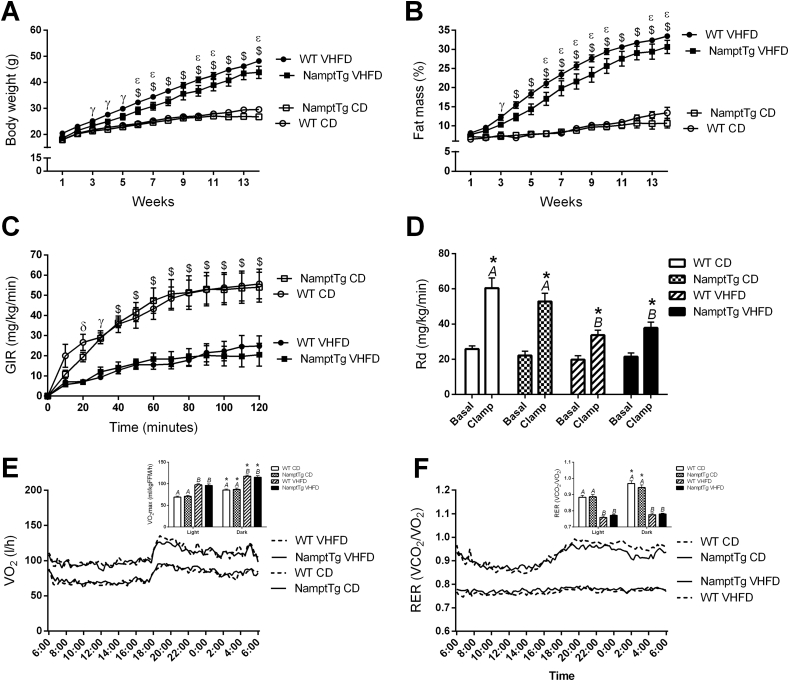
3.3. NAMPT overexpression in skeletal muscle partially protected against body weight gain but not against insulin resistance, without behavioral changes in mice fed VHFD
Mice fed VHFD had a larger increase in body weight and fat mass percentage than mice fed CD (p < 0.001, Figure 4A–B). NamptTg mice were partially protected against increases in body weight and fat mass percentage due to VHFD feeding, with 9.0% lower body weight (p < 0.001, Figure 4A) and 8.5% lower fat mass percentage (p < 0.001, Figure 4B) versus WT mice. No difference in body weight or fat mass percentage was observed between NamptTg and WT mice fed CD (Figure 4A–B, p > 0.050). Average glucose infusion rate, insulin stimulated glucose disposal rate (Rd), 24 h oxygen consumption (VO2), and respiratory exchange ratio (RER) was not different between NamptTg and WT mice on the same diet (p > 0.050, Figure 4C–F). Daily food intake was not different between mice fed the same diet (p > 0.050, Figure S2A). Daily water intake and daily ambulatory activity was similar across groups (p > 0.050, Figures S2B–C).
Figure 4.
NAMPT overexpression in skeletal muscle partially protected against body weight gain but not against insulin resistance, without behavioral changes in mice fed VHFD. (A) body weight over time, (B) fat mass percentage over time, (C) glucose infusion rate (GIR) during the hyperinsulinemic-euglycemic clamp after 16 weeks fed very high fat diet (VHFD) or chow diet (CD), (D) glucose rate of disposal (Rd) during the hyperinsulinemic-euglycemic clamp after 16 weeks fed very high fat diet (VHFD) or chow diet (CD, (E) oxygen consumption (VO2) over 24 h, and (F) respiratory exchange ratio (RER) over 24 h after 16 weeks fed VHFD or CD. WT CD = Wild Type Chow Diet; NamptTg CD = NamptTg Chow Diet; WT VHFD = Wild Type Very High Fat Diet; NamptTg VHFD = NamptTg Very High Fat Diet. Letters indicate significant differences between groups (p < 0.050). γ p < 0.050 WT VHFD versus WT CD and NamptTg CD; $ p < 0.050 all VHFD versus all CD; ε p < 0.050 WT VHFD versus NamptTg VHFD; δ p < 0.050 WT CD versus WT VHFD and NamptTg VHFD; *p < 0.050 basal versus clamp and light versus dark within the same group. Data are mean ± SEM.
3.4. NAMPT overexpression in skeletal muscle coupled with voluntary exercise training increased exercise endurance capacity
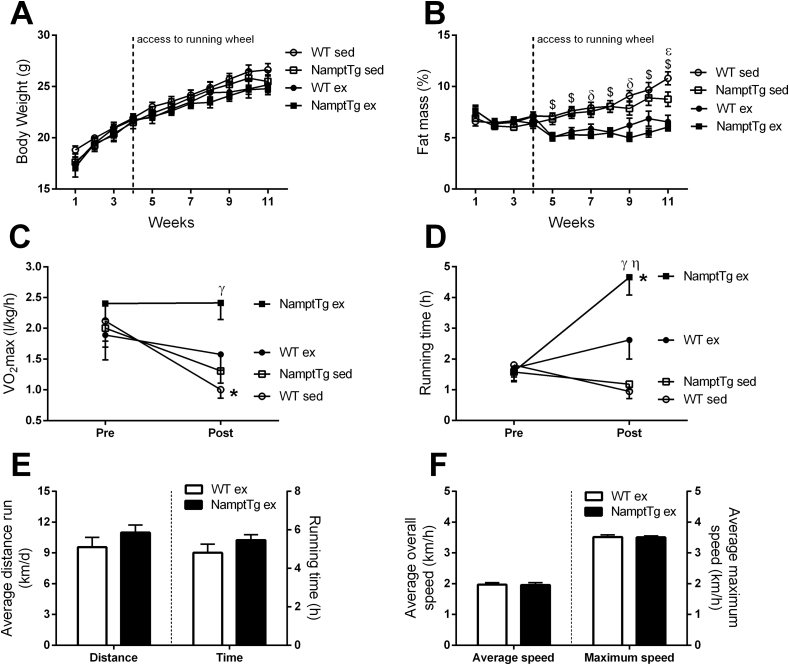
NamptTg and WT mice were housed without (sedentary) or with (exercised) running wheels in their home cages for 7 weeks. Body weight was not different between WT and NamptTg mice at the start of the protocol (p > 0.050, Figure 5A). Fat mass percentage remained similar in WT and NamptTg mice during the first 4 weeks of the study (p > 0.050, Figure 5B). However, access to running wheels resulted in lower fat mass percentage in exercised mice versus sedentary mice (p < 0.001, Figure 5B). There was no difference in fat mass percentage between NamptTg and WT mice subjected to the same voluntary exercise protocol (p > 0.050, Figure 5B). VO2max (p > 0.050, Figures 5C and S3A) and exercise endurance (p > 0.050, Figure 5D) were not different across groups before placement of running wheels. VO2max significantly decreased over time in sedentary WT mice (p < 0.050, Figures 5C and S3A). Sedentary NamptTg mice did not decrease VO2max significantly, suggesting that skeletal muscle NAMPT overexpression might provide some protection (p > 0.050, Figures 5C and S3A). Interestingly, VO2max post-intervention was significantly higher in exercised NamptTg mice versus sedentary WT mice (p < 0.050, Figures 5C and S3A) and sedentary NamptTg mice (p < 0.050, Figures 5C and S3A). Forced running times using a treadmill apparatus (to measure exercise endurance capacity) were comparable in sedentary WT, sedentary NamptTg, and exercised WT mice (p > 0.050, Figure 5D). However, exercised WT mice ran longer in the treadmill test after 7-week exposure to running wheels than sedentary WT mice (p = 0.024, Figure 5D). Interestingly, exercised NamptTg mice increased forced running time by ∼2.9-fold compared to baseline (p < 0.001, Figure 5D) and ran longer post-intervention than any of the other groups (p < 0.010, Figure 5D). The higher endurance displayed by exercised NamptTg mice was not attributable to higher levels of voluntary exercise training, as there were no differences in the average values for distance run per day, running time, speed, or maximal speed versus exercised WT mice (p > 0.050, Figure 5E–F). Skeletal muscle fiber type ratio changed with exercise training but was similar between NamptTg and WT mice that underwent the same intervention (Figure S3B). Heart weight was higher in exercised mice compared to sedentary mice but was not different between NamptTg and WT mice that were either sedentary or performed exercise training (Figure S3C). Skeletal muscle and liver glycogen content was not different across groups (Figure S3D–E).
Figure 5.
NAMPT overexpression in skeletal muscle coupled with voluntary exercise training increased exercise endurance capacity. (A) body weight over time, (B) fat mass percentage over time, (C) aerobic capacity (VO2max) pre- and post-voluntary exercise training or sedentary conditions, (D) endurance capacity (running time) pre- and post-voluntary exercise training or sedentary conditions, (E) running distance and total running time per 24 h, (F) average running speed and maximum running speed per 24 h. WT sed = sedentary Wild Type; NamptTg sed = sedentary NamptTg; WT ex = exercised Wild Type; NamptTg ex = exercised NamptTg. $ p < 0.050 all ex versus all sed; ε p < 0.050 WT sed versus NamptTg sed; δ p < 0.050 WT sed versus WT ex and NamptTg ex; γ p < 0.050 NamptTg ex versus WT sed and NamptTg sed; η p < 0.050 WT ex versus WT sed; *p < 0.050 pre-versus post-within the same group. Data are mean ± SEM.
3.5. NAMPT overexpression in skeletal muscle and voluntary exercise training increased mitochondrial gene expression but not SIRT1 gene expression or complex III protein content
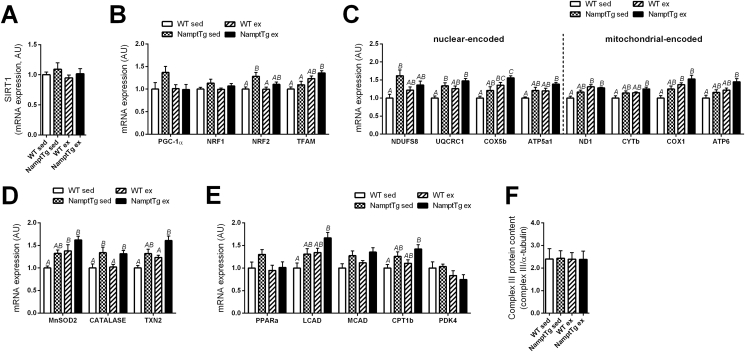
Gene expression and complex III protein content were analyzed in red and white quadriceps muscle of sedentary and exercised mice. SIRT1 gene expression was not different across groups in red quadriceps muscle (Figure 6A). In white quadriceps muscle, SIRT1 gene expression was lower in exercised NamptTg mice versus sedentary WT and sedentary NamptTg mice (p < 0.050, Fig. S4A). Differences in mitochondrial gene expression were observed in red quadriceps muscle (Figure 6B–E). Four genes (NRF2, NDUFS8, UQCRC1, CATALASE; p < 0.050, Figure 6B–E) showed higher expression in sedentary NamptTg mice in comparison to sedentary WT mice, and another 4 genes (COX5b, ND1, COX1, MnSOD2; p < 0.050, Figure 6B–E) showed higher expression in exercised WT mice in comparison to sedentary WT mice. A total of 13 genes (TFAM, UQCRC1, COX5b, ATP5a1, ND1, CYTb, COX1, ATP6, MnSOD2, CATALASE, TXN2, LCAD, CPT1b; p < 0.050, Figure 6B–E) showed higher expression in exercised NamptTg mice in comparison to sedentary WT mice. Complex III protein content in red quadriceps muscle was not significantly different across groups (Figure 6F). There were no remarkable differences displayed in mitochondrial gene expression, and complex III protein content by white quadriceps muscle from WT and NamptTg mice, despite the latter having markedly increased NAMPT protein levels (Figure S4A–F).
Figure 6.
NAMPT overexpression in skeletal muscle and voluntary exercise training increased mitochondrial gene expression but not SIRT1 gene expression and complex III protein content. Gene expression of (A) SIRT1 and genes related to (B) mitochondrial biogenesis, (C) oxidative phosphorylation, (D) oxidative stress modulation, and (E) substrate selection in red quadriceps. (F) Complex III protein content in red quadriceps. WT sed = sedentary Wild Type; NamptTg sed = sedentary NamptTg; WT ex = exercised Wild Type; NamptTg ex = exercised NamptTg. Letters indicate significant differences between groups (p < 0.050). Data are normalized to WT sed, and are mean ± SEM.
4. Discussion
Our data reveal that NAMPT overexpression in skeletal muscle of C57BL6/J mice increased skeletal muscle NAMPT enzyme activity by approximately 7-fold and, in turn, elevated skeletal muscle NMN and NAD+ levels. NamptTg mice fed VHFD were partially-protected from body weight gain, however, they developed insulin resistance that was comparable to WT mice fed VHFD. After prolonged access to a running wheel in the home cages, NamptTg mice exhibited a 3-fold higher exercise endurance capacity than WT mice despite equivalent bouts of voluntary running wheel activity.
This is the first study to report tissue levels of NAMPT enzyme activity. The 7-fold increase in skeletal muscle NAMPT activity due to expression of the NAMPT transgene resulted in only a 1.6-fold increase in skeletal muscle NAD+ levels. This relatively modest increase of NAD+ is comparable to other published studies that used various strategies to boost tissue NAD+ levels [27], [28], [29], [30]. Activation of NAD+ consuming pathways or the contribution of other homeostatic mechanisms probably serves to place a ceiling on the maximal NAD+ level. For instance, physiological levels of NAD+ can inhibit NAMPT activity thus posing a powerful feedback inhibition loop that blunts the impact of elevated NAMPT activity [31].
Skeletal muscle NAMPT overexpression increased expression of genes related to mitochondrial metabolism in the skeletal muscle of mice fed LFD and HFD. The increase in mitochondrial gene expression was especially clear in mice fed HFD; NamptTg mice fed HFD displayed higher gene expression for 8 genes related to mitochondrial metabolism in comparison to WT mice fed HFD (compared to 2 genes when fed LFD). The higher gene expression that was observed when mice were fed HFD might suggest that NamptTg mice have a better oxidative profile, which, in turn, might protect against metabolic complications that arise from HFD feeding. Nicotinamide riboside (NR) administration to mice fed VHFD enhanced mitochondrial gene expression in skeletal muscle when compared to mice that were not dosed with NR, and this was accompanied by partial protection against body weight and fat mass gain [27]. Similarly, NamptTg mice fed VHFD were partially protected against body weight gain as compared to WT mice fed VHFD. A moderate reduction in body weight has also been observed in older mice that had life-long overexpression of NAMPT in skeletal muscle when compared to age-matched WT mice [32].
Despite the lower body weight gain in the present study, NamptTg mice fed VHFD still developed insulin resistance comparable to WT mice on the same diet. The Baur group reported that skeletal muscle NAMPT overexpression did not protect from metabolic consequences after 24 weeks of VHFD feeding [28], and temporal overexpression of SIRT1 in skeletal muscle had no effect on insulin sensitivity in adult mice [33]. In contrast, increasing whole-body NAD+ levels in rodents using NAD+-precursors such as NMN and NR [27], [30], [34] attenuated the glucose and insulin intolerance, which developed in response to HFD feeding. Thus, the exclusive elevation of NAD+ in skeletal muscle in the NamptTg mouse might not be enough to improve the metabolic profile. On the other hand, NR or NMN administration will increase the NAD+ concentrations in other organs (such as adipose tissue and liver) that might play a pivotal role in preserving metabolic health.
A striking observation in our study was the 3-fold higher endurance capacity of exercised NamptTg mice versus exercised WT mice, despite similar duration and intensity of the recorded voluntary (running wheel) exercise activity. We showed previously that skeletal muscle NAMPT protein content was higher in athletes than in sedentary persons, and positively correlated with VO2max [9]. The investigation reported herein is the first to probe the impact of (voluntary) exercise training in the setting of increased skeletal muscle NAD+ concentrations. We further observed partial protection from decreases in VO2max over time in sedentary NamptTg mice when compared to sedentary WT mice. Cantó et al. reported a slight increase in VO2max and endurance capacity after 12 weeks of NR supplementation when compared to control mice [27]. Moreover, a recent study showed decreased endurance capacity by skeletal muscle-specific NAMPT knock-out mice [32]. Consequently, our data suggest that higher skeletal muscle NAD+ concentrations themselves might be beneficial for exercise performance. This is supported by earlier findings in older mice that had life-long overexpression of NAMPT in skeletal muscle, and showed protection against aging-induced decreases in exercise endurance [32].
The enhanced endurance capacity in the exercised NamptTg mice was concordant with gene expression changes evident in skeletal muscle derived from these animals. Thirteen out of 20 genes that govern mitochondrial metabolism showed higher gene expression in exercised NamptTg mice versus sedentary WT mice. While some of these genes were also elevated in sedentary NamptTg mice or exercised WT mice when compared to sedentary WT mice, 7 genes were uniquely elevated in exercised NamptTg mice, suggesting that when NAMPT is upregulated exercise training has a larger effect. These gene expression changes were seen in red muscle tissue, but not in white muscle tissue. Red muscle tissue predominantly consists of slow-twitch oxidative muscle fibers and is typically considered the specialized skeletal muscle type for endurance activity [35], which fits the enhanced endurance capacity profile observed of the exercised NamptTg mice.
While gene expression for genes related to mitochondrial metabolism was elevated in exercised NamptTg mice (versus sedentary WT mice), SIRT1 gene expression was not elevated. A previous study found no significant increases in protein levels of SIRT1 in gastrocnemius muscle of Sprague–Dawley rats that performed 12-weeks of exercise training [36]. There is evidence that exercise might increase SIRT1 enzyme activity via elevation of its necessary co-substrate NAD+, rather than increasing SIRT1 expression or protein content per se [12], [13]. We did not analyze SIRT1 enzyme activity in the present study, however, Cantó et al. concluded that AMP-kinase (AMPK) – a critical regulator of mitochondrial biogenesis and function in response to energy deprivation [37] – increased SIRT1 enzyme activity by increasing the intracellular levels of NAD+ [12]. Moreover, exercise training increased SIRT1 enzyme activity but not protein content in skeletal muscle of young and old male Wistar rats [13].
In conclusion, our study has revealed a fascinating interaction between elevated NAMPT activity in skeletal muscle and voluntary exercise that was manifest as a striking improvement of exercise endurance. While we have yet to unveil the precise molecular mechanism for this interaction, there are several fascinating possibilities to explore. Hence, there are tantalizing clues gleaned from published studies to be pursued as we seek to more fully elucidate the pivotal role played by NAMPT in health and fitness.
Acknowledgment
We would like to acknowledge the contributions of Estrellita Bermudez, Diana Albarado, and Dillon Chustz (Pennington Biomedical Research Center) for mouse colony management and tissue collection; Carmen Ruiz and Ginger Ku (Pennington Biomedical Research Center) for technical assistance with UPLC-MS/MS; Rochelle Holt, Samuel Goldberg, and Madhumita Jambhekar (Sanford-Burnham Medical Research Institute) for tissue processing and mouse colony management; and Emily King and Rochelle Holt of the Sanford-Burnham Cardiometabolic Phenotyping Core for in vivo metabolic phenotyping experiments.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.molmet.2017.10.012.
Author contribution
S.R.C. designed the study, performed experiments, analyzed the data and critically reviewed and edited the manuscript, B.B. performed experiments, analyzed the data and wrote the manuscript, M.E.H. performed experiments and critically reviewed the manuscript, L.M.S. interpreted results of experiments and critically reviewed and edited the manuscript, M.D. performed experiments and critically reviewed the manuscript, A.P.G. performed experiments and critically reviewed the manuscript, H.H.C. interpreted results of experiments and critically reviewed the manuscript, C.P. performed experiments and critically reviewed the manuscript, P.P. performed experiments and critically reviewed the manuscript, H.X. performed statistical analysis and critically reviewed the manuscript, F.Y. performed statistical analysis and critically reviewed the manuscript, G.A.W. interpreted results of experiments and critically reviewed the manuscript, T.F.O. interpreted results of experiments and critically reviewed the manuscript, D.A.S. interpreted results of experiments and critically reviewed the manuscript, R.L.M. interpreted results of experiments and critically reviewed the manuscript, J.E.A. interpreted results of experiments and critically reviewed the manuscript, S.J.G. designed the study, performed experiments, analyzed the results and critically reviewed and edited the manuscript, S.R.S. designed the study, performed experiments, analyzed the data and critically reviewed and edited the manuscript. S.R.S. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Funding
This work was supported by a Pennington Biomedical Research Center Pilot and Feasibility grant (SRC); NIH R01 (1 R01 AG030226-01; SRS); USDA (2005-34323-15741; SRS); and the PBRC Nutrition Obesity Research Unit [NORC] (NIH 5 P30 DK072476). The transgenic core at the Pennington Biomedical Research Center was partially supported by the NORC grant (P30 DK072476-12) and COBRE grant (P30 GM118430). The Metabolomics Core at SBP is a partner within the Southeast Center for Integrated Metabolomics (SECIM) and is partially supported by the NIH Common Fund (1 U24 DK097209-01A1; SJG, SRS).
Conflict of interest
The authors have no relevant conflict of interest to disclose.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Imai S., Guarente L. NAD+ and sirtuins in aging and disease. Trends in Cell Biology. 2014;24:464–471. doi: 10.1016/j.tcb.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mori V., Amici A., Mazzola F., Di Stefano M., Conforti L., Magni G. Metabolic profiling of alternative NAD biosynthetic routes in mouse tissues. PLoS One. 2014;9 doi: 10.1371/journal.pone.0113939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins P.B., Chaykin S. The management of nicotinamide and nicotinic acid in the mouse. The Journal of Biological Chemistry. 1972;247:778–783. [PubMed] [Google Scholar]
- 4.Revollo J.R., Grimm A.A., Imai S. The regulation of nicotinamide adenine dinucleotide biosynthesis by Nampt/PBEF/visfatin in mammals. Current Opinion in Gastroenterology. 2007;23:164–170. doi: 10.1097/MOG.0b013e32801b3c8f. [DOI] [PubMed] [Google Scholar]
- 5.Wang T., Zhang X., Bheda P., Revollo J.R., Imai S., Wolberger C. Structure of Nampt/PBEF/visfatin, a mammalian NAD+ biosynthetic enzyme. Nature Structural & Molecular Biology. 2006;13:661–662. doi: 10.1038/nsmb1114. [DOI] [PubMed] [Google Scholar]
- 6.Hirschey M.D., Shimazu T., Jing E., Grueter C.A., Collins A.M., Aouizerat B. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Molecular Cell. 2011;44:177–190. doi: 10.1016/j.molcel.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Revollo J.R., Grimm A.A., Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. The Journal of Biological Chemistry. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 8.Rongvaux A., Shea R.J., Mulks M.H., Gigot D., Urbain J., Leo O. Pre-B-cell colony-enhancing factor, whose expression is up-regulated in activated lymphocytes, is a nicotinamide phosphoribosyltransferase, a cytosolic enzyme involved in NAD biosynthesis. European Journal of Immunology. 2002;32:3225–3234. doi: 10.1002/1521-4141(200211)32:11<3225::AID-IMMU3225>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 9.Costford S.R., Bajpeyi S., Pasarica M., Albarado D.C., Thomas S.C., Xie H. Skeletal muscle NAMPT is induced by exercise in humans. American Journal of Physiology, Endocrinology and Metabolism. 2010;298:E117–E126. doi: 10.1152/ajpendo.00318.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez-Marcos P.J., Auwerx J. Regulation of PGC-1alpha, a nodal regulator of mitochondrial biogenesis. American Journal of Clinical Nutrition. 2011;93:884S–890S. doi: 10.3945/ajcn.110.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gurd B.J. Deacetylation of PGC-1alpha by SIRT1: importance for skeletal muscle function and exercise-induced mitochondrial biogenesis. Applied Physiology, Nutrition, and Metabolism. 2011;36:589–597. doi: 10.1139/h11-070. [DOI] [PubMed] [Google Scholar]
- 12.Canto C., Jiang L.Q., Deshmukh A.S., Mataki C., Coste A., Lagouge M. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metabolism. 2010;11:213–219. doi: 10.1016/j.cmet.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koltai E., Szabo Z., Atalay M., Boldogh I., Naito H., Goto S. Exercise alters SIRT1, SIRT6, NAD and NAMPT levels in skeletal muscle of aged rats. Mechanisms of Ageing and Development. 2010;131:21–28. doi: 10.1016/j.mad.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright D.C., Han D.H., Garcia-Roves P.M., Geiger P.C., Jones T.E., Holloszy J.O. Exercise-induced mitochondrial biogenesis begins before the increase in muscle PGC-1alpha expression. The Journal of Biological Chemistry. 2007;282:194–199. doi: 10.1074/jbc.M606116200. [DOI] [PubMed] [Google Scholar]
- 15.Lira V.A., Benton C.R., Yan Z., Bonen A. PGC-1alpha regulation by exercise training and its influences on muscle function and insulin sensitivity. American Journal of Physiology, Endocrinology and Metabolism. 2010;299:E145–E161. doi: 10.1152/ajpendo.00755.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hogan B.C.F., Lacy E. Cold Spring Harbor Laboratory; Cold Spring Harbor, New York: 1986. Manipulating the mouse embryo: a laboratory manual. [Google Scholar]
- 17.Sparks L.M., Xie H., Koza R.A., Mynatt R., Hulver M.W., Bray G.A. A high-fat diet coordinately downregulates genes required for mitochondrial oxidative phosphorylation in skeletal muscle. Diabetes. 2005;54:1926–1933. doi: 10.2337/diabetes.54.7.1926. [DOI] [PubMed] [Google Scholar]
- 18.Zhang R.Y., Qin Y., Lv X.Q., Wang P., Xu T.Y., Zhang L. A fluorometric assay for high-throughput screening targeting nicotinamide phosphoribosyltransferase. Analytical Biochemistry. 2011;412:18–25. doi: 10.1016/j.ab.2010.12.035. [DOI] [PubMed] [Google Scholar]
- 19.Ayala J.E., Bracy D.P., Hansotia T., Flock G., Seino Y., Wasserman D.H. Insulin action in the double incretin receptor knockout mouse. Diabetes. 2008;57:288–297. doi: 10.2337/db07-0704. [DOI] [PubMed] [Google Scholar]
- 20.Ayala J.E., Bracy D.P., James F.D., Burmeister M.A., Wasserman D.H., Drucker D.J. Glucagon-like peptide-1 receptor knockout mice are protected from high-fat diet-induced insulin resistance. Endocrinology. 2010;151:4678–4687. doi: 10.1210/en.2010-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ayala J.E., Bracy D.P., James F.D., Julien B.M., Wasserman D.H., Drucker D.J. The glucagon-like peptide-1 receptor regulates endogenous glucose production and muscle glucose uptake independent of its incretin action. Endocrinology. 2009;150:1155–1164. doi: 10.1210/en.2008-0945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burmeister M.A., Ferre T., Ayala J.E., King E.M., Holt R.M., Ayala J.E. Acute activation of central GLP-1 receptors enhances hepatic insulin action and insulin secretion in high-fat-fed, insulin resistant mice. American Journal of Physiology, Endocrinology and Metabolism. 2012;302:E334–E343. doi: 10.1152/ajpendo.00409.2011. [DOI] [PubMed] [Google Scholar]
- 23.Debodo R.C., Steele R., Altszuler N., Dunn A., Bishop J.S. On the hormonal regulation of carbohydrate metabolism; studies with C14 glucose. Recent Progress in Hormone Research. 1963;19:445–488. [PubMed] [Google Scholar]
- 24.Steele R., Wall J.S., De Bodo R.C., Altszuler N. Measurement of size and turnover rate of body glucose pool by the isotope dilution method. American Journal of Physiology. 1956;187:15–24. doi: 10.1152/ajplegacy.1956.187.1.15. [DOI] [PubMed] [Google Scholar]
- 25.Bloemberg D., Quadrilatero J. Rapid determination of myosin heavy chain expression in rat, mouse, and human skeletal muscle using multicolor immunofluorescence analysis. PLoS One. 2012;7 doi: 10.1371/journal.pone.0035273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson J.E., Wold B.J., Hauschka S.D. Muscle creatine kinase sequence elements regulating skeletal and cardiac muscle expression in transgenic mice. Molecular and Cellular Biology. 1989;9:3393–3399. doi: 10.1128/mcb.9.8.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Canto C., Houtkooper R.H., Pirinen E., Youn D.Y., Oosterveer M.H., Cen Y. The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metabolism. 2012;15:838–847. doi: 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frederick D.W., Davis J.G., Davila A., Jr., Agarwal B., Michan S., Puchowicz M.A. Increasing NAD synthesis in muscle via nicotinamide phosphoribosyltransferase is not sufficient to promote oxidative metabolism. The Journal of Biological Chemistry. 2015;290:1546–1558. doi: 10.1074/jbc.M114.579565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li L., Muhlfeld C., Niemann B., Pan R., Li R., Hilfiker-Kleiner D. Mitochondrial biogenesis and PGC-1alpha deacetylation by chronic treadmill exercise: differential response in cardiac and skeletal muscle. Basic Research in Cardiology. 2011;106:1221–1234. doi: 10.1007/s00395-011-0213-9. [DOI] [PubMed] [Google Scholar]
- 30.Yoshino J., Mills K.F., Yoon M.J., Imai S. Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metabolism. 2011;14:528–536. doi: 10.1016/j.cmet.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burgos E.S., Schramm V.L. Weak coupling of ATP hydrolysis to the chemical equilibrium of human nicotinamide phosphoribosyltransferase. Biochemistry. 2008;47:11086–11096. doi: 10.1021/bi801198m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frederick D.W., Loro E., Liu L., Davila A., Jr., Chellappa K., Silverman I.M. Loss of NAD homeostasis leads to progressive and reversible degeneration of skeletal muscle. Cell Metabolism. 2016;24:269–282. doi: 10.1016/j.cmet.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Svensson K., LaBarge S.A., Martins V.F., Schenk S. Temporal overexpression of SIRT1 in skeletal muscle of adult mice does not improve insulin sensitivity or markers of mitochondrial biogenesis. Acta Physiologica. 2017;221:193–203. doi: 10.1111/apha.12897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang S.J., Choi J.M., Kim L., Park S.E., Rhee E.J., Lee W.Y. Nicotinamide improves glucose metabolism and affects the hepatic NAD-sirtuin pathway in a rodent model of obesity and type 2 diabetes. The Journal of Nutritional Biochemistry. 2014;25:66–72. doi: 10.1016/j.jnutbio.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 35.Schiaffino S., Reggiani C. Fiber types in mammalian skeletal muscles. Physiological Reviews. 2011;91:1447–1531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- 36.Huang C.C., Wang T., Tung Y.T., Lin W.T. Effect of exercise training on skeletal muscle SIRT1 and PGC-1alpha expression levels in rats of different age. International Journal of Medical Sciences. 2016;13:260–270. doi: 10.7150/ijms.14586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zong H., Ren J.M., Young L.H., Pypaert M., Mu J., Birnbaum M.J. AMP kinase is required for mitochondrial biogenesis in skeletal muscle in response to chronic energy deprivation. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15983–15987. doi: 10.1073/pnas.252625599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.