SUMMARY
Germinal centers (GCs) are the primary sites of clonal B cell expansion and affinity maturation, directing the production of high-affinity antibodies. This response is a central driver of pathogenesis in autoimmune diseases, such as systemic lupus erythematosus (SLE), but the natural history of autoreactive GCs remains unclear. Here, we present a novel mouse model where the presence of a single autoreactive B cell clone drives the TLR7-dependent activation, expansion, and differentiation of other autoreactive B cells in spontaneous GCs. Once tolerance was broken for one self-antigen, autoreactive GCs generated B cells targeting other self-antigens. GCs became independent of the initial clone and evolved towards dominance of individual clonal lineages, indicating affinity maturation. This process produced serum autoantibodies to a breadth of self-antigens, leading to antibody deposition in the kidneys. Our data provide insight into the maturation of the self-reactive B cell response, contextualizing the epitope spreading observed in autoimmune disease.
Keywords: Germinal Center, Autoreactive B Cell, Systemic Lupus Erythematosus, Epitope Spreading, Autoantibody, Break-of-Tolerance
INTRODUCTION
Systemic lupus erythematosus (SLE) is characterized by the production of antibodies to nucleic acid antigens (Ags) (Rahman and Isenberg, 2008), with >75% of patients having serum autoantibodies to double-stranded DNA (compared to ~0.5% of healthy controls), which typically appear a few years before SLE is diagnosed (Arbuckle et al., 2003). SLE patients at or after disease onset drift in their serological reactivities towards a variety of nuclear, nucleolar, and protein-DNA complexes: a process known as epitope spreading. Although the mechanism is not well understood, epitope spreading is thought to be driven by chronic immune responses causing inclusion of new autoreactive B cell clones (Arbuckle et al., 2003; Cornaby et al., 2015; Vanderlugt and Miller, 2002).
Affinity-matured antibodies (Abs) arise in germinal centers (GCs), wherein B cell clones cycle between division, somatic hypermutation (SHM), and selection based on Ag affinity (Victora et al., 2010). This process of random mutation can generate BCRs that recognize self-Ags (Mietzner et al., 2008; Tiller et al., 2007); however, it has long been thought that GCs are able to limit the affinity maturation of autoreactive cells (Han et al., 1995; Pulendran et al., 1995; Shokat and Goodnow, 1995; Vinuesa et al., 2009). In fact, display of self-Ag by follicular dendritic cells (FDCs) within GCs can delete autoreactive cells (Yau et al., 2013), and autoreactive B cells can mutate away from autoreactivity (Reed et al., 2016; Sabouri et al., 2014). Therefore, it has remained questionable whether autoreactive B cells in GCs follow the same rules of Ag engagement with FDCs, clonal evolution, and affinity maturation as those in foreign-Ag-elicited GCs.
Many autoimmune mouse models have spontaneous GC formation (Luzina et al., 2001), but as these models are based on genetically modified B or T cells or have uncharacterized, complex genetic backgrounds, they are poorly suited to studying natural autoreactive GC behavior. Here, we developed a chimera mouse model that has spontaneous autoreactive GCs composed of self-reactive B cells from the wild-type (WT) repertoire in a genetically normal context. Surprisingly, we find that there is no limit on the evolution of autoreactive GCs – that is, they generate B cells targeting other self-Ags once tolerance is broken.
RESULTS
564Igi Mice Display Spontaneous GCs, VDJ diversification, SHM, and CSR
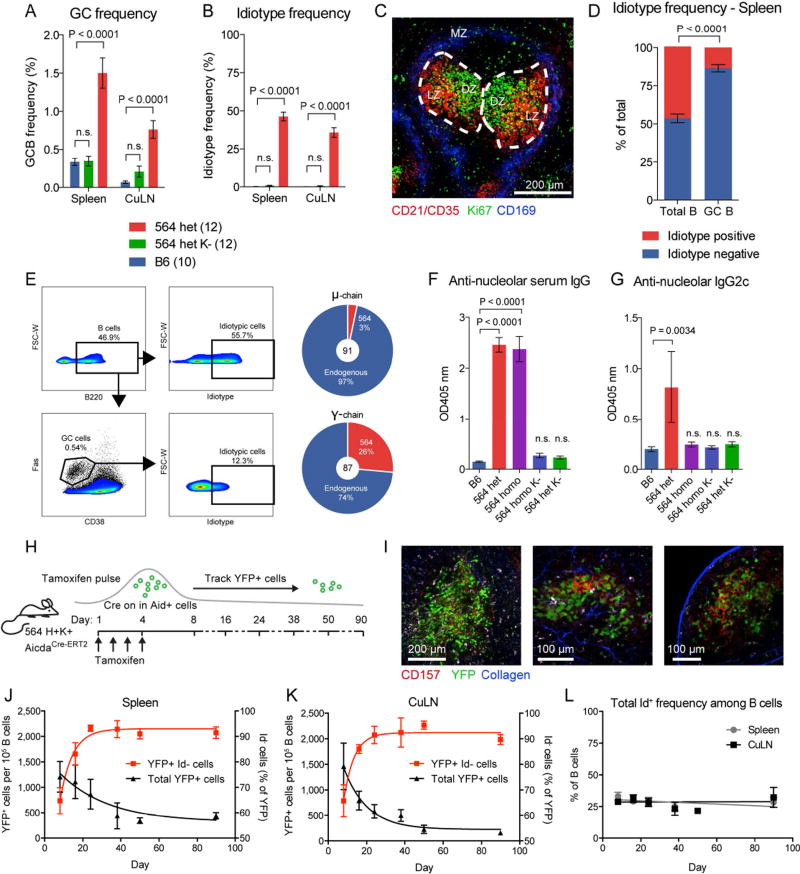
The 564Igi mouse is a murine model of SLE on C57BL/6 (B6) background, generated by knock-in of the heavy (H) and kappa light (K) chain of an autoreactive B cell clone targeting ribonuclear complexes (Berland et al., 2006). In heterozygous 564Igi mice, which carry a single copy of the knock-in H and K chain, ~50% of circulating B cells express the knock-in BCR (identified by anti-idiotype [Id] Ab staining). The remaining half are Id negative (Id−) due to receptor editing or allelic inclusion (Berland et al., 2006; Chatterjee et al., 2013; Das et al., 2017; Luning Prak et al., 2011). The mice have high titers of IgG against nuclear-associated Ags, but do not exhibit disease until late in life.
Already at 6 weeks of age, heterozygous 564Igi mice (564het) harbored robust GC B cell populations in spleen and cutaneous lymph nodes (LN) (Figure 1A). Littermates that had the H chain knock-in but lacked the K chain (564het K−) had baseline GC B cell frequencies comparable to B6 controls (Figure 1A). The presence of spontaneous GC B cells correlated with the presence of circulating Id+ (knock-in BCR) cells (Figure 1B and Figure S1A, Spearman correlation, p=0.0002 and p<0.0001, for spleen and LN, respectively). As reported previously, GC structures were found in the spleens of heterozygous 564Igi mice (Chatterjee et al., 2013) (Figure 1C).
Figure 1. Spontaneous, Chronic GCs in 564Igi Mice Display VDJ diversification, SHM, and CSR.
(A) GC frequency within the B220+ gate of B6 (n=10), heterozygous 564Igi K− (n=12), and heterozygous 564Igi mice (n=12). Mean±SEM, multiplicity-adjusted P values for two-way ANOVA with Dunnett’s posttest.
(B) Id frequency within the B220+ gate as in (A).
(C) Representative confocal IF microscopy of a 10-µm splenic section from a heterozygous 564Igi mouse, displaying GC structures identified by staining for CD21/35 (dim on B cells, bright on FDC in the light zone [LZ], red) and centroblasts (dividing cells in the dark zone [DZ], Ki67 positive cells, green), and staining of the marginal zone (MZ, CD169+ metallophilic macrophages, blue) indicating the border of the red and white pulp.
(D) Frequency of Id+ versus Id− cells in the total versus GC B cell gate in spleen for the group of mice represented in (A) and (B). Mean±SEM, multiplicity-adjusted P-value for repeated-measures two-way ANOVA with Sidak’s posttest.
(E) FACS plots showing frequency of Id+ cells within the total B220+ pool (top right panel) versus the GC population (bottom right panel) defined as Fas+CD38− B cells. Sequencing of the mu chain (top) and gamma chain (bottom) of FACS-sorted GC B cells of heterozygous 564Igi mice.
(F) Anti-nucleolar serum IgG of B6 (n=4), heterozygous 564Igi (n=10), homozygous 564Igi (n=7), heterozygous 564Igi K− (n=8), and homozygous 564Igi K− mice (n=3), as measured by ELISA. Mean±SEM, multiplicity-adjusted P values for one-way ANOVA with Dunnett’s posttest.
(G) Anti-nucleolar serum IgG2c, as in panel (F).
(H) Overview of experimental approach.
(I) Representative two-photon imaging of explanted spleen at low (left panel) and high (middle panel) magnification, and of explanted inguinal LN at high magnification (right panel), showing YFP+ cells (green), CD157+ FDC (red), and second harmonics generation (blue).
(J) A pulsed GC population followed in the spleen over 90 days. Mean±SEM for 6 (Day 8, 16, 24), 2 (Day 38 and 50) and 3 (Day 90) mice from 3 independent cohorts.
(K) Similar to (J), but for cutaneous LN.
(L) Overall Id frequencies in spleen and LN over the course of the experiment.
Strikingly, Id+ cells were underrepresented in GCs compared to the circulating repertoire (Figure 1D). GC B cells were sorted from the spleens of two heterozygous 564Igi mice, and sequencing their BCR repertoire revealed that ~95% of the IgM and ~75% of the IgG sequences were not derived from the knock-in allele (Figure 1E, Table S1), but were derived by V(D)J recombination of the other allele or by receptor editing of the knock-in allele (essentially recombining it again). These “WT-derived B cells” had undergone SHM, as judged by the mutations in their heavy chain VDJ (Figure S1B), and the mutation frequency was higher in IgG sequences than in IgM sequences (two-tailed Mann-Whitney test, p<0.0001), suggesting affinity maturation and class-switch recombination (CSR), but it was unclear whether these WT-derived clones were autoreactive.
We next assessed the reactivity of WT-derived serum antibodies to nucleolar autoAg. The 564Igi heavy chain locus is derived from the IgH-1a allotype (which encodes IgG2a), whereas the endogenous B6 heavy chain locus is IgH-1b (which encodes IgG2c). Therefore, we can distinguish antibodies produced by 564Igi B cells versus WT based on their respective IgG2a and IgG2c allotypes. We screened for IgG2c antibodies against nucleolar autoAg in the sera of heterozygous 564Igi mice. Samples from B6 (IgG2c only), heterozygous 564Igi kappa negative (564het K−) (IgG2a and IgG2c), and homozygous 564Igi (IgG2a only) mice were included as controls. As expected, heterozygous and homozygous 564Igi mice harbored IgG antibodies targeting nucleoli, whereas B6 and 564het K− littermates did not (Figure 1F). Strikingly, heterozygous 564Igi mice harbored IgG2c Ab targeting nucleoli (Figure 1G), meaning that a subset of their WT-derived B cells had become autoreactive.
To investigate the trajectory of Id frequencies in spontaneous GCs, the 564Igi strain was crossed to an Aid-CreERT2 EYFP reporter (Dogan et al., 2009). In this model, GC B cells will constitutively express YFP upon tamoxifen administration, allowing us to track these cells and their progeny (Figure 1H). Following tamoxifen injection, YFP+ cells represented 2/3 to 3/4 of the total GC B cell population (day 8, ~1,500 YFP+ cells per 105 B cells, with most YFP+ cells being GC B cells, compared with a total GC B cell frequency of ~2,000 per 105, Figure 1 I, J, and K, Figure S1C, Movie S1 and S2). The fidelity and tamoxifen dependence of the YFP expression were verified in control experiments (Figure S1D–J).
The YFP+ GC population and its progeny were followed for 3 months, showing an overall steady decrease in YFP+ cells with an increase in Id− YFP+ cells (Figure 1J and K), despite circulating Id frequencies remaining constant (Figure 1L). Bulk sorting and sequencing of YFP+ cells 90 days after tamoxifen revealed that >80% of IgM sequences and >90% of IgG sequences were not derived from the 564 knock-in allele (Figure S1K), confirming that high numbers of WT-derived B cells were included in the GC and downstream plasma and memory cell population over time. These cells had undergone SHM, with more mutations in IgG sequences than in IgM sequences (two-tailed Mann-Whitney test, p<0.0001; Figure S1L), supporting affinity maturation and CSR. SHM is classically associated with GC responses, but also occurs in extrafollicular foci (Herlands et al., 2008; William et al., 2002). However, the observed mutation rates of the YFP cells were an order of magnitude above those expected from extrafollicular foci. Taken together, these results demonstrate that in heterozygous 564Igi mice, autoreactive WT-derived cells are maturing in spontaneous GCs, thus undergoing affinity maturation, SHM, and CSR.
564Igi Mixed Chimeras Have Spontaneous GCs Composed of WT B Cells
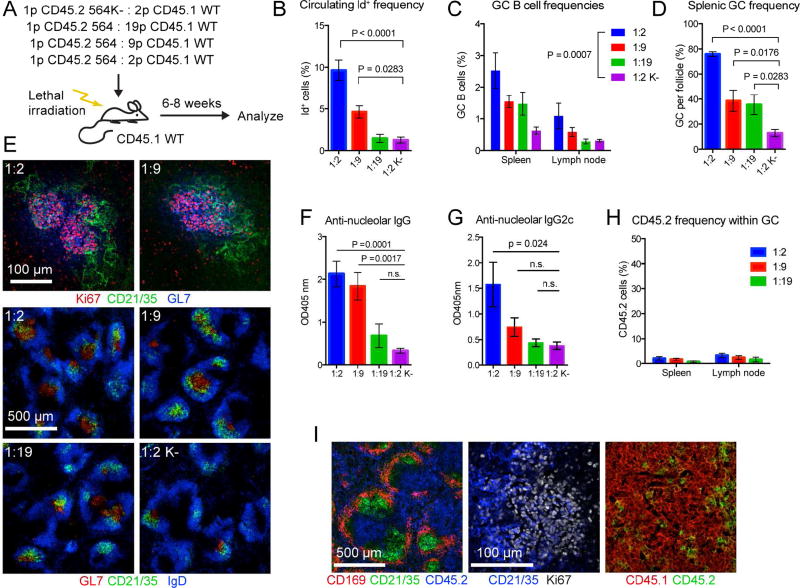
In heterozygous 564Igi mice, spontaneous GCs were predominantly composed of GC B cells with WT-derived BCRs. However, all developing B cells in these mice harbor the knock-in allele, thus they are not strictly WT B cells. To follow true WT cells in spontaneous GCs, we used a mixed chimera approach. WT B6 mice were irradiated, ablating their immune system, and reconstituted with a mixture of bone marrows (BM) from WT B6 and homozygous 564Igi donors. In this scenario, the 564Igi cells still kick-start autoimmunity, but they are ‘locked in’, as they are bi-allelically pre-rearranged, and they are genetically segregated from the WT B cell response (Figure S2A). The cells were followed using two allotypes of leukocyte marker CD45 (CD45.1 for WT cells and CD45.2 for 564Igi cells) (Figure 2A). For controls, we used mixed chimeras in which homozygous 564Igi donors were K-chain negative (CD45.2 564K−) (Figure 2A). T cells should be unaffected by the knock-in status, as they do not express the BCR, and all chimeras had the predicted ratios of CD45.1 to CD45.2 T cells.
Figure 2. An Autoreactive B Cell Clone is Sufficient to Induce Spontaneous GCs Composed Predominantly of WT-derived B Cells.
(A) Overview of experimental approach for generating and analyzing 564 CD45.1 mixed chimeras.
(B) Id frequencies in chimera blood. Mean±SEM for 1:2 (n=7), 1:9 (n=7), 1:19 (n=5), and 1:2K− (n=5) chimeras from 3 independent cohorts. Multiplicity-adjusted P value for one-way ANOVA using Holm-Sidak’s posttest.
(C) GC B cell frequencies in spleen and cutaneous LN analyzed by flow cytometry. As for (B), but using two-way ANOVA with Holm-Sidak’s posttest.
(D) Frequency of GC per follicle assessed by confocal IF microscopy. Two slices per mouse listed in (B). Mean±SEM, multiplicity-adjusted P value for one-way ANOVA using Holm-Sidak’s posttest.
(E) Representative confocal IF microscopy of 10-µm spleen sections. Top: representative spleen sections showing T-dependent GC defined by GL7 (blue) and Ki67 (red) bounded by FDC networks (green); Bottom: representative spleen sections at lower magnification showing GL7+ GC (red) and FDC networks (green) within follicular areas containing IgD positive B cells (blue).
(F) Serum anti-nucleolar IgG titers for a subset of mice in (B). Mean±SEM for 1:2 (n=6), 1:9 (n=4), 1:19 (n=5), and 1:2K− (n=6) chimeras from 3 independent cohorts. Multiplicity-adjusted P value for one-way ANOVA using Holm-Sidak’s posttest.
(G) Serum anti-nucleolar IgG2c titers, as in (F).
(H) CD45.2 frequencies within GC of panel (C).
(I) Left panel: confocal IF microscopy of spleen section showing distribution of 564 BM-derived cells (CD45.2, blue) in relation to the marginal zone (CD169, red) and FDC networks (CD21/35, green). Right two panels: distribution of CD45.2 (green) vs. CD45.1 (red) cell populations within the GC defined by FDC (light zone, CD21/35 in blue) and dividing centroblasts (dark zone, Ki67 in white).
See also Figure S2.
The mixed chimeras had fewer-than-expected 564Igi B cells (CD45.2) when compared to 564Igi T cells, likely because the autoreactive 564Igi cells were selected against (Figure S2B–E). A smaller, but significant negative selection was observed in the BM of 564K− mixed chimeras, likely because of higher tonic BCR signal in the double-heavy-chain-expressing 564K− B cells (Figure S2B). However, in spleen, skin-draining LN, and blood, the reduced frequency of 564Igi B cells was more drastic than for 564K− B cells (Figure S2C–E). This finding agrees with the previously reported negative selection of self-reactive B cells at the transitional stage observed in 564Igi mice (Berland et al., 2006; Chatterjee et al., 2013). The chimeras displayed normal and consistent total B cell frequency (Figure S2F–I).
Despite negative selection against 564Igi B cells, the input BM ratios had a titration effect on circulating Id frequencies (Figure 2B). Weaker effects on GCs were also observed in spleen and skin-draining LN (Figure 2C–E). The frequency of Id+ cells in blood correlated with GC B cell frequencies in spleen (Spearman r=0.73, p<0.0001) and skin-draining LN (Spearman r=0.65, p=0.0006) (Figure S2J). Furthermore, the ratios impacted the level of circulating anti-nucleolar IgG and IgG2c (produced by WT cells) (Figure 2F and G). Strikingly, at 6–8 weeks post reconstitution, GCs were almost exclusively composed of WT-derived cells (Figure 2H and I), while the 564K− chimera controls had no GCs. Because the observed phenotypes were most robust in the 1:2 chimeras, this BM ratio was chosen for most subsequent experiments.
B Cells of Spontaneous GCs in 564Igi Mixed Chimeras are Autoreactive and Converge on Similar BCRs
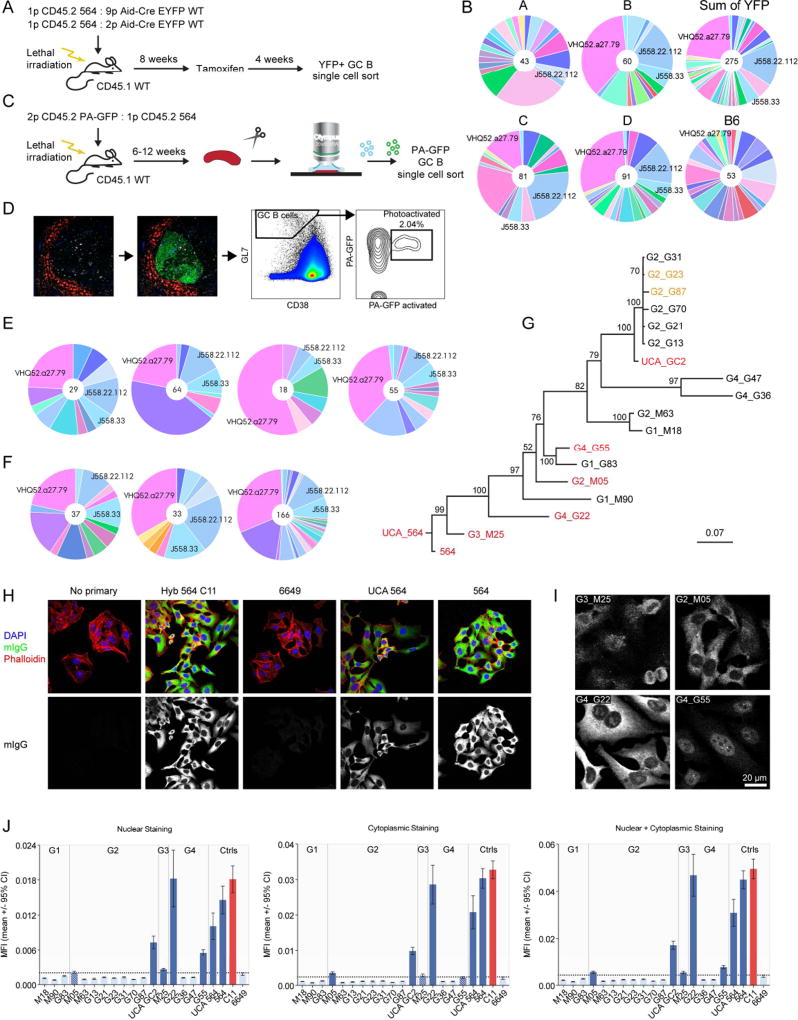
To analyze the WT-derived GC B cells further, mixed chimeras were generated using a mixture of BM from Aid-CreERT2 EYFP WT and homozygous 564Igi donors transferred into irradiated WT recipients (Figure 3A). Following reconstitution, mice were pulsed with tamoxifen, and 4 weeks later, the BCR repertoire of the YFP+ GC B cells was sequenced (Figure 3A and B), capturing a total of 275 heavy chains from 4 chimeras, representing a total of 46 distinct V segments (the V segment determines most of the Ag specificity). The most prevalent V segment accounted for over 1/5 of the total sequences, while the second- and third-most accounted for ~1/6 and ~1/15, respectively. The most prevalent and the second-most prevalent V segment were observed in 3 and 4 out of 4 mice, respectively (Figure 3B), indicating that the WT-derived GC B cells converged on particular heavy chain V segments, suggesting they were targeting similar Ags. The sequences had many mutations, indicating a significant degree of SHM, as compared to naïve mature B cells from B6 mice (Kruskal-Wallis test with Dunn’s multiple comparison, P<0.0001 for pairwise comparison of mouse A–D with B6) (Figure S3C).
Figure 3. WT GC Cells Converge on Stereotypic Autoreactive Sequence Elements.
(A) Overview of experimental setup for single-cell sequence analysis of temporal global GC populations.
(B) Single-cell sequence analysis of temporal global GC populations from 4 mice (two 1:9 [A+B] and two 1:2 chimeras [C+D]), the sum of all observed YFP-derived sequences (from A–D) and a control B6. A unique color is assigned to each unique V element, and colors are consistent across graphs in Figure 3 and Figure S3.
(C) Schematic overview of experimental setup for single GC-level single GC B cell analysis.
(D) Example of photoactivation and sort gates for single GC-level single GC B cell sorting. Leftmost panel: the marginal zone is labeled with anti-CD169-PE and based on tingible-body macrophage morphology; follicles containing GC are delineated for photoactivation. Second panel from left: GFP signal following photoactivation. Second panel from right: GC B cell gate defined as CD38lo, GL7hi. Rightmost panel: signal for photoactivated GFP vs. unactivated PA-GFP, and indication of terminal sort gate for photoactivated GC B cells.
(E) Clonal composition of four splenic GC from the same mouse, as defined by heavy chain V-segments.
(F) Sum of observed Vh-segments in four splenic GC each, for 3 independent mice.
(G) Phylogenetic tree for the 17 synthesized PA-GFP clones, 564, and the 564 UCA, based on the nucleotide sequences of heavy+light chains. Clones identified as autoreactive in HEp-2 assay, nucleolar ELISA, and autoAg array are highlighted in red, whereas clones displaying limited autoreactivity in autoAg array only are in orange. The tree was resampled 100 times, and the resulting confidence of branchpoint determination is indicated.
(H) Representative results for confocal analysis of human epithelial (HEp-2) cells stained with (from left to right): no primary; 564 Ab derived from a hybridoma (clone C11); cloned heavy and light chain pairs from control anti-influenza HA head Ab (6649); reconstructed 564 unmutated common ancestor (564 UCA); and 564. Development for murine IgG (green), counterstained with DAPI (blue) and phalloidin (red). Overlay of all three channels (top row) or mIgG channel alone (bottom row).
(I) Representative examples of the different staining patterns observed for cloned sequences from single GC: perinuclear (G1_M25), predominantly cytoplasmic (G2_M05), cytoplasmic+nucleolar (G4_G22), and nuclear stain with nucleolar exclusion (G4_G55). Channel intensity was adjusted to facilitate visualization of the pattern.
(J) Quantified staining intensities based on CellProfiler analysis of raw images. Nuclear staining, cytoplasmic staining, and the sum of the two (left to right). Cloned antibodies are grouped according to GC origin as indicated. The threshold for background signal, defined as the upper limit of the 95% confidence interval for the mean signal in the negative control (6649), is indicated with a horizontal dotted line. Measurements for which the limit of the 95% confidence interval for the mean did not overlap with threshold were considered positive. Positive measurements are indicated in dark blue, while negative are light blue, and borderline are patterned. The positive control, hybridoma-derived 564 C11, is indicated in red.
See also Figure S3.
This analysis is a gross under-sampling of a vast pool of B cells and may not adequately reflect clonal selection processes at the level of a single GC. To improve the analysis, chimeras were generated based on mixing WT PA-GFP BM with homozygous 564Igi BM in WT recipients (Figure 3C). Following reconstitution, single GCs from fresh spleen explants were photoactivated as previously described (Victora et al., 2010) and sorted into single cells (Figure 3C and D). Again, clonal expansion and selection were observed in the heavy-chain-derived V segments (Figure 3E and F and Figure S3A and B). Specifically, the VHQ52.a27.79 (IGHV2-9*02) and J558.33 (IGHV1S81*02) segments occurred in 3/4, 4/4, and 4/4 GCs analyzed in 3 independent chimeras (Figure S3B). Surprisingly, these sequence elements were shared, not only among clones of distinct GC within the same mice, but also among clones of different mice. Again, mutation analysis indicated a significant degree of SHM, both when comparing all sequences from each PA-GFP mouse to naïve mature B cells from B6 mice (Kruskal-Wallis test with Dunn’s multiple comparison, P<0.0001 for pairwise comparison of E–G with B6) (Figure S3D) and when comparing sequences of individual GC with those from B6 (Kruskal-Wallis test with Dunn’s multiple comparison, P≤0.0001 for pairwise comparison of G1–G4 with B6) (Figure S3E).
Of note, the observed V segments were also prominent in Aid-CreERT2 EYFP chimeras. Furthermore, we identified similar sequences in YFP+ cells from tamoxifen-pulsed Aid-CreERT2 EYFP heterozygous 564Igi mice, indicating that this convergence across independent animals was not an artifact of the chimera approach. An unbiased BLASTP search of CDR3 amino acid sequences (the CDR3 region being the most critical for Ag recognition) of clones carrying the VHQ52.a27.79 (IGHV2-9*02), J558.22.112 (IGHV1-22*01) and J558.33 (IGHV1S81*02) elements yielded hits for anti-DNA binding antibodies (including the original 564 VH clone) and anti-phosphocholine antibodies. These findings indicate that WT-derived GC B cells converged on specific autoreactive sequence elements.
To definitively determine whether BCRs from WT-derived clones were autoreactive, paired H and K chains from 16 clones and 1 inferred unmutated common ancestor (UCA) from 4 GCs of a PA-GFP 564Igi mixed chimera were synthesized, cloned, and expressed (Table S2). As controls, we cloned and expressed the original 564 Ab, the inferred 564 UCA, and an influenza hemagglutinin (HA)–specific Ab (6649). An overview of the sequence diversity of the 17 PA-GFP-derived clones and the 564 clone is shown in the phylogram in Figure 3G, which was rooted on the 564 UCA. The 20 total recombinant antibodies, and the control 564 Ab derived from a hybridoma, were assayed for cross-reactivity with cellular components in human epithelial (HEp-2) cells (Figure 3H–J). Representative examples of the different staining patterns observed for cloned sequences from single GCs are shown in Figure 3I: perinuclear (G3_M25), predominantly cytoplasmic (G2_M05), cytoplasmic+nucleolar (G4_G22), and nuclear stain with nucleolar exclusion (G4_G55). Staining intensities were quantified in CellProfiler and are shown in Figure 3J.
Out of 4 GCs analyzed, autoreactivity was confirmed in 3. We further validated our findings in nucleolar ELISA-type assays (Figure S3F). The analysis was extended and refined by screening with autoAg arrays (Ayoglu et al., 2016), where each clone was assayed against 241 Ags (227 unique targets) and 30 controls (10 unique antibodies) (Table S3), and overall, results were congruent with the HEp-2 and nucleolar ELISA results (Figure S3G–K). The 564 clone, 564 UCA, G4_G22, and UCA GC2 were strongly reactive with single-stranded (ss)DNA (Figure S3H). G2_M05 also reacted to ssDNA, but reacted most strongly with macrophage-derived chemokine (MDC, CCL22). Clones G2_G87, G2_G23, and G3_M25 displayed unique reactivity, albeit weaker, to this target (Figure S3I). These differences suggested a link between subcellular targeting and finer Ag specificity, because in the HEp-2-assay, G4_G22 and G2_M05 stained nucleolar+cytoplasmic and cytoplasmic, respectively, whereas G3_M25 had perinuclear staining. Also, G4_G55, which had nuclear staining with nucleolar exclusion, was strongly reactive with Smith D2 Ag (Figure S3J). Interestingly, UCA_GC2 had additional reactivity to TGFβ-RII, not present in the other ssDNA-reactive clones (Figure S3K). The finding that the inferred UCA of the IgG-switched clones in GC2 was autoreactive suggests that germline precursors are autoreactive and enter GCs as such. Furthermore, there was CSR, as both autoreactive IgMs and IgGs were observed. Taken together, these results confirm that the spontaneous WT-derived GC B cells were autoreactive and were the likely source of the WT-derived, affinity-matured IgG2c autoantibodies observed in the chimeras.
GCs of 564Igi Mixed Chimeras Are T-Dependent with a Prominent Follicular Helper T Cell Population
GCs that form in response to foreign Ag depend on T cell help. To test whether those in 564Igi mice and 564Igi mixed chimeras do as well, we treated mice with anti-CD40L Ab to block T cell interaction. Similar to foreign Ag GCs, CD40L blockade ablated the GCs in spleen, cutaneous LN, and mesenteric LN of both heterozygous 564Igi mice and 564Igi mixed chimeras (Figure S4A–D). Splenic Id frequencies were unaffected, whereas those of cutaneous and mesenteric LN were marginally decreased. T follicular helper (Tfh, CXCR5hiPD-1hi) cell populations were present in both heterozygous 564Igi mice and 564Igi mixed chimeras, and they were similarly ablated by CD40L blockade, as previously shown (Durie et al., 1993; Yusuf et al., 2014). In contrast, overall CD4 T cell levels were unaffected (Figure S4E–H).
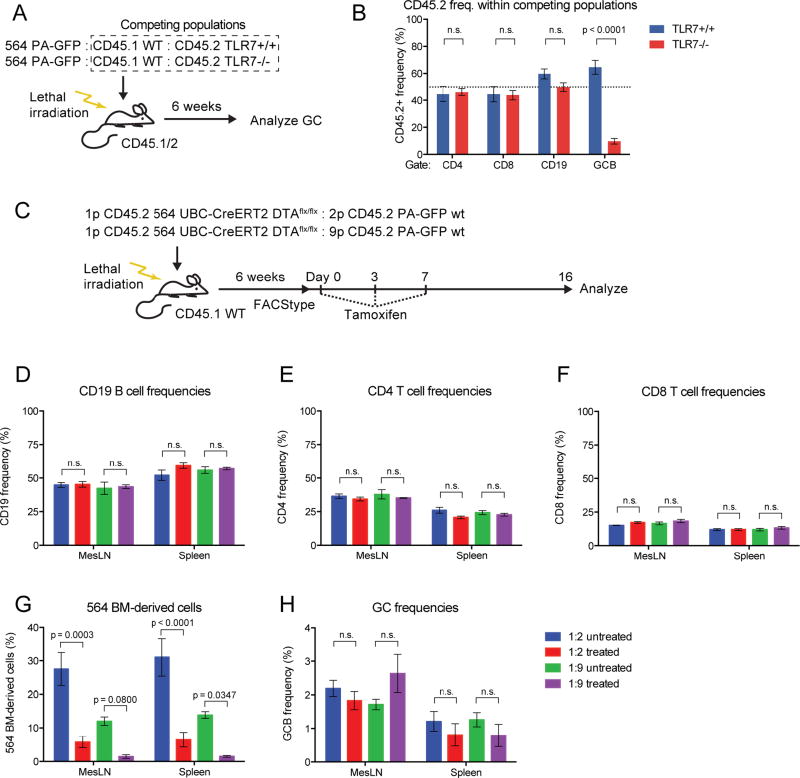
WT-derived GC B Cells In 564Igi Mixed Chimeras Depend on TLR7 Signals
B cell–intrinsic TLR7 signaling is a driver of escape of tolerance for self-reactive B cells, as observed for Id+ cells in the 564Igi model (Berland et al., 2006; Green et al., 2012; Lau et al., 2005; Leadbetter et al., 2002). To test whether the WT-derived GC B cells in 564Igi mixed chimeras similarly depended on intrinsic TLR7 signaling, three-part mixed chimeras were generated using irradiated CD45.1/2 recipients. One part was 564Igi BM (identified by CD45.2 and PA-GFP congenic markers), one part was WT (CD45.1) BM, and the last part was either TLR7−/− or TLR7+/+ BM (both CD45.2) (Figure 4A). Both TLR7−/− and TLR7+/+ BM mixtures reconstituted mice equally, as demonstrated by near-expected (50:50) ratios of CD45.1 to CD45.2 T and B cells after excluding CD45.1/2 double-positive (recipient-derived) and PA-GFP-positive cells (564Igi-derived) (Figure S5A–H). Normal and comparable B and T cell frequencies were confirmed (Figure S5I–K). Robust and comparable GC frequencies were observed in both spleen and mesenteric LN for TLR7−/− and TLR7+/+ chimeras, both in terms of total GC B cells and GC B cells after exclusion of PA-GFP and CD45.1/2 cells (Figure S5L and M).
Figure 4. WT GCs Depend on B Cell–Intrinsic TLR7 Signaling and Become Self-Sustained.
(A) Overview of experimental approach for generation and analysis of TLR7+/+ and 564 TLR7−/− mixed 564 chimeras.
(B) Splenic CD45.2 frequencies in the competition gate within CD4, CD8, CD19 and GC B populations of 564 TLR7+/+ (n=3) and 564 TLR7−/− (n=4) mixed chimeras. Mean±SEM, multiplicity-adjusted P values for two-way ANOVA with Dunnett’s posttest. Dotted line indicates the expected 50% frequency.
(C) Overview of experimental layout for generating and analyzing 564 DTA mixed chimeras.
(D) CD19 frequencies of untreated vs. treated 1:2 and 1:9 564 DTA mixed chimeras. Mean±SEM for 3 mice per group, multiplicity-adjusted P values for 2-way ANOVA with Tukey’s posttest, n.s.=not significant.
(E) Similar to panel (D), but showing CD4 frequencies.
(F) Similar to panel (D), but showing CD8 frequencies.
(G) Frequencies of 564 BM-derived cells in mesenteric LN and spleen of untreated vs. treated 1:2 and 1:9 564 DTA chimeras.
(H) As panel I, but showing GC B cell frequencies.
See also Figures S4 and S5.
There were significantly fewer TLR7−/− GC B cells than TLR7+/+ GC B cells in the spleens of mixed chimeras (Figure 4B), meaning that WT B cells needed intrinsic TLR7 signaling for entry or persistence in autoreactive GCs. In mesenteric LN, there was a less dramatic, albeit significant, reduction in TLR7−/− cell frequency (Figure S5N). The reduced dependence on the TLR7 pathway in intestinal LN GCs is likely due to multiple other TLR ligands originating from gut flora or food Ags.
Spontaneous GCs of 564Igi Mixed Chimeras Become Self-Propagating
Because of the low frequency of 564-derived cells in the autoreactive GCs of mixed chimeras, we asked whether the GCs were dependent on 564Igi cells for their persistence or whether the epitope spreading was sufficient to maintain them. We crossed 564Igi mice with UBC-CreERT2 mice and a conditional attenuated-diphtheria toxin (DTA) line (564 UBC-CreERT2 DTA). The BM from these mice (CD45.2) were mixed with BM from PA-GFP WT mice (CD45.2) and transferred to irradiated WT recipients (CD45.1) (Figure 4C). This set-up allowed specific ablation of 564-derived cells in a tamoxifen-induced manner. Following reconstitution, a high degree of chimerism was verified, and the expected frequencies of donor and normal B cells were observed (Figure S5O–R).
Half the cohort was treated with tamoxifen and mice were analyzed two weeks later. As expected, overall CD4 T cell, CD8 T cell, and CD19 B cell frequencies were unaffected (Figure 4D–F). Because the frequency of 564-derived B cells was already low and therefore harder to quantify, CD8 T cells were used to measure the ablation strategy’s effect on the 564-derived hematopoietic compartment. In the 1:2 chimeras, the 564-derived cell frequency was reduced approximately 80% (from ~30% down to ~6%); whereas in the 1:9 chimeras, it was reduced ~90% (from ~12% down to ~1.2%) (Figure 4G). Despite this dramatic reduction in 564-derived cells, splenic and mesenteric GC B cell frequencies were only marginally reduced (Figure 4H). Therefore, 564-derived cells were not required once autoreactive GCs had been established.
Autoreactive GCs Evolve Towards Pauciclonality
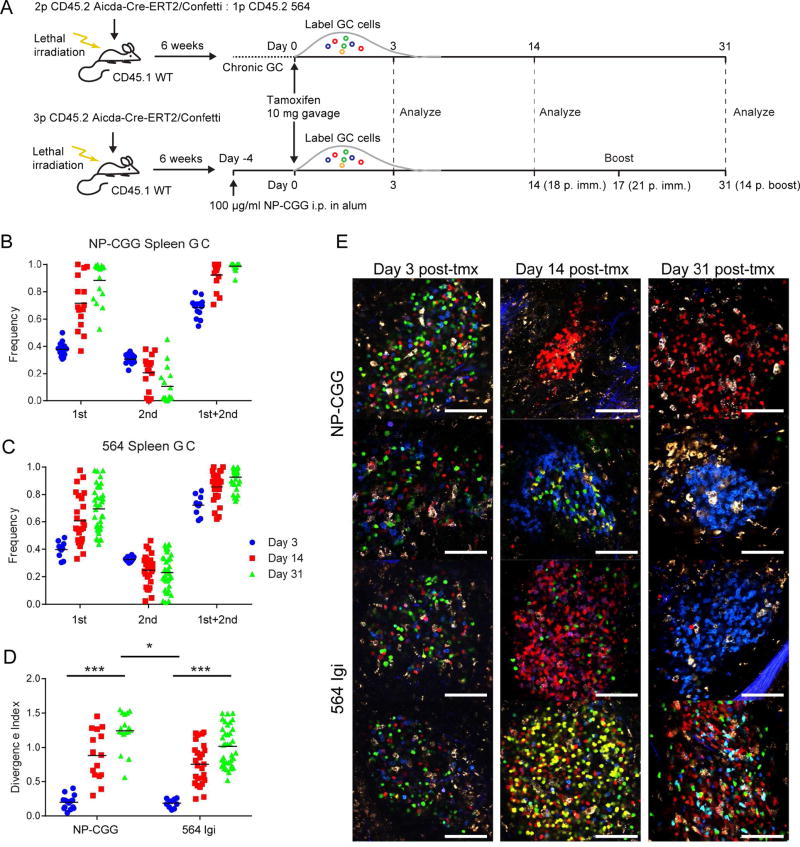
To analyze autoreactive GC dynamics globally, we generated mixed 564Igi and Aid-CreERT2 Confetti BM chimeras. In this set-up, 564-driver cells elicit autoreactive GCs largely composed of WT-derived Aid-CreERT2 Confetti B cells. Upon tamoxifen induction, Aid+ (GC) cells and their progeny express 1 of 10 different color combinations, allowing the visualization of clonal selection, as previously done for foreign Ag (Tas et al., 2016).
Robust induction of autoreactive GCs was observed, and the autoreactive GCs were composed of WT clones as evidenced by Confetti positivity (Movie S3 and S4). As an internal control, “Confetti only” chimeras without 564 driver BM were generated and immunized with a foreign Ag (NP-CGG) analogous to that of Tas et al. (Figure 5A). Over 4 weeks, foreign-Ag-elicited GCs lost color diversity, as evidenced by the frequencies of the most dominant and second-most dominant color, and the sum of the two relative to the remainder of the GC (Figure 5B and E). Unlike for Tas et al., these data do not account for unrecombined cells; and therefore, the results are not directly comparable.
Figure 5. Clonal Evolution in Autoreactive GCs in the Aid-Confetti Model.
(A) Overview of experimental setup for generating and analyzing Aid-Confetti mixed 564 chimeras.
(B) Frequency of most (1st), second most (2nd) and their sum observed in individual splenic GC at Day 3, 14, and 31 post-tamoxifen induction of Confetti recombination in NP-CGG-immunized Confetti chimeras. The mean of an average of 7.8 GC per spleen for 2 mice per time point is indicated by the bar.
(C) As panel (B), but for 564 Confetti mixed chimeras. The mean of an average of 9.2 GC per spleen for 3 mice per time point is indicated by the bar.
(D) Divergence index for the GC represented in (B) and (C). Significance indicated for two-way ANOVA with Tukey’s posttest (*=P<0.05, **=P<0.001).
(E) Representative images used for quantification of color dominance. The scale bars indicate 100 µm.
However, we found that the autoreactive GCs evolved in a similar manner as foreign-Ag GCs, losing color diversity at only a slightly decreased rate (Figure 5C and E), which was confirmed by comparing the divergence index for the two groups (Figure 5D). Of note, whereas foreign-Ag-induced GCs are synchronized and timed, the 564Igi GCs were spontaneous and chronic, making it difficult to determine GC age. However, the effect of GC age could be controlled for by comparing to mesenteric LN GCs, which are chronic, in both groups. Notably, we observed no difference between the mesenteric LN GCs of foreign-Ag-immunized Confetti-only mice and those of 564Igi chimeric mice (Figure S6A–I), indicating that there is no effect from GC age. We conclude that autoreactive GCs, once tolerance has been broken, evolve towards pauciclonality - that is, the dominance of a single or a few clones and their progeny.
Mixed 564Igi Chimeras Undergo Epitope Spreading
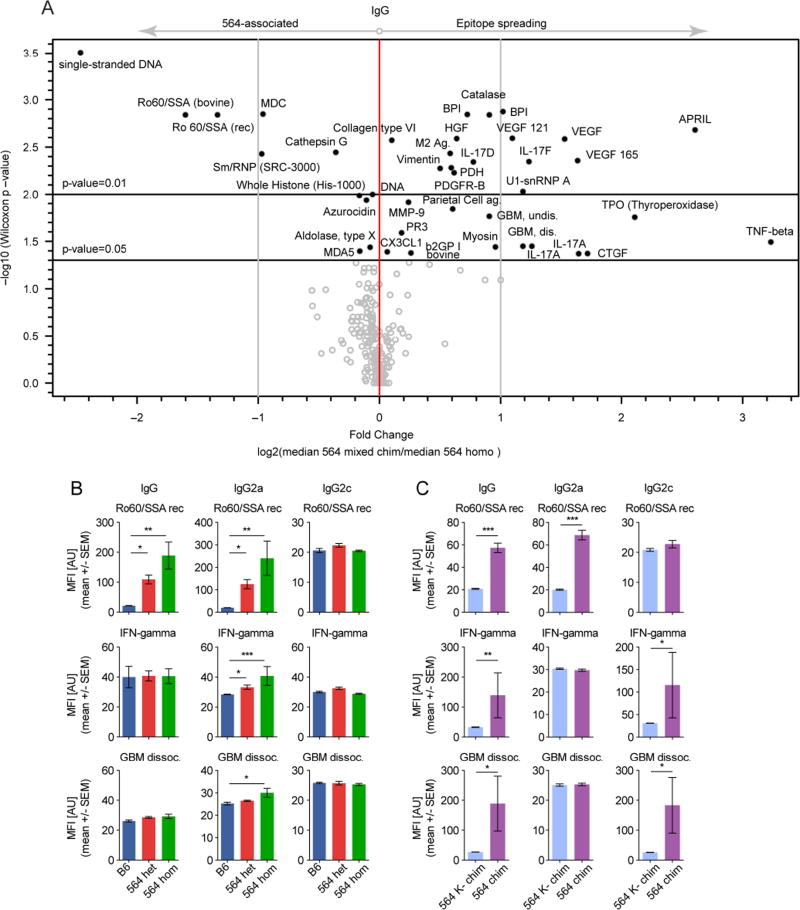
Given the differential Vh usage and clonal selection of WT-derived GC B cells, we hypothesized that the WT-derived response may target more self-Ags than the original 564Igi clone. IgG (total), IgG2a (564-derived), and IgG2c (WT-derived) from 9 heterozygous 564Igi, 7 homozygous 564Igi, 6 1:2 564 mixed chimeras, 2 1:9 564 mixed chimeras, 6 1:2 564K− mixed chimeras, and 5 B6 mice were assayed for binding to 241 Ags (227 unique targets) and 30 controls (10 unique Abs) (Figure S7A–C, Table S3). Individual mice within each group varied in reactivity, but the IgG antibodies of heterozygous 564Igi mice, homozygous 564Igi mice, and 564Igi mixed chimeras had stronger reactivity across a broad range of self-Ags than did IgG from 564K− mixed chimeras and B6 mice (Figure S7A). IgG2a (564-derived) was strongly reactive in homozygous 564Igi and to a lesser extent heterozygous 564Igi mice (Figure S7B), whereas IgG2c (WT-derived) was strongly reactive in 564Igi mixed chimeras (Figure S7C), agreeing with the preceding experiments. A shift, or broadening, of reactivities was notable when comparing 564Igi mixed chimeras and homozygous 564Igi mice, which have B cells that are doubly pre-arranged and so cannot easily acquire new targets (Figure 6A). TNFβ, GBM (glomerular basement membrane), and numerous other targets were significantly overrepresented in the mixed chimeras (upper right quadrant, Figure 6A). Many of these targets are well-established autoimmune targets (GBM, Collagen Type VI, U1-snRNP A etc.) including cytokines (APRIL, VEGF, IL-17 etc.) (Cappellano et al., 2012).
Figure 6. The Convergence on Stereotypic Autoreactive Sequence Elements is Mirrored Serologically by Functional Epitope Spreading.
(A) Volcano plot comparison of median IgG signal intensities in 564 mixed chimeras (n=8) relative to homozygous 564Igi mice (n=7), with indication of log2-fold change on the x-axis and significance level on the y-axis.
(B) Bar graph representations of IgG (left column), IgG2a (middle column) and IgG2c (right column) Ab towards Ro60/SSA (top row), IFNγ (middle row) and dissociated GBM (bottom row) in sera of B6 (n=5), heterozygous (n=9), and homozygous (n=7) 564Igi mice. Mean±SEM, multiplicity-adjusted significance for one-way ANOVA with Dunn’s posttest.
(C) Bar graph representations of IgG (left column), IgG2a (middle column), and IgG2c (right column) Ab towards Ro60/SSA (top row), IFNγ (middle row) and dissociated GBM (bottom row) in sera of 564 mixed chimeras (n=8) and 564K− mixed chimeras (n=6). Mean±SEM, multiplicity-adjusted significance for two-tailed Mann-Whitney test.
See also Figure S7.
The original 564 clone did not react with IFNγ, GBM, SmD2 (Smith D2), or TNFβ, suggesting that these are targets of WT-derived autoreactive B cells (IgG and IgG2c) (Figure 6B and C, and Figure S7D and E). The 564 clone reacts with ssDNA (Berland et al., 2006; Gavalchin et al., 1987) (compare also with Figure S3), so not surprisingly, ssDNA and Ro60/SSA were the most notable 564-associated targets (upper left quadrant, Figure 6A). Homozygous 564Igi and to a lesser extent heterozygous 564Igi mice had a strong anti-ssDNA profile for IgG and IgG2a (564-derived), but not for IgG2c (WT-derived) (Figure S7D). 564Igi mixed chimeras had a significant but lower level of anti-ssDNA IgG and IgG2a than homozygous and heterozygous 564Igi mice, whereas 564K− mixed chimeras had none (Figure S7E). A similar pattern was noted for Ro60/SSA (Figure 6B and C), in line with the polyreactive nature of the parental 564 clone. Taken together, these results suggest that inclusion of WT B cells in chronic autoreactive GCs drives epitope spreading.
Mixed 564Igi Chimeras Have Ab Deposits in Their Kidneys
The reactivity of 564Igi mixed chimeras towards GBM and collagen type IV suggested autoAb deposition in their kidneys, as those antigens are often targeted in autoimmune renal disease. We found that aged (9–12 months old) heterozygous 564Igi mice and 564Igi mixed chimeras had prominent IgG2c (WT-derived) deposits in the kidney; whereas, homozygous 564Igi and homozygous 564Igi K− mice did not (Figure 7A and B). As expected, IgG2a (knock-in allele) deposits were prominent in homozygous 564Igi mice, as well as 564Igi mixed chimeras, but were not detected in the homozygous 564Igi K− mice (Figure 7A and C). This IgG2c deposition suggests a contribution of the epitope spreading process to pathogenesis.
Figure 7. WT-Derived Autoantibodies Are Deposited in Kidneys of 564Igi mice and 564Igi mixed chimeras.
(A) Representative immunofluorescence analysis of IgG2a (top) and IgG2c (bottom) deposition in glomeruli of kidneys from a homozygous 564Igi K− control, a homozygous 564Igi, a homozygous 564Igi mouse, and a 564Igi mixed chimera. Glomerular vessels defined by CD31 (red), nuclei counterstained with Hoechst (blue), Ab deposits in green.
(B) Quantification of IgG2c Ab deposits within the glomerular area defined by CD31 staining. The analysis was based on an average of 2.5 images from each mouse for 564 homo K− (n=1), 564 homo (n=1), 564het (n=2), and 564 mixed chimeras (n=3). Statistical significance for Kruskal-Wallis test with Dunn’s posttest.
(C) Similar to B, but for IgG2a.
DISCUSSION
We made the unexpected observation of widespread epitope spreading in the 564Igi model. In heterozygous 564Igi mice, upwards of half of circulating naïve mature B cells carried an affinity-matured autoreactive BCR derived from the knock-in. Yet, these cells failed to win in the GC reaction, giving way to WT-derived cells. This was surprising, because in mice that have a knock-in BCR specific for foreign Ag, the knock-in B cells dominate the GC response. For example, in B1–8 mice, 5% of the pool harbor a BCR specific for the hapten NP, and upon NP-carrier immunization, these cells dominate the response. However, this is arguably an oversimplified system as the hapten response is very narrow. Our findings in the autoreactive setting may reflect those of Kuraoka and colleagues, whereby the process of SHM increases the breadth of the response to complex Ag (Kuraoka et al., 2016).
Although the maturation of autoreactive BCRs is generally restricted by T cell support, this check may be bypassed by linked recognition. In fact, autoreactive IgG memory antibodies in patients with SLE may arise from nonreactive and polyreactive precursors (Mietzner et al., 2008). However, mechanisms may exist to prevent the emergence of autoreactive B cells in GCs (Reed et al., 2016; Sabouri et al., 2014), perhaps relying in part on FDC presentation of autoAg (Yau et al., 2013). The nature of the self-Ag may be an important factor, as many nuclear self-Ags, such as the 564Igi Ag, trigger FDCs to secrete IFNα via endosomal TLR7 (Das et al., 2017), potentially leading to increased recruitment of self-reactive B cells from the immature repertoire.
To circumvent artifacts that might arise from transgenic B cells, we developed a mixed chimera model with spontaneous, autoreactive GCs composed of WT B cells with 564Igi cells initiating autoimmunity. We found that these autoreactive GCs are largely composed of WT B cells and are dependent on intrinsic TLR7 signaling, similarly to 564Igi B cells (Berland et al., 2006). Moreover, we found that the GCs became self-sustained and gained independence from the initial 564Igi trigger, explaining how autoreactive GCs become chronic and how disease is propagated.
We found that autoreactive GCs are polyclonal, but evolve towards pauciclonality at varying rates, agreeing with the observation that affinity maturation can occur in the absence of homogenizing selection, with many clones maturing in parallel within the same GC (Tas et al., 2016). This ‘clonal permissiveness’ is in line with earlier reports that transient foreign-Ag-driven GCs are dynamic and open structures (Schwickert et al., 2009; 2007). We found that clonal evolution was largely similar between foreign Ag and autoAg GCs. Our observations suggest that once tolerance is broken, an increasingly broader array of self-Ags will be targeted.
We observed clonal convergence and selection leading to the predominance of autoreactive V segments in the WT-derived GC B cell population, similar to previous observations in autoimmunity and cancer (Hershberg and Luning Prak, 2015; Shlomchik et al., 1987). Serum antibodies displayed epitope spreading, further supported by our finding of WT-derived Ab deposits (IgG2c) in the kidneys of 564Igi mixed chimeras.
Our findings support that autoreactive GCs are at the heart of the autoimmune response. The chronic, progressive nature of epitope spreading calls for early interventions, even before disease onset. In light of the failure of CD40L blockade in humans, alternative GC checkpoints should be explored. We have focused here on the evolution of the B cell response, but questions remain. Although we demonstrated T cell dependence in our model, the extent and nature of T cell involvement has not been addressed. Future experiments will determine whether the T cell pool is not only necessary, but also sufficient, to sustain and confer autoreactivity.
We conclude that self-reactive 564Igi B cells are sufficient to break tolerance and induce the formation of autoreactive GCs, which are predominantly composed of WT-derived cells. We dub this novel tool, which can be used to study WT epitope spreading and autoreactive GC responses, the “ARTEMIS model” to signify Autoreactive B cell driven T-dependent Epitope Migration towards Immunity to Self.
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Michael C. Carroll (Michael.Carroll@childrens.harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
C57BL/6J, B6.SJL (CD45.1), Photo-Activatable (PA)-GFP transgenic mice (Victora et al., 2010) (B6.Cg-Ptprca Tg(UBC-PA-GFP)1Mnz/J), and TLR7 knockouts (B6.129S1-Tlr7tm1Flv/J) were obtained from Jackson Laboratories. AicdaCreERT2 flox-stop-flox-EYFP mice (Dogan et al., 2009) were from Claude-Agnès Reynaud and Jean-Claude Weill (Institut Necker). The UBC-CreERT2 strain (B6.Cg-Tg(UBC-cre/ERT2)1Ejb/J) was kindly made available by Arlene Sharpe. The attenuated DTA line (B6;129-Gt(ROSA)26Sor<tm1(DTA)Mrc>/J) was kindly provided by Fred Alt (Boston Children’s Hospital). 564Igi mice (Berland et al., 2006) were originally provided by Theresa Imanishi-Kari (Tufts University) and were maintained in-house. All mice were bred and maintained in the AAALAC-accredited facility at Harvard Medical School. AicdaCreERT2-Rosa26Confetti (Tas et al., 2016) donors were from Gabriel D. Victora (Whitehead Institute). Mice were specific pathogen-free (SPF) and maintained under a 12-hr light/dark cycle with standard chow diet. Both male and female mice were used. All animal experiments were conducted in accordance with the guidelines of the Laboratory Animal Center of National Institutes of Health. The Institutional Animal Care and Use Committee of Harvard Medical School approved all animal protocols (protocol numbers IS00000095 and IS00000111).
Cells
For the nucleolar purification protocol and for expression of IgG’s, FreeStyle 293-F HEK cells were purchased from Invitrogen and grown in FreeStyle 293 Expression Medium at 37°C with 8% CO2. For IF staining, HEp-2 cells were obtained from ATCC and cultured in EMEM containing 10% FBS at 37°C with 5% CO2.
METHOD DETAILS
Antibodies and staining reagents
Commercial antibodies and staining reagents, from BioLegend: GL7-PacBlue, GL7-PE, GL7-A647, B220-PerCP-Cy5.5, B220-APC-Cy7, IgMb-FITC, IgMa-PE, CD45.2-FITC, CD45.2-APC, CD45.1-FITC, CD45.1-PE, IgD-PB, CD21/35 (7E9)-PE, CD138-PE, CD38-PE-Cy7, CD31-A647, CD157-PE, Streptavidin-PE/Cy7; from eBioscience: CD95 (APO-1/Fas)-PE (clone 15A7) and viability dye Fixable live/dead stain Efluor780; from ThermoFisher Scientific: Rabbit-anti-Goat-A488, Hoechst 33342, DAPI (4',6-Diamidino-2-Phenylindole, Dihydrochloride), Phalloidin-A568; from Southern Biotech: AP-Goat-anti-Mouse IgG, AP-Goat-anti-Mouse IgG2a, AP-Goat-anti-Mouse IgG2c; from Rockland Immunochemicals: Rabbit polyclonal anti-B-Phycoerythrin; from DAKO: Rabbit polyconal anti-Mouse Immunoglobulins-biotin; from Perkin-Elmer: Europium-labeled streptavidin. In-house generated anti-idiotypic Ab, clone 9D11, conjugated to Alexa Fluor 647 or Alexa Fluor 568; in-house generated Rabbit polyclonal anti-C3b conjugated to Alexa Fluor 633.
Genotyping and FACStyping
Genotyping was performed using the primers and reaction conditions indicated in Table S1. For FACStyping mice were bled retroorbitally. Using heparinized capillary tubes, approximately 60–100 µl blood was drawn into Eppendorf tubes containing 30 µl acid-citrate-dextrose solution. Following collection, the stabilized blood was briefly spun down, then underlayered with 1 ml of Lymphocyte Separation Medium, and spun for 25 min, 1,600 RPM at RT. The mononuclear cell layer was aspirated and transferred into 1 ml ice-cold FACSbuffer, mixed, then pelleted at 200 g for 5 min. Cells were resuspended in FACSbuffer and processed for flow cytometric analysis as described further below. FACStyping of 564 mice was performed using B220, anti-IgMa, anti-IgMb, and 9D11. PA-GFP mice were FACStyped for PA-GFP positivity based on signal in the AmCyan/Pacific Orange channel.
Chimeras
Recipients were irradiated with 950 rad, then immediately placed on water containing antibiotics (sulfamethoxazole/trimethoprim) to prevent opportunistic infections during the reconstitution phase. Femurs and tibia were extracted from BM donors, mechanically cleaned and rinsed through several rounds of HBSS supplemented with 10 mM HEPES, pH 7.2, 1 mM EDTA and 2% heat-inactivated FBS (BM buffer). The bones were subsequently crushed in a mortar, and the cell extract was passed through a 70 µm cell strainer (Corning). An aliquot was subjected to RBC lysis and counted in a standard hemacytometer (Neubauer chamber). Based on cell counts, appropriate ratios of mixed BM were calculated to achieve final desired donor ratios. Cells were pelleted by centrifugation (200 g, 5 min) and resuspended at 1 * 108 cells per ml, and 100 µl was injected i.v. into each irradiated recipient ~10–12 hours post irradiation.
Additional procedures
To generate foreign-Ag-elicited GCs, mice were immunized intraperitoneally with 100 µg of chicken gamma globulin (Rockland Immunochemicals) precipitated in an equal volume of Imject Alum (ThermoScientific). Recombination of the Rosa26Confetti allele in AicdaCreERT2 mice was induced by a single gavage of 10 or 15 mg of tamoxifen (Sigma) dissolved in sunflower oil at 20 or 30 mg/ml. For in vivo-labeling of FDC networks, mice were injected intraperitoneally with 100 µg of rabbit polyclonal anti-B-PE (Rockland) two days prior to sacrifice, and subsequently intravenously with 10 µg B-PE (Molecular Probes/ThermoFisher) or 1 mg SA-PE-Cy7 (BioLegend) the day before sacrifice. Alternative FDC labeling approaches included i.v. injection of 10 µg A633-conjugated rabbit polyclonal-anti-C3b (in-house generated) or PE-conjugated anti-CD157 Ab (BioLegend) 1–2 days before sacrifice.
Mutiphoton imaging and photoactivation of explants
Spleen and LNs were harvested at different times post-immunization/tamoxifen. Adipose tissue was carefully removed under a dissecting microscope. Either thick transversal sections (~1 mm) were manually cut from spleens using surgical scissors, or longitudinal thick sections (~2–3 mm) were prepared by clamping the spleen between Styrofoam holders and manually slicing with a surgical scalpel. LNs and spleen sections were placed in FACS buffer (PBS, pH 7.2, 1 mM EDTA and 2% heat-inactivated FBS) in vacuum-grease chambers on microscope slides. Chambers were coverslipped and kept on ice before and immediately following imaging/photoactivation. All imaging was performed on an upright Olympus FV1200 MPE multiphoton system microscope fitted with either a 20× 0.95NA Plan water-immersion objective or a 25× 1.05NA Plan IR optimized water-immersion objective, a MaiTai HP DeepSee Ti-Sapphire laser (Spectraphysics), and 4 non-descanned detectors (2 GaAsP and 2 regular PMTs). Imaging of Confetti alleles was performed using λ=940 nm excitation. Fluorescence emission was collected in three channels, using the following filter sets: a pair of CFP (480/40 nm) and YFP (525/50 nm) filters, separated by a 505 nm dichroic mirror, for CFP/GFP/YFP detection, and a third filter (605/70 nm) for RFP detection. Alexa 633-conjugated Ab was imaged using 830 nm excitation and a 665/40 nm emission filter. Imaging of PA-GFP explants was performed at λ=930–940 nm, photoactivation was performed at λ=830 nm. For live imaging, mice were anaesthetized using an isoflurane vaporizer, with 1.5–2% isofluorane for the induction phase, followed by 0.5–1% for maintenance. Adequate surgical plane of anesthesia was frequently verified throughout the procedure by testing the pedal withdrawal reflex. The mice were prepared using standard surgical technique.
Confetti analyses
Acquired stacks were rendered in Fiji (ImageJ). Counting was performed on 3–4 z planes from stacks acquired at 5 µm steps through the GC. Slices were at least 15–20 µm apart to preclude the possibility of double counting. Very sparse, <50% occupation GC were discarded prior to analysis, as they were considered to harbor at least one expanded dark clone. Typically, greater than 250 GC B cells were quantified for a single GC per time point. Observers were blinded to animal status and inter-observer reliability was confirmed by independent counts of 15 GC with a concordance of more than 95% between 2 observers. Raw counts were converted to relative frequencies for each observable color to simplify analysis. Clonal dominance was calculated as the frequency of the most dominant clone/color at a given time point. Clonal divergence score was reported previously (Tas et al., 2016) and is calculated at a given time point by comparing the observed distribution of colors at time t to the expected frequencies resulting from random recombination. Expected frequencies for each of 10 possible colors was derived using explant data from day 3 post tamoxifen treatment where color distribution most closely mimics random recombination and prior to any selection events:
Descriptors of GC behavior and clonality were graphed using GraphPad Prism 6.
Flow cytometry
Spleens and LNs were harvested into ice cold FACS buffer and mechanically dissociated using pestles in 1.5 ml Eppendorf tubes. Samples were filtered through 70 µm cell strainers (Corning) and spun down at ~200 g for 5 min. LN samples were resuspended in FACS buffer. Spleen samples were resuspended in RBC lysis buffer (155 mM NH4Cl, 12 mM NaHCO3, 0.1 mM EDTA), incubated for 2–3 minutes then spun down as before and resuspended in FACS buffer. Femurs and tibia were harvested, mechanically cleaned using coarse paper towels and placed into ice cold FACS buffer. Bones were processed by maceration in a mortar and the resulting BM cell suspensions were collected and passed through 70 µm cell strainers. BM cell suspensions were spun down and subjected to RBC lysis as for the spleen, then spun again and resuspended in FACS buffer. Peyer’s patches were snipped off the outer surface of the small intestine, collected into ice cold FACS buffer and subsequently washed through 5–10 changes of FACS buffer to remove contaminating fecal matter. Peyer’s patches were then mechanically dissociated using pestles in 1.5 ml Eppendorf tubes as for the spleens and LNs, filtered through 70 µm cell strainers, spun down and finally resuspended in FACS buffer. Samples were added to wells of 96-well round-bottom plates, spun down and resuspended in 100 µl staining mix (appropriate Ab and viability dye cocktail in FACS buffer). Staining was performed for 30 min on ice, followed by addition of 150 µl wash buffer. Plates were subsequently centrifuged 200 g for 5 min, and supernatants flicked out of the plates. For two-step staining procedures secondary staining mix was added (appropriate secondary Ab mix and viability dye in FACS buffer), and the process repeated. Following the last wash, samples were resuspended in 200 µl FACS buffer and transferred to FACS tubes. Flow cytometric analyses were performed on one of 3 different instruments (BD Biosciences), depending on the experiment: a four color, six parameter FACSCalibur equipped with 488 nm and 633 nm lasers; a standard 3 lasers-configuration (405 nm, 488 nm and 633 nm) FASCSCanto2, with 8-color and 10 parameter analytical capabilities; and a 5-laser, 20-parameter FACSAria 2 SORP (lasers: 355 nm, 488 nm, 633 nm, 405 nm, 594 nm) equipped with a PMT option for FSC, enhanced optics and digital focusing and a 300 mW 488 nm laser from Coherent.
Bulk sequence analysis
Cell suspensions were stained as for flow cytometry, washed, and resuspended in FACS buffer. Bulk sort was performed on a 5-laser FACSARIA II Special Order system (355, 405, 488, 640, 594 nm laser lines; FCS-PMT module and enhanced optics and digital focusing), using a single-cell sort mask. Cells were sorted directly into RNA lysis buffer (Zymo Research) in FACS tubes, spun down briefly following completion of collection, and frozen at −80°C until further processing. RNA was purified using the Zymo Quick RNA Miniprep kit (Zymo Research) following the manufacturer’s instructions, and RNA yields determined using a NanoDrop1000. Complementary DNA was generated using M-MLV Reverse Transcriptase (ThermoFisher Scientific) at standard conditions. Amplification of V(D)J segments was performed using consensus primers based on Tiller et al., and Busse et al., (see also Table S1) and GoTaq DNA polymerase (Promega) under the conditions described herein (Busse et al., 2013; Tiller et al., 2009). Amplicons were TOPO-TA cloned into the pCR2.1-TOPO vector and used for transformation of DH5α, which were subsequently plated on LB-agar containing kanamycin, X-gal and IPTG. Following incubation overnight at 37°C, single defined white colonies were picked into 10 µl of autoclaved distilled water. From each sample, 1 µl was used as input for amplicon PCR using M13 Forward and Reverse primers and GoTaq DNA polymerase. From 20 µl reactions, 2–5 µl were run on analytical agarose gels, and the remainder was precipitated by addition of 1/10th volume 3 M sodium acetate (pH 5.2) and 2.5 volumes 96% ethanol. Following overnight incubation at −20°C, samples were spun at 3,500 rpm for 30 min, the supernatant flicked off, and the pellet washed with 96% ethanol. Pellets were subsequently dried and resuspended in 12 µl 10 mM Tris buffer, pH 8.5. Nucleic acid concentration and purity was measured on NanoDrop1000, and samples were submitted to sequencing at the DFCI Sequencing Core using either T7 or M13 Reverse primer. Sequence traces were oriented with V(D)J in forward orientation and truncated based on the msVHE and mu- or gamma-inner primer sequences. Files were imported into CLC Sequence Viewer 7, the resulting sequences extracted and grouped, then exported as sequence lists in fasta format. Sequences were submitted to IgBlast against the full NCBI V(D)J dataset with standard settings (Ye et al., 2013). Output was imported into Microsoft Excel, filtered to include only full-length sequences, then tabulated using the PivotTable function. Resulting VH segment usage and mutation tables were rendered in GraphPad Prism 6.
Single cell sequence analysis
Cell suspensions were stained as for flow cytometry, washed, and resuspended in FACS buffer. Single cell sort was performed on a 5-laser FACSARIA II Special Order system (355, 405, 488, 640, 594 nm laser lines; FCS-PMT module and enhanced optics and digital focusing), using a single-cell sort mask. Single cells were sorted directly into wells of 96-well semi-skirted PCR plates containing 5 µl buffer RLT (Qiagen) supplemented with 1% beta-mercaptoethanol. Following collection, plates were sealed with PCR tape, spun briefly, and immediately placed at −80°C. Plates were processed separately in a laminar flow hood following wipe-down and UV illumination. All reagents were prepared fresh for each experiment, based on pre-aliquots and stock solutions. Plates were thawed on ice, and to each well was added 2.2 vol. RNA-SPRI beads (11 µl per well, Agencourt RNAClean XP). Samples and beads were mixed well by pipetting, followed by incubation 10 min at RT. The plates were then incubated 5 min on a plate magnet, and the supernatant was removed using an 8× multichannel and following the direction of the magnet. Beads were washed 3 times with 100 µl 80% EtOH, in each round by displacing and repositioning wells from one magnet bar to a neighboring one several times. Washed beads were air dried for 10 min, and RNA was eluted in 4.5 µl of Mix1 (1.1 µM anchored Oligo-dT, 2.2 mM dNTP, and 1.1 U/µl of RNAseOut) per well by dispensing it directly over the beads and by pipetting up and down. Plates were spun briefly, incubated 3 min at 72°C, followed by 1 min on ice, then to each well was added 5.5 µl of Mix2 (1.8 U/µl RNAseOUT, 3.6 U/µl Maxima H- Reverse Transcriptase, 0.9 M Betaine, 5.5 mM MgCl2, and containing 2 µl 5× RT buffer per reaction). Plates were again spun briefly, and RT reactions were then run on a thermocycler, 50°C for 1h, 4°C hold. For PCRs, again all reagents were prepared fresh for each experiment, based on pre-aliquots and stock solutions. The 10 µl cDNA reactions were diluted 5-fold with 40 µl autoclaved MilliQ water, and from this 5 µl was used per 20 µl 1st round PCR reaction. From the 1st round PCR, 1 µl was used as input for 2nd round (semi-nested) PCR reaction. Primers and PCR conditions were as described above for the bulk sequence analysis. Amplicons were precipitated and analyzed as described, then sent for sequencing using Mu-inner and Gamma-inner, for mu and gamma heavy chain amplicons respectively. Sequence traces were oriented with V(D)J in forward orientation and truncated based on the msVHE primer sequence. Files were imported into CLC Sequence Viewer 7, the resulting sequences extracted and grouped, then exported as sequence lists in fasta format. Sequences were submitted to IgBlast against the full NCBI V(D)J dataset with standard settings (Ye et al., 2013). Output was imported into Microsoft Excel, filtered to include only full-length sequences, then tabulated using the PivotTable function. Resulting VH segment usage and mutation tables were rendered in GraphPad Prism 6.
Clonal lineage determination and inference
The program Cloanalyst (Kepler, 2013) was used to annotate isolated paired heavy-light VDJ/VJ sequences, and to infer intermediates and the unmutated common ancestor (UCA). The phylogenetic tree of the clonal lineages was then reconstructed.
Cloning, expression and purification of IgGs
The 38 genes encoding the Ab VH, VL and the UCA were synthesized (Integrated DNA Technologies) and cloned into the modified pVRC8400 vector between a tissue plasminogen activation (TPA) signal sequence and the constant domains of the mouse IgG1 CH1-CH3 and CL (Kuraoka et al., 2016). Mouse IgG1,k were produced by transient transfection of 293T adapted for suspension culture, using polyethylenimine (PEI, Polysciences) as the transfection reagent. The supernatant was harvested 6 days after transfection and clarified from cellular debris by centrifugation at 4,200 rpm for 20 min. IgGs were purified using Protein G agarose (Thermo) and dialyzed in PBS. Purified IgGs were concentrated and stored at 4°C until need ed.
Nucleolar ELISA/TRIFMA
Nucleoli were purified from cultured HEK293F FreeStyle cells (Invitrogen). The cells were grown to a density of ~1 * 106 cells per ml, spun down and resuspended in fresh pre-warmed medium the day before, and on the day of isolation added 1/10 volume fresh medium 1 hr before commencement of the protocol. For the isolation, ~108 cells were spun down at ~200 g (~1000 rpm, Beckman GS-6 centrifuge, GH-3.8 rotor) for 5 min, the supernatant decanted, and they were resuspended in 5 ml ice-cold Buffer A (10 mM Hepes, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT). After incubation on ice for 5 min, the cells were transferred to a pre-cooled 7 ml Dounce homogenizer (Wheaton Scientific) and subjected to 10 strokes using a tight pestle (“A” specification: 0.0010" – 0.0030" clearance), while keeping the homogenizer on ice. An aliquot of the homogenate was checked under a phase contrast microscope, and the procedure repeated as necessary, until >90% of the cells were burst, leaving intact nuclei, with various amounts of cytoplasmic material attached. The homogenate was then centrifuged 200 g for 5 min at 4°C. Following aspiration of the supernatant, the pellet, containing enriched but not highly pure nuclei, was resuspended in 3 ml S1 solution (0.25 M Sucrose, 10 mM MgCl2). The resuspended pellet was layered over 3 ml of S2 solution (0.35 M Sucrose, 0.5 mM MgCl2) and centrifuged at ~1,400 g (2,500 rpm) for 5 min at 4°C. The pellet was resuspended in 3 ml of S 2 solution and the resulting nuclear suspension was sonicated for 6 × 10 second bursts (with 10 second rest intervals between each burst) using a Misonix XL 2020 sonicator fitted with a microtip probe, and set at power setting 5. The sonicated nuclei were periodically checked under a phase contrast microscope to ensure the virtual absence of intact cells and nuclei, and confirm the presence of free nucleoli observable as dense refractile bodies. The sonicated sample was layered over 3 ml of S3 solution (0.88 M Sucrose, 0.5 mM MgCl2) and centrifuged at 3,000 g (3,500 rpm) for 10 min at 4°C. The nucleoplasmic fraction was aspirated and the nucleolar pellet was resuspended in 0.5 ml of S2 solution, followed by centrifugation at 1,430 g (2,500 rpm) for 5 min at 4°C and resuspension in 0.5 ml of S2 solution. Each batch was checked carefully under a phase contrast microscope to ensure presence of highly purified nucleoli without any other material, and stored at −80°C until use. For quantification of nucleoli 10-f old dilution series were prepared and loaded in a standard hemacytometer, allowed to settle, and then counted using phase-contrast. We routinely obtained approximately ~108 nucleoli, i.e., 1 nucleolus per 1 cell input, indicating a recovery of 25–33% (on average 3–4 nucleoli per HEK293F cell), and we occasionally observed partially intact nuclei or fragments hereof (around 1/40 to 1/50), indicating a purity of >97.5%. Fluoronunc microtiter plates were added nucleoli, 2 × 106/ml in PBS, 100 µl per well, spun at 2,500 rpm (~1,300 g), 10 min at 4°C, then left to coat overnight at 4°C. Wells were emptied and blocked by incubation with BSA, 1 mg/ml PBS, for 1 hr at RT. Plates were washed three times with TBS/Tw (10 mM Tris-HCl, 145 mM NaCl, pH 7.4, containing 0.05% v/v Tween-20), then added standard, controls and samples: standard, 564 C11 mAb in sqrt10 dilution series from ~4000 ng/ml to ~3.9 ng/ml, plus "0" (buffer only); samples and controls, serum diluted 1/100; all in duplicate, 100 µl per well in TBS/Tw, incubated 2 hrs at RT, then washed 3× with TBS/Tw. This was followed by incubation with secondary AP-conjugated Ab (total IgG, IgG2a, IgG2c, all from Southern Biotech), 1/2,000 in TBS/Tw, 100 µl/well, for 1 hr at RT. Finally, wells were washed 3 × with TBS/Tw and developed with phosphatase substrate, 1 mg/ml AP substrate buffer (1 M diethanolamine, pH 9.8, with 0.5 mM MgCl2), reading out on a SPECTRAmax 340 Microplate Reader running SoftMax Pro v. 5.4. In a time-resolved immunofluorometric assay (TRIFMA) variation of the assay, two-step development was performed by adding in-house biotiniylated rabbit anti-mouse IgG Ab (DAKO) followed by Eu3+-labeled streptavidin (0.1 mg/ml in TBS/Tw containing 25 µM EDTA). After the final wash, enhancement solution (Perkin-Elmer) was added and time-resolved fluorescence was read on a Victor3 (Perkin-Elmer).
Immunofluorescence confocal microscopy
Freshly harvested tissue was embedded in OCT (TissueTek) and immediately frozen at −80°C. Tissue blocks were equilibrated at cutting temperature (−16 to −20°C depending on tissue type) and 6–10 µm thick sections were cut on a cryostat. Tissue sections were mounted on SuperFrost+ slides (Fisher Scientific) and fixed using either ice-cold acetone or freshly thawed 4% PFA, for 5–10 min. PFA-fixed slides were incubated with TBS to block residual primary amine reactivity. Slides were then rinsed with PBS and incubated with block/perm buffer (PBS, 2% FBS, 0.1% NaN3, and 0.1% Triton-X100) for 30 min. This was followed by incubation with primary Ab mixture in staining buffer (PBS, 2% FBS, 0.1% NaN3), overnight at 4°C. For two step staining procedures, the slides were washed 3 times with PBS, 0.01% Tween-20, then added secondary Ab mixture in staining buffer, and incubated for 2 hrs at RT. At the end of either one- or two-step staining procedures, slides were washed once with staining buffer for 5 min, then 3 times 5 min with PBS, 0.01% Tween-20. Slides were spot-dried, then mounted in Fluoro-Gel (Electron Microscopy Sciences) and coverslipped. Imaging was performed using a Fluoview FV1000 inverted Olympus IX 81 confocal microscope, equipped with 6 laser lines (405, 457, 488, 515, 559, 635 nms) and 4 fluorescence + 1 transmission detector (PMTs).
HEp-2 Immunofluorescence assay
HEp-2 cells (ATCC) were split 1/3 in T175 on day −2 and seeded to glass coverslips at day −1 (100K/well, 24 well). Cells were fixed with 1%PFA for 20 min at RT. Cells were then blocked and permeabilized with block/perm buffer (2% FBS, 0.1% Tx-100 in PBS). After 30 min, cells were stained o/n in block/perm buffer with indicated antibodies at 10 ug/ml, 1 ug/ml or 0.1 ug/ml (C11 and 564 only). The next day, coverslips were washed 3 times and incubated for one hour with Goat anti Mouse IgG (H+L) Alexa488 (Life Technologies), phalloidin Alexa568 (Life Technologies) and DAPI. Coverslips were then washed 4 times and mounted on slides with Fluorogel as mounting medium. Detectors were set to no primary Ab control. Anti HA Ab 6649 was used as negative control. C11, the original hybridoma from which the 564 BCR was created was used as positive control in addition to re-cloned 564.
Image analysis
Images were quantified using CellProfiler (Lamprecht et al., 2007). For kidney sections, glomerulus specific masks were generated based on CD31 signal and the mean pixel intensity was measured within each mask. For HEp-2 slides, quantifications were done based on the DAPI (nucleus) and Phalloidin staining. Cell outlines were detected using the phalloidin staining and as a function of the detected nuclei. A cytoplasmic mask was generated by subtracting the nucleus mask from the cell mask. Each bar represents a mean of individual objects (for instance a mean of MFI from 20 nuclei).
Autoantigen arrays
Autoantigen arrays were generated and assays on the arrays were performed as described previously (Ayoglu et al., 2016). Unless stated otherwise in Table S3, 6 µg of each Ag and a three-point dilution series of control mouse and anti-mouse antibodies (0.25, 1, 4 µg) were diluted in PBS and transferred into 3 × 96-well plates. Analytes were coupled to 1×106 carboxylated magnetic beads per ID (MagPlex-C, Luminex Corp.). Beads were distributed into 3 × 96-well plates (Greiner BioOne), washed and re-suspended in phosphate buffer (0.1 M NaH2PO4, pH 6.2) using a plate washer (Biotek). Bead surface was activated by addition of 100 µl of phosphate buffer containing 0.5 mg of 1-ethyl-3(3-dimethylamino-propyl)carbodiimide (Pierce) and 0.5 mg N-hydroxysuccinimide (Pierce). After 20 min incubation on a shaker (Grant Bio), beads were washed and re-suspended in activation buffer (0.05 M MES, pH 5.0). Diluted Ags and control antibodies were incubated with beads for 2 h at RT. The beads were washed 3× in 100 µl PBS-T, re-suspended in 60 µl storage buffer (Blocking reagent for ELISA, Roche) and stored in plates at 4°C overnight . Immobilization of the Ags were confirmed and the assay conditions were optimized by analysis of several mouse monoclonal antibodies, such as anti-La/SSB (Santa Cruz), anti-Ro52 (Santa Cruz), anti-His6 tag (Invitrogen), anti-Scl70 (Immunovision), anti-SSB (Immunovision), anti-IL17A (eBiosciences), and a rabbit polyclonal Ab, anti-H2b (abcam), all at 1 µg/ml (data not shown). In addition, dilution series (1:50 to 1:300) of various autoimmune disease state human plasma for ds-DNA, Scl-70, SSA, SSB, cardiolipin, whole histones and RNP (Immunovision), as well as normal human sera (Immunovision) and previously in-house characterized MRL/lpr sera pool and BALB/c sera pool were used to optimize the assay conditions prior to analysis (data not shown).
Based on the optimized assay conditions, the mouse sera were diluted 1:60 in an assay buffer of 0.05% PBS-T supplemented with 3% (w/v) BSA (Sigma) and 5% (w/v) blotting-grade nonfat dry milk (BioRad) and transferred into a 96-well plate. The bead array was distributed into a 384-well plate (Greiner BioOne) by transfer of 5 µl bead array per well. Using a liquid handler (SELMA, CyBio), 45 µl of the 1:60 diluted sera were aliquoted and transferred into each of the three different quadrants of a 384-well plate. Samples were incubated for 90 min on a shaker (Grant Bio) at RT. The beads were washed with 3 × 60 µl PBS-T on a plate washer (EL406, Biotek). Using a liquid handler (SELMA, CyBio), 50 µl of R-PE conjugated Fcγ fragment specific goat anti-mouse IgG (Jackson), R-PE conjugated Fcγ subclass 2a specific goat anti-mouse IgG (Jackson) and R-PE conjugated Fcγ subclass 2c specific goat anti-mouse IgG (Jackson) were transferred to the three different quadrants of the 384-well plate, allowing for a parallel detection different isotypes. All secondary antibodies were diluted 1:500 in a buffer consisting of 3% BSA in 0.05% PBS-T. After incubation with the secondary antibodies for 45 min, the plate was washed with 3 × 60 µl PBS-T and re-suspended in 60 µl PBS-T for read-out in a FlexMap3D instrument (Luminex Corp.). Cloned antibodies were analyzed following the same protocol, where all the antibodies were diluted to 20 µg/ml in 3% (w/v) BSA and detected with the 1:500 diluted R-PE conjugated Fcγ fragment specific goat anti-mouse IgG (Jackson). At least 100 events per bead ID were counted and binding events were displayed as median fluorescence intensity (MFI). Data was analyzed using R. Heatmaps were rendered using the pheatmap package following log2-transformation.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analyses were performed in GraphPad Prism version 6, as defined throughout the text and figure legends.
DATA AND SOFTWARE AVAILABILITY
The data presented in this manuscript are tabulated in the main paper and in the supplementary materials. The sequences used for production of recombinant antibodies are compiled in Table S2. The Heavy chain Ig VDJ sequences have been deposited in GenBank under ID codes MF429952 – MF430833. The Kappa chain Ig VJ sequences have been deposited in GenBank under ID codes MF430834 – MF430850.
Supplementary Material
(A) Correlation plot for Id+ cell frequency versus GC B cell frequency in spleen or cutaneous LN, across the cohort presented in Figure 1A–C. Each dot represents one mouse, for a total of 34 mice analyzed.
(B) Mutation analysis for the heavy chains (IgM or IgG) of bulk-sorted GC B cells from two heterozygous 564Igi mice. Each dot represents one heavy chain. The mean number of mutations is indicated. P-value given for two-tailed Mann-Whitney test.
(C) GC B cell frequency measured within the YFP+ population for the cohort shown in Figure 1 H–L. Mean±SEM is shown for 6 (Day 8, 16, 24), 2 (Day 38 and 50) and 3 (Day 90) mice from 3 independent cohorts.
(D) Total GC B and CD138+ (plasma) cells in spleen of tamoxifen pulsed 564 Aid-Cre EYFP (n=8), non-pulsed 564 Aid-Cre EYFP (n=2) and 564 K− controls (n=4). Mean±SEM is given.
(E) as in (D), but for cutaneous LN.
(F) Frequencies of YFP+CD138− (GC and memory) and YFP+CD138+ (plasma) cells in spleen of the cohort represented in (D).
(G) As in (F), but for cutaneous LN.
(H) As in (F), but for BM.
(I) Id frequencies of untreated, tamoxifen in sunflower oil-treated, sunflower oil only-treated, and peanut oil-treated B6, as well as untreated 564het and 564het K− mice (n=1 per group, except for 564het with n=2, mean±SEM).
(J) GC B cell frequencies of the same cohort as in (I).
(K) FACS plots showing the frequency of YFP+ cells in a Day 90 tamoxifen pulsed 564Igi Aid-Cre EYFP mouse (top panel) versus a negative control (bottom panel). Results of sequencing of the mu-chain (left) and gamma chain (right) of FACS sorted GC cells of 564Igi mice (top) and Day 90 tamoxifen pulsed 564Igi Aid-Cre EYFP mice (bottom).
(L) Mutation analysis for the heavy chains (IgM or IgG) of bulk-sorted YFP+ cells from two Day 90 tamoxifen pulsed 564Igi Aid-Cre EYFP mice. Each dot represents one heavy chain. The mean number of mutations is indicated. P-value given for two-tailed Mann-Whitney test.
(A) Illustration of conceptual difference between intra- and inter-B cell BCR competition occurring in 564hets and 564 mixed BM chimeras, respectively. In the 564hets, “WT cells” arise through inactivation of the 564 locus and rearrangement of the endogenous Ig locus. In the mixed chimeras, use of 564 homozygous donors “locks in” the 564 cell fate, whereas the wild type repertoire is entirely normal.
(B) CD45.2 (564 BM-derived) cell frequencies in the B220− (triangles) and the B220+ (circles) population of the lymphocyte gate in BM of 564 CD45.1 mixed chimeras. Mean±SD are also indicated.
(C) As (B), but for spleen.
(D) As (B), but for skin-draining LN.
(E) As (B), but for blood.
(F) B220+ cell frequencies in the lymphocyte gate in BM of 564 CD45.1 mixed chimeras. Mean±SD are also indicated.
(G) As (F), but for spleen.
(H) As (F), but for skin-draining LN.
(I) As (F), but for blood.
(J) Correlation plot for Id+ cell frequency versus GC B cell frequency in spleen or cutaneous LN, across the cohort presented in Figure 2. Each dot represents one mouse, for a total of 24 mice analyzed.
(A) Heavy chain V segments observed in B6, sum of all YFP, and sum of all PA-GFP, clones. Colors congruent with Figure 3.
(B) Heavy chain V segments observed in 4 GC of 3 independent mice analyzed. Colors congruent with Figure 3.
(C) Mutation analysis for the heavy chains (IgM and IgG) of single-sorted YFP+ GC B cells from 4 different (A–D) Aid-Cre EYFP WT 564Igi mixed chimeras. Each dot represents one heavy chain. The mean number of mutations is indicated. P-value given for Kruskal-Wallis test with Dunn’s posttest comparing to naïve follicular B cells from B6.
(D) Mutation analysis for the heavy chains (IgM and IgG) of single-sorted photoactivated GC B cells of 4 GC (1–4) of 3 different (E–G) PA-GFP WT 564Igi mixed chimeras. Each dot represents one heavy chain. The mean number of mutations is indicated. P-value given for Kruskal-Wallis test with Dunn’s posttest comparing to naïve follicular B cells from B6.
(E) Mutation analysis for the heavy chains (IgM and IgG) of single-sorted photoactivated GC B cells of 4 GC (1–4) of one (G) of the PA-GFP WT 564Igi mixed chimeras. Each dot represents one heavy chain. The mean number of mutations is indicated. P-value given for Kruskal-Wallis test with Dunn’s posttest comparing to naïve follicular B cells from B6.
(F) Results of nucleolar TRIFMA titrations of cloned antibodies from Figure 3. Positive or borderline antibodies from HEp-2 screen are indicated in dark blue or mixed, while negative antibodies are in light blue. The positive control, hybridoma-derived 564 C11, is displayed in red.
(G) Clustered, unscaled heatmap of autoAg reactivities of cloned antibodies from Figure 3. Clone identity is indicated on the righthand side. Identity of individual Ags from left to right is indicated in Table S3. The scale is indicated in the top righthand corner.
(H) Bar graph representation of data from panel G for ssDNA reactivity.
(I) Bar graph representation of data from panel G for MDC reactivity.
(J) Bar graph representation of data from panel G for SmD2 reactivity.
(K) Bar graph representation of data from panel G for TGFβ-RII reactivity.
Epitope Spread GCs and Associated TfH are Ablated by CD40L Blockade. (A) GC B cell frequencies across spleen, cutaneous LN and mesenteric LN of untreated vs. anti-CD40L Ab treated heterozygous 564Igi mice and 564 mixed BM chimeras. Mean±SEM is given for 2 mice per group. Multiplicity-adjusted P values for two-way ANOVA with Tukey’s posttest.
(B) Frequencies of GC per follicle as quantified by confocal IF analysis of spleen sections from treated vs. untreated heterozygous 564Igi mice. Mean±SEM is indicated. Multiplicity-adjusted P-values given for one-way ANOVA with Dunnett’s posttest.
(C) as panel (B), but for 564 mixed chimeras.
(D) As panel (A), but showing Id frequencies. P values given for two-way ANOVA with Sidak’s posttest.
(E) Tfh (CXCR5hiPD-1hi) cells in spleen, skin-draining LN and mesenteric LN of untreated and CD40L-treated heterozygous 564Igi mice. P-values given for two-way ANOVA with Sidak’s posttest.
(F) As panel (E), but showing CD4 T cell frequencies.
(G) As panel (E), but for 564 mixed chimeras.
(H) As panel (E), but showing CD4 T cell frequencies for 564 mixed chimeras.
(A) Schematic overview of exclusion and competition gates in the experiment, with indication of the different compartments, their congenic markers and BCR identities.
(B) Total frequencies of 45.1 positive, 45.1/2 double positive, and 45.2 positive cells (as indicated on the x-axis) within the CD45 gate in blood of 564 TLR7+/+ (circles) or 564 TLR7−/− (squares) chimeras. Mean±SEM is indicated.
(C) as panel (B), but for the T cell gate.
(D) as panel (B), but for the B cell gate.
(E) PA-GFP+ (564 BM-derived) cell frequencies within the total lymphocyte gate, the B cell gate, and the T cell gate (as indicated on the x-axis) within the CD45 gate in blood of 564 TLR7+/+ (circles) or 564 TLR7−/− (squares) chimeras. Mean±SEM is indicated.
(F) Relative frequencies of CD45.1 and CD45.2 cells in the CD45 gate, within the competition gate, in blood of 564 TLR7 mixed chimeras. Mean±SEM is shown.
(G) as panel (F), but for the T cell gate.
(H) as panel (F), but for the B cell gate.
(I) Total B and T cell frequencies in blood of 564 TLR7+/+ (circles) and 564 TLR7−/− (squares) mixed chimeras. Mean±SEM is indicated.
(J) Total splenic CD4, CD8 and CD19 frequencies (as indicated on the x-axis) within the lymphocyte gate for 564 TLR7+/+ (blue) and 564 TLR7−/− (red) mixed chimeras. Mean±SEM is shown.
(K) as panel (J), but for mesenteric LN.
(L) Total GC B frequencies in spleen and mesenteric LN.
(M) GC B frequencies within the competition gate.
(N) CD45.2 frequencies within the competition gate for CD4, CD8, CD19 and GCB populations (as indicated on the x-axis) in mesenteric LN. Mean±SEM shown, with multiplicity adjusted P-values for two-way ANOVA using Sidak’s posttest. The dotted line indicates the expected 50% frequency.
(O) Frequency of donor-derived (CD45.2) vs. recipient derived (CD45.1) cells within the lymphocyte gate in blood of 1:2 (circles) and 1:9 (squares) 564 DTA chimeras. Mean±SD is indicated.
(P) Similar to panel (O), but for the B cell subgate.
(Q) B220 frequencies in 1:2 and 1:9 564 DTA chimeras, compared to B6 mice. Mean±SD is indicated.
(R) Frequency of PA-GFP+ (wt BM-derived) cells after exclusion of recipient-derived CD45.1+ cells, within the lymphocyte gate, B220+ gate and B220− gate of 1:2 (circles) and 1:9 (squares) 564 DTA mixed chimeras. Mean±SD is indicated.
Clonal Evolution in GCs of Mesenteric LN. (A) Frequency of most (1st), second most (2nd) and the sum of the most and second most predominant colors observed in individual mesenteric LN GC at Day 3, 14 and 31 post tamoxifen induction of Confetti recombination in NP-CGG immunized Confetti chimeras. The mean of an average of 3.7 GC per mouse in mesenteric LN of 2 mice per time point is indicated by the bar.
(B) As panel (A), but for 564 Confetti mixed chimeras. The mean of an average of 3.7 GC per mouse in mesenteric LN of 3 mice per time point is indicated by the bar.
(C) Divergence index for the GC represented in (A) and (B). P values given for two-way ANOVA with Tukey’s posttest.
(D) Line graph showing evolution of average frequencies of most, second most, and the sum of the most and second most predominant colors over time. Data as in (A).
(E) as in panel (D), but for 564 data as in (B).
(F) Divergence index data as in (C), but with a side-by-side comparison of NP-CGG and 564 chimeras at each time point.
(G) For NP-CGG immunized Confetti chimeras, the relative frequency of GC out of total mesenteric GC, plotted as a function of frequency of the most dominant color within each GC, demonstrating the shift over time from a high frequency of no-dominance GC (most dominant color=~0.35) to a higher frequency of color-dominant GC (most dominant color frequency increases above 50%, i.e. 0.5, and beyond).
(H) as for (G), but for 564 Confetti mixed chimeras.
(I) Representative images used for quantification of color.
(A) Overall patterns of reactivities of IgG in sera from 9 heterozygous 564Igi, 7 homozygous 564Igi, 6 1:2 564 mixed chimeras, 2 1:9 564 mixed chimeras, 6 1:2 564K− mixed chimeras, and 5 B6, assayed for 245 Ags. Each row represents a mouse, and mice are grouped as indicated in the legend. Each column represents an Ag (for a list of Ags in order left to right, please refer to Table S3). The log2-transformed signal intensity scale is indicated in the legend, going from high signal (red) to low signal (blue).
(B) as A, but for IgG2a development.
(C) as A, but for IgG2c development.
(D) Bar graph representations of IgG (left column), IgG2a (middle column) and IgG2c (right column) Ab towards single-stranded DNA (top row), SmD2 (middle row) and TNF-beta (bottom row), in sera of B6 (n=5), heterozygous (n=9), and homozygous (n=7) 564Igi mice. Mean±SEM and multiplicity adjusted significance for one-way ANOVA with Dunn’s posttest.
(E) Bar graph representations of IgG (left column), IgG2a (middle column) and IgG2c (right column) Ab towards single-stranded DNA (top row), SmD2 (middle row) and TNF-beta (bottom row), in sera of 564 mixed chimeras (n=8) and 564K− mixed chimeras (n=6). Mean±SEM and multiplicity adjusted significance for two-tailed Mann-Whitney test.
YFP+ GC in tamoxifen-treated 564 Aid-Cre EYFP reporter. Five fps video of an 85 µm stack through a GC in an explanted inguinal LN, moving at a step size of 5 µm. Second harmonics generation from collagen rendered in blue, YFP+ cells rendered in green and FDC network labeled with anti-CD157 Ab in red.
Intravital imaging of YFP+ GC in tamoxifen-treated 564 Aid-Cre EYFP reporter. Fifteen fps intravital video of a GC over 1 hr at an interval of 15 s per frame. Second harmonics generation from collagen rendered in blue, YFP+ cells rendered in green and FDC network labeled with PE-ICs in red.
High resolution 3-dimensional reconstruction of a mesenteric GC from an explanted mesenteric LN 3 days following oral tamoxifen treatment. Slices were obtained at 1 um steps; collagen second harmonics generation rendered in blue, confetti recombined cells are apparent in a variety of colors including GFP, YFP, RFP, CFP, and combinations of these, throughout the GC.
Intravital imaging of Confetti+ GC Day 3 post tamoxifen treatment. Twenty-five fps intravital video of a GC over 45 min at an interval of 20.5 s per frame. Brainbow colors as indicated.
Genotyping (primers and protocols) and IgPCRs (primers and protocols).
Sequences of cloned antibodies.
Overview of antigens and control analytes used in autoantigen arrays.
Acknowledgments
SED was supported by the Benzon Foundation and by a Marie Curie International Outgoing Fellowship within the 7th European Community Framework Programme (PIOF-GA-2012-330134). CEvdP was supported by a fellowship from GSK. DJF was supported by the HHMI Medical Research Fellows Program. BA was supported by the Knut and Alice Wallenberg Foundation Postdoctoral Scholarship Program (KAW 2014.0412). This research was supported by NIH grants R21AI117737, R01AI039246, R21AI117737 (MCC), 1R01AI119006 (GDV), U19-AI110491, R01 AI125197-01 (PJU), by the Donald E. and Delia B. Baxter Foundation (Career Development Award to PJU), the Henry Gustav Floren Trust (gift to PJU), and a gift from Elizabeth F. Adler (to PJU). F. Alt provided the attenuated DTA strain and A. Sharpe the UBC-CreERT2 line. We thank H. Leung of the Optical Microscopy Core and N. Barteneva and K. Ketman of the Flow and Imaging Cytometry Resource at the PCMM for technical assistance. We also thank the SciLifeLab Autoimmunity Profiling Facility for access to FlexMap3D instrument. We are grateful to E. Alicot for technical assistance, D. Weseman for helpful discussion of the topic, and C. Usher for help editing the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
SED conceived the project. SED, CEvdP, DJF, BA, FAAQ, and GB planned and performed experiments and analyzed data. C-EvdP and DJF contributed equally. LM, CAR, J-CW, PJU, and GDV provided crucial reagents and expertise. PJU, GDV, and MCC oversaw the project. SED wrote the manuscript and all authors provided critical feedback.
References
- Arbuckle MR, McClain MT, Rubertone MV, Scofield RH, Dennis GJ, James JA, Harley JB. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N Engl J Med. 2003;349:1526–1533. doi: 10.1056/NEJMoa021933. [DOI] [PubMed] [Google Scholar]
- Ayoglu B, Mitsios N, Kockum I, Khademi M, Zandian A, Sjöberg R, Forsström B, Bredenberg J, Lima Bomfim I, Holmgren E, et al. Anoctamin 2 identified as an autoimmune target in multiple sclerosis. Proceedings of the National Academy of Sciences. 2016;113:2188–2193. doi: 10.1073/pnas.1518553113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berland R, Fernandez L, Kari E, Han J-H, Lomakin I, Akira S, Wortis HH, Kearney JF, Ucci AA, Imanishi-Kari T. Toll-like receptor 7-dependent loss of B cell tolerance in pathogenic autoantibody knockin mice. Immunity. 2006;25:429–440. doi: 10.1016/j.immuni.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Busse CE, Czogiel I, Braun P, Arndt PF, Wardemann H. Single-cell based high-throughput sequencing of full-length immunoglobulin heavy and light chain genes. Eur J Immunol. 2013;44:597–603. doi: 10.1002/eji.201343917. [DOI] [PubMed] [Google Scholar]
- Cappellano G, Orilieri E, Woldetsadik AD, Boggio E, Soluri MF, Comi C, Sblattero D, Chiocchetti A, Dianzani U. Anti-cytokine autoantibodies in autoimmune diseases. Am J Clin Exp Immunol. 2012;1:136–146. [PMC free article] [PubMed] [Google Scholar]
- Chatterjee P, Agyemang AF, Alimzhanov MB, Degn S, Tsiftsoglou SA, Alicot E, Jones SA, Ma M, Carroll MC. Complement C4 maintains peripheral B-cell tolerance in a myeloid cell dependent manner. Eur J Immunol. 2013;43:2441–2450. doi: 10.1002/eji.201343412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornaby C, Gibbons L, Mayhew V, Sloan CS, Welling A, Poole BD. B cell epitope spreading: mechanisms and contribution to autoimmune diseases. Immunol. Lett. 2015;163:56–68. doi: 10.1016/j.imlet.2014.11.001. [DOI] [PubMed] [Google Scholar]
- Das A, Heesters BA, Bialas A, O’Flynn J, Rifkin IR, Ochando J, Mittereder N, Carlesso G, Herbst R, Carroll MC. Follicular Dendritic Cell Activation by TLR Ligands Promotes Autoreactive B Cell Responses. Immunity. 2017;46:106–119. doi: 10.1016/j.immuni.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogan I, Bertocci B, Vilmont V, Delbos F, Mégret J, Storck S, Reynaud C-A, Weill J-C. Multiple layers of B cell memory with different effector functions. Nat Immunol. 2009;10:1292–1299. doi: 10.1038/ni.1814. [DOI] [PubMed] [Google Scholar]
- Durie FH, Fava RA, Foy TM, Aruffo A, Ledbetter JA, Noelle RJ. Prevention of collagen-induced arthritis with an antibody to gp39, the ligand for CD40. Science. 1993;261:1328–1330. doi: 10.1126/science.7689748. [DOI] [PubMed] [Google Scholar]
- Gavalchin J, Seder RA, Datta SK. The NZB X SWR model of lupus nephritis. I. Cross-reactive idiotypes of monoclonal anti-DNA antibodies in relation to antigenic specificity, charge, and allotype. Identification of interconnected idiotype families inherited from the normal SWR and the autoimmune NZB parents. J. Immunol. 1987;138:128–137. [PubMed] [Google Scholar]
- Green NM, Moody K-S, Debatis M, Marshak-Rothstein A. Activation of autoreactive B cells by endogenous TLR7 and TLR3 RNA ligands. J Biol Chem. 2012;287:39789–39799. doi: 10.1074/jbc.M112.383000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Zheng B, Dal Porto J, Kelsoe G. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. IV. Affinity-dependent, antigen-driven B cell apoptosis in germinal centers as a mechanism for maintaining self-tolerance. J Exp Med. 1995;182:1635–1644. doi: 10.1084/jem.182.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlands RA, Christensen SR, Sweet RA, Hershberg U, Shlomchik MJ. T cell-independent and toll-like receptor-dependent antigen-driven activation of autoreactive B cells. Immunity. 2008;29:249–260. doi: 10.1016/j.immuni.2008.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg U, Luning Prak ET. The analysis of clonal expansions in normal and autoimmune B cell repertoires. Philos Trans R Soc Lond, B, Biol Sci. 2015;370 doi: 10.1098/rstb.2014.0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepler TB. Reconstructing a B-cell clonal lineage. I. Statistical inference of unobserved ancestors. F1000Res. 2013;2:103. doi: 10.12688/f1000research.2-103.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraoka M, Schmidt AG, Nojima T, Feng F, Watanabe A, Kitamura D, Harrison SC, Kepler TB, Kelsoe G. Complex Antigens Drive Permissive Clonal Selection in Germinal Centers. Immunity. 2016;44:542–552. doi: 10.1016/j.immuni.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamprecht M, Sabatini D, Carpenter A. CellProfiler™: free, versatile software for automated biological image analysis. Biotech. 2007;42:71–75. doi: 10.2144/000112257. [DOI] [PubMed] [Google Scholar]
- Lau CM, Broughton C, Tabor AS, Akira S, Flavell RA, Mamula MJ, Christensen SR, Shlomchik MJ, Viglianti GA, Rifkin IR, et al. RNA-associated autoantigens activate B cells by combined B cell antigen receptor/Toll-like receptor 7 engagement. J Exp Med. 2005;202:1171–1177. doi: 10.1084/jem.20050630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leadbetter EA, Rifkin IR, Hohlbaum AM, Beaudette BC, Shlomchik MJ, Marshak-Rothstein A. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature. 2002;416:603–607. doi: 10.1038/416603a. [DOI] [PubMed] [Google Scholar]
- Luning Prak ET, Monestier M, Eisenberg RA. B cell receptor editing in tolerance and autoimmunity. Ann N Y Acad Sci. 2011;1217:96–121. doi: 10.1111/j.1749-6632.2010.05877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzina IG, Atamas SP, Storrer CE, daSilva LC, Kelsoe G, Papadimitriou JC, Handwerger BS. Spontaneous formation of germinal centers in autoimmune mice. J Leukoc Biol. 2001;70:578–584. [PubMed] [Google Scholar]
- Mietzner B, Tsuiji M, Scheid J, Velinzon K, Tiller T, Abraham K, Gonzalez JB, Pascual V, Stichweh D, Wardemann H, et al. Autoreactive IgG memory antibodies in patients with systemic lupus erythematosus arise from nonreactive and polyreactive precursors. Proc Natl Acad Sci USA. 2008;105:9727–9732. doi: 10.1073/pnas.0803644105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B, Kannourakis G, Nouri S, Smith KGC, Nossal GJV. Soluble antigen can cause enhanced apoptosis of germinal-centre B cells. Nature. 1995;375:331–334. doi: 10.1038/375331a0. [DOI] [PubMed] [Google Scholar]
- Rahman A, Isenberg DA. Systemic Lupus Erythematosus. New England Journal of Medicine. 2008;358:929–939. doi: 10.1056/NEJMra071297. [DOI] [PubMed] [Google Scholar]
- Reed JH, Jackson J, Christ D, Goodnow CC. Clonal redemption of autoantibodies by somatic hypermutation away from self-reactivity during human immunization. Journal of Experimental Medicine. 2016;213:1255–1265. doi: 10.1084/jem.20151978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabouri Z, Schofield P, Horikawa K, Spierings E, Kipling D, Randall KL, Langley D, Roome B, Vazquez-Lombardi R, Rouet R, et al. Redemption of autoantibodies on anergic B cells by variable-region glycosylation and mutation away from self-reactivity. Proceedings of the National Academy of Sciences. 2014;111:E2567–E2575. doi: 10.1073/pnas.1406974111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwickert TA, Alabyev B, Manser T, Nussenzweig MC. Germinal center reutilization by newly activated B cells. Journal of Experimental Medicine. 2009;206:2907–2914. doi: 10.1084/jem.20091225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwickert TA, Lindquist RL, Shakhar G, Livshits G, Skokos D, Kosco-Vilbois MH, Dustin ML, Nussenzweig MC. In vivo imaging of germinal centres reveals a dynamic open structure. Nature. 2007;446:83–87. doi: 10.1038/nature05573. [DOI] [PubMed] [Google Scholar]
- Shlomchik MJ, Marshak-Rothstein A, Wolfowicz CB, Rothstein TL, Weigert MG. The role of clonal selection and somatic mutation in autoimmunity. Nature. 1987;328:805–811. doi: 10.1038/328805a0. [DOI] [PubMed] [Google Scholar]
- Shokat KM, Goodnow CC. Antigen-induced B-cell death and elimination during germinal-centre immune responses. Nature. 1995;375:334–338. doi: 10.1038/375334a0. [DOI] [PubMed] [Google Scholar]
- Tas JMJ, Mesin L, Pasqual G, Targ S, Jacobsen JT, Mano YM, Chen CS, Weill J-C, Reynaud C-A, Browne EP, et al. Visualizing antibody affinity maturation in germinal centers. Science. 2016;351:1048–1054. doi: 10.1126/science.aad3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T, Busse CE, Wardemann H. Journal of Immunological Methods. J Immunol Methods. 2009;350:183–193. doi: 10.1016/j.jim.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Tiller T, Tsuiji M, Yurasov S, Velinzon K, Nussenzweig MC, Wardemann H. Autoreactivity in Human IgG+ Memory B Cells. Immunity. 2007;26:205–213. doi: 10.1016/j.immuni.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderlugt CL, Miller SD. Epitope spreading in immune-mediated diseases: implications for immunotherapy. Nat Rev Immunol. 2002;2:85–95. doi: 10.1038/nri724. [DOI] [PubMed] [Google Scholar]
- Victora GD, Schwickert TA, Fooksman DR, Kamphorst AO, Meyer-Hermann M, Dustin ML, Nussenzweig MC. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 2010;143:592–605. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinuesa CG, Sanz I, Cook MC. Dysregulation of germinal centres in autoimmune disease. Nat Rev Immunol. 2009;9:845–857. doi: 10.1038/nri2637. [DOI] [PubMed] [Google Scholar]
- William J, Euler C, Christensen S, Shlomchik MJ. Evolution of autoantibody responses via somatic hypermutation outside of germinal centers. Science. 2002;297:2066–2070. doi: 10.1126/science.1073924. [DOI] [PubMed] [Google Scholar]
- Yau IW, Cato MH, Jellusova J, Hurtado de Mendoza T, Brink R, Rickert RC. Censoring of Self-Reactive B Cells by Follicular Dendritic Cell-Displayed Self-Antigen. J Immunol. 2013;191:1082–1090. doi: 10.4049/jimmunol.1201569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Ma N, Madden TL, Ostell JM. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41:W34–W40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusuf I, Stern J, McCaughtry TM, Gallagher S, Sun H, Gao C, Tedder T, Carlesso G, Carter L, Herbst R, et al. Germinal Center B Cell Depletion Diminishes CD4+ Follicular T Helper Cells in Autoimmune Mice. PLoS ONE. 2014;9:e102791–17. doi: 10.1371/journal.pone.0102791. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Correlation plot for Id+ cell frequency versus GC B cell frequency in spleen or cutaneous LN, across the cohort presented in Figure 1A–C. Each dot represents one mouse, for a total of 34 mice analyzed.
(B) Mutation analysis for the heavy chains (IgM or IgG) of bulk-sorted GC B cells from two heterozygous 564Igi mice. Each dot represents one heavy chain. The mean number of mutations is indicated. P-value given for two-tailed Mann-Whitney test.
(C) GC B cell frequency measured within the YFP+ population for the cohort shown in Figure 1 H–L. Mean±SEM is shown for 6 (Day 8, 16, 24), 2 (Day 38 and 50) and 3 (Day 90) mice from 3 independent cohorts.
(D) Total GC B and CD138+ (plasma) cells in spleen of tamoxifen pulsed 564 Aid-Cre EYFP (n=8), non-pulsed 564 Aid-Cre EYFP (n=2) and 564 K− controls (n=4). Mean±SEM is given.
(E) as in (D), but for cutaneous LN.
(F) Frequencies of YFP+CD138− (GC and memory) and YFP+CD138+ (plasma) cells in spleen of the cohort represented in (D).
(G) As in (F), but for cutaneous LN.
(H) As in (F), but for BM.
(I) Id frequencies of untreated, tamoxifen in sunflower oil-treated, sunflower oil only-treated, and peanut oil-treated B6, as well as untreated 564het and 564het K− mice (n=1 per group, except for 564het with n=2, mean±SEM).
(J) GC B cell frequencies of the same cohort as in (I).
(K) FACS plots showing the frequency of YFP+ cells in a Day 90 tamoxifen pulsed 564Igi Aid-Cre EYFP mouse (top panel) versus a negative control (bottom panel). Results of sequencing of the mu-chain (left) and gamma chain (right) of FACS sorted GC cells of 564Igi mice (top) and Day 90 tamoxifen pulsed 564Igi Aid-Cre EYFP mice (bottom).
(L) Mutation analysis for the heavy chains (IgM or IgG) of bulk-sorted YFP+ cells from two Day 90 tamoxifen pulsed 564Igi Aid-Cre EYFP mice. Each dot represents one heavy chain. The mean number of mutations is indicated. P-value given for two-tailed Mann-Whitney test.
(A) Illustration of conceptual difference between intra- and inter-B cell BCR competition occurring in 564hets and 564 mixed BM chimeras, respectively. In the 564hets, “WT cells” arise through inactivation of the 564 locus and rearrangement of the endogenous Ig locus. In the mixed chimeras, use of 564 homozygous donors “locks in” the 564 cell fate, whereas the wild type repertoire is entirely normal.
(B) CD45.2 (564 BM-derived) cell frequencies in the B220− (triangles) and the B220+ (circles) population of the lymphocyte gate in BM of 564 CD45.1 mixed chimeras. Mean±SD are also indicated.
(C) As (B), but for spleen.
(D) As (B), but for skin-draining LN.
(E) As (B), but for blood.
(F) B220+ cell frequencies in the lymphocyte gate in BM of 564 CD45.1 mixed chimeras. Mean±SD are also indicated.
(G) As (F), but for spleen.
(H) As (F), but for skin-draining LN.
(I) As (F), but for blood.
(J) Correlation plot for Id+ cell frequency versus GC B cell frequency in spleen or cutaneous LN, across the cohort presented in Figure 2. Each dot represents one mouse, for a total of 24 mice analyzed.
(A) Heavy chain V segments observed in B6, sum of all YFP, and sum of all PA-GFP, clones. Colors congruent with Figure 3.
(B) Heavy chain V segments observed in 4 GC of 3 independent mice analyzed. Colors congruent with Figure 3.
(C) Mutation analysis for the heavy chains (IgM and IgG) of single-sorted YFP+ GC B cells from 4 different (A–D) Aid-Cre EYFP WT 564Igi mixed chimeras. Each dot represents one heavy chain. The mean number of mutations is indicated. P-value given for Kruskal-Wallis test with Dunn’s posttest comparing to naïve follicular B cells from B6.
(D) Mutation analysis for the heavy chains (IgM and IgG) of single-sorted photoactivated GC B cells of 4 GC (1–4) of 3 different (E–G) PA-GFP WT 564Igi mixed chimeras. Each dot represents one heavy chain. The mean number of mutations is indicated. P-value given for Kruskal-Wallis test with Dunn’s posttest comparing to naïve follicular B cells from B6.
(E) Mutation analysis for the heavy chains (IgM and IgG) of single-sorted photoactivated GC B cells of 4 GC (1–4) of one (G) of the PA-GFP WT 564Igi mixed chimeras. Each dot represents one heavy chain. The mean number of mutations is indicated. P-value given for Kruskal-Wallis test with Dunn’s posttest comparing to naïve follicular B cells from B6.
(F) Results of nucleolar TRIFMA titrations of cloned antibodies from Figure 3. Positive or borderline antibodies from HEp-2 screen are indicated in dark blue or mixed, while negative antibodies are in light blue. The positive control, hybridoma-derived 564 C11, is displayed in red.
(G) Clustered, unscaled heatmap of autoAg reactivities of cloned antibodies from Figure 3. Clone identity is indicated on the righthand side. Identity of individual Ags from left to right is indicated in Table S3. The scale is indicated in the top righthand corner.
(H) Bar graph representation of data from panel G for ssDNA reactivity.
(I) Bar graph representation of data from panel G for MDC reactivity.
(J) Bar graph representation of data from panel G for SmD2 reactivity.
(K) Bar graph representation of data from panel G for TGFβ-RII reactivity.
Epitope Spread GCs and Associated TfH are Ablated by CD40L Blockade. (A) GC B cell frequencies across spleen, cutaneous LN and mesenteric LN of untreated vs. anti-CD40L Ab treated heterozygous 564Igi mice and 564 mixed BM chimeras. Mean±SEM is given for 2 mice per group. Multiplicity-adjusted P values for two-way ANOVA with Tukey’s posttest.
(B) Frequencies of GC per follicle as quantified by confocal IF analysis of spleen sections from treated vs. untreated heterozygous 564Igi mice. Mean±SEM is indicated. Multiplicity-adjusted P-values given for one-way ANOVA with Dunnett’s posttest.
(C) as panel (B), but for 564 mixed chimeras.
(D) As panel (A), but showing Id frequencies. P values given for two-way ANOVA with Sidak’s posttest.
(E) Tfh (CXCR5hiPD-1hi) cells in spleen, skin-draining LN and mesenteric LN of untreated and CD40L-treated heterozygous 564Igi mice. P-values given for two-way ANOVA with Sidak’s posttest.
(F) As panel (E), but showing CD4 T cell frequencies.
(G) As panel (E), but for 564 mixed chimeras.
(H) As panel (E), but showing CD4 T cell frequencies for 564 mixed chimeras.
(A) Schematic overview of exclusion and competition gates in the experiment, with indication of the different compartments, their congenic markers and BCR identities.
(B) Total frequencies of 45.1 positive, 45.1/2 double positive, and 45.2 positive cells (as indicated on the x-axis) within the CD45 gate in blood of 564 TLR7+/+ (circles) or 564 TLR7−/− (squares) chimeras. Mean±SEM is indicated.
(C) as panel (B), but for the T cell gate.
(D) as panel (B), but for the B cell gate.
(E) PA-GFP+ (564 BM-derived) cell frequencies within the total lymphocyte gate, the B cell gate, and the T cell gate (as indicated on the x-axis) within the CD45 gate in blood of 564 TLR7+/+ (circles) or 564 TLR7−/− (squares) chimeras. Mean±SEM is indicated.
(F) Relative frequencies of CD45.1 and CD45.2 cells in the CD45 gate, within the competition gate, in blood of 564 TLR7 mixed chimeras. Mean±SEM is shown.
(G) as panel (F), but for the T cell gate.
(H) as panel (F), but for the B cell gate.
(I) Total B and T cell frequencies in blood of 564 TLR7+/+ (circles) and 564 TLR7−/− (squares) mixed chimeras. Mean±SEM is indicated.
(J) Total splenic CD4, CD8 and CD19 frequencies (as indicated on the x-axis) within the lymphocyte gate for 564 TLR7+/+ (blue) and 564 TLR7−/− (red) mixed chimeras. Mean±SEM is shown.
(K) as panel (J), but for mesenteric LN.
(L) Total GC B frequencies in spleen and mesenteric LN.
(M) GC B frequencies within the competition gate.
(N) CD45.2 frequencies within the competition gate for CD4, CD8, CD19 and GCB populations (as indicated on the x-axis) in mesenteric LN. Mean±SEM shown, with multiplicity adjusted P-values for two-way ANOVA using Sidak’s posttest. The dotted line indicates the expected 50% frequency.
(O) Frequency of donor-derived (CD45.2) vs. recipient derived (CD45.1) cells within the lymphocyte gate in blood of 1:2 (circles) and 1:9 (squares) 564 DTA chimeras. Mean±SD is indicated.
(P) Similar to panel (O), but for the B cell subgate.
(Q) B220 frequencies in 1:2 and 1:9 564 DTA chimeras, compared to B6 mice. Mean±SD is indicated.
(R) Frequency of PA-GFP+ (wt BM-derived) cells after exclusion of recipient-derived CD45.1+ cells, within the lymphocyte gate, B220+ gate and B220− gate of 1:2 (circles) and 1:9 (squares) 564 DTA mixed chimeras. Mean±SD is indicated.
Clonal Evolution in GCs of Mesenteric LN. (A) Frequency of most (1st), second most (2nd) and the sum of the most and second most predominant colors observed in individual mesenteric LN GC at Day 3, 14 and 31 post tamoxifen induction of Confetti recombination in NP-CGG immunized Confetti chimeras. The mean of an average of 3.7 GC per mouse in mesenteric LN of 2 mice per time point is indicated by the bar.
(B) As panel (A), but for 564 Confetti mixed chimeras. The mean of an average of 3.7 GC per mouse in mesenteric LN of 3 mice per time point is indicated by the bar.
(C) Divergence index for the GC represented in (A) and (B). P values given for two-way ANOVA with Tukey’s posttest.
(D) Line graph showing evolution of average frequencies of most, second most, and the sum of the most and second most predominant colors over time. Data as in (A).
(E) as in panel (D), but for 564 data as in (B).
(F) Divergence index data as in (C), but with a side-by-side comparison of NP-CGG and 564 chimeras at each time point.
(G) For NP-CGG immunized Confetti chimeras, the relative frequency of GC out of total mesenteric GC, plotted as a function of frequency of the most dominant color within each GC, demonstrating the shift over time from a high frequency of no-dominance GC (most dominant color=~0.35) to a higher frequency of color-dominant GC (most dominant color frequency increases above 50%, i.e. 0.5, and beyond).
(H) as for (G), but for 564 Confetti mixed chimeras.
(I) Representative images used for quantification of color.
(A) Overall patterns of reactivities of IgG in sera from 9 heterozygous 564Igi, 7 homozygous 564Igi, 6 1:2 564 mixed chimeras, 2 1:9 564 mixed chimeras, 6 1:2 564K− mixed chimeras, and 5 B6, assayed for 245 Ags. Each row represents a mouse, and mice are grouped as indicated in the legend. Each column represents an Ag (for a list of Ags in order left to right, please refer to Table S3). The log2-transformed signal intensity scale is indicated in the legend, going from high signal (red) to low signal (blue).
(B) as A, but for IgG2a development.
(C) as A, but for IgG2c development.
(D) Bar graph representations of IgG (left column), IgG2a (middle column) and IgG2c (right column) Ab towards single-stranded DNA (top row), SmD2 (middle row) and TNF-beta (bottom row), in sera of B6 (n=5), heterozygous (n=9), and homozygous (n=7) 564Igi mice. Mean±SEM and multiplicity adjusted significance for one-way ANOVA with Dunn’s posttest.
(E) Bar graph representations of IgG (left column), IgG2a (middle column) and IgG2c (right column) Ab towards single-stranded DNA (top row), SmD2 (middle row) and TNF-beta (bottom row), in sera of 564 mixed chimeras (n=8) and 564K− mixed chimeras (n=6). Mean±SEM and multiplicity adjusted significance for two-tailed Mann-Whitney test.
YFP+ GC in tamoxifen-treated 564 Aid-Cre EYFP reporter. Five fps video of an 85 µm stack through a GC in an explanted inguinal LN, moving at a step size of 5 µm. Second harmonics generation from collagen rendered in blue, YFP+ cells rendered in green and FDC network labeled with anti-CD157 Ab in red.
Intravital imaging of YFP+ GC in tamoxifen-treated 564 Aid-Cre EYFP reporter. Fifteen fps intravital video of a GC over 1 hr at an interval of 15 s per frame. Second harmonics generation from collagen rendered in blue, YFP+ cells rendered in green and FDC network labeled with PE-ICs in red.
High resolution 3-dimensional reconstruction of a mesenteric GC from an explanted mesenteric LN 3 days following oral tamoxifen treatment. Slices were obtained at 1 um steps; collagen second harmonics generation rendered in blue, confetti recombined cells are apparent in a variety of colors including GFP, YFP, RFP, CFP, and combinations of these, throughout the GC.
Intravital imaging of Confetti+ GC Day 3 post tamoxifen treatment. Twenty-five fps intravital video of a GC over 45 min at an interval of 20.5 s per frame. Brainbow colors as indicated.
Genotyping (primers and protocols) and IgPCRs (primers and protocols).
Sequences of cloned antibodies.
Overview of antigens and control analytes used in autoantigen arrays.