Abstract
Aim
Homeobox (HOX) genes and their protein products have been found to function as oncogenes in the progression of many cancers. But the role of Homeobox C10 (HOXC10) in osteosarcoma (OS) still remains less understood. In this study, we firstly determine the biologic functions of HOXC10 in OS.
Materials and methods
We examined the expression of HOXC10 in OS tissues by quantitative real-time polymerase chain reaction and Western blot assays. We investigated the effects of HOXC10 on cell proliferation, apoptosis and caspase 3 activity in three OS cell lines by RNA interference, Cell Counting Kit-8, flow cytometry and colorimetric assays.
Results
We found that HOXC10 was elevated in OS tissues. Silencing HOXC10 significantly inhibited cell proliferation, induced cell apoptosis and increased the expression and activity of caspase 3. The resistance assay further suggested that HOXC10 affected cell growth and apoptosis through regulating the expression and activity of caspase 3.
Conclusion
HOXC10 might function as an oncogene in OS by regulating the expression and activity of caspase 3.
Keywords: apoptosis, caspase 3, HOXC10, osteosarcoma, proliferation
Introduction
Osteosarcoma (OS) is an aggressive bone malignancy. Mesenchymal stem cells and committed osteoblast precursors have been suggested as the cell origin of OS.1,2 OS most commonly occurs at sites of bone growth in children and adolescents, such as the proximal end of tibia or humerus or the distal end of femur.3,4 Surgical removal of the malignant lesion is the mainstay therapy for OS. Neoadjuvant chemotherapy combined with limb-sparing surgery has effectively increased the survival rates of OS. However, about 20% of OS patients have metastatic spread when it is firstly diagnosed. The survival rate of these patients still remains between 15% and 30%. Also, current clinical therapy is helpless for metastatic patients.5–7 Therefore, novel targets that can advance the development of OS therapy are still urgently needed.8
Homeobox (HOX) genes are identified as a group of evolutionarily conserved genes that control the cell differentiation and embryonic development.9 The protein products of HOX gene act as transcription factors by binding to the promoters of various target genes and regulating their expression. In humans, four HOX clusters (A–D) are located on four chromosomes (7, 17, 12 and 2, respectively). On the basis of sequence similarities and location within the clusters, HOX genes are divided into 13 paralogous groups. Homeobox A10 (HOXA10), Homeobox C10 (HOXC10) and Homeobox D10 (HOXD10) are three paralogous genes, inactivation of which may affect motor neuron patterning and endometrial differentiation.10,11 In recent years, more and more evidence has indicated that HOX genes and their protein products are associated with carcinogenesis.12 For example, HOXA10 was found to be frequently upregulated in various human cancers, such as leukemia, lung cancer, epithelial ovarian cancer and glioma.13–16 López et al also suggested that expression of HOXC10 was elevated in cervical cancer cells, which was involved in the invasiveness of cervical cancer cells.17 Lower HOXD10 mRNA levels were significantly associated with higher grade breast cancer.18
In this study, we assessed the expression level of HOXC10 in OS. Also, we selected two OS cell lines combined with primary OS cells to analyze the biologic functions and mechanisms of HOXC10 in tumor progression. Our data collectively established an important role for HOXC10 in OS and highlight HOXC10 as a potential therapeutic target for OS patients.
Materials and methods
Tissue samples
OS and normal bone tissues were obtained from 45 patients with OS (Enneking’s stage II) and 15 patients with other diseases, respectively treated at the Department of Orthopedics, The Second Affiliated Hospital of Zhejiang University. All these tissues were stored at −80°C until being analyzed. This study was approved by the Ethics Committee of The Second Affiliated Hospital of Zhejiang University. Written informed consent was obtained from all patients, according to the guidelines of the Ethics Committee.
Quantitative real-time polymerase chain reaction analysis
Total RNA was extracted with Trizol reagent (Invitrogen) and reverse transcribed using cDNA Synthesis Kit (Fermentas). Real-time polymerase chain reaction (PCR) was carried out using a standard SYBR Green PCR kit, as previously described.19 The cycle conditions were: 10 min at 95°C followed by 40 cycles of 15 s at 95°C and 45 s at 60°C. The real-time PCR data were analyzed using ABI Prism 7300 SDS software. GAPDH was used as an internal control. The following real-time PCR primers were used: HOXC10 (NM_0,17,409.3), 5′-TGACTTCAATTGCGGGGTGA-3′ and 5′-ACTAGGTGGGTAGGAGCAGG-3′; caspase 3 (NM_0,04,346.3), 5′-AACTGGACTGTGGCATTGAG-3′ and 5′-ACAAAGCGACTGGATGAACC-3′; GAPDH (NM_00,12,56,799.1), 5′-CACCCACTCCTCCACC TTTG-3′ and 5′-CCACCACCCTGTTGCTGTAG-3′.
Western blot assay
Total protein was extracted by using radioimmunoprecipitation buffer. Samples were then separated on sodium dodecyl sulphate-polyacrylamide gel electrophoresis and electrophoretically transferred to a nitrocellulose membrane. After blocking with 5% skimmed milk, the blots were incubated with primary antibodies, followed by incubation with secondary antibody (Beyotime). The signal was visualized using enhanced chemiluminescence (EMD Millipore). The band intensity was quantified with ImageJ Software. The primary antibodies used were as follows: HOXC10 (Ab153904, 1:1500; Abcam), caspase 3 (Ab44976, 1:500; Abcam) and GAPDH (#5174, 1:2000; Cell Signaling Technology).
Cell isolation and cell culture
Primary OS cells were isolated from available OS tissues of three patients. Briefly, the OS tissues were cut into 0.5–1 mm3 segments and transferred into a centrifuge tube with 1.5 mL tryspin (10.25%) and 3 mL collagenase (4%). After 2 h of incubation at 37°C, the cell suspension was centrifuged at 500 rpm for 5 min. The supernatant was collected and centrifuged at 3000 rpm for 10 min. The supernatant was discarded and the cells were resuspended in Dulbecco’s Modified Eagle Medium (DMEM)/F12 medium. Cell passage was performed once the cell achieved 70%–80% confluence.
Five OS cell lines, OS-732, MG63, U-20S, SaoS2 and 143B, were purchased from American Type Culture Collection. OS-732 and MG63 cells were cultured in Minimum Essential Medium. U-20S, SaoS2 and 143B cells were maintained in DMEM culture medium. All culture media were supplemented with 10% fetal bovine serum, 100 U/mL penicillin and 100 μg/mL streptomycin. All the OS cells were maintained at 37°C under 5% CO2/95% air. The cell lines used here were between passage 5 and 15.
RNA interference
siRNA transfection was performed to silence the HOXC10 expression in OS cells. siRNA targeting human HOXC10 mRNA (734–752: CGGATAACGAAGCGAAAGA) was synthesized and transfected into OS cells using Lipofectamine 2000 (Invitrogen). A nonspecific control siRNA (NC) sequence was also synthesized which served as the negative control. All procedures done were according to the instructions of the manufacturer. RNAi efficiency was evaluated by detecting the mRNA and protein levels of HOXC10 at 48 h after siRNA transfection.
Proliferation analysis
Cell Counting Kit-8 (CCK-8) assay was performed to measure the cell growth of OS cells according to the instructions of the manufacturer. U-20S, SaoS2 and primary OS cells were seeded into 96-well plates (5×103). After allowing adherence overnight, these cells were transfected with HOXC10 siRNA or NC siRNA and CCK-8 solution was added to each well at 0, 24, 48 and 72 h after siRNA transfection. Following incubation for 1 h at 37°C, the optical density at 450 nm (OD450) was determined to evaluate cell growth.
Cell apoptosis
OS cells were harvested at 48 h after RNA interference. Cell apoptosis was measured using Annexin V-allophycocyanin/propidium iodide Apoptosis Detection kit (KeyGEN BioTECH, Nanjing, China) according to the manufacturer’s instructions. Cells were examined by a flow cytometry (BD Biosciences) and fluorescence of at least 10,000 events was examined.
Caspase 3 activity assay
Caspase 3 activity assay was performed using the caspase 3 activity kit (NanJing JianCheng Bioengineering Institute). OS cells were collected and disrupted in lysis buffer (containing 0.5 μL dithiothreitol/50 μL). The lysates were then incubated with acetyl-Asp-Glu-Val-Asp-p-nitroanilide (Ac-DEVD-pNA) for >4 h at 37°C. The OD value at 400 nm was measured with a microplate reader. The activity of caspase 3 was assessed with the ratio of inducer OD value and control OD value.
Resistance assay
To further reveal the functional mechanism of HOXC10 in OS, we performed resistance experiment in MG63 cells using a Caspase 3-like proteases inhibitor (Ac-DEVD-CHO). The groups were as follows: 1) NC group: MG63 cells were transfected with a NC sequence; 2) siRNA group: MG63 cells were transfected with HOXC10-siRNA sequence; 3) Ac-DEVD-CHO group: MG63 cells were transfected with a nonspecific scramble siRNA sequence and treated with 40 μM Ac-DEVD-CHO and 4) siRNA-Ac-DEVD-CHO group: MG63 cells were transfected with HOXC10-siRNA sequence and treated with 40 μM Ac-DEVD-CHO. CCK-8 was performed at 0, 24, 48 and 72 h after treatment. After 48 h, the cells were harvested to perform flow cytometry and Western blot assays.
Statistical analysis
Data analysis was conducted with GraphPad Prism software (GraphPad Software, Inc., San Diego, CA, USA). Values were presented as mean ± SD. Differences were determined by analysis of variance test. P<0.05 was considered to be statistically significant.
Results
HOXC10 expression was elevated in OS tissues
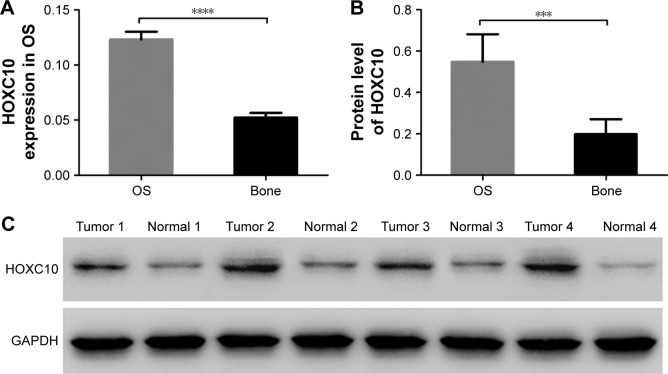
We assessed the expression level of HOXC10 in OS tissues (n=45) and normal bone tissues (n=15). As shown in Figure 1, both the mRNA and protein levels of HOXC10 were significantly increased in OS tissues compared to normal bone tissues. These data indicate that HOXC10 expression was abnormally elevated in OS.
Figure 1.
HOXC10 was overexpressed in OS tissues.
Notes: (A) mRNA expression of HOXC10 was highly expressed in OS tissues (n=45) compared to normal bone tissues (n=15). (B and C) The protein expression of HOXC10 was obviously increased in OS tissues (n=45) compared to normal bone tissues (n=15). Data shown as mean ± SD, ***P<0.001, ****P<0.0001.
Abbreviations: HOXC10, Homeobox C10; OS, osteosarcoma.
HOXC10 expression was decreased in OS cells after RNA interference
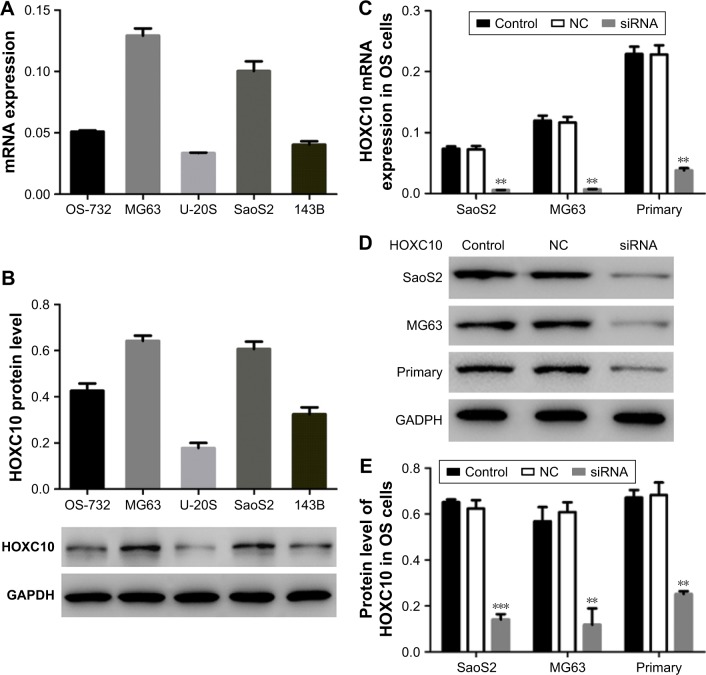
We then used quantitative real-time polymerase chain reaction and Western blot to analyze HOXC10 expression level in five OS cell lines, including OS-732, MG63, U-20S, SaoS2 and 143B. We found that HOXC10 expression was much higher in MG63 and SaoS2 cells than in other three cell lines (Figure 2A and B). So, MG63 and SaoS2 cells were selected to perform further assays. Primary OS cells (Figure S1) were also isolated from OS tissues for the following experiments.
Figure 2.
The expression of HOXC10 was decreased after RNA interference.
Notes: (A and B) mRNA and protein expression of HOXC10 in five OS cell lines (n=3). HOXC10 was overexpressed in SaoS2 and MG63 cell lines. These two cell lines were selected to perform further assays. (C) mRNA of HOXC10 was remarkably silenced after siRNA transfected in three OS cell lines (n=3). (D and E) The protein level of HOXC10 was obviously decreased after RNA interference in three OS cell lines (n=3). Data shown as mean ± SD, **P<0.01, ***P<0.001 (compared with negative controls).
Abbreviations: HOXC10, Homeobox C10; NC, nonspecific control siRNA; OS, osteosarcoma.
To explore the role of HOXC10 in OS progression, we transfected siRNA targeting human HOXC10 mRNA into SaoS2, MG63 and primary OS cells to silence the expression of HOXC10. As illustrated in Figure 2C–E, the mRNA and protein levels of HOXC10 were remarkably decreased in siRNA groups as compared to NC and control groups.
HOXC10 silencing reduced cell proliferation and induced cell apoptosis in OS cells
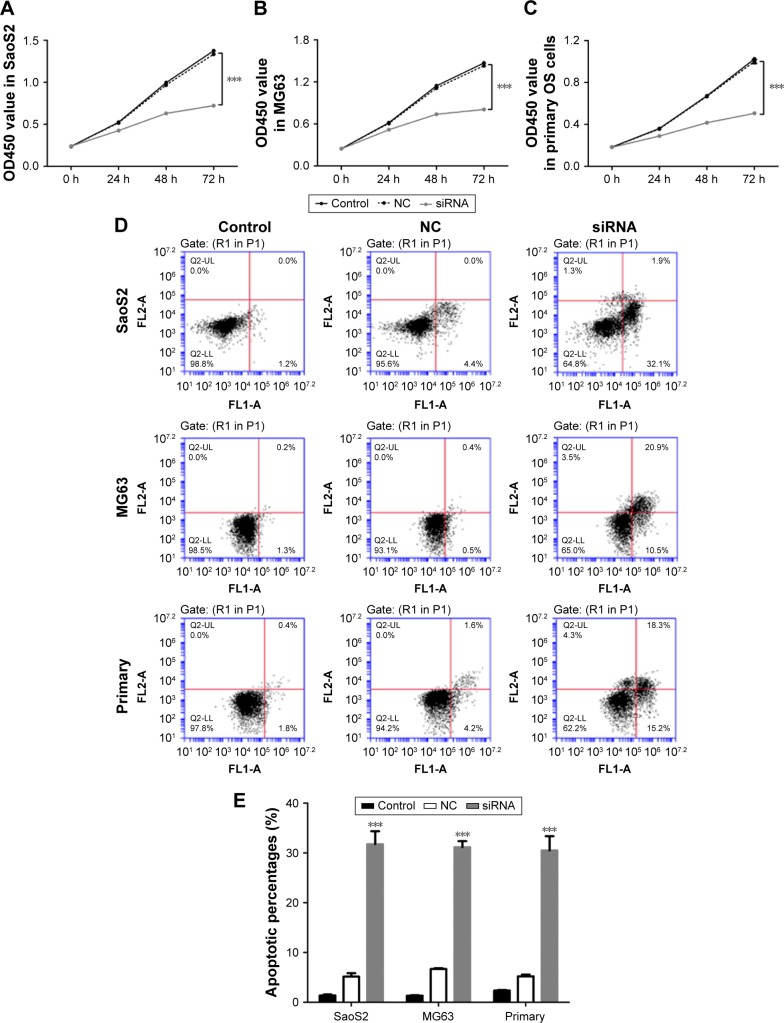
Then, we analyzed cell proliferation in three OS cell lines at 0, 24, 48 and 72 h after RNA interference using CCK-8 assay. As shown in Figure 3A–C, cell growth in SaoS2, MG63 and primary OS cells was obviously inhibited at 24, 48 and 72 h after siRNA transfected. These results proved that knockdown of HOXC10 suppressed proliferation in OS cell lines.
Figure 3.
Depletion of HOXC10 significantly inhibited cell proliferation and induced cell apoptosis in OS cell lines.
Notes: (A–C) Cell proliferation of three cell lines was examined at 0, 24, 48 and 72 h after siRNA transfection (n=3). The cell growth was obviously decreased in siRNA groups compared to NC group. (D and E) Cell apoptosis was examined at 48 h after RNA interference by flow cytometry (n=3). HOXC10 knockdown significantly increased the percentages of apoptotic cells. Data shown as mean ± SD, ***P<0.001 (compared with negative controls).
Abbreviations: HOXC10, Homeobox C10; NC, nonspecific control siRNA; OS, osteosarcoma.
We also performed Annexin V-fluorescein isothiocyanate/propidium iodide staining assay to evaluate the function of HOXC10 in OS cell apoptosis. As shown in Figure 3D and E, flow cytometry analysis indicated that knockdown of HOXC10 in SaoS2, MG63 and primary OS cells significantly induced cell apoptosis, compared with NC and control groups.
Knockdown of HOXC10 repressed the expression and activity of caspase 3 in OS cell lines
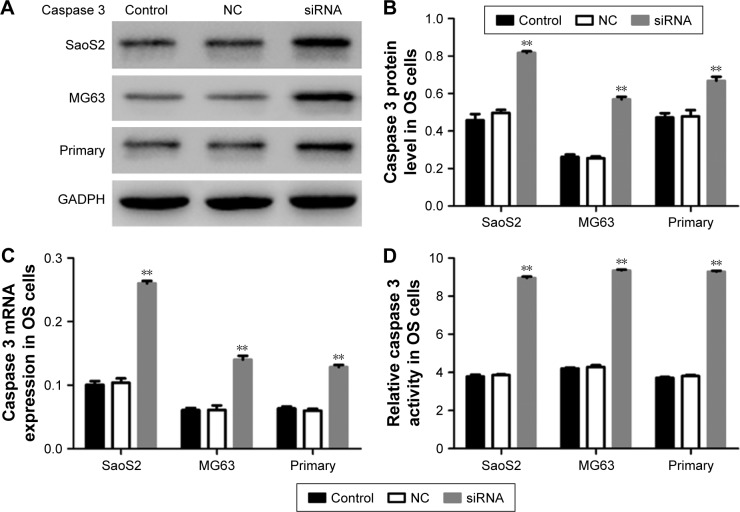
As silencing HOXC10 promoted cell apoptosis in three cell lines, we then examined the endogenous expression and activity of caspase 3 at 48 h after siRNA transfection. As shown in Figure 4A–C, the protein and mRNA levels of caspase 3 were significantly increased in SaoS2, MG63 and primary OS cell lines. The intracellular caspase 3 activity was also obviously elevated in siRNA transfected cells compared to NC transfected cells (Figure 4D). Taken together, these data showed that silencing HOXC10 significantly promoted the expression and activity of caspase 3 in OS cells.
Figure 4.
Depletion of HOXC10 significantly increased the expression and activity of caspase 3 in OS cells.
Notes: (A and B) The protein level of caspase 3 in three cell lines (n=3). (C) mRNA expression of caspase 3 in OS cells (n=3). (D) Intracellular activity of caspase 3 in OS cell lines (n=3). Data shown as mean ± SD, **P<0.01 (compared with negative controls).
Abbreviations: HOXC10, Homeobox C10; NC, nonspecific control siRNA; OS, osteosarcoma.
HOXC10 affected cell proliferation and apoptosis through regulating caspase 3
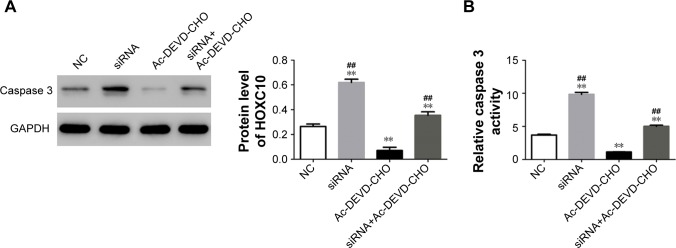
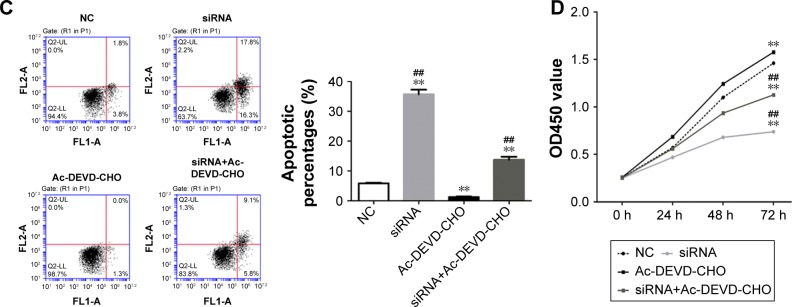
In resistance assay, we used Ac-DEVD-CHO to downregulate the expression of caspase 3 in MG63 cells. As shown in Figure 5A and B, we found that Ac-DEVD-CHO treatment significantly decreased the intracellular protein level and activity of caspase 3. But the effects of HOXC10 knockdown on the expression and activity of caspase 3 were partially relieved by Ac-DEVD-CHO treatment. The antiproliferative and proapoptotic effects of HOXC10 knockdown were also partially repressed by Ac-DEVD-CHO (as illustrated in Figure 5C and D). These results suggested that, HOXC10 might affect cell growth and apoptosis by regulating the expression and activity of caspase 3.
Figure 5.
Resistant assay with a caspase 3-like proteases inhibitor suggested HOXC10 affected cell proliferation and apoptosis through regulating caspase 3.
Notes: MG63 cells were randomly divided into four groups: NC group, cells were transfected with an NC; siRNA group, cells were transfected with HOXC10-siRNA (siRNA); Ac-DEVD-CHO group, cells were transfected with NC and treated with 40 μM Ac-DEVD-CHO; and, siRNA-Ac-DEVD-CHO group, cells were transfected with siRNA and treated with 40 μM Ac-DEVD-CHO. (A) The protein level of caspase 3 was determined by Western blot assay (n=3). (B) Intracellular activity of caspase 3 was assessed (n=3). (C) Cell apoptosis was evaluated by flow cytometry analysis (n=3). (D) Cell proliferation by CCK-8. Data shown as mean ± SD, **P<0.01 (compared with negative controls); ##P<0.01 (compared with Ac-DEVD-CHO group).
Abbreviations: CCK-8, Cell Counting Kit-8; HOXC10, Homeobox C10; NC, nonspecific control siRNA; OS, osteosarcoma.
Discussion
HOX genes have been well identified as important players in development and morphogenesis. In recent years, more and more evidence has suggested that HOX genes may also function in carcinogenesis. HOXC10 overexpression has been proved in breast cancer, which may be associated with cancer progression.20 Zhai et al indicated that HOXC10 functioned as a key mediator in the invasion of cervical squamous cell carcinoma.21 Upregulation also promotes progression of human thyroid cancer and indicates poor survival outcome.22 Therefore, we aimed to determine the biologic effects of HOXC10 on OS progression. We found that HOXC10 was abnormally overexpressed in OS tissues compared to normal bone tissues. Moreover, silencing HOXC10 expression significantly repressed the proliferation and promoted apoptosis in all three OS cell lines. All these data were consistent with a recent report23 and suggested that HOXC10 might promote the progression of OS.
Caspase 3 plays a critical role in the execution of apoptosis. It can be activated by diverse death-inducing signals.24 The regulation of caspase 3 is closely involved in tumorigenesis.25–27 To reveal the oncogene functions of HOXC10 in OS cells, we then analyzed the mRNA and protein levels of caspase 3 in SaoS2, MG63 and primary OS cells after RNA interference. We found that depletion of HOXC10 remarkably increased the intracellular level of caspase 3 in the three cell lines. The results of caspase 3 activity assay also showed obvious promotion induced by HOXC10 knockdown in OS cells. We further performed resistant assay in MG63 cells using a caspase 3-like proteases inhibitor. We found that HOXC10 affected cell proliferation and apoptosis through regulating the expression and activity of caspase 3 in OS cells. The results indicated that HOXC10 functioned as an oncogene by inhibiting the expression and activity of caspase 3 in OS.
To summarize, our results revealed HOXC10 was overexpressed in OS. Depletion of HOXC10 inhibits proliferation and promotes apoptosis, and HOXC10 may affect these biologic progresses through regulating the expression and intracellular activity of caspase 3. Our study provides valuable information for further investigation toward a comprehensive understanding of HOXC10.
Supplementary material
Primary OS cells.
Abbreviation: OS, osteosarcoma.
Footnotes
Disclosure
The authors alone are responsible for the content and writing of the paper. The authors report no conflicts of interest in this work.
References
- 1.Mutsaers AJ, Walkley CR. Cells of origin in osteosarcoma: mesenchymal stem cells or osteoblast committed cells? Bone. 2014;62:56–63. doi: 10.1016/j.bone.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Ottaviani G, Jaffe N. Pediatric and Adolescent Osteosarcoma. Springer; NY: 2009. The epidemiology of osteosarcoma; pp. 3–13. [DOI] [PubMed] [Google Scholar]
- 3.Biondi A, Pizzo PA, Poplack DG, editors. Principles and practice of pediatric oncology. Ann Oncol. 2003;14(4):661–662. [Google Scholar]
- 4.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2010;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 5.Avnet S, Longhi A, Salerno M, et al. Increased osteoclast activity is associated with aggressiveness of osteosarcoma. Int J Oncol. 2008;33(6):1231–1238. [PubMed] [Google Scholar]
- 6.Man TK, Chintagumpala M, Visvanathan J, et al. Expression profiles of osteosarcoma that can predict response to chemotherapy. Cancer Res. 2005;65(18):8142–8150. doi: 10.1158/0008-5472.CAN-05-0985. [DOI] [PubMed] [Google Scholar]
- 7.Provisor AJ, Ettinger LJ, Nachman JB, et al. Treatment of nonmetastatic osteosarcoma of the extremity with preoperative and postoperative chemotherapy: a report from the Children’s Cancer Group. J Clin Oncol. 1997;15(1):76–84. doi: 10.1200/JCO.1997.15.1.76. [DOI] [PubMed] [Google Scholar]
- 8.Akiyama T, Dass CR, Choong PF. Novel therapeutic strategy for osteosarcoma targeting osteoclast differentiation, bone-resorbing activity, and apoptosis pathway. Mol Cancer Ther. 2008;7(11):3461–3469. doi: 10.1158/1535-7163.MCT-08-0530. [DOI] [PubMed] [Google Scholar]
- 9.Mcginnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68(2):283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 10.Duboule D. The vertebrate limb: a model system to study the Hox/HOM gene network during development and evolution. Bioessays. 1992;14(6):375–384. doi: 10.1002/bies.950140606. [DOI] [PubMed] [Google Scholar]
- 11.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276(5688):565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 12.Grier DG, Thompson A, Kwasniewska A, Mcgonigle GJ, Halliday HL, Lappin TR. The pathophysiology of HOX genes and their role in cancer. J Pathol. 2005;205(2):154–171. doi: 10.1002/path.1710. [DOI] [PubMed] [Google Scholar]
- 13.Calvo R, West J, Franklin W, et al. Altered HOX and WNT7A expression in human lung cancer. Proc Natl Acad Sci U S A. 2000;97(23):12776–12781. doi: 10.1073/pnas.97.23.12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrando AA, Armstrong SA, Neuberg DS, et al. Gene expression signatures in MLL-rearranged T-lineage and B-precursor acute leukemias: dominance of HOX dysregulation. Blood. 2003;102(1):262–268. doi: 10.1182/blood-2002-10-3221. [DOI] [PubMed] [Google Scholar]
- 15.Murat A, Migliavacca E, Gorlia T, et al. Stem cell–related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26(18):3015–3024. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 16.Cheng W, Liu J, Yoshida H, Rosen D, Naora H. Lineage infidelity of epithelial ovarian cancers is controlled by HOX genes that specify regional identity in the reproductive tract. Nat Med. 2005;11(5):531–537. doi: 10.1038/nm1230. [DOI] [PubMed] [Google Scholar]
- 17.López R, Garrido E, Vázquez G, et al. A subgroup of HOX Abd-B gene is differentially expressed in cervical cancer. Int J Gynecol Cancer. 2006;16(3):1289–1296. doi: 10.1111/j.1525-1438.2006.00603.x. [DOI] [PubMed] [Google Scholar]
- 18.Sekar P, Bharti JN, Nigam JS, Sharma A, Soni PB. Evaluation of p53, HoxD10, and E-cadherin status in breast cancer and correlation with histological grade and other prognostic factors. J Oncol. 2014;2014:702527. doi: 10.1155/2014/702527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Z, Li S, Liu J, et al. RNAi-mediated silencing of AQP1 expression inhibited the proliferation, invasion and tumorigenesis of osteosarcoma cells. Cancer Biol Ther. 2015;16(9):1332–1340. doi: 10.1080/15384047.2015.1070983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ansari KI, Hussain I, Kasiri S, Mandal SS. HOXC10 is overexpressed in breast cancer and transcriptionally regulated by estrogen via involvement of histone methylases MLL3 and MLL4. J Mol Endocrinol. 2012;48(1):61–75. doi: 10.1530/JME-11-0078. [DOI] [PubMed] [Google Scholar]
- 21.Zhai Y, Kuick R, Nan B, et al. Gene expression analysis of preinvasive and invasive cervical squamous cell carcinomas identifies HOXC10 as a key mediator of invasion. Cancer Res. 2007;67(21):10163–10172. doi: 10.1158/0008-5472.CAN-07-2056. [DOI] [PubMed] [Google Scholar]
- 22.Feng X, Li T, Liu Z, Shi Y, Peng Y. HOXC10 up-regulation contributes to human thyroid cancer and indicates poor survival outcome. Mol Biosyst. 2015;11(11):2946–2954. doi: 10.1039/c5mb00253b. [DOI] [PubMed] [Google Scholar]
- 23.Xiong W, Zhou Q, Liu G, Liu XS, Li XY. Homeodomain-containing gene 10 inhibits cell apoptosis and promotes cell invasion and migration in osteosarcoma cell lines. Tumor Biol. 2017;39(5) doi: 10.1177/1010428317697566. 1010428317697566. [DOI] [PubMed] [Google Scholar]
- 24.Porter AG, Jänicke RU. Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 1999;6(2):99–104. doi: 10.1038/sj.cdd.4400476. [DOI] [PubMed] [Google Scholar]
- 25.Devarajan E, Sahin AA, Chen JS, et al. Down-regulation of caspase 3 in breast cancer: a possible mechanism for chemoresistance. Oncogene. 2002;21(57):8843–8851. doi: 10.1038/sj.onc.1206044. [DOI] [PubMed] [Google Scholar]
- 26.Huang Q, Li F, Liu X, et al. Caspase 3-mediated stimulation of tumor cell repopulation during cancer radiotherapy. Nat Med. 2011;17(7):860–866. doi: 10.1038/nm.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandlekar S, Yu R, Tan TH, Kong AN. Activation of caspase-3 and c-Jun NH2-terminal kinase-1 signaling pathways in tamoxifen-induced apoptosis of human breast cancer cells. Cancer Res. 2000;60(21):5995–6000. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primary OS cells.
Abbreviation: OS, osteosarcoma.