Abstract
Cyclospora spp. in nonhuman primates are most closely related to Cyclospora cayetanensis, an emerging human pathogen causing outbreaks of cyclosporiasis in North America. Studies thus far indicate the possible existence of host specificity in Cyclospora spp. In this study, 411 fecal specimens from free-range rhesus monkeys (Macaca mulatta) were collected and examined for Cyclospora by sequence analysis of the small subunit rRNA gene. A novel Cyclospora species was identified in 28 (6.8 %) specimens and named Cyclospora macacae based on morphologic and molecular characterizations. The oocyst of C. macacae is spherical and measures 8.49±0.55× 8.49 ± 0.49 μm in diameter. Phylogenetic analysis grouped this species together with the other four Cyclospora species infecting primates, including C. cayetanensis in humans, forming a monophyletic group closely related to avian Eimeria species. In addition, C. cayetanensis was detected in one specimen, although whether rhesus monkeys can serve as a natural reservoir host of C. cayetanensis needs further investigation.
Keywords: Cyclospora, Cyclospora macacae, Rhesus monkeys, SSU rRNA gene
Introduction
Cyclospora cayetanensis, an important human pathogen causing acute diarrhea, is commonly found in some developing countries such as Haiti, Guatemala, Peru, and Nepal and rarely found in developed countries. However, it has caused numerous outbreaks in the USA and Canada since the mid 1990s (Hall et al. 2012; Herwaldt 2000; Ortega and Sanchez 2010; CDC 1996), including recent multi-state outbreaks in summer 2013 in the USA (CDC 2013). Most of these outbreaks are foodborne and associated with imported fresh produce (Hall et al. 2012; Herwaldt 2000; Ortega and Sanchez 2010; Shields and Olson 2003). Ingestion of contaminated water has also been implicated as a risk factor for C. cayetanensis infections (Hall et al. 2011; Ortega and Sanchez 2010). Because of the scarcity of human C. cayetanensis specimens and lack of animal models, studies in nonhuman primates have been used to improve the understanding of the biology of Cyclospora spp. (Eberhard et al. 1999a; Eberhard et al. 2014; Perez Cordon et al. 2008; Smith et al. 1996; Zhao et al. 2013).
Morphologically, oocysts of C. cayetanensis are spherical and 8–10 μm in diameter and differ significantly from other Cyclospora spp. in insectivores and rodents (Ortega et al. 1994; Ortega et al. 1993). Phylogenetic analysis based on the small subunit rRNA (SSU rRNA) gene suggests that C. cayetanensis and other Cyclospora spp. in primates are related to members of the Eimeria genus (Relman et al. 1996). Thus far, of 19 known species of Cyclospora, three species, including C. cercopitheci, C. colobi, and C. papionis from nonhuman primates (African green monkeys, colobus monkeys, and baboons, respectively), are most closely related to C. cayetanensis based on morphologic and molecular characterization (Eberhard et al. 1999a; Lopez et al. 1999). Moreover, these four Cyclospora species from primates (including humans) are probably host specific, even though their nonhuman primate hosts have overlapping ranges (Eberhard et al. 2001; Ortega and Sanchez 2010).
In November 2010, we collected fecal samples from free-range rhesus monkeys (Macaca mulatta) in a public park in Guizhou province in Southwestern China to conduct an epidemiological survey of cryptosporidiosis, giardiasis, and microsporidiosis (Ye et al. 2012). Organisms similar to Cyclospora spp. were identified in some specimens. In the present study, we conducted morphologic and molecular characterization of Cyclospora sp. in these animals and describe the occurrence of a parasite unique to macaque monkeys. We have proposed the name Cyclospora macacae n. sp. for this newly identified parasite from rhesus monkeys.
Materials and methods
Specimens
A total of 411 fecal specimens collected in November 2010 from rhesus monkeys (M. mulatta) were used in this study. They consisted of fresh stools picked up from grounds frequented by the animals. The rhesus monkeys were free-range animals in a public park in Guiyang City, Guizhou Province, China. Twenty-three water samples collected from a lake where these monkeys bathed were also examined. Permission for specimen collection was obtained from the park management prior to the execution of the study. The detailed specimen collection was described in a previous publication (Ye et al. 2012). The fresh specimens were stored in 2.5 % aqueous potassium dichromate at 4 °C prior to DNA extraction.
DNA extraction and PCR
After washing 200 μl of fecal specimens or water concentrates twice with distilled water by centrifugation at 3000×g for 10 min, DNA was extracted from them using the FastDNA SPIN Kit for Soil (MP Biomedicals, Irvine, CA). Cyclospora spp. in the specimens were genetically characterized by nested PCR amplification of a 680-bp fragment of the SSU rRNA gene (Li et al. 2011). The near full-length (1677 bp) SSU rRNA gene of the unique Cyclospora sp. was amplified by nested PCR analysis of three overlapping fragments (fragments 1, 2, and 3) using the following primers. The primers for fragment-1 (680 bp) were previously described (Li et al. 2011). The primers for fragment-2 (580 bp) were CYCP2F1 (5′-TGTAAAACCCTTCCAGAGAAC-3′) and CYCP2R1 (5′-AGAAGTGATGCGGAAACCAAA-3′) in primary PCR and CYCP2F2 (5′-TGTCGTGGTCATCCGGCC-3′) and CYCP2R2 (5′-ACCTGGTGAGTTTCCCCG-3′) in secondary PCR. The primers for fragment-3 (676 bp) were CYCP3F1 (5′-AACCTGGTTGATCCTGCCAG-3′) and CYCP3R1 (5′-TGATCCTTCTGCAGGTTCACCTA-3′) in primary PCR and CYCP3F2 (5′-AATCAAAGTCTCTGGGTTCTGGG-3′) and CYCP3R2 (5′-CGTGTTACGACTTT TGCATCCTT-3′) in secondary PCR. The PCR condition for fragment-2 and fragment-3 was similar to that used in PCR for fragment-1, except that the annealing temperatures of 55 and 60 °C were used, respectively, in primary and secondary PCR for fragment-2 and 53 and 55 °C, respectively, in primary and secondary PCR for fragment-3.
Sequence analysis
The secondary PCR products were sequenced in both directions on an ABI 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA). The sequences obtained were assembled using ChromasPro (http://www.technelysium.com.au/ChromasPro.html), edited using BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html), and aligned using ClustalX (http://bips.ustrasbg.fr/fr/Documentation/ClustalX/). To determine the taxonomic identity of the newly identified Cyclospora sp., a neighbor-joining tree based on near-complete sequences of the SSU rRNA gene was constructed using genetic distances from the Kimura two-parameter model and the software Mega 6 (http://www.megasoftware.net/). The reliability of cluster formation was evaluated by the bootstrap method with 1000 replicates.
Morphological assessment
Oocysts of the new Cyclospora sp. were purified by discontinuous sucrose and cesium chloride gradient centrifugation (Arrowood and Donaldson 1996). The purified oocysts were examined as wet mounts by differential interference contrast microscopy and epifluorescence microscopy with a 330–380-nm ultraviolet excitation filter (Zhou et al. 2011).
Results
Occurrence of Cyclospora spp. in rhesus monkeys
Of the 411 fecal specimens from rhesus monkeys, 28 (6.8 %) were positive for Cyclospora spp. by PCR analysis of the ~680-bp fragment. There were no obvious differences in fecal consistency between positive and negative specimens. DNA sequence analysis identified two types of sequences of the SSU rRNA gene. One sequence was identical to the reference sequence (AF111183) for C. cayetanensis, whereas the other 27 sequences were identical to the 621-bp SSU rRNA sequence (KC441080) from a Cyclospora sp. recently detected in a crab-eating macaque (Macaca fascicularis) in China (Ye et al. 2014). Of the 23 water samples from the lake where the monkeys bathed, four samples (17.4 %) were identified as positive for the new Cyclospora sp.
Sequence characteristics of SSU rRNA gene of Cyclospora sp
Efforts were made to acquire the near-complete sequence of the SSU rRNA gene of the newly identified Cyclospora sp. Altogether, a 1677-bp sequence, except for the 5′ and 3′ ends of the gene, was obtained and deposited in GenBank with the accession number KP335196. Pair-wise comparisons of the SSU rRNA sequence between Cyclospora sp. and known Cyclospora spp. in human and nonhuman primates (GenBank accession no. AF111183, AF111184, AF111185, AF111186, and AF111187) showed sequence differences of 1.5, 0.9, 1.0, and 1.0 % from C. cayetanensis, C. colobi, C. cercopitheci, and C. papionis, respectively.
Phylogenetic relationship of Cyclospora spp
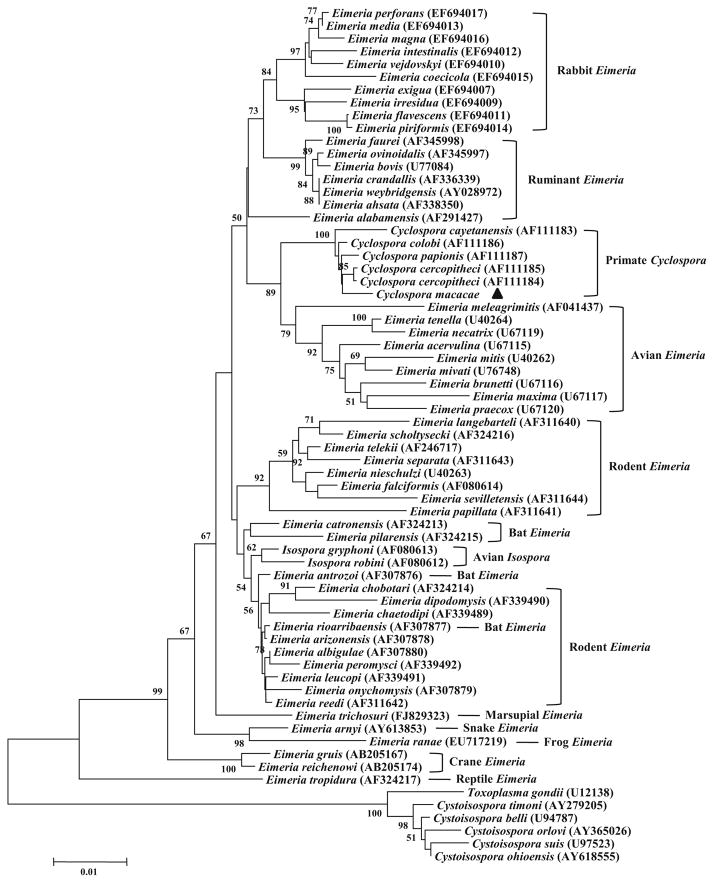
To establish the relationship between the newly identified Cyclospora species and other related apicomplexans, the obtained SSU rRNA sequence (1677 bp) was aligned with reference sequences in GenBank, including those from Cyclospora, Eimeria, Isospora, Cystoisospora, and Toxoplasma. In a neighbor-joining analysis, the novel Cyclospora sp. from rhesus monkeys grouped with other Cyclospora spp. from nonhuman primates (C. colobi, C. cercopitheci, and C. papionis) and was relatively distant from the human pathogen C. cayetanensis (Fig. 1). These five Cyclospora species from primates formed a monophyletic group with 100 % bootstrap support. This primate Cyclospora group clustered with a clade containing avian Eimeria species. They were more distant from bovine, rabbit, and rodent Eimeria species. In addition, the avian Isospora species formed a cluster within various Eimeria groups and were more related to the rodent Eimeria species.
Fig. 1.
Phylogenetic relationship among common Cyclospora, Eimeria, Isospora, Cystoisospora, and Toxoplasma species as inferred by a neighbor-joining analysis of the SSU rRNA sequence, based on genetic distances calculated by Kimura two-parameter model. Bootstrap values greater than 50 % from 1000 replicates are shown. The novel Cyclospora species C. macacae is indicated by a filled triangle
Morphometrics of oocysts of Cyclospora sp
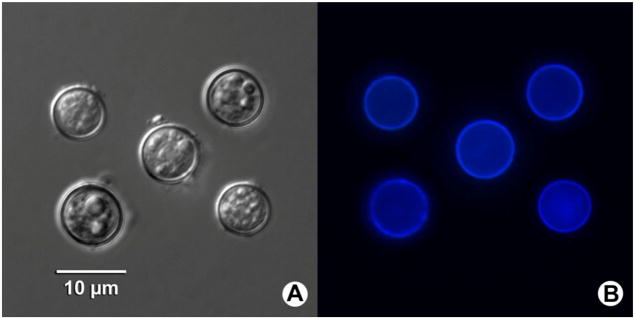
Oocysts of the novel Cyclospora sp. were almost perfectly spherical in microscopy. They measured 8.49±0.55×8.49± 0.49 μm in size, with a length/width shape index of 1.02 (n=11, Fig. 2a), and showed typical blue autofluorescence under an epifluorescence microscope with a 330–380-nm excitation filter (Fig. 2b). Both unsporulated and partially sporulated oocysts could be seen by microscopy (Fig. 2a).
Fig. 2.
Photomicrographs of Cyclospora macacae oocysts isolated from feces of rhesus monkeys (Macaca mulatta) in China. (a) Oocysts under differential interference contrast microscopy of wet mount. (b) Typical blue autofluorescence of the oocysts is observed under epifluorescence microscopy using a 330–380-nm ultraviolet excitation filter
Description of C. macacae n. sp
Based on the unique genetic feature of the novel Cyclospora and the apparent restriction of the parasite to macaque monkeys, we propose to name the Cyclospora sp. identified in this study as C. macacae n. sp.
Diagnosis—oocysts measure 8.49±0.55×8.49±0.49 μm with a length/width shape index of 1.02 (n=11). Two sporocysts are seen in each oocyst.
Host type—Rhesus monkeys (M. mulatta).
Other natural hosts—crab - eating macaques (M. fascicularis).
Type locality—Guiyang, Guizhou Province, China.
Prevalence—found in 6.6 % of rhesus monkeys sampled.
Site of infection—unknown, oocysts collected from feces.
Material deposited—the SSU rRNA sequence of this species has been deposited in GenBank under accession number KP335196.
Etymology—the species name of C. macacae was derived from the genus name of its host (M. mulatta) from which this parasite was recovered.
Discussion
To date, only four Cyclospora spp. from primates have been identified in previous studies using a combination of morphologic observations and molecular characterizations, including the human-pathogenic species C. cayetanensis and three non-human primate species: C. cercopitheci, C. colobi, and C. papionis from monkeys and baboons (Eberhard et al. 1999a; Ortega et al. 1994). In a few reports, some organisms resembling the primate Cyclospora species were also observed in fecal samples of monkeys, baboons, and mandrills, although the taxonomic status of these Cyclospora-like organisms has not been clearly established because of the lack of either morphologic or molecular data (Eberhard et al. 2014; Perez Cordon et al. 2008; Smith et al. 1996; Zhao et al. 2013). In this study, we identified a novel Cyclospora species from rhesus monkeys in southwestern China and named it C. macacae based on morphologic and molecular characterizations.
The oocysts of C. macacae under microscopy (spherical, mean 8.5 μm in diameter) are similar to morphologic descriptions of other Cyclospora spp. in primates, such as C. cayetanensis (spherical, mean 8.6 μm in diameter), C. cercopitheci (spherical, mean 9.2 μm in diameter), C. colobi (spherical, mean 8.3 μm in diameter), and C. papionis (spherical, mean 8.8 μm in diameter) (Eberhard et al. 1999a; Ortega et al. 1994). This observation suggests that all five known Cyclospora species infecting primates have small and spherical oocysts, in contrast to those Cyclospora species that infect insectivores and rodents, which have large and oblong oocysts (Eberhard et al. 1999a). Although these Cyclospora spp. infecting primates are morphologically similar and may not be easily defined at the light-microscopy level, the sequence and phylogenetic analysis based on the SSU rRNA gene can distinguish C. macacae from the other four primate Cyclospora species. Data from this and previous studies further support the conclusion of host specificity in Cyclospora species, although similar primate species, such as two species of macaque monkeys in the case of C. macacae and two species of colobus moneys in the case of C. colobi, can be infected naturally with the same Cyclospora species (Eberhard et al. 1999a; Eberhard et al. 2001; Eberhard et al. 2014; Zhao et al. 2013).
For the specimens used in this study, previous genetic characterization of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi indicated that the rhesus monkeys in close contact with humans in the public park are commonly infected with human-pathogenic protist species/genotypes or subtypes, an indication of cross-species transmission of enteric pathogens between monkeys and humans (Ye et al. 2012). In this study, we conducted an examination of Cyclospora infections in these monkeys and found 27 monkeys infected with C. macacae and one monkey infected with C. cayetanensis. Here, C. macacae has been reported to infect rhesus monkeys for the first time, and it has never been reported in humans thus far. In the lake where these monkeys frequently bathed, 4/23 water samples were identified to contain C. macacae, indicating that drinking water or recreational water might have played a role in the transmission of C. macacae among monkeys. Surprisingly, C. cayetanensis was also detected in one fecal specimen of a rhesus monkey by PCR analysis. C. cayetanensis is known to be human pathogenic, and humans are probably the only natural host (Aksoy et al. 2014; Ortega and Sanchez 2010). Attempts have been made to identify nonhuman hosts or reservoirs for C. cayetanensis and to establish experimental infection model in multiple laboratory animals (also including rhesus monkeys), but none of the tested animals became infected (Eberhard et al. 1999b; Eberhard et al. 2000). However, results of this and a previous study have identified the presence of C. cayetanensis in the fecal samples of two rhesus monkeys in China and Nepal, suggesting that C. cayetanensis may under rare occasions infect rhesus monkeys (Chu et al. 2004). Because of limited numbers of specimens and the absence of tissue analysis, whether rhesus monkeys can serve as a natural reservoir host of C. cayetanensis needs further investigation.
Our phylogenetic analysis based on the 1677-bp sequence of the SSU rRNA gene showed that C. macacae from rhesus monkeys is more closely related to Cyclospora spp. from non-human primates than to C. cayetanensis from humans, probably reflecting the host differences between humans and lower primates. The tree also places the primate Cyclospora group between various Eimeria groups, forming a cluster with avian Eimeria species rather than to mammalian (rabbit, bovine, and rodent) Eimeria species. This is consistent with previous taxonomic descriptions of coccidia (Eberhard et al. 1999a; Li et al. 2007; Morrison et al. 2004; Zhao et al. 2013). In addition to the placement of the primate Cyclospora within the Eimeria clade, two avian Isospora species are placed there as well, whereas the mammalian Cystoisospora species form sister groups with Toxoplasma gondii as reported previously (Morrison et al. 2004). However, the true taxonomic placement of Cyclospora spp. will require genetic characterization of diverse parasites at other loci.
The identification and description of C. macacae in rhesus monkeys extends our knowledge on the taxonomy and transmission of Cyclospora spp. in primates. With further characterization, the C. macacae-rhesus monkey relationship may serve as a useful model to improve our understanding of the biology of C. cayetanensis. In the era of genomics, whole genome sequencing is needed to better understand the genetic relationship between Cyclospora spp. and Eimeria spp. and among various Cyclospora species, and the genetic basis for host specificity of C. cayetanensis.
Acknowledgments
This work was supported in part by the National Natural Science Foundation of China (31425025 and 31302078), China Postdoctoral Science Foundation (2014 M560310), Open Funding Project of the State Key Laboratory of Veterinary Etiological Biology, Lanzhou, China (SKLVEB2014KFKT008), and Fundamental Research Funds for the Central Universities, China. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Contributor Information
Na Li, State Key Laboratory of Bioreactor Engineering, School of Resources and Environmental Engineering, East China University of Science and Technology, Shanghai, China.
Jianbin Ye, State Key Laboratory of Bioreactor Engineering, School of Resources and Environmental Engineering, East China University of Science and Technology, Shanghai, China.
Michael J. Arrowood, Division of Foodborne, Waterborne, and Environmental Diseases, National Center for Emerging and Zoonotic Infectious Diseases, Centers for Disease Control and Prevention, Atlanta, GA, USA
Jingbo Ma, State Key Laboratory of Bioreactor Engineering, School of Resources and Environmental Engineering, East China University of Science and Technology, Shanghai, China.
Lin Wang, State Key Laboratory of Bioreactor Engineering, School of Resources and Environmental Engineering, East China University of Science and Technology, Shanghai, China.
Hailing Xu, State Key Laboratory of Bioreactor Engineering, School of Resources and Environmental Engineering, East China University of Science and Technology, Shanghai, China.
Yaoyu Feng, State Key Laboratory of Bioreactor Engineering, School of Resources and Environmental Engineering, East China University of Science and Technology, Shanghai, China.
Lihua Xiao, Division of Foodborne, Waterborne, and Environmental Diseases, National Center for Emerging and Zoonotic Infectious Diseases, Centers for Disease Control and Prevention, Atlanta, GA, USA.
References
- Aksoy U, Marangi M, Papini R, Ozkoc S, Bayram Delibas S, Giangaspero A. Detection of Toxoplasma gondii and Cyclospora cayetanensis in Mytilus galloprovincialis from Izmir Province coast (Turkey) by real time PCR/high-resolution melting analysis (HRM) Food Microbiol. 2014;44:128–135. doi: 10.1016/j.fm.2014.05.012. [DOI] [PubMed] [Google Scholar]
- Arrowood MJ, Donaldson K. Improved purification methods for calf-derived Cryptosporidium parvum oocysts using discontinuous sucrose and cesium chloride gradients. J Eukaryot Microbiol. 1996;43:89S. doi: 10.1111/j.1550-7408.1996.tb05015.x. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Update: outbreaks of Cyclospora cayetanensis infection—United States and Canada, 1996. Morb Mortal Wkly Rep. 1996;45:611–612. [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Outbreaks of cyclosporiasis—United States, June–August 2013. Morb Mortal Wkly Rep. 2013;62:862. [PMC free article] [PubMed] [Google Scholar]
- Chu DM, Sherchand JB, Cross JH, Orlandi PA. Detection of Cyclospora cayetanensis in animal fecal isolates from Nepal using an FTA filter-base polymerase chain reaction method. Am J Trop Med Hyg. 2004;71:373–379. [PubMed] [Google Scholar]
- Eberhard ML, da Silva AJ, Lilley BG, Pieniazek NJ. Morphologic and molecular characterization of new Cyclospora species from Ethiopian monkeys: C. cercopitheci sp.n., C. colobi sp.n., and C. papionis sp.n. Emerg Infect Dis. 1999a;5:651–658. doi: 10.3201/eid0505.990506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard ML, Nace EK, Freeman AR. Survey for Cyclospora cayetanensis in domestic animals in an endemic area in Haiti. J Parasitol. 1999b;85:562–563. [PubMed] [Google Scholar]
- Eberhard ML, Ortega YR, Hanes DE, Nace EK, Do RQ, Robl MG, Won KY, Gavidia C, Sass NL, Mansfield K, Gozalo A, Griffiths J, Gilman R, Sterling CR, Arrowood MJ. Attempts to establish experimental Cyclospora cayetanensis infection in laboratory animals. J Parasitol. 2000;86:577–582. doi: 10.1645/0022-3395(2000)086[0577:ATEECC]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Eberhard ML, Njenga MN, DaSilva AJ, Owino D, Nace EK, Won KY, Mwenda JM. A survey for Cyclospora spp. in Kenyan primates, with some notes on its biology. J Parasitol. 2001;87:1394–1397. doi: 10.1645/0022-3395(2001)087[1394:ASFCSI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Eberhard ML, Owens JR, Bishop HS, de Almeida ME, da Silva AJ. Cyclospora spp. in drills, Bioko Island, Equatorial Guinea. Emerg Infect Dis. 2014;20:510–511. doi: 10.3201/eid2003.131368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall RL, Jones JL, Herwaldt BL. Surveillance for laboratory-confirmed sporadic cases of cyclosporiasis—United States, 1997–2008. Morb Mortal Wkly Rep. 2011;60:1–11. [PubMed] [Google Scholar]
- Hall RL, Jones JL, Hurd S, Smith G, Mahon BE, Herwaldt BL. Population-based active surveillance for Cyclospora infection—United States, Foodborne Diseases Active Surveillance Network (FoodNet), 1997–2009. Clin Infect Dis. 2012;54(Suppl 5):S411–S417. doi: 10.1093/cid/cis049. [DOI] [PubMed] [Google Scholar]
- Herwaldt BL. Cyclospora cayetanensis: a review, focusing on the outbreaks of cyclosporiasis in the 1990s. Clin Infect Dis. 2000;31:1040–1057. doi: 10.1086/314051. [DOI] [PubMed] [Google Scholar]
- Li G, Xiao S, Zhou R, Li W, Wadeh H. Molecular characterization of Cyclospora-like organism from dairy cattle. Parasitol Res. 2007;100:955–961. doi: 10.1007/s00436-006-0380-z. [DOI] [PubMed] [Google Scholar]
- Li W, Kiulia NM, Mwenda JM, Nyachieo A, Taylor MB, Zhang X, Xiao L. Cyclospora papionis, Cryptosporidium hominis, and human-pathogenic Enterocytozoon bieneusi in captive baboons in Kenya. J Clin Microbiol. 2011;49:4326–4329. doi: 10.1128/JCM.05051-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez FA, Manglicmot J, Schmidt TM, Yeh C, Smith HV, Relman DA. Molecular characterization of Cyclospora-like organisms from baboons. J Infect Dis. 1999;179:670–676. doi: 10.1086/314645. [DOI] [PubMed] [Google Scholar]
- Morrison DA, Bornstein S, Thebo P, Wernery U, Kinne J, Mattsson JG. The current status of the small subunit rRNA phylogeny of the coccidia (Sporozoa) Int J Parasitol. 2004;34:501–514. doi: 10.1016/j.ijpara.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Ortega YR, Sanchez R. Update on Cyclospora cayetanensis, a food-borne and waterborne parasite. Clin Microbiol Rev. 2010;23:218–234. doi: 10.1128/CMR.00026-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega YR, Sterling CR, Gilman RH, Cama VA, Diaz F. Cyclospora species—a new protozoan pathogen of humans. N Engl J Med. 1993;328:1308–1312. doi: 10.1056/NEJM199305063281804. [DOI] [PubMed] [Google Scholar]
- Ortega YR, Gilman RH, Sterling CR. A new coccidian parasite (Apicomplexa: Eimeriidae) from humans. J Parasitol. 1994;80:625–629. [PubMed] [Google Scholar]
- Perez Cordon G, Hitos Prados A, Romero D, Sanchez Moreno M, Pontes A, Osuna A, Rosales MJ. Intestinal parasitism in the animals of the zoological garden “Pena Escrita” (Almunecar, Spain) Vet Parasitol. 2008;156:302–309. doi: 10.1016/j.vetpar.2008.05.023. [DOI] [PubMed] [Google Scholar]
- Relman DA, Schmidt TM, Gajadhar A, Sogin M, Cross J, Yoder K, Sethabutr O, Echeverria P. Molecular phylogenetic analysis of Cyclospora, the human intestinal pathogen, suggests that it is closely related to Eimeria species. J Infect Dis. 1996;173:440–445. doi: 10.1093/infdis/173.2.440. [DOI] [PubMed] [Google Scholar]
- Shields JM, Olson BH. Cyclospora cayetanensis: a review of an emerging parasitic coccidian. Int J Parasitol. 2003;33:371–391. doi: 10.1016/s0020-7519(02)00268-0. [DOI] [PubMed] [Google Scholar]
- Smith HV, Paton CA, Girdwood RW, Mtambo MM. Cyclospora in non-human primates in Gombe, Tanzania. Vet Rec. 1996;138:528. [PubMed] [Google Scholar]
- Ye J, Xiao L, Ma J, Guo M, Liu L, Feng Y. Anthroponotic enteric parasites in monkeys in public park, China. Emerg Infect Dis. 2012;18:1640–1643. doi: 10.3201/eid1810.120653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Xiao L, Li J, Huang W, Amer SE, Guo Y, Roellig D, Feng Y. Occurrence of human-pathogenic Enterocytozoon bieneusi, Giardia duodenalis and Cryptosporidium genotypes in laboratory macaques in Guangxi, China. Parasitol Int. 2014;63:132–137. doi: 10.1016/j.parint.2013.10.007. [DOI] [PubMed] [Google Scholar]
- Zhao G, Cong M, Bian Q, Cheng W, Wang R, Qi M, Zhang L, Lin Q, Zhu X. Molecular characterization of Cyclospora-like organisms from golden snub-nosed monkeys in Qinling Mountain in Shaanxi province, Northwestern China. PLoS One. 2013;8:e58216. doi: 10.1371/journal.pone.0058216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Lv B, Wang Q, Wang R, Jian F, Zhang L, Ning C, Fu K, Wang Y, Qi M, Yao H, Zhao J, Zhang X, Sun Y, Shi K, Arrowood MJ, Xiao L. Prevalence and molecular characterization of Cyclospora cayetanensis, Henan, China. Emerg Infect Dis. 2011;17:1887–1890. doi: 10.3201/eid1710.101296. [DOI] [PMC free article] [PubMed] [Google Scholar]