Abstract
Introduction:
Stereotactic hypofractionated radiotherapy is an effective treatment for brain metastases in oligometastatic patients. Its planning is however time-consuming because of the number of organs at risk to be manually segmented. This study evaluates 2 automated segmentation commercial software.
Methods:
Patients were scanned in the treatment position. The computed tomography scan was registered on a magnetic resonance imaging and volumes were manually segmented by a clinician. Then 2 automated segmentations were performed (with iPlan and Smart Segmentation). RT STRUCT files were compared with Aquilab’s Artistruct segment comparison module. We selected common segmented volume ratio as the main judging criterion. Secondary criteria were Dice-Sørensen coefficients, overlap ratio, and additional segmented volume.
Results:
Twenty consecutive patients were included. Agreement between manual and automated contouring was poor. Common segmented volumes ranged from 7.71% to 82.54%, Dice-Sørensen coefficient ranged from 0.0745 to 0.8398, overlap ratio ranged from 0.0414 to 0.7275, and additional segmented volume ranged from 9.80% to 92.25%. Each software outperformed the other on some organs while performing worse on others.
Conclusion:
No software seemed clearly better than the other. Common segmented volumes were much too low for routine use in stereotactic hypofractionated brain radiotherapy. Manual editing is still needed.
Keywords: radiation therapy, automated segmentation, brain, radiosurgery
Introduction
Stereotactic radiosurgery (SRS) or stereotactic hypofractionated radiation therapy is a highly effective local treatment for brain metastases. It allows the delivery of high doses of radiation per fraction while sparing normal tissues by using numerous beams and entry points.1 This technique is becoming more and more prevalent throughout the world thanks to the availability of newer linear accelerators as well as improvements made to the cobalt-based Gamma Knife. It has been suggested that SRS may increase survival in young patients with few metastases and a favorable disease-specific graded prognostic assessment.2 Avoiding whole-brain radiation therapy may also improve the quality of life.3
This technique is often based on inverse planning, with various regions of interest being segmented by the clinician and dose constraints being set.1 As this technique aims to allow steep dose gradients between the target and the organs at risk (OARs), the accuracy of segmentation is extremely important to get the best possible dosimetry for the patient.1
The high number of OAR to be taken into account for treatment planning (lenses, cochleae, eyes, hippocampi, brain stem, chiasma, pituitary gland, and so on) can make the segmentation process time-consuming. While manual segmentation is currently viewed as the gold standard, it is subject to interobserver variation and allows fatigability to come into play at the risk of lowering accuracy.4,5
A potential way to spare time would be automatic segmentation, with numerous commercial and homemade software being developed.6-10 Very few of them, especially commercial ones, have been evaluated in clinical practice, however, for brain segmentation, especially in the high-precision context of stereotactic hypofractionated radiation therapy, and their accuracy is not known. The aim of this study was to evaluate 2 different segmentation software as compared to the current gold standard of manual segmentation.
Methods
Data Creation
This study was conducted on the treatment planning computed tomography (CT) scans of 20 consecutive patients referred to our institution for brain metastases. All patients treated in our institution are informed on admission about the potential use of their collected data for future research. Need for written informed consent from the participants was waived by our institutional review board, but oral consent was required in accordance with French laws (data, data collection, and freedom law, CNIL, January, 6, 1978). For this study, no personal data were collected beyond age and sex and all patient’s data were anonymized by the physicians in charge of the patients before being used for analysis.
The patients were treated on a CyberKnife M6 system (Accuray, Sunnyvale, California) either exclusively or as an adjuvant postsurgery strategy. Each patient was positioned (head-first supine position, arms resting on each side of the body). A thermoformed contention mask was modeled. They were then scanned on a LightSpeed CT (GE Healthcare, Little Chalfont, United Kingdom): helical acquisition, contiguous slices of 1.25 mm, 512 × 512 × 512 matrix, 120 kV, Tube Current - Exposure Time Product > 350 mA.s speed: 3.75, mode: 0.75, no contrast injection, software version 0.7MW11.10.
The obtained digital imaging and communications in medicine (DICOM) data were used as samples for analysis. Manual segmentation was performed after registration of the CT scan with a 3-D, T1-weighted, contrast-enhanced magnetic resonance imaging (MRI; most often using the diagnostic MRI performed outside of our institution, voxels of 0.25-1.25 mm3). Structures were manually segmented by a first clinician and subsequently verified and edited by a second clinician on Eclipse V.13 (Varian Medical Systems, Palo Alto, California).
The 2 commercial treatment planning systems used for this study were iPlan RT Image v. 4.1.1 (Brainlab AG, Munich, Germany) and Eclipse v. 13’s Smart Segmentation. The former uses a library of atlases which are included in the software at the time of purchase. The latter lets the clinician choose from a series of example cases and increment the library with new cases.11 This potentially allows an automated segmentation that is closer to that of the clinician who created the library.
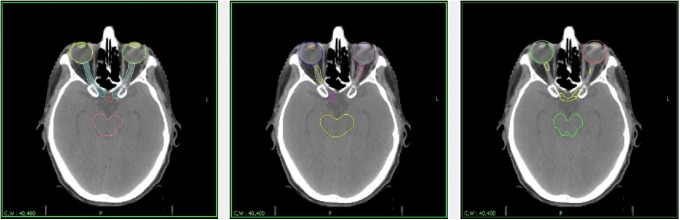
As the Smart Segmentation software did not include a brain expert case, a new atlas was created using a CT scan and an MRI from an archived patient data set. The patient selected for the atlas had no brain lesion, so as to avoid skewing automated segmentation results. The newly created atlas was then used for automatic segmentation on each CT scan. The images from each patient’s planning CT scan and MRI were also transferred on an iPlan planning console where an automated segmentation was also performed, using the MRI images and then resampled on the CT scan. Automated segmentation was performed using the full width of Hounsfield units, as the width and level did not impact iPlan automated segment but impacted Smart Segmentation results. We also ensured that repeated segmentation attempts yielded the same results, both on the index case used for creating the atlas and the new cases (data not shown). As the aim of the study was to perform an evaluation of each software in replacing the clinician, no editing was made to the segmentation (Figure 1).
Figure 1.
Automated segmentation results.
Figure 1 shows the same slice, segmented by the authors (left), Smart Segmentation (center), and iPlan (right). As can be seen here, both automated software failed to correctly segment the lenses and the optical nerves, instead including portions of adjacent tissue compared to anatomical boundaries.
Statistical Analysis
Computed tomography scan, MRI images, and structures were then exported as DICOM files (DICOM image and DICOM for radiation therapy—DICOM RT STRUCT). Each automated set was compared to the gold standard set (manual segmentation) via commercial software Artiview 4 (Aquilab, Loos Les Lille, France), which was used as a segment comparison module. The module allows comparison of DICOM RT STRUCT files and is able to compute each of the following indices (with A being the reference volume and B the automated volume):
Dice-Sørensen coefficient (DSC): DSC = 2 (A ∩ B)/(A + B), where 0.7 and above is considered good.12
Overlap ratio (OR): OR = (A ∩ B)/(A U B), much like the DSC, it should be as close to 1 as possible.
Common segmented volume (CSV): CSV = (A ∩ B)/A, which should be as close to 100% as possible.
Additional segmented volume (ASV): ASV = (B − A)/B, which should be as close to 0% as possible.
These data were calculated for each patient’s brain stem, lenses, eyes, optic chiasm, and optic nerves. We chose the CSV as our main judging criterion to reflect the accuracy of the automated segmentation of the structure. Data were analyzed using open-source R ×64 3.3.1. Mean values were calculated for each relevant index alongside standard deviations (SDs) with the open-source prettyR package.
We conducted a paired, nonparametric statistical test, that is, a Wilcoxon signed rank paired test, for each comparison. This test is more conservative than the t test and allows comparisons of matched smaller samples when one cannot assume that the samples are normally distributed, such as in small samples.13 We also applied a Bonferroni correction to the risk α to take the multiple comparisons performed in this work into account and keep a global α risk below 0.05.14 The threshold for statistical significance was thus lowered to 0.05/32 = 0.0015625.
Results
Patient Characteristics
Twenty patients were included in the study, aged between 38 and 86 years (median: 61 years). Their clinical characteristics are reported in Table 1. They had between 1 and 4 metastases (median: 1) and were treated with stereotactic radiation therapy either alone or as an adjunct. The biggest target volume was 81.75 cm3 (postoperatively), with an average target volume of 7.79 cm3 (SD: 16.28). None of the lesions invaded any of the structures to be segmented.
Table 1.
Clinical Characteristics.a
| Age | Gender | Number of Lesions | Volume (cm3) | Strategy | Lesion | |
|---|---|---|---|---|---|---|
| Patient #1 | 55 | Female | 1 | 10.414 | Exclusive | Posterior upper right frontal lobe |
| Patient #2 | 62 | Male | 1 | 0.141 | Exclusive | Anterior horn of the right ventricle |
| Patient #3 | 38 | Female | 1 | 81.75 | Adjuvant | Posterior horn of the left ventricle |
| Patient #4 | 54 | Male | 3 | 0.268 | Exclusive | Posterior left parafalcorial |
| 0.212 | Anterior upper left frontal lobe | |||||
| 5.699 | Posterior lower left frontal lobe | |||||
| Patient #5 | 55 | Female | 1 | 3.025 | Exclusive | Upper anterior frontal lobe |
| Patient #6 | 61 | Male | 3 | 0.141 | Exclusive | Middle upper left frontal lobe |
| 8.556 | Left occipital lobe | |||||
| 0.384 | Posterior left parafalcorial | |||||
| Patient #7 | 80 | Female | 4 | 0.282 | Exclusive | Anterior upper left frontal lobe |
| 0.834 | Middle left frontal lobe | |||||
| 0.059 | Middle right frontal lobe | |||||
| 0.748 | Right occipital lobe | |||||
| Patient #8 | 65 | Male | 1 | 34.544 | Exclusive | Upper anterior right frontal lobe |
| Patient #9 | 59 | Male | 1 | 9.839 | Exclusive | Lower left cerebellar hemisphere |
| Patient #10 | 49 | Female | 1 | 0.31 | Exclusive | Upper left cerebellar hemisphere |
| Patient #11 | 86 | Female | 1 | 3.419 | Exclusive | Middle upper left frontal lobe |
| Patient #12 | 40 | Male | 2 | 0.177 | Exclusive | Middle left cerebellar hemisphere |
| 2.626 | Posterior upper left frontal lobe | |||||
| Patient #13 | 66 | Female | 1 | 13.388 | Exclusive | Posterior horn of the right ventricle |
| Patient #14 | 51 | Female | 1 | 2.243 | Exclusive | Middle upper left frontal lobe |
| Patient #15 | 78 | Female | 1 | 2.087 | Exclusive | Posterior right parafalcorial |
| Patient #16 | 64 | Female | 1 | 15.712 | Exclusive | Left occipital lobe |
| Patient #17 | 67 | Male | 1 | 4.111 | Exclusive | Left temporal lobe |
| Patient #18 | 66 | Male | 1 | 0.62 | Exclusive | Middle right cerebellar hemisphere |
| Patient #19 | 61 | Male | 1 | 13.038 | Exclusive | Posterior horn of the right ventricle |
| Patient #20 | 56 | Female | 1 | 3.525 | Adjuvant | Middle left frontal lobe |
aReported here are the main clinical characteristics of each segmented case.
Automated segmentation took less than 5 minutes to perform and performed similarly in all patients aside from 1 (patient 3) who had a voluminous lesion surgically removed before being referred to our institution. The resulting structural deformation led to a poorer performance for segmentation. Five (25%) patients had their lenses removed (for cataract treatment) prior to treatment, but neither Smart Segmentation nor iPlan was able to identify the lenses as missing and therefore still segmented both regions of interest. We had planned to compare the data for the cochlea as well; however, Smart Segmentation failed to segment it in all patients, and as such, no comparison was made.
Common Segmented Volume
As can be seen in Table 2, Smart Segmentation performed significantly better for the eyes (P < .001), with a trend toward a better performance for the optical nerves, while iPlan performed better for the right lens (P < .001), with a trend toward a better performance for the other lens and the brain stem.
Table 2.
Common Segmented Volume.a
| Organ | Brain Stem | Chiasm | Left Eye | Right Eye | Left Lens | Right Lens | Left Optical Nerve | Right Optical Nerve |
|---|---|---|---|---|---|---|---|---|
| CSV smart segmentation (%) | 73.10 | 45.51 | 82.54 | 80.99 | 7.71 | 11.37 | 64.02 | 59.50 |
| Standard deviation | 8.72 | 32.43 | 8.03 | 7.75 | 9.18 | 11.22 | 16.45 | 16.57 |
| CSV iPlan (%) | 79.48 | 48.10 | 72.85 | 75.06 | 46.17 | 50.31 | 46.15 | 52.07 |
| Standard deviation | 9.24 | 31.23 | 12.87 | 11.13 | 31.47 | 31.76 | 22.42 | 21.89 |
| P value | .0020 | .5075 | .0002 | .0004 | .0039 | .0004 | .0017 | .4091 |
Abbreviation: CSV, common segmented volume.
aReported here are average percentages by organ of voxels belonging to each organ at risk on the manually segmented computed tomography (CT) scan, which were rightly included in the organ at risk region of interest by the automated segmentation software.
Overall performance was rather poor, however with at most 82.54% and 80.99% agreement in the eyes (Smart Segmentation) and 79.48% for the brain stem (iPlan). It went as low as 7.71% and 11.37% in the lenses (Smart Segmentation). Other small structures such as the chiasm and optical nerves were only about 50% correct.
Additional Segmented Volume
Additional segmented volume, that is, voxels mistakenly identified as belonging to the OAR, may lead to worse target coverage. The values given in Table 3 are relative to the organ’s volume, meaning the absolute additional eye volume is greater than that of the lenses. Smart Segmentation performed better than iPlan in both optical nerves and the brain stem. Other organs did not reach statistical significance.
Table 3.
Additional Segmented Volume.a
| Organ | Brain Stem | Chiasm | Left Eye | Right Eye | Left Lens | Right Lens | Left Optical Nerve | Right Optical Nerve |
|---|---|---|---|---|---|---|---|---|
| ASV smart segmentation (%) | 14.94 | 78.32 | 16.43 | 17.28 | 92.25 | 92.16 | 73.94 | 56.83 |
| Standard deviation | 7.63 | 23.40 | 16.02 | 8.82 | 12.02 | 8.02 | 8.28 | 11.64 |
| ASV iPlan (%) | 20.47 | 76.76 | 9.80 | 14.22 | 74.61 | 72.65 | 85.02 | 83.71 |
| Standard deviation | 7.19 | 23.25 | 7.68 | 8.56 | 18.32 | 20.57 | 11.79 | 12.17 |
| P value | .0008 | .7404 | .0826 | .4304 | .0280 | .0067 | .0009 | .0002 |
Abbreviation: ASV, additional segmented volume.
aReported here are average percentages by organ of voxels belonging to each organ at risk on the automated segmented computed tomography (CT) scan, which were mistakenly included in the organ-at-risk region of interest by the automated segmentation software.
Dice-Sørensen Coefficients and ORs
As reported in Table 4, iPlan performed better for the right lens, while Smart Segmentation performed better for the optical nerves for both indices. However, only the biggest structures such as the eyes and the brain stem reached over 0.7. Agreement in smaller structures was much poorer and not acceptable for inverse planning.
Table 4.
Dice-Sørensen Coefficients and Overlap Ratios.a
| Organ | Brain Stem | Chiasm | Left Eye | Right Eye | Left Lens | Right Lens | Left Optical Nerve | Right Optical Nerve |
|---|---|---|---|---|---|---|---|---|
| OR smart segmentation | 0.64 | 0.14 | 0.73 | 0.69 | 0.04 | 0.05 | 0.23 | 0.33 |
| Standard deviation | 0.05 | 0.12 | 0.08 | 0.08 | 0.06 | 0.05 | 0.06 | 0.09 |
| OR iPlan | 0.66 | 0.14 | 0.67 | 0.66 | 0.20 | 0.24 | 0.14 | 0.14 |
| Standard deviation | 0.07 | 0.09 | 0.09 | 0.08 | 0.15 | 0.19 | 0.12 | 0.08 |
| P value | .478 | .9058 | .0251 | .1850 | .0101 | .0002 | .0083 | .0001 |
| DSC smart segmentation | 0.78 | 0.22 | 0.84 | 0.81 | 0.07 | 0.09 | 0.37 | 0.48 |
| Standard deviation | 0.04 | 0.18 | 0.05 | 0.05 | 0.10 | 0.08 | 0.09 | 0.11 |
| DSC iPlan | 0.79 | 0.24 | 0.80 | 0.79 | 0.32 | 0.35 | 0.20 | 0.23 |
| Standard deviation | 0.06 | 0.15 | 0.07 | 0.06 | 0.22 | 0.25 | 0.10 | 0.13 |
| P value | .4980 | .7946 | .0215 | .1650 | .0120 | .0002 | .0003 | .0005 |
Abbreviations: DSC, Dice-Sørensen coefficients; OR, overlap ratios.
aReported here are indices of segmentation accuracy.
Exploratory Analyses
Exploratory analyses were carried out to evaluate whether the size of the lesions or the absence of lenses had any further impact on segmentation. Overall, software performance was roughly the same (±2% for CSV and ASV, ±0.025 for DSC and OR). Excluding patients whose lenses had been previously removed did lower the power of the statistical analysis, leading to a lack of statistical significance in most cases. Nonstatistically significant results remained nonsignificant after exclusion (data not shown), except for the left optical nerve’s DSC and OR (P < .001 for all analyses).
Discussion
Both automated segmentation algorithms performed similarly when considering CSV, each performing better on several organs, with no clear shape or size reasoning. iPlan may not have performed as well as Smart Segmentation for the eyes and optical nerves as it was used on the MRI, in which they may be positioned differently from the planning CT scan. iPlan did however trend toward a better performance for the lenses which are not necessarily easier to view on an MRI than on a CT scan.
We chose the CSV as our main criterion because this percentage allows to evaluate how much of each OAR is correctly segmented, with the best possible value being 100%. In the context of inverse planning, each missed pixel can lead to highly damaging consequences, especially if the organ is close to the treated volume. As stereotactic radiation’s survival benefit is somewhat unclear, we felt that the need to lower the toxicity was the most important parameter. Indeed, the report of American Association of Physicists in Medecine (AAPM) Task Group 101 on stereotactic body radiation therapy lists dose constraints for volumes as low as 0.2 cm3 (optical pathway) and 0.5 cm3 (brain stem), going as far as 0.035 cm3 for point doses.1 Here, the software missed over 35% of each optical nerve (about 0.1 cm3) and 20% of the brain stem (about 5 cm3), which could severely impact planning with such precise dose constraints if the lesion were close to the OAR.
Furthermore, the only acceptable DSCs produced by the commercial software were those of the brain stem and the eyes. Every other OAR ranked below 0.7, which is viewed as acceptable in practice.10 Isambert et al evaluated another platform for head-and-neck conformational radiation therapy, where many of the OARs are similar. Using DSC as a measurement, it proved to be useful for bigger structures. In smaller organs, the performance was poor,7 which is consistent with the results of our study. One should note, however, that DSC is a relative volumetric measurement. The bigger the organ, the smaller the relative error becomes. Here, for instance, the DSC was higher for the eyes and the brain stem than the lenses, chiasm, and optical nerves. Again, in the context of hypofractionated radiation therapy, where dose constraints have been established on small volumes,1,15,16 such a comparison did not seem appropriate.
It is important to note that accuracy is viewed on a matrix made of discrete voxels instead of a continuous volume. Likewise, a voxel can only be classified as either correct or incorrect with no in-between. A change in the size of the voxels in the matrix will almost always automatically move a slight part of the volume from inside to outside of the structure, from correct to incorrect or vice versa.
As the automated segmentation was performed on 2 different software, the volume of each structure from the DICOM RT STRUCT may vary in part simply because of the matrix and not because of an actual change in size. As such, the various data reported here are the closest possible approximations of the truth using each of the software’s voxel matrixes. This imprecision is however often minute compared to the size of the entire organ as it only affects the voxels on the border of the structure (long narrow organs such as the optic nerves may be the most affected ones). It is also unavoidable as each planning image itself is acquired as a voxel matrix.
The exploratory analyses seemed to show that the software performed in the same way regardless of the size of the lesions and the removal of the lenses, as all stayed nonstatistically significant but one, which may simply be linked to sample fluctuation.
A limitation of this study is that the Eclipse Smart Segmentation software used only 1 atlas from a patient, which can explain why it did not perform as well as iPlan’s automated segmentation software in some regards. A way to improve the segmentation would probably have been the use of multiple atlases. Van de Velde et al evaluated the number of atlases necessary for automated segmentation using the simultaneous truth and performance level estimation (STAPLE) and found that 4 were necessary and as many as 9 were best.17
While there have been studies in patients with large metastases8,9 and in other body regions such as head and neck and pelvis,7,18-22 to our knowledge, this study is the first to compare multiple commercial segmentation platforms in the context of intracranial stereotactic hypofractionated radiation therapy, using both an user-generated and a commercial software. In this context, tumors are often smaller and as such induce less displacement, making it easier to use an automated segmentation software. This is evidenced in our study by the fact that Smart Segmentation and iPlan both performed the most poorly in the patient with the biggest lesions.
Despite the predictable reproducibility of this situation, as intracranial structures are subject to less internal movement and interpersonal variations than other parts of the human body, automated segmentation was a rather ineffective tool. Manual editing, often even manual segmentation altogether, remained mandatory. This warrants further studies and cooperation between vendors and radiation oncologists to improve those platforms before they can be used in routine practice.
One could also view automated segmentation as a first step, followed by manual editing of the structures, which could allow a time gain while retaining human control. Deeley et al showed that both automated software and experts were challenged for tubular structure segmentation in the brain (ie, chiasm and optic nerves), as compared to a STAPLE-generated ground truth.8 Using atlas-based registration to segment the eyes and the brain stem and the atlas-navigated optimal medial axis and deformable model algorithm for other structures followed by manual editing, they then showed in another study that it could reduce interobserver variation, improve accuracy, and reduce segmentation time by about 60%.9
Another limitation of our study is that we used a homemade atlas for the automated segmentation in Smart Segmentation. On the other hand, our atlas was put together by 2 different clinicians, which has been shown to reduce interobserver variation as well.23 One should also note that Smart Segmentation only allows elastic registration from an atlas derived from a single case and does not allow to create a STAPLE-type ground truth as had been studied by other authors.8,11,24 There are other tools such as segmentation by region or by contours,25 which have been studied by Özsavaş et al.26 On a sample of cases, it took us slightly more time to edit the case than to do a straight manual contouring (46 vs 41 minutes, not statistically significant). The time gain with automated segmentation has been debated in the literature, with some reporting gains in efficiency while others note a longer time, depending on the organ and the segmenting tools and who is segmenting the structures.22,23,27
While a variety of contouring atlases have been published in the past few years,28-31 often sharing DICOM RT files for institutional use,28,31 both for target volumes and for OAR, none of these have been made in the context of hypofractionated stereotactic brain radiation therapy. Dose constraints in stereotactic radiation therapy remain mostly empiric and defined according to trial or institutional guidelines that may vary. While recent efforts have been made to make them somewhat more consensual,1,15,16 clear segmentation guidelines are needed to make these constraints as accurate as they should be. The next step would be to assess the dosimetric consequences of these contouring discrepancies in order to predict their potential clinical effects.
Conclusion
Each of the automated segmentation software has its strengths and weaknesses. If automated segmentation alone was accurate, this could reduce segmentation time. However, the physician is still needed to further segment structures,4,9 especially in the context of inverse-planned hypofractionated radiation therapy. Using both an automated software and manual editing may be the current best of both worlds to deliver safe treatment to our patients.
Abbreviations
- ASV
additional segmented volume
- CSV
common segmented volume
- CT
computed tomography
- DICOM RT
digital imaging and communications in medicine for radiation therapy
- DSC
Dice-Sørensen coefficients
- MRI
magnetic resonance imaging
- OARs
organs at risk
- OR
overlap ratio
- SRS
stereotactic radiosurgery
- STAPLE
simultaneous truth and performance level estimation
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflict of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
References
- 1. Benedict SH, Yenice KM, Followill D, et al. Stereotactic body radiation therapy: the report of AAPM task group 101. Med Phys. 2010;37(8):4078–4101. [DOI] [PubMed] [Google Scholar]
- 2. Halasz LM, Uno H, Hughes M, et al. Comparative effectiveness of stereotactic radiosurgery versus whole-brain radiation therapy for patients with brain metastases from breast or non-small cell lung cancer: WBRT vs SRS for brain metastases. Cancer. 2016;122(13):2091–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sahgal A, Aoyama H, Kocher M, et al. Phase 3 trials of stereotactic radiosurgery with or without whole-brain radiation therapy for 1 to 4 brain metastases: individual patient data meta-analysis. Int J Radiat Oncol Biol Phys. 2015;91(4):710–717. [DOI] [PubMed] [Google Scholar]
- 4. Ramkumar A, Dolz J, Kirisli HA, et al. User interaction in semi-automatic segmentation of organs at risk: a case study in radiotherapy. J Digit Imaging. 2016;29(2):264–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Park SH, Gao Y, Shi Y, Shen D. Interactive prostate segmentation using atlas-guided semi-supervised learning and adaptive feature selection. Med Phys. 2014;41(11):111715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pointreau Y, Bera G, Barillot I. Aide à la délinéation: quels outils pratiques? Cancer Radiother. 2009;13(6-7):600–605. [DOI] [PubMed] [Google Scholar]
- 7. Isambert A, Dhermain F, Bidault F, et al. Evaluation of an atlas-based automatic segmentation software for the delineation of brain organs at risk in a radiation therapy clinical context. Radiother Oncol. 2008;87(1):93–99. [DOI] [PubMed] [Google Scholar]
- 8. Deeley MA, Chen A, Datteri R, et al. Comparison of manual and automatic segmentation methods for brain structures in the presence of space-occupying lesions: a multi-expert study. Phys Med Biol. 2011;56(14):4557–4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deeley MA, Chen A, Noble J, et al. Segmentation editing improves efficiency while reducing inter-expert variation and maintaining accuracy for normal brain tissues in the presence of space-occupying lesions. Phys Med Biol. 2013;58(12):4071–4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Anders LC, Stieler F, Siebenlist K, Schäfer J, Lohr F, Wenz F. Performance of an atlas-based autosegmentation software for delineation of target volumes for radiotherapy of breast and anorectal cancer. Radiother Oncol. 2012;102(1):68–73. [DOI] [PubMed] [Google Scholar]
- 11. Food and Drug Administration. Premarket Notification [510(k)] Summary Smart Segmentation Knowledge Based Contouring. December 29, 2011. https://www.accessdata.fda.gov/cdrh_docs/pdf11/K112778.pdf. Accessed December 17, 2017.
- 12. Zijdenbos AP, Dawant BM, Margolin RA, Palmer AC. Morphometric analysis of white matter lesions in MR images: method and validation. IEEE Trans Med Imaging. 1994;13(4):716–724. [DOI] [PubMed] [Google Scholar]
- 13. Wilcoxon F. Individual comparisons by ranking methods. Biom Bull. 1945;1(6):80. [Google Scholar]
- 14. Miller RG. Simultaneous Statistical Inference [Internet] New York, NY: Springer New York; 1981. . Springer Series in Statistics http://link.springer.com/10.1007/978-1-4613-8122-8. Accessed December 17, 2017. [Google Scholar]
- 15. Grimm J, LaCouture T, Croce R, Yeo I, Zhu Y, Xue J. Dose tolerance limits and dose volume histogram evaluation for stereotactic body radiotherapy. J Appl Clin Med Phys. 2011;12(2):3368 http://jacmp.org/index.php/jacmp/article/viewArticle/3368. Accessed December 17, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Emami B. Tolerance of normal tissue to therapeutic radiation. Rep Radiother Oncol. 2013;1(1). http://journals.sbmu.ac.ir/rro/article/download/4316/3851. Accessed December 17, 2017. [DOI] [PubMed] [Google Scholar]
- 17. Van de Velde J, Wouters J, Vercauteren T, et al. Optimal number of atlases and label fusion for automatic multi-atlas-based brachial plexus contouring in radiotherapy treatment planning. Radiat Oncol. 2016;11:1 http://www.ro-journal.com/content/11/1/1. Accessed December 17, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Greenham S, Dean J, Fu CK, et al. Evaluation of atlas-based auto-segmentation software in prostate cancer patients. J Med Radiat Sci. 2014;61(3):151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Beyer GP, Velthuizen RP, Murtagh FR, Pearlman JL. Technical aspects and evaluation methodology for the application of two automated brain MRI tumor segmentation methods in radiation therapy planning. Magn Reson Imaging. 2006;24(9):1167–1178. [DOI] [PubMed] [Google Scholar]
- 20. Tao C-J, Yi J-L, Chen N-Y, et al. Multi-subject atlas-based auto-segmentation reduces interobserver variation and improves dosimetric parameter consistency for organs at risk in nasopharyngeal carcinoma: a multi-institution clinical study. Radiother Oncol. 2015;115(3):407–411. [DOI] [PubMed] [Google Scholar]
- 21. Hoang Duc AK, Eminowicz G, Mendes R, et al. Validation of clinical acceptability of an atlas-based segmentation algorithm for the delineation of organs at risk in head and neck cancer. Med Phys. 2015;42(9):5027–5034. [DOI] [PubMed] [Google Scholar]
- 22. Gambacorta MA, Valentini C, Dinapoli N, et al. Clinical validation of atlas-based auto-segmentation of pelvic volumes and normal tissue in rectal tumors using auto-segmentation computed system. Acta Oncol. 2013;52(8):1676–1681. [DOI] [PubMed] [Google Scholar]
- 23. Walker GV, Awan M, Tao R, et al. Prospective randomized double-blind study of atlas-based organ-at-risk autosegmentation-assisted radiation planning in head and neck cancer. Radiother Oncol. 2014;112(3):321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bondiau P-Y, Malandain G, Chanalet S, et al. Atlas-based automatic segmentation of MR images: validation study on the brainstem in radiotherapy context. Int J Radiat Oncol. 2005;61(1):289–298. [DOI] [PubMed] [Google Scholar]
- 25. Pasquier D, Peyrodie L, Denis F, Pointreau Y, Bera G, Lartigau E. Segmentation automatique des images pour la planification dosimetrique en radiotherapie. Cancer Radiother. 2010;14(suppl 1):S6–S13. [DOI] [PubMed] [Google Scholar]
- 26. Özsavaş EE, Telatar Z, Dirican B, Sağer Ö, Beyzadeoğlu M. Automatic segmentation of anatomical structures from CT scans of thorax for RTP. Comput Math Methods Med. 2014;2014:472890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reed VK, Woodward WA, Zhang L, et al. Automatic segmentation of whole breast using atlas approach and deformable image registration. Int J Radiat Oncol. 2009;73(5):1493–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brouwer CL, Steenbakkers RJHM, Bourhis J, et al. CT-based delineation of organs at risk in the head and neck region: DAHANCA, EORTC, GORTEC, HKNPCSG, NCIC CTG, NCRI, NRG Oncology and TROG consensus guidelines. Radiother Oncol. 2015;117(1):83–90. [DOI] [PubMed] [Google Scholar]
- 29. Baldini EH, Wang D, Haas RL, et al. Treatment guidelines for preoperative radiation therapy for retroperitoneal sarcoma: preliminary consensus of an international expert panel. Int J Radiat Oncol. 2015;92(3):602–612. [DOI] [PubMed] [Google Scholar]
- 30. Gaffney DK, King B, Viswanathan AN, et al. Consensus recommendations for radiation therapy contouring and treatment of vulvar carcinoma. Int J Radiat Oncol. 2016;95(4):1191–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Offersen BV, Boersma LJ, Kirkove C, et al. ESTRO consensus guideline on target volume delineation for elective radiation therapy of early stage breast cancer. Radiother Oncol. 2015;114(1):3–10. [DOI] [PubMed] [Google Scholar]