Abstract
Background
Since the discovery of giant viruses infecting amoebae in 2003, many dogmas of virology have been revised and the search for these viruses has been intensified. Over the last few years, several new groups of these viruses have been discovered in various types of samples and environments.In this work, we describe the isolation of 68 giant viruses of amoeba obtained from environmental samples from Brazil and Antarctica.
Methods
Isolated viruses were identified by hemacolor staining, PCR assays and electron microscopy (scanning and/or transmission).
Results
A total of 64 viruses belonging to the Mimiviridae family were isolated (26 from lineage A, 13 from lineage B, 2 from lineage C and 23 from unidentified lineages) from different types of samples, including marine water from Antarctica, thus being the first mimiviruses isolated in this extreme environment to date. Furthermore, a marseillevirus was isolated from sewage samples along with two pandoraviruses and a cedratvirus (the third to be isolated in the world so far).
Conclusions
Considering the different type of samples, we found a higher number of viral groups in sewage samples. Our results reinforce the importance of prospective studies in different environmental samples, therefore improving our comprehension about the circulation anddiversity of these viruses in nature.
Electronic supplementary material
The online version of this article (doi: 10.1186/s12985-018-0930-x) contains supplementary material, which is available to authorized users.
Keywords: Giant viruses, Prospection, Brazil, Antarctica, Pandoravirus, Cedratvirus, Marseillevirus, Mimivirus
Background
The discovery of Acanthamoeba polyphaga mimivirus (APMV) in 2003, the first isolated giant virus infecting amoebas, interested the scientific community due to its size and genome content, which culminated in the search for and isolation of new giant viruses [1, 2]. The giant amoebal viruses have many phenotypic and genomic features which had never been seen in other viruses before, like large viral particles presenting up to 1.5 μm in length and large double-stranded DNA genomes ranging from 350 kb in Marseilleviridae members to 2500 kb for pandoravirus [3, 4]. These genes encode many hypothetical proteins, uncharacterized, or with functions that have never orrarely been observed before in other viruses, such as those related to translation and DNA repair [5–7]. Common characteristics shared by giant and large DNA viruses permitted their incorporation into a supposedly viral monophyletic group, named nucleocytoplasmic large DNA viruses (NCLDV), created in 2001 [8]. When the NCLDV group was proposed, it was composed of families Poxviridae (e.g. Vaccinia virus, Crocodilepox virus), Asfarviridae (e.g. African swine fever virus) Iridoviridae (e.g. Frog virus 3) and Phycodnaviridae (e.g. Emiliania huxleyi virus 86, Aureococcus anophagefferens virus) [8].
Subsequently, viruses belonging to the Mimiviridae, Marseilleviridae, Ascoviridae family and also the pandoravirus, faustovirus, pithovirus, mollivirus, kaumoebavirus, cedratvirus and pacmanvirus were also incorporated to NCLDV group [9–17]. Recent prospective studies have shown that giant viruses are ubiquitous, as are their protozoa hosts [2, 18, 19]. The use of high-throughput techniques and different species of amoebae in culture for viral isolation has allowed the discovery of a large variety of new viruses and new lineages in recent years. They have been detected and/or isolated in all continents of Earth. Metagenomic studies have indicated an outstanding profile of giant virus distribution and diversity in natural environments and organisms, including water, soil, invertebrates and mammals [14, 20–27]. It is important to note that mimiviruses and marseilleviruses have also been isolated from human samples, raising questions abouttheir possible role as pathogenic agents of diseases, but this possibility still under investigation, and these viruses may be components of healthy humans virome [24, 25, 28–31].
Despite the advances made in the techniques used to isolate new giant viruses, which have increased the success of detection and the isolation of these viruses in different environments around the world, the diversity, distribution and role of these viruses in nature is still far from completely understood. Therefore, in order to better understand the diversity and distribution of giant viruses in the environment, this work aimed at the isolation and identification of giant viruses obtained from clinical and environmental samples from different regions of Brazil and Antarctica. A total of 976 samples were analyzed and 68 viruses were isolated. Taken together, our results reinforcethat giant viruses, in particular mimiviruses, are ubiquitous and may play an important role in the control of amoebal populations, both in natural and anthropogenic-affected environments.
Methods
Samples collection and treatment
In this work, a collection composed of 976 clinical and environmental samples was analyzed: 495 soil samples (mean weight was 3 g of each sample), 124 water samples (10 mL of each sample), 140 sewage samples (10 mL of each sample), 200 human nasopharyngeal aspirate samples (1,5 mL of each sample) and 17 capybara samples (mean weight was 2 g of each sample) (Table 1 and Fig. 1). All collections were collected in different locations using sterile tubes.
Table 1.
Collections and locations of samples analyzed
| Collections | Type of sample | Collection site | Date of collection |
|---|---|---|---|
| Serra do Cipó | |||
| 13 samples | Freshwater | Serra do Cipó, MG, Brazil | Jan.2015 |
| 47 samples | Soil | Serra do Cipó, MG, Brazil | Jan. 2015 |
| Sewage creeks Pampulha | |||
| 110 samples | Sewage | Pampulha Creeks, Belo Horizonte, MG, Brazil | Oct.2016 |
| Farm Sewage | |||
| 30 samples | Sewage | Itaúna, MG, Brazil | Nov. 2016 |
| Water treatment station | |||
| 50 samples | Freshwater | COPASA, Belo Horizonte, MG, Brazil | Dec. 2016 |
| Antarctic | |||
| 7 samples | Marine Water | Antarctic | Dec. 2014 |
| Capybara Stool | |||
| 17 samples | Stool | Serra do Cipó, MG, Brazil Pampulha, MG, Brazil Serro, MG, Brazil Pantanal, MS, Brazil |
Dec. 2012 Dec. 2012 Dec. 2012 |
| Minas Gerais Soil | |||
| 470 samples | Soil | MG, Brazil | Jan. 2014 |
| Pantanal soil | |||
| 12 samples | Soil | Pantanal, MT, Brazil | Mar. 2015 |
| Human nasopharyngeal aspirate | |||
| 200 samples | Human nasopharyngeal aspirate | Laboratório Central do Estado do Rio Grande do Sul, Brazil | Nov. 2014 |
| Bromeliads Water | |||
| 10 samples | Freshwater | Maceió, AL, Brazil | Set. 2015 |
| Mangrove water | |||
| 10 samples | Mangroove water Marine water |
ES, Brazil | Feb. 2015 |
Fig. 1.
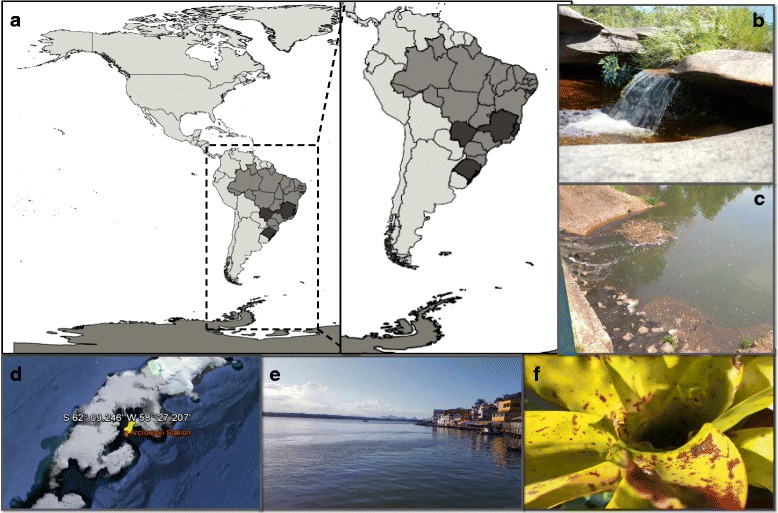
Locations where the environmental samples were collected. Schematic map (a) indicating in dark gray the location of collections tested and pictures from representative areas represented by letters (b-f). River at Serra do Cipó, MG, Brazil (b); Sewage creeks, Belo Horizonte, MG, Brazil (c); Location of collection of one of marine water samples in Antarctica (d); Location of collection of mangrove water, ES, Brazil (e). Bromeliads at Serra da Saudinha, AL, Brazil (f)
The samples of human nasopharyngeal aspirate were used under approval of the ethics committee of Universidade Federal de Ciências da Saúde de Porto Alegre (protocol number 1774/12, register 928/12). After collection, all samples were stored at 4 °C until inoculation procedures were performed.
Initially, the samples were divided into two groups, one with sediment-free water, including human clinical samples and other with a high concentration of sediment and soil. Samples with only water and no sediment were directly inoculated onto amoebalcultures. The soil samples were transferred to conical tubes of 15 mL and treated with 5 mL of phosphate buffered saline (PBS). The system was left for 24 h for sediment decantation and then the supernatants were collected and inoculated onto amoebal cultures.
Culture procedures
For viral isolation, we used Acanthamoeba polyphaga (ATCC 30461), Acanthamoeba castellanii (ATCC 30234) kindly provided by the Laboratório de Amebíases (Departamento de Parasitologia, ICB/UFMG) and Vermamoeba vermiformis (ATCC CDC19), kindly provided by Professor Bernard La Scola from Aix Marseille University. Amoeba were grown in 75 cm2 Nunc™ Cell Culture Treated Flasks with Filter Caps (Thermo Fisher Scientific, USA) with 30 mL of peptone-yeast extract-glucose (PYG) medium supplemented with 0,14 mg/mL penicillin (Sigma-Aldrich, USA), 50 mg/mL gentamycin (Thermo Fisher Scientific, USA) and 2.5 mg/mL amphotericin (Bristol-Myers- Squibb, New York, USA) at 32 °C.
For co-culture, amoeba were re-suspended in 10 mL of PYG supplemented with an antibiotic mix containing 0,004 mg/mL ciprofloxacin (Cellofarm, Brazil), 0,004 mg/mL vancomycin (Sigma-Aldrich, U.S.A), and 0,020 mg/mL doxycycline (Sigma-Aldrich, U.S.A). The suspension was then diluted 1:10 in PBS and then inoculated in96-well plates containing 4 × 104 cells per well. The plates were incubated for 7 days at 32 °C and observation of the cytopathic effect was done daily using an inverted optical microscope. The well contents were then collected, frozen and thawed three times to lyse the bacterial and fungal cells that may be present in the samples and thereby decrease the chance of co-culture contamination and also helps release the viruses of amoeba cells not lysed. Posteriorly, the samples were re-inoculated for two new sub-cultures on fresh amoeba, as described above (blind passages). The contents of wells with cytopathic effect were collected and inoculated in a new 25 cm2 Nunc™ Cell Culture Treated Flasks with Filter Caps (Thermo Fisher Scientific, USA) culture containing 1 million cells, the cytopathic effect was confirmed and this culture was centrifuged 10,000 rpm for 10 min (Centrifuge Sigma 1–14) for lysate clearance and were further analyzed for giant viruses. Negative controls with no sample inoculated amoeba were used in all microplates.
DNA extraction and PCR
After the identification of cultures with a cytopathic effect, screening was done to identify which giant virus was present in samples using PCR with specific targets for some giant virus groups (Table 2). For this, 200 μL of each positive suspension was used for DNA extraction. DNA was extracted using the phenol-chloroform method [32] and used at the concentration 50 μg/μg as a template for PCR assays. The genes targeted in the PCR assays were: helicase of mimivirus lineage A; DNA polymerase B of mimivirus lineage B; DNA polymerase B of mimivirus lineage C; the major protein of the capsid of the family Mimiviridae (generic reaction targeting lineages A, B and C), Marseilleviridae, pandoravirus and cedratvirus. The primers were designed using a freely available primer design tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) at the National Center for Biotechnology Information, U.S.A (NCBI); the sequences are described in Table 2. The primers and reactions were designed and standardized considering all analyzed viruses available on GenBank to avoid cross-amplification. PCR assays were performed using 1 μL of extracted DNA (~ 50 nanograms) in an amplification reaction mix containing 5 μL of SYBR Green Master Mix and 0.4 μL (10 μM) of forward and reverse primers. The final volume of the reaction was adjusted with ultrapure waterto 10 μL. The conditions of the StepOne thermal cycler reactions (Applied Biosystem, USA) were: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min, which was followed by a final step of 95 °C for 15 s, 60 °C for 1 min and 95 °C for 15 s. Positive samples in the PCR were those that amplified, showing the specific melting temperature, using the primers listed in Table 2, whereas the negative samples did not amplify in the PCR. As negative controls we used DNA extracted from non-inoculated amoebas with purified viruses or samples, and as a positive control we used DNA from amoebae infected with purified virus. Samples that were not possible to identify using the PCR assay were identified by electron microscopy and/or hemacolor staining.
Table 2.
Primer sequences used for specific PCR
| Target genes | Forward sequence | Reverse sequence |
|---|---|---|
| Helicase of mimivirus lineage A | 5’-ACCTGATCCACATCCCATAACTAAA-3′ | 5’-GGCCTCATCAACAAATGGTTTCT-3′ |
| DNA polymerase beta of mimivirus lineage B | 5’-AGTTCATCCGCACTTGGAGA-3′ | 5’-TCAACGGATAAAATCCCTGGTACT-3′ |
| DNA polymerase beta of mimivirus lineage C | 5′- TCCGAATTCTATGAGGGAGAGA-3′ | 5’-TGTTCCTTTTTGGGAGAACCA-3′ |
| Main protein of the capsid of the family Mimiviridae | 5’-ACTTTATTATCATTATCAGCGAATA-3’ | 5’-GCTCTTAACCCTGAAGAACA-3’ |
| Main protein of the capsid of the family Marseilleviridae | 5’-CTTTTGCACCTGCTTCATGA-3’ | 5’-GCGGTAACCCTCCCACTTAT-3’ |
| Main protein of the capsid of pandoravirus | 5’-GGATGGCTCGACGTCTCTT-3’ | 5’-CCTYGGTRAGCAMAGGCAAC-3’ |
| Main protein of the capsid of cedratvirus | 5′- AGAGTATGCTCGCAACCACC-3’ | 5’-CACGTTAAGGCCGGGGTAAT −3’ |
Sequencing validation and phylogeny
Four isolates were selected for sequencing validation. The genome of two pandoraviruses, the cedratvirus and one mimiviruses of lineage B positive samples were sequenced using the Illumina MiSeq instrument (Illumina Inc., San Diego, CA, USA) with the paired-end application. The sequenced reads were imported to CLC_Bio software and assembled into contigs by the de novo method. The prediction of open reading frame (ORF) sequences was carried out using the Fgenes V tool. ORFs smaller than 100aa were excluded from the annotation. Paralogous groups of genes were predicted by OrthoMCL program. The ORFs were functionally annotated using similarity analyses with sequences in the NCBI data base using BLAST tools. One fragment of 327 amino acid of DNA polymerase B gene sequence of the samples was aligned with sequences from other giant viruses, previously deposited in GenBank, using the ClustalW program. After the alignment analysis, phylogeny reconstruction was performed using the Neighbor-joining method implemented by the MEGA7 software.
Viral stock production and titration
For seed pool production, A. castellanii or A. polyphaga cells were cultivated and infected with 500 μL of isolates. After observation of a cytopathic effect, the titer was obtained by end-point method [33]. Stocks were kept at − 80 °C freezers.
Hemacolor staining
A. castellanii and A. polyphaga cells were infected with isolates at a M.O.I of 0.01 following the procedures described above. After approximately 18 h, amoeba became rounded, so 10 μL of the previously inoculated suspension was spread on a histological slide and fixed with methanol. The virus factories and viral particles were observed after hemacolor (Renylab, Brazil) or crystal violet (Labsynth, Brazil) staining, respectively. After, slides were analyzed under an optical microscope (OlympusBX41, Japan) with 1000X zoom.
Electron microscopy
For transmission electron microscopy (TEM), A. castellanii and A. polyphaga cells were cultivated until the observation of 80–90% confluence and infected with the isolates in an M.O.I of 0.01. The samples were prepared as described previously [34]. Briefly, 12 h post-infection, when approximately 50% of the cells were presenting a cytopathic effect, the medium was discarded and the monolayer gently washed twice with 0.1 M phosphate buffer. Glutaraldehyde 2.5% (v/v) was added to the system, followed by incubation for 1 h at room temperature for fixation. The cells were then collected, centrifuged at 3000 rpm for 10 min, the medium discarded and the cells stored at 4 °C in phosphate buffer until electron microscopy analyses.
For the scanning electron microscopy (SEM) assay, the isolates were prepared onto round glass blades covered by poly-L-lysine and fixed with glutaraldehyde 2.5% in 0.1 M cacodylate buffer for 1 h at room temperature. Samples were then washed three times with 0.1 M cacodylate buffer and post-fixed with 1.0% osmium tetroxide for 1 h at room temperature. After a second fixation, the samples were washed three times with 0.1 M cacodylate buffer and immersed in 0.1% tannic acid for 20 min. Samples were then washed in cacodylate buffer and dehydrated by serial passages in ethanol solutions with concentrations ranging from 35% to 100%. They were dried at the critical CO2 point, transferred into stubs and metalized with a 5 nm gold layer. The analyses were completed with scanning electronic microscopy (FEG Quanta 200 FEI) at the Center of Microscopy of UFMG, Brazil.
Results
Here, we report the screening of 976 environmental and clinical samples collected between 2014 and 2017 and the isolation of 68 giant viruses (6.97% isolation rate). Among all of the isolated viruses, 17 (25%) were isolated in A. polyphaga and 51 were isolated in A. castellanii (75%). No virus was isolated in V. vermiformes (Additional file 1: Table S1).
The PCR, hemacolor staining and electron microscopy assays showed that 22 samples were positive for mimivirus lineage A, 17 were positive for mimivirus lineage B and 2 were positive for mimivirus lineage C. In addition, 2 samples were positive for pandoravirus, 1 for cedratvirus and 1 for marseillevirus (Fig. 2). Twenty-three other samples were identified as mimiviruses by PCR (capsid gene, generic reaction), by hemacolor staining or by electron microscopy, but it was not possible to discriminate the lineage of these viruses using the specific PCR (Additional file 1: Table S1).
Fig. 2.
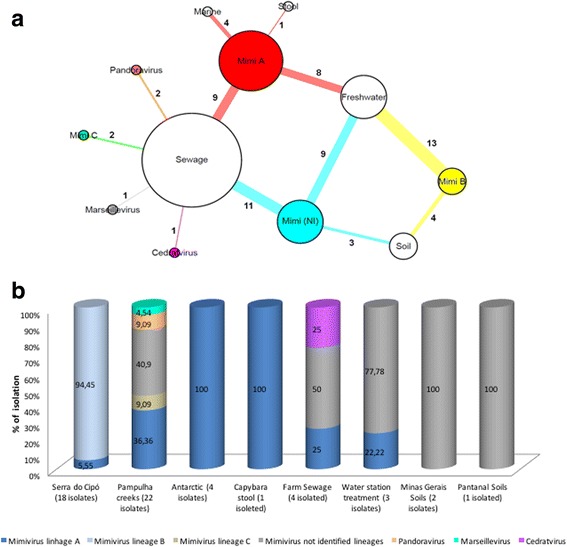
Diversity of isolated giant virus by type of sample and collections. Network graph showing the viral groups isolated and identified by PCR and electron microscopy assays in different samples. Each node represents a type of sample (white nodes) or viral group (colored nodes). The node diameter is proportional to the edge degree. The numbers of isolated viruses in each sample are shown on the respective edge. The layout was generated using a force based algorithm followed by manual rearrangement for a better visualization of the connections (a). A total of 7 viral groups are represented. Isolation rate of each virus groups by collections (b)
Twelve samples were positive in PCR for mimivirus lineage B and for marseillevirus. Four isolates were selected for genome sequencing and phylogenetic analyzes performed with the DNA polymerase gene of these viruses confirmed the identification by PCR (Fig. 3). In order to investigate the occurrence of co-infections, these samples were analyzed via hemacolor staining, and two of them were randomly selected for diagnosis via TEM. The samples tested showed only particles with morphology similar to the mimivirus, showing no marseillevirus-like particles (Additional file 1: Table S1). In addition, no marseillevirus-like factories were observed by hemacolor staining, just mimiviruses-like ones.
Fig. 3.
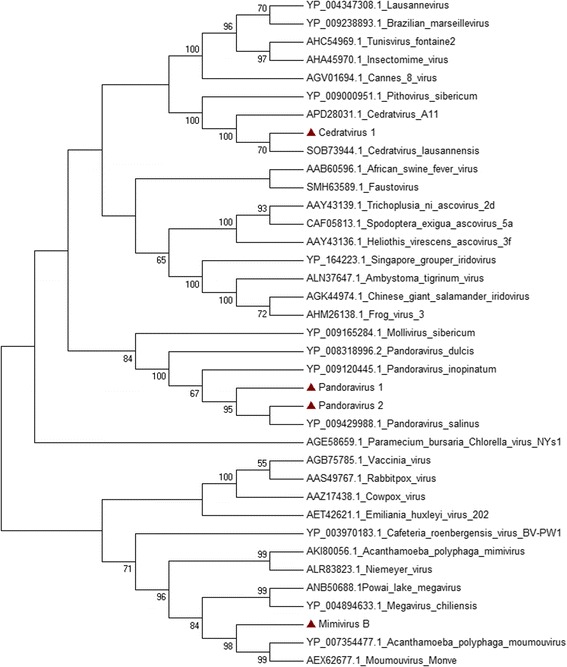
Phylogenetic tree of isolates. A neighbor-joining phylogenetic tree constructed using a 327 amino acid fragment of the DNA polymerase B gene. Tree was constructed by using MEGA version 7.0 (www.megasoftware.net) on the basis of the amino acids sequences with 1000 bootstrap replicates. Bootstrap values > 40% are shown. Nucleotide sequences were obtained from GenBank. The isolates are highlighted with red triangle. Scale bar indicates rate of evolution
The highest isolation percentages (27.42%) were obtained from the water samples, with 34 isolates from 124 samples. Of these, 4 were isolated from 7 seawater samples (57.14% isolation rate) and 30 were isolated from 117 freshwater samples (isolation rate of 25.64%) (Fig. 2).
In addition, with an isolation success of 18.57%, 26 viruses were obtained from 140 sewage samples, followed by samples of capybara feces (5.88%), with an isolate obtained from 17 samples, and soil samples (1.41%), with 7 isolates from 495 samples. In addition, 200 samples of human nasopharyngeal aspirate were tested and no isolates were obtained from these samples (Fig. 2).
Although water samples have shown the highest number of isolated virus, sewage samples presented the highest diversity of viruses groups isolated (Fig. 2). In the fresh and marine water samples, only Mimiviridae family viruses (12 of lineage A, 13 of lineage B and 9 unidentified) were identified, while besides Mimiviridae (9 of lineage A, 2 of lineage C and 11 unidentified), 1 marseillevirus, 1 cedratvirus and 2 pandoraviruses (Fig. 2) were found in the sewage samples. Soil and stool samples also showed only viruses of Mimiviridae family (4 of lineage B and 3 unidentified in soil, and 1 of lineage A in stool) (Fig. 2).
Comparing the percentage of isolates per collection region, we can observe that the isolation was higher in the Antarctica collection with a 57% isolation success rate; however, this collection has few samples (4 isolates from 7 samples). The collections of Serra do Cipó and sewage creeks appear with 30% (18 isolates from 60 samples) and 20% (22 isolates from 110 samples) positivity, respectively. The collections of farm sewage and Pantanal soils presented a percentage of isolations of 13.33% (4 isolates from 30 samples) and 8.33% (1 isolate from 12 samples), respectively (Fig. 2).
The collections of water station treatment, capybara stool and Minas Gerais soils, showed percentages of isolation of 8% (4 isolates from 50 samples), 5.88% (1 isolate from 17 samples) and 0,4% (2 isolates from 470 samples), respectively. Collections from bromeliad, mangrove, and human nasopharyngeal aspirate, showed no viral isolates (Fig. 2). The collection of creek sewages showed the greatest viral diversity, with isolates of Mimiviridae, Marseilleviridae and Pandoravirus groups; followed by the collection of farm sewage, with isolates of mimivirus and cedratvirus. In the remaining collection, only mimiviruses were identified (Fig. 2).
Electron microscopy assays showed that two isolated samples of Mergulhão (Fig. 4b-c) and Bom Jesus (Fig. 4e) sewage creek show pandoravirus-like morphology, with particles having an average length of 1 μm, as described by Philippe and colleagues in 2013. Antarctica isolates showed a mimivirus-like morphology, with particles of about 750 nm, as described by La Scola and colleagues in 2003 (Fig. 4d and g). SEM analyses of a sample of sewage farm collection showed cedratvirus-like morphology with particles of approximately 1.2 μm, as described by Andreani and colleagues in 2016 (Fig. 4a and d). The images obtained from another isolate from Bom Jesus creek showed particles with marseillevirus-like morphology, apparently with icosahedral symmetry and dimensions of about 200 nm (Fig. 4f).
Fig. 4.
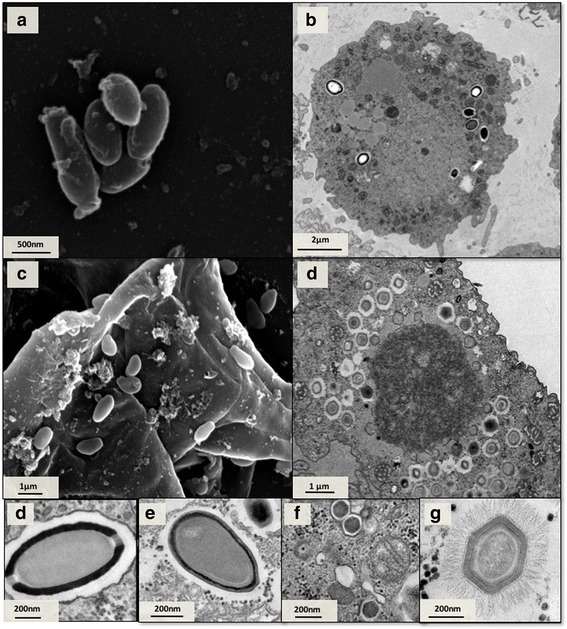
Electron microscopy images of viruses isolated. SEM of Cedratvirus isolated from sewage farm of MG (a) TEM (b) and SEM (c) of Pandoravirus isolate from Mergulhão sewage creek. TEM of Mimivirus isolated from Antarctica (c) TEM of Cedratvirus isolated from sewage farm of MG (d) TEM of Pandoravirus isolate from Bom Jesus sewage creek (e) marseillevirus isolated from Bom Jesus sewage creek (f). TEM mimivirus particle detail that was isolated from Antarctica (g). Scale Bars: (a-d) 500 nm; (e) 50 nm
Discussion
The search for giant viruses in environmental and clinical samples from different regions of Brazil and Antarctica resulted in 68 isolates, reinforcing the results obtained in other prospective studies involving environmental Brazilian samples, in which a large variety of giant viruses, specially mimiviruses, were isolated [18, 35–39]. The present work corroborates those studies, since 64 out of 68 viruses isolated (94.11%) were identified as mimiviruses; however, for the first time many mimiviruses of lineages B and C were isolated in Brazil.
Although Brazil is one of the most exploited countries regarding the presence of giant viruses, only two viruses of the Marseilleviridae family had been isolated in this territory to date. Brazilian marseillevirus and Golden marseillevirus presented a high genomic diversity, thus suggesting that these isolates form two new lineages within the family [37, 38]. This study presents the third marseillevirus isolated from Brazil. The genomic characterization of this isolate (in progress) may expand even more the plethora of marseillevirus lineages.
Regarding pandoraviruses, since their discovery in 2013, only 4 isolates have been described worldwide [18, 40–42]. These viruses form a new group among the NLCDVs, known as new TRUC (an acronym for Things Resisting Uncompleted Classification) members [10]. Here, we add two members to this club, providing the possibility of a wider study of this virus biology. In addition, this study reports for the first time the isolation of a giant virus from capybara feces. This type of sample had not yet been explored for the presence of giant viruses although DNA from poxvirus, another member of the NCLDV group, was previously detected in this collection [43].
The isolation and detection rates of giant viruses vary in the different studied samples, with noisolation in human nasopharyngeal aspirate samples and higher rates in water and sewage, followed by stool and soil samples. These results corroborate other studies in which giant viruses are more abundant in water and sewage than in soil samples [18, 19, 34, 39]. Furthermore, we demonstrated that giant viruses are not commonly found in nasopharyngeal clinical samples, as reported elsewhere [44–46]. Considering the amoebas used in this study, A. castellanii was shown to be more effective in the isolation of a greater diversity of giant viruses, as demonstrated by Dornas and colleagues in 2015.
The difficulty in amplifying some preserved lineage-specific regions by PCR can be explained by the high genetic diversity among these viruses [36]. This may be one of the reasons why we could not identify the strains of all mimiviruses isolated in this work. It is also important to consider that among these, there may be new strains which have not yet been described. In addition, two samples that were PCR positive for mimivirus lineage B and marseillevirus revealed only mimiviruses particles or factories when analyzed by electron microscopy or hemacolor staining. This finding also reinforces the importance ofusing a set of techniques for the identification of giant viruses, as performedin this study.
Metagenomic studies have indicated that the presence of the giant virus gene marker is common in all continents including Antarctica, a region with extreme environmental conditions [20, 21, 47, 48]. Virophages have already been isolated from this region, which is an additional indicative of the presence of giant viruses [22, 49]. However, to our knowledge, there has been no description of mimiviruses isolated in this continent to date. Nevertheless, we report the first giant amoebal viruses in Antarctica, confirming some previous expectations and the ubiquity of these microorganisms.
Altogether, our results lead us one step further into knowledge about the giant virus diversity and ecology, but important questions were raised. What could be the role of giant viruses in an extreme environment such as Antarctica? Will the host spectrum of these viruses be the same, or are they capable of infecting other more well-adapted hosts at extreme conditions? In-depth investigations regarding genetic and biological aspects of these isolates might provide some answers. Moreover, new prospecting studies, exploring different isolation strategies in environments that have never been explored around the globe, will bring insights about the ecology of giant viruses and completely new NCLDV members could be brought to light, boosting our knowledge about the diversity of this complex group within the virosphere.
Conclusions
This work presented the isolation of different giant virus species from the prospecting study of a large collection of environmental samples, providing the isolation of viruses never previously isolated in Brazil and Antarctica. The findings of this study reinforce the idea that giant viruses are ubiquitous and open the door to further study of the biology of these isolates, which contributes to an understanding of the diversity of these viruses.
Acknowledgments
We thank our colleagues from Gepvig and the Laboratório de Vírus fortheir excellent technical support. We thank CNPq, CAPES and FAPMEIG for scholarship and the Center of Microscopy of UFMG. To Drs. Ulysses Lins, Fernanda de Ávila Abreu, Pedro Leão, Tatiana Schaffer Gregianini, Ana Beatriz Gorini da Veiga, for crucial participation on sample collection. To the financial and logistic support from the Brazilian Antarctic Program, PROANTAR, as part of the IPY Activity no. 403 ‘MIDIAPI Microbial Diversity of Terrestrial and Maritime ecosystems in Antarctic Peninsula’ (520194/2006-3) is acknowledged. The Brazilian National Science and Technology Instituteon Antarctic Environmental Research (INCT-APA, CNPq 574018/2008) is also acknowledged. JSA, EGK, GST are CNPq researchers.
Funding
The scholarships were provided by CNPq, CAPES and FAPMEIG. The financial and logistic support were provided by The Brazilian National Science and Technology Institute on Antarctic Environmental Research (INCT-APA, CNPq 574018/2008) and Brazilian Antarctic Program, PROANTAR, as part of the IPY Activity no. 403 ‘MIDIAPI Microbial Diversity of Terrestrial and Maritime ecosystems in Antarctic Peninsula’ (520194/2006–3). JSA, EGK, GST are CNPq researchers.
Availability of data and materials
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
Abbreviations
- AL
Alagoas
- APMV
Acanthamoeba polyphaga mimivirus
- ES
Espírito Santo
- LACEN/RS
Laboratório Central do Estado do Rio Grande do Sul
- M.O.I
Multiplicity of infection
- MG
Minas Gerais
- NCBI
National Center for Biotechnology Information
- NCLDV
Nucleocytoplasmic large DNA viruses
- PBS
Phosphate buffered saline
- PCR
Polymerase chain reaction
- SEM
Scanning electron microscopy
- TEM
Transmission electron microscopy
Additional file
Identification and locations of viruses isolated. (DOCX 34 kb)
Authors’ contributions
ACSPA: performed experiments, wrote the manuscript, microscopical assays, designed the experiments; TSA: performed experiments; RALR: performed experiments, TBM: performed experiments; IAB: performed experiments; LALD: performed experiments FPD: microscopical assays; JSA: designed the experiments; EGK: designed the experiments; DBO: designed the experiments; GST: samples collections; GA: samples collections; JRC: samples collections; WA: samples collections; IB: samples collections; LGAB: samples collections; CF: samples collections; MFL: samples collections. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Two hundred samples of human nasopharyngeal aspiratewere also kindly provided by Laboratório Central do Estado do Rio Grande do Sul (LACEN/RS) (Table 1). These samples were used underapproval of the ethics committee of Universidade Federal de Ciências da Saúde de Porto Alegre (protocol number 1774/12, register 928/12).
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (doi: 10.1186/s12985-018-0930-x) contains supplementary material, which is available to authorized users.
References
- 1.La Scola B, Audic S, Robert C, Jungang L, de Lamballerie X, Drancourt M, et al. A giant virus in amoebae. Science. 2003;299:2033. doi: 10.1126/science.1081867. [DOI] [PubMed] [Google Scholar]
- 2.Aherfi S, Colson P, La Scola B, Raoult D. Giant viruses of amoebas: an update. Front Microbiol. 2016;7:1–14. doi: 10.3389/fmicb.2016.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyer M, Yutin N, Pagnier I, Barrassi L, Fournous G, Espinosa L, et al. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc Natl Acad Sci. 2009;106:21848–21853. doi: 10.1073/pnas.0911354106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Legendre M, Bartoli J, Shmakova L, Jeudy S, Labadie K, Adrait A, et al. Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc Natl Acad Sci. 2014;111:4274–4279. doi: 10.1073/pnas.1320670111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abrahão JS, Araújo R, Colson P, La Scola B. The analysis of translation-related gene set boosts debates around origin and evolution of mimiviruses. PLoS Genet. 2017;16:1–12. doi: 10.1371/journal.pgen.1006532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colson P, La Scola B, Levasseur A, Caetano-Anollés G, Raoult D. Mimivirus: leading the way in the discovery of giant viruses of amoebae. Nat Rev Microbiol. 2017;4:243–254. doi: 10.1038/nrmicro.2016.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, et al. The 1.2-megabase genome sequence of mimivirus. Science. 2004;306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- 8.Iyer LM, Aravind L, Koonin EV. Common origin of four diverse families of large eukaryotic DNA viruses. J Virol. 2001;75:11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma V, Colson P, Pontarotti P, Raoult D. Mimivirus inaugurated in the 21st century the beginning of a reclassification of viruses. Curr Opin Microbiol. 2016;31:16–24. doi: 10.1016/j.mib.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 10.Sharma V, Colson P, Chabrol O, Scheid P, Pontarotti P, Raoult D. Welcome to pandoraviruses at the “Fourth TRUC” club. Front Microbiol. 2015;6:1–11. doi: 10.3389/fmicb.2015.00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma V, Colson P, Chabrol O, Pontarotti P, Raoult D. Pithovirus sibericum, a new bona fide member of the “Fourth TRUC” club. Front Microbiol. 2015;6:1–9. doi: 10.3389/fmicb.2015.00722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benamar S, DGI R, Bandaly V, Labas N, Raoult D, La Scola B. Faustoviruses: comparative genomics of new megavirales family members. Front Microbiol. 2016;7:1–9. doi: 10.3389/fmicb.2016.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andreani J, Bou Khalil JY, Sevvana M, Benamar S, Di Pinto F, Bitam I, et al. Pacmanvirus, a new giant icosahedral virus at the crossroads between Asfarviridae and Faustoviruses. J Virol. 2017;91:212–7. [DOI] [PMC free article] [PubMed]
- 14.Legendre M, Lartigue A, Bertaux L, Jeudy S, Bartoli J, Lescot M, et al. In-depth study of Mollivirus sibericum, a new 30,000-y-old giant virus infecting Acanthamoeba. Proc Natl Acad Sci. 2015;112:5327–5335. doi: 10.1073/pnas.1510795112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bajrai LH, Benamar S, Azhar EI, Robert C, Levasseur A, Raoult D, et al. Kaumoebavirus, a new virus that clusters with Faustoviruses and Asfarviridae. Viruses. 2016;8:1–10. doi: 10.3390/v8110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andreani J, Aherfi S, JYB K, Di Pinto F, Bitam I, Raoult D, et al. Cedratvirus, a double-cork structured giant virus, is a distant relative of pithoviruses. Viruses. 2016;8:1–11. doi: 10.3390/v8110300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertelli C, Mueller L, Thomas V, Pillonel T, Jacquier N, Greub G. Cedratvirus lausannensis - digging into Pithoviridae diversity. Environ Microbiol. 2017;10:4022–4034. doi: 10.1111/1462-2920.13813. [DOI] [PubMed] [Google Scholar]
- 18.Dornas FP, JYB K, Pagnier I, Raoult D, Abrahão J, La Scola B. Isolation of new Brazilian giant viruses from environmental samples using a panel of protozoa. Front Microbiol. 2015;6:1–9. doi: 10.3389/fmicb.2015.01086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.La Scola B, Campocasso A, N’Dong R, Fournous G, Barrassi L, Flaudrops C, et al. Tentative characterization of new environmental giant viruses by MALDI-TOF mass spectrometry. Intervirology. 2010;53:344–353. doi: 10.1159/000312919. [DOI] [PubMed] [Google Scholar]
- 20.Ghedin E, Claverie J-MM. Mimivirus relatives in the Sargasso sea. Virol J. 2005;2:1–62. doi: 10.1186/1743-422X-2-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang W, Zhou J, Liu T, Yu Y, Pan Y, Yan S, et al. Four novel algal virus genomes discovered from Yellowstone Lake metagenomes. Sci Rep. 2015;5:1–13. doi: 10.1038/srep15131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerepesi C, Grolmusz V. The “Giant Virus Finder” discovers an abundance of giant viruses in the Antarctic dry valleys. Arch Virol. 2017;162:1–6. doi: 10.1007/s00705-017-3286-4. [DOI] [PubMed] [Google Scholar]
- 23.Boughalmi M, Pagnier I, Aherfi S, Colson P, Raoult D, La Scola B. First isolation of a marseillevirus in the diptera syrphidae eristalis tenax. Intervirology. 2013;56:386–394. doi: 10.1159/000354560. [DOI] [PubMed] [Google Scholar]
- 24.Popgeorgiev N, Colson P, Thuret I, Chiarioni J, Gallian P, Raoult D, et al. Marseillevirus prevalence in multitransfused patients suggests blood transmission. J Clin Virol. 2013;58:722–725. doi: 10.1016/j.jcv.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 25.Saadi H, DGI R, Colson P, Aherfi S, Minodier P, Pagnier I, et al. Shan virus: a new mimivirus isolated from the stool of a tunisian patient with pneumonia. Intervirology. 2013;56:424–429. doi: 10.1159/000354564. [DOI] [PubMed] [Google Scholar]
- 26.Dornas FP, Rodrigues FP, PVM B, LCF S, PCP F, Bonjardim CA, et al. Mimivirus circulation among wild and domestic mammals, Amazon Region, Brazil. Emerg Infect Dis. 2014;20:469–472. doi: 10.3201/eid2003.131050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reteno DG, Benamar S, Khalil JB, Andreani J, Armstrong N, Klose T, et al. Faustovirus, an Asfarvirus-related new lineage of giant viruses infecting amoebae. J Virol. 2015;89:6585–6594. doi: 10.1128/JVI.00115-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colson P, Aherfi S, La Scola B, Raoult D. The role of giant viruses of amoebas in humans. Curr Opin Microbiol. 2016;31:199–208. doi: 10.1016/j.mib.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 29.Aherfi S, Colson P, Raoult D. Marseillevirus in the pharynx of a patient with neurologic disorders. Emerg Infect Dis. 2016;22:2008–2010. doi: 10.3201/eid2211.160189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aherfi S, Colson P, Audoly G, Nappez C, Xerri L, Valensi A, et al. Marseillevirus in lymphoma: a giant in the lymph node. Lancet Infect Dis. 2016;1:225–234. doi: 10.1016/S1473-3099(16)30051-2. [DOI] [PubMed] [Google Scholar]
- 31.Popgeorgiev N, Michel G, Lepidi H, Raoult D, Desnues C. Marseillevirus adenitis in an 11-month-old child. J Clin Microbiol. 2013;51:4102–4105. doi: 10.1128/JCM.01918-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Russell DW. Molecular Cloning-Sambrook & Russel. 3rd ed. New York: Cold Spring Harbor Laboratory Press; 2001.
- 33.Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 34.Campos RK, Boratto PV, Assis FL, Aguiar ER, Silva LC, Albarnaz JD, et al. Samba virus: a novel mimivirus from a giant rain forest, the Brazilian Amazon. Virol J. 2014;11:95. doi: 10.1186/1743-422X-11-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andrade KR, Boratto PPVM, Rodrigues FP, LCF S, Dornas FP, Pilotto MR, et al. Oysters as hot spots for mimivirus isolation. Arch Virol. 2014;160:477–482. doi: 10.1007/s00705-014-2257-2. [DOI] [PubMed] [Google Scholar]
- 36.Assis FL, Bajrai L, Abrahao JS, Kroon EG, Dornas FP, Andrade KR, et al. Pan-genome analysis of Brazilian lineage a amoebal mimiviruses. Viruses. 2015;7:3483–3499. doi: 10.3390/v7072782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.dos Santos RN, Campos FS, Medeiros de Albuquerque NR, Finoketti F, Côrrea RA, Cano-Ortiz L, et al. A new marseillevirus isolated in Southern Brazil from Limnoperna fortunei. Sci Rep. 2016;6:35237. doi: 10.1038/srep35237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dornas FP, Assis FL, Aherfi S, Arantes T, Abrahão JS, Colson P, et al. A Brazilian marseillevirus is the founding member of a lineage in family marseilleviridae. Viruses. 2016;8:76. doi: 10.3390/v8030076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.PVM B, Arantes TS, LCF S, Assis FL, Kroon EG, La Scola B, et al. Niemeyer virus: a new mimivirus group A isolate harboring a set of duplicated aminoacyl-tRNA synthetase genes. Front Microbiol. 2015;6:1–11. doi: 10.3389/fmicb.2015.01256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Antwerpen MH, Georgi E, Zoeller L, Woelfel R, Stoecker K, Scheid P. Whole-genome sequencing of a pandoravirus isolated from keratitis-inducing acanthamoeba. Genome Announc. 2015;3:1–2. doi: 10.1128/genomeA.00136-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Philippe N, Legendre M, Doutre G, Coute Y, Poirot O, Lescot M, et al. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science. 2013;341:281–286. doi: 10.1126/science.1239181. [DOI] [PubMed] [Google Scholar]
- 42.Scheid P. A strange endocytobiont revealed as largest virus. Curr Opin Microbiol. 2016;31:58–62. doi: 10.1016/j.mib.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 43.Dutra LAL, de Freitas Almeida GM, Oliveira GP, Abrahão JS, Kroon EG, de Trindade GS. Molecular evidence of Orthopoxvirus DNA in capybara (Hydrochoerus hydrochaeris) stool samples. Arch Virol. 2017;162:439–448. doi: 10.1007/s00705-016-3121-3. [DOI] [PubMed] [Google Scholar]
- 44.JYB K, Robert S, Reteno DG, Andreani J, Raoult D, La Scola B. High-throughput isolation of giant viruses in liquid medium using automated flow cytometry and fluorescence staining. Front Microbiol. 2016;7:1–9. doi: 10.3389/fmicb.2016.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dare RK, Chittaganpitch M, Erdman DD. Screening pneumonia patients for Mimivirus1. Emerg Infect Dis. 2008;14:465–467. doi: 10.3201/eid1403.071027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saadi H, Pagnier I, Colson P, Cherif JK, Beji M, Boughalmi M, et al. First isolation of Mimivirus in a patient with pneumonia. Clin Infect Dis. 2013;57:127–134. doi: 10.1093/cid/cit354. [DOI] [PubMed] [Google Scholar]
- 47.Kerepesi C, Grolmusz V. Giant viruses of the Kutch Desert. Arch Virol. 2016;161:721–724. doi: 10.1007/s00705-015-2720-8. [DOI] [PubMed] [Google Scholar]
- 48.Santini S, Jeudy S, Bartoli J, Poirot O, Lescot M, Abergel C, et al. Genome of Phaeocystis globosa virus PgV-16T highlights the common ancestry of the largest known DNA viruses infecting eukaryotes. Proc Natl Acad Sci. 2013;110:10800–10805. doi: 10.1073/pnas.1303251110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yau S, Lauro FM, MZ DM, Brown MV, Thomas T, Raftery MJ, et al. Virophage control of antarctic algal host-virus dynamics. Proc Natl Acad Sci. 2011;108:6163–6168. doi: 10.1073/pnas.1018221108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.


