Abstract
Background
Auxin response factors (ARFs) in auxin signaling pathway are an important component that can regulate the transcription of auxin-responsive genes involved in almost all aspects of plant growth and development. To our knowledge, the comprehensive and systematic characterization of ARF genes has never been reported in Prunus sibirica, a novel woody biodiesel feedstock in China.
Results
In this study, we identified 14 PsARF genes with a perfect open reading frame (ORF) in P. sibirica by using its previous transcriptomic data. Conserved motif analysis showed that all identified PsARF proteins had typical DNA-binding and ARF domain, but 5 members (PsARF3, 8 10, 16 and 17) lacked the dimerization domain. Phylogenetic analysis of the ARF proteins generated from various plant species indicated that ARFs could be categorized into 4 major groups (Class I, II, III and IV), in which all identified ARFs from P. sibirica showed a closest relationship with those from P. mume. Comparison of the expression profiles of 14 PsARF genes in different developmental stages of Siberian apricot mesocarp (SAM) and kernel (SAK) reflected distinct temporal or spatial expression patterns for PsARF genes. Additionally, based on the expressed data from fruit and seed development of multiple plant species, we identified 1514 ARF-correlated genes using weighted gene co-expression network analysis (WGCNA). And the major portion of ARF-correlated gene was characterized to be involved in protein, nucleic acid and carbohydrate metabolic, transport and regulatory processes.
Conclusions
In summary, we systematically and comprehensively analyzed the structure, expression pattern and co-expression network of ARF gene family in P. sibirica. All our findings provide theoretical foundation for the PsARF gene family and will pave the way for elucidating the precise role of PsARF genes in SAM and SAK development.
Electronic supplementary material
The online version of this article (10.1186/s12870-017-1220-2) contains supplementary material, which is available to authorized users.
Keywords: Prunus Sibirica, Auxin response factor, Expression profile, Co-expression analysis
Background
Siberian apricot (Prunus sibirica L.), belonging to family Rosaceae, is diploid plant: 2n = 2× = 16 [29]. Recently, Siberian apricot has become a novel and important woody oilseed plant. In China, the total area of Siberian apricot is approximately 1.7 million ha, and the annual harvest of seeds is nearly 192,500 tons [48]. Siberian apricot is a multipurpose tree species with ecological and economic value. The Siberian apricot mesocarp (SAM) can be eaten either dried or fresh. The Siberian apricot kernel (SAK) was identified with high content of oil (over 50%), and the SAK oil was determined to be suitable for biodiesel production based on evaluation of cold filter plugging point, cetane number, oxidative stability and flash point [49]. Recently, the investigations of morphological characteristics and oil contents in developing SAK revealed that the development of SAK is a dynamic process involving a complex series of specific respond to different developmental signals [11, 32].
Increasing evidences suggested the ubiquitous involvement of auxin in transcriptional regulation of various genes, which may function in most aspects of growth and development processes, such as embryogenesis, organogenesis, tissue differentiation, apical dominance, root initiation, flower, fruit and seed development [6, 7, 25]. Numerous studies in Arabidopsis and other plant species elucidated that these auxin-inducible genes contain auxin response element (AuxRE: TGTCTC) in their promoters, and auxin response factors (ARFs) involved in auxin perception and signaling could specifically bind AuxRE [18, 20, 24, 25, 42, 47, 55]. The typical ARF proteins contain 3 representative components, an N-terminal DNA-binding domain (DBD), a middle domain (MD) and a carboxy-terminal dimerization domain (CTD) [6]. The DBD domain, as a plant-specific B3-type protein domain, is responsible for the recognition of AuxRE in the promoter of auxin-responsive genes. The CTD domain, consisting of two highly conserved dimerization domains III and IV, similar to those found in auxin/indole-3-acetic acid (Aux/IAA) proteins [17, 18]. The MD, in the middle region between the DBD and CTD, functions as an activation domain (AD) or repression domain (RD) to regulate the expression of downstream genes [12]. AD is enriched in glutamine (Q) residues, while RD is enriched in proline (P), serine (S) and threonine (T) residues [6].
Since, cloning of the first ARF1 gene from Arabidopsis [46], genome-wide analyses have identified ARF gene family from 29 plant species [6]. For example, 23 genes from Arabidopsis [18], 35 genes from Gossypium raimondi [42], 22 genes from Citrus sinensis [24], 17 genes from Eucalyptus grandis [55], 15 genes from Cucumis sativus [27], 36 genes from Zea mays [28], 25 genes from Oryza sativa [47] and 39 genes from Populus trichocarpa [20] have been defined. The quantity of ARF genes in different plant species were obviously different, which could be due to extensive duplication and diversification in the evolution of these plants [3]. Interestingly, most ARF proteins from these plant species are nuclear proteins, and maintain consistency of conserved domains as above described [6, 25]. Although wide analysis of ARFs function, expression, and regulation has been performed in annual herbaceous plants, relatively few reports focus on perennial woody plants [25].
To date, biochemical and genetic approaches have enabled identifying ARF gene functions in development of plant seeds and fruits. For example, mutation analyses revealed that AtARF2 is a general repressor of cell division to regulate seed size and weight [38]. The mutation of AtARF5 in Arabidopsis suggested that the AtARF5 gene influences embryo pattern formation as well as vascular development by mediating axialized behavior of plant cells in response to auxin cues [19]. T-DNA insertion alleles demonstrated that AtARF8 is an important regulator of fruit initiation and that the disruption of its normal function induces parthenocarpy in Arabidopsis [14]. In tomato (Solanum lycopersicon), SlARF4 was demonstrated to be involved in sugar metabolism and cell wall architecture during tomato fruit development [36, 37]. The transgenic antisense-OsARF1 rice showed extremely low growth, poor vigor, short curled leaves and sterility, suggesting that the OsARF1 is essential for growth in vegetative organs and seed development [1]. Together, these studies have shown that the ARF gene family is widely involved in regulating growth and development of seeds and fruits.
Here, we identified ARF genes from the previous transcriptomic data of Siberian apricot, and analyzed their phylogenetic relationship, gene structure and protein motifs. To understand the contribution of PsARFs in SAM and SAK development, we also characterized the expression profiles and co-expression networks of PsARF genes. Our present work is a necessary step in formulating further studies of the function of PsARF genes and the ARF-mediated auxin signaling pathway in growth and development of Siberian apricot.
Methods
Plant material
The different developmental stages of Siberian apricot were obtained from the same tree located at the Beijing Forestry University experimental station, Beijing, China. The developmental processes of Siberian apricot from flowering to seed maturity were observed from May to July 2017. Flowers with the same anthesis were marked, and then fruits were respectively harvested at 10, 30, 50, 60, and 70 days after flowering (DAF), based on our previous report [31]. The SAM and SAK in different development periods were immediately separated and frozen in liquid nitrogen, and stored at −80 °C until use.
PsARF sequence retrieval and conserved motif analysis
Our data were obtained from the previous transcriptomic studies of SAM and SAK at 10, 30, 50, 60, and 70 DAF by Illumina sequencing (PRJNA260249) [31], and a mixture of buds, leaves, stems, flowers, fruits by 454 pyrosequencing (SRX339392) [9]. After removal of the adapter sequences, the low-quality sequences (reads with ambiguous bases ‘N’) and reads with more than 10% Q < 20 bases, the clean reads were assembled into unigenes with the Trinity program [53]. A total of 124,070 unigenes (N50: 1603 bp) with the mean length of 829.62 bp was obtained. The Siberian apricot unigenes were annotated using BLASTX alignment with an E-value cut-off of 10−5 against the following protein databases: Arabidopsis proteome (www.arabidopsis.org), NCBI nonredundant (https://www.ncbi.nlm.nih.gov/) and SwissProt (www.uniprot.org). Based on the annotated results, the unigenes with ARF family domain were filter out for the prediction of open reading frame (ORF) by NCBI ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/). All non-overlapping PsARF genes with a perfect ORF were validated by PCR amplification and gene sequencing, and also were used for further analysis. The primer sequences of PCR amplification are shown in Additional file 1: Table S1.
To exhibit the structural divergence of PsARF proteins, the conserved motifs were performed with Multiple Expectation Maximization for Motif Elicitation (MEME) 4.11.2 online program [2]. The following parameters were employed in the analysis: the maximum number of motifs 20; minimum motif width 6; and maximum motif width 50. A phylogenetic tree of PsARF proteins was constructed with the MEGA 7.0 software.
Phylogenetic analysis PsARF genes
For phylogenetic reconstruction of the ARF gene family, we downloaded ARF genes from the sequenced genomes, including Arabidopsis (23) [18], Brassica rapa (31) [30], Citrus sinensis (19) [24] and P. mume (17) [41]. Amino acid sequences of the PsARF genes were aligned using ClustalW with default options [43], and the alignment was manually corrected at both ends to eliminate regions of poor alignment by using Jalview [51]. Phylogenetic trees were constructed by the neighbor-joining method using program MEGA 7.0 [21]. The parameters of the constructed trees were: phylogeny test and options, bootstrap (1000 replicates); gaps/missing data, complete deletion; model, amino acid; Poisson correction; substitutions to include, all; pattern among lineages, same (homogeneous); and rates among sites, uniform rates.
Co-expression analysis of PsARF
To precisely determine genes whose expression tightly correlates with that of ARF genes in fruit and seed development, a series of expression data was analyzed derived from Elaeis guineensis [4], P. sibirica [31], B. napus, Ricinus communis, and Euonymus alatus [44]. The expression data contains 5, 5, 4, 4, 4, 5 and 5 developmental phases of oil palm mesocarp, date palm mesocarp, B. napus seed, R. communis seed, E. alatus seed, SAM and SAK, respectively. Automatic construction of the gene network and identification of modules were conducted by using the R package of weighted gene co-expression network analysis (WGCNA) [22]. By pickSoftThreshold analysis, the power 12 was selected to amplify the strong connections between genes and penalize the weaker connections. Here, we used a convenient one-step network construction and module detection, and chose a relatively large minimum module size of 30 and a medium sensitivity (DeepSplit = 2) to cluster splitting [22]. The resulting genes (threshold weight ≥ 0.1) involved in PsARFs was analyzed with BGI WEGO (http://wego.genomics.org.cn/) to assign gene ontology (GO) terms [54]. Cytoscape was implemented for visualizing interaction networks [39].
Expressed analysis by FPKM and qRT-PCR
The recent development of the RNA-Seq method provides information on wide and sensitive gene expression. In our RNA-seq data, the expression levels were calculated by Fragment Per Kilobase of exon model per Million mapped reads (FPKM). From these results, we extracted the expression data related to PsARF gene family, and conducted further analysis.
The equal weight of three biological samples of SAM and SAK in different developmental stages was mixed. And then total RNA of SAM and SAK was separately extracted from the mixture using RNeasy Plant Mini Kits (Qiagen) according to the manufacturer’s protocol. Three biological repetitions were performed for each RNA extraction. The fist-strand cDNA was synthesized by using oligo d(T) primers and reverse transcription System (Promega). The amplification primers were designed using PrimerQuest (http://www.idtdna.com/PrimerQuest/Home/Index) software with melting temperatures at 62 °C, and the absence of secondary structures was verified by the UNAFold program (http://eu.idtdna.com/UNAFold). According to our previous studies, cyclophilin and ubiquitin-conjugating enzyme were used as internal controls [33]. The qRT-PCR was performed using the SYBR Premix Ex Taq Kit (TaKaRa) according to the manufacturer’s protocol. Negative controls consisting of nuclease-free water instead of template, and reverse transcriptase controls prepared by substituting reverse transcriptase for nuclease-free water in the cDNA synthesis step were included in all analyses for each primer pair. Three technical repetitions were performed for qRT-PCR. The qRT-PCR primer sequences are shown in Additional file 1: Table S1. The statistical analysis of expressed data was performed using SPSS software.
Results
Identification of PsARF gene families in P. sibirica
To comprehensively identify the PsARF genes in P. sibirica, we integrated the transcriptomic data of P. sibirica form different developing kernels by Illumina/Solexa sequencing [31] and different tissues by 454 GS FLX Titanium sequencing [9]. Simultaneously, the 23 protein sequences of ARFs from the TAIR 10 (The Arabidopsis Information Resource) database were used as queries to search PsARFs in this work. After PCR amplification and gene sequencing (Additional file 2: Figure S1), we characterized 14 PsARF genes with a perfect ORF (Table 1). The minimum ORF length of PsARFs is 1803 bp (PsARF17) encoding 600 amino acid residues (66.23 kDa), and the maximum ORF length of PsARFs is 3489 bp (PsARF19) encoding 1162 amino acid residues (130.01 kDa) (Table 1). It is worth noting that the theoretical PI values of all PsARF proteins were less than 7 (Table 1), suggesting that those PsARF genes encoded weakly acidic proteins. The nomenclature system for PsARFs in the present study was given to the homologies of Arabidopsis ARFs.
Table 1.
ARF genes in P. sibirica
| Genes | NCBI ID |
ORF length (bp) | Deduced polypeptide | TIGR locus ID | E-value | ||
|---|---|---|---|---|---|---|---|
| Length (aa) | MW (kDa) | PI | |||||
| PsARF1 | MF373590 | 2037 | 678 | 75.47 | 6.00 | AT1G59750 | 0 |
| PsARF2 | MF373591 | 2540 | 846 | 94.26 | 6.55 | AT5G62000 | 0 |
| PsARF3 | MF373592 | 2160 | 719 | 78.50 | 6.19 | AT2G33860 | e-174 |
| PsARF4 | MF373593 | 2406 | 801 | 88.47 | 5.96 | AT5G60450 | 0 |
| PsARF5 | MF373594 | 2865 | 954 | 104.29 | 5.27 | AT1G19850 | 0 |
| PsARF6 | MF373595 | 2670 | 889 | 98.46 | 6.11 | AT1G30330 | 0 |
| PsARF7 | MF373596 | 3423 | 1140 | 126.22 | 6.19 | AT5G20730 | 0 |
| PsARF8 | MF373597 | 2172 | 723 | 80.56 | 6.16 | AT5G37020 | 0 |
| PsARF9 | MF373598 | 2046 | 681 | 75.34 | 6.20 | AT4G23980 | 0 |
| PsARF10 | MF373599 | 2025 | 674 | 74.69 | 6.46 | AT2G28350 | 0 |
| PsARF11 | MF373600 | 2097 | 698 | 77.41 | 6.37 | AT2G46530 | 0 |
| PsARF16 | MF373601 | 2061 | 686 | 75.94 | 6.66 | AT4G30080 | 0 |
| PsARF17 | MF373602 | 1803 | 600 | 66.23 | 6.52 | AT1G77850 | e-145 |
| PsARF19 | MF373603 | 3489 | 1162 | 130.01 | 6.20 | AT1G19220 | 0 |
Conserved motifs and domains in PsARF proteins
By using MEME online program, a total of 14 motifs was characterized from 14 PsARF proteins (Fig. 1a and Additional file 3: Figure S2). Most ARF proteins contain a highly conserved N terminal DBD composed of an unknown subdomain, a plant specific B3-type subdomain, an ARF subdomain and nuclear localization signal (NLS) [55]. In the present work, motif 1–4, 6–9, 12, and 13 correspond to the N-terminal DBD in all of 14 PsARFs. Among these motifs, the biological significance of motif 3 and 4 remains uncharacterized (Additional file 3: Figure S2). However, motif 2–1-9 and 6–13–8-12-7 orderly constituted the B3-type and ARF subdomain, respectively (Fig. 1a and b). Interestingly, 3 out of the 14 PsARFs (PsARF10, 16 and 17) exhibited an additional short segment of amino acids between the B3-type and ARF subdomain (Fig. 1a). At the end of the DBD domain, all of the PsARFs contain a motif 11, which correspond to a conserved putative NLS (Fig. 1a). In addition, the motif 14, 10 and 5 orderly formed the CTD region consisting of subdomain III and IV (Fig. 1b).The predicted protein structures of PsARF3, 8 and 17 are lacking dimerization subdomain III and IV (Fig. 1a), while PsARF10 and 16 have a truncated CTD (only segmental subdomain III) (Fig. 1a and Additional file 3: Figure S2). It was reported that these non-conserved MD in at least some Arabidopsis ARFs function as transcriptional activation (Q-rich) or repression domains (P, S and T-rich) [45]. By component analysis in the amino acid of 14 PsARF proteins, the middle region of PsARF1–6, 10–11, and 16–17 is rich in P or S, whereas the middle region of PsARF7 and 19 is rich in Q (Fig. 1a and Additional file 4: Table S2). Additionally, the middle region of PsARF 8 and 9 is enriched in QS and Asparagine (N), respectively (Fig. 1a). Overall, the conserved motifs were found in PsARF proteins, indicating a strongly functional constraint during the course of evolution.
Fig. 1.
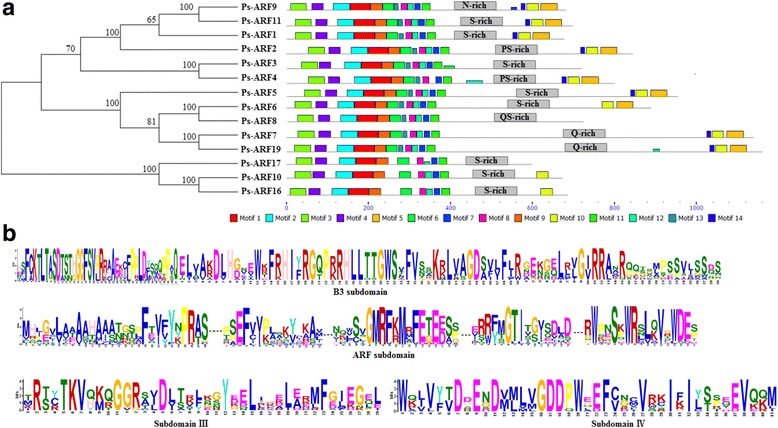
Diagrams and predicted sequences of PsARFs in P. sibirica. a Schematic diagrams of 14 PsARFs. b Amino acid sequence alignments of B3-type subdomain, ARF subdomain, subdomain III and subdomain IV
Phylogenetic analysis of PsARF proteins
To investigate the evolutionary relationship between PsARFs and those in other species, a neighbor-joining tree was generated based on alignments of the complete protein sequences of 14 PsARFs, 23 AtARFs, 31 BrARFs, 19 CsARFs, and 17 PmARFs. The results showed that all the ARFs could be grouped into four major clusters (I, II, III, and IV) based on their phylogenetic relationship (Fig. 2). 14 PsARFs were distributed among the four clusters, for example, Cluster I included PsARF1 and 2; Cluster II included PsARF9 and 11; Cluster III included PsARF3, 4, 5, 6, 7, 8, and 19; and Cluster IV included PsARF10, 16, and 17 (Fig. 2). Interestingly, our phylogenetic analysis unambiguously established the orthologous relationship between PsARF and PmARF proteins, indicating that the organization of the PsARF proteins is very similar to that of the PmARF proteins. These results imply that PsARF and PmARF proteins are close relative, and indeed P. mume belongs to the same family Rosaceae with our experimental material (P. sibirica).
Fig. 2.
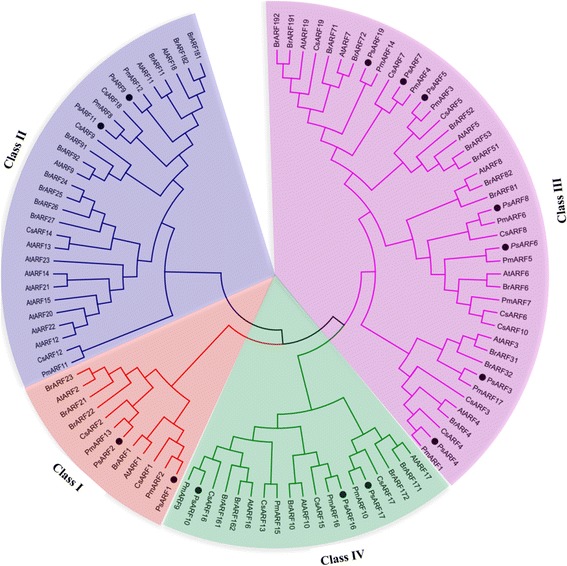
Phylogenetic analysis of ARFs from Arabidopsis, B. rapa, C. sinensis, P. mume, and P. sibirica by using Neighbor-joining method. Groups of genes are represented by color arcs. At: Arabidopsis thaliana, Br: Brassica rapa, Cs:Citrus sinensis, Pm: Prunus mume, Ps: Prunus sibirica
Expression of PsARF genes in developing mesocarp and kernel
To better understand the expressional characteristics of each PsARF gene in developmental process of SAM and SAK, the expression profiles of PsARF genes were investigated by qRT-PCR and FPKM. In this study, the expressions of all 14 PsARF genes could be detected in both the SAM and SAK (Figs. 3 and 4).
Fig. 3.
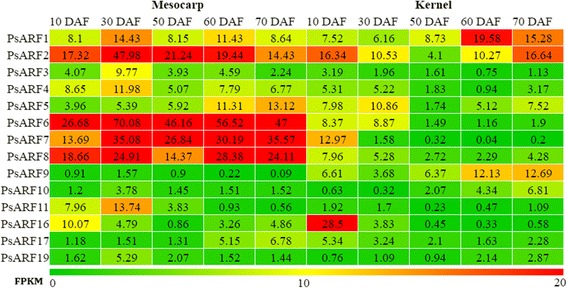
The expression levels (FPKM) of PsARF genes in developing SAM and SAK. The data in heatmap showed FPKM values
Fig. 4.
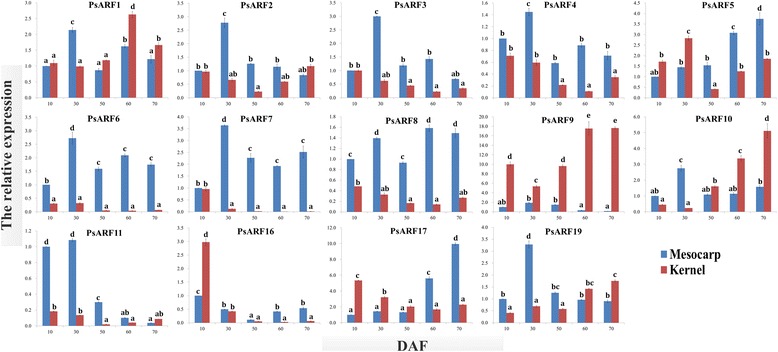
The qRT-PCR analysis of PsARF genes in developing SAM and SAK. The relative expression levels were calculated as 2-△△Ct, and the expression data of SAM at 10 DAF served as control. Data are means ± SE of three separate measurements
During the development of SAM, 14 PsARF genes presented complicated expression patterns (Figs. 3 and 4). 10 PsARF genes (PsARF1–8, 11 and 16) were highly induced (value >10) at specific stages of SAM development, while PsARF9, 10, 17 and 19 showed low expression (value <10) at all stages of SAM development (Fig. 3). Among the highly expressed genes, most of PsARF genes, such as PsARF1–4, 11 and 16, showed transcript accumulation before 30 DAF, whereas only PsARF5 were significantly up-regulated at late development (60–70 DAF) of SAM (Fig. 3), as was in accordance with our qRT-PCR results (Fig. 4). Notably, PsARF2 and 6–8 exhibited high expression levels (value >10) at all stages of mesocarp development (Fig. 3).
As for the development of kernel, PsARF1, 2, 5, 7, 9 and 16 demonstrated significant induction (value >10) at specific stages of SAK development (Fig. 3). Our qRT-PCR analysis revealed that PsARF7 and 16 were significantly up-expressed at 10 DAF, whereas PsARF1 and 9 were abundantly transcribed at 60–70 DAF (Fig. 4). Additionally, PsARF2 and 5 showed down-regulated transcript at 50 DAF (Figs. 3 and 4). It is worthy to note that more PsARF genes, including PsARF3, 4, 6, 8, 10, 11, 17 and 19, showed low expression (value <10) at all developmental stages of SAK in comparison to SAM (Fig. 3).
In a comparison of the expression profiles of PsARF genes at different developmental stages of SAM and SAK, PsARF4, 6, 7, 8, and 11 exhibited specifically higher expression in SAM, while PsARF9 were transcribed more strongly in SAK (Figs. 3 and 4). Also, PsARF 10, 17 and 19 showed low expressions (value <10) at all stages of SAM and SAK development (Fig. 3). Interestingly, PsARF16 shared a similar pattern of mRNA accumulation in developing SAM and SAK (Figs. 3 and 4), implying that PsARF16 may play similar roles in early development of SAM and SAK. Together, our expression data showed a high variability in transcript abundance of the PsARF genes between SAM and SAK (Figs. 3 and 4), probably indicating the diversified functions of the PsARF genes in controlling SAM and SAK development.
Co-expression network analysis of PsARF genes
By using previous expression data from E. guineensis [4], P. sibirica [31], B. napus, R. communis, and E. alatus [44], we attempted to establish the co-expression network of ARF genes. A total of 8 ARF genes (homologous PsARF1, 2, 6, 8, 9, 11, 16 and 17) were shared by the above 5 plant species, suggesting that these ARF genes may be constitutively expressed. Using WGCNA software, the resulting 79, 1265, 9, 35, 2 and 148 genes were identified to be involved in the co-expression network of the homologous PsARF1, 8, 9, 11, 16 and 19, respectively (Additional file 5: Table S3). To fully explore the biological functions of these ARF-correlated genes, GO annotation was performed. The functional terms of 1514 ARF-correlated genes, covering 41 subcategories, were assigned to 3 main GO categories, for example, “GO:0044464 cell part”, “GO:0008152 metabolic process” and “GO:0003824 catalytic activity” had the highest frequencies in cellular components, biological processes and molecular function, respectively (Fig. 5a and Additional file 6: Table S4). Notably, most of ARF-correlated genes were identified to be related to cell metabolism, and thus we further explored the detailed category. “GO:0044260 protein metabolic process”, “GO:0006139 nucleobase, nucleoside, nucleotide and nucleic acid metabolic process”, “GO:0006810 transport”, “GO:0010467 gene expression” and “GO:0005975 carbohydrate metabolic process” were the top 5 subcategories (Fig. 5b). Moreover, these ARF-correlated genes participated in phosphorus, ketone, lipid, amino acid derivative and other metabolic processes (Fig. 5b), suggesting that ARFs may play a crucial role in several metabolic processes.
Fig. 5.
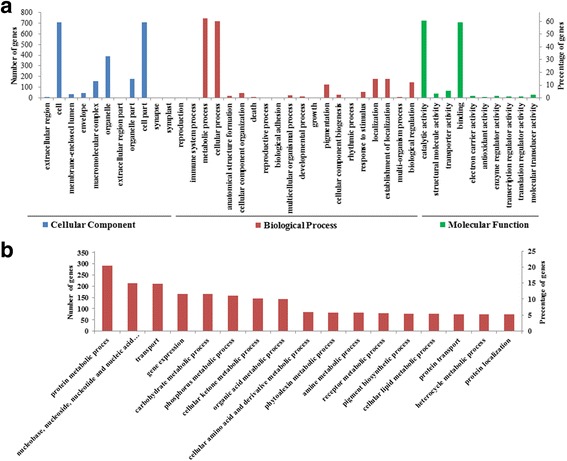
Histogram presentation of Gene Ontology classification. a The results are summarized in three main categories: biological process, cellular component, and molecular function. b The major subcategories in metabolic process. The y-axis on the left indicates the number of genes, and the y-axis on the right means the percent of genes in a category
Discussion
It is known that auxin is a key signaling molecule for most organogenesis and patterning processes during plant development. In plants, the ARFs could directly bind to the AuxRE of down-stream target genes and mediate their transcription involved in various biological processes [6, 25]. Recently, the characterization of ARF genes has been reported in Arabidopsis [18], O. sativa [47], Z. mays [28], P. trichocarpa [20] and Gossypium raimondii [42], but not in Siberian apricot, a novel and important woody oilseed species. In this study, 14 ARF transcription factor genes with a complete ORF were identified in P. sibirica according to transcriptomic data form different developing fruits [31] and different tissues [9].
Generally, the detailed information about the protein domains is helpful for understanding the function of the corresponding gene. It has been reported that most ARF proteins consist of an N-terminal B3-type DBD, a C-terminal CTD, and a variable MD that functions as an AD (rich in Q) or RD (rich in PST) [12]. As a typical ARF-type structure, the DBD domain is important for the recognition of AuxRE (TGTCTC) in the promoters of auxin-responsive genes [55]. Indeed, the DBD was identified in all 14 PsARF proteins (Fig. 1a), suggesting that DBD may be essential region for the function of PsARF proteins. It was reported that some ARFs exhibited an additional short segment of amino acids between the B3 and ARF subdomain in Arabidopsis [18] and P. trichocarpa [20]. Such a feature has also been observed in PsARF10, 16 and 17 proteins (Fig. 1a). At the end of the DBD domain, all the PsARF proteins contain a monopartite NLS, which has been experimentally confirmed to be able to direct the ARF proteins into the nucleus by a synthetic green fluorescent protein fusion assay in rice [40]. For the conserved structure of CTD, the region is directly involved in the hetero-dimerization of ARF and Aux/IAA proteins [23]. In this study, 5 PsARFs (PsARF3, 8, 10, 16 and 17), which lack a partial or complete CTD (Fig. 1a), should consequently be insensitive to auxin [13]. However, loss of CTD could also have consequences on the interaction of ARFs with other transcription factors [12]. The amino acid composition analysis of MD sequence in all 14 PsARF proteins revealed that PST rich regions were found in the middle regions of PsARF1–6, 10–11 and 16–17 (Fig. 1a), suggesting that these PsARFs might function as transcriptional repressors. In addition, PsARF7 and 19 harbour a Q-rich middle region (Fig. 1a), implying that PsARF7 and 19 are possibly transcriptional activators. Certainly, the specific regulatory role of PsARFs is misleading if without further experiments of functional verification.
We also built a phylogenetic tree to analyze the relationship of ARF families between Arabidopsis, B. rapa, C. sinensis, P. mume, and P. sibirica (Fig. 2). The result revealed that ARFs are distributed into four major clusters (I, II, III, and IV), which was similar to the previously phylogenetic classifications of ARFs in E. grandis [55], banana [52] and Arabidopsis [34]. Generally, the close relative species should expect to have similar structures in the same protein family. Indeed, ARFs from the P. sibirica were more closely related to those from P. mume than those from Arabidopsis, B. rapa and C. sinensis (Fig. 2).
Fruit (including mesocarp or seed) development is a complex interplay of cell division, differentiation and expansion that occurs in a temporally and spatially coordinated manner in the reproductive organs [15]. To assess the function of the PsARF genes in developing SAM and SAK, we investigated the expression profiles of PsARF genes by FPKM and qRT-PCR. Although the expressions of all 14 PsARF genes could be detected in both the SAM and SAK, different PsARF showed distinct temporal and spatial expression pattern in developing SAM and SAK (Figs. 3 and 4). In Arabidopsis, ARF1 may act with ARF2 to control aspects of maturation and senescence [10]. The homologous PsARF1 was significantly expressed at 60–70 DAF of SAK (Fig. 4). This finding, together with the involvement of highly PsARF1-correlated ETHYLENE INSENSITIVE 4 gene in growth and development of Arabidopsis [26], suggested that PsARF1 might play crucial roles in late SAK development, such as the acquisition of dormancy and desiccation tolerance. In addition, the high expression of PsARF 9 was characteristically transient at 60–70 DAF of SAK (Figs. 3 and 4). It has been reported that AtARF9 act in suspensor cells to mediate hypophysis specification in Arabidopsis [35]. One of PsARF9-correlated genes was also identified to code a RCD1-like protein involved in cell differentiation [50].
The mutation analyses revealed that AtARF2 is a general repressor of cell division to regulate seed size and weight [38]. Indeed, PsARF2 exhibited high expression levels (value >10) at all developmental stages of SAM (Fig. 3), implying potential influence of PsARF2 in fruit size. Additionally, high expression levels (value >10) of PsARF6, 7 and 8 at all developmental stages of SAM (Fig. 3) suggested that these genes might be involved in mesocarp development, as was reported that ARF8 regulate fertilization and fruit development in Arabidopsis [14]. By GO annotation in biological process, we identified most genes correlated with PsARF8 genes were widely involved in cellular metabolic and regulatory process, including carbohydrate, amino acid, organic acid and lipid metabolism. Interestingly, the SlARF4 that is highly homologous to PsARF8 was identified to be involved in the control of sugar metabolism during tomato fruit development [36]. Thus, it is tempting to speculate that PsARF8 may be a master transcriptional factor participated in the regulation of carbohydrate metabolism during SAM development.
Previous investigation in stem cell of Arabidopsis showed the responsible of AtARF16 for pattern specification process that results in the creation of defined areas or spaces within an organism to which cells respond and eventually are instructed to differentiate [8]. In this study, specifically transcriptional accumulation of PsARF16 was found at early development (10 DAF) of SAM and SAK (Figs. 3 and 4). Remarkably, PsARF16-correlated REVEILLE 6 gene, as one of circadian rhythm regulator, could inhibit plant growth in Arabidopsis [16]. Thus, the abundantly similar transcripts of PsARF16 at early development of SAM and SAK may help to a series of programmed cell divisions and the basic architecture of the plant. In addition, the high expression of PsARF11 was characteristically transient at early SAM development (Figs. 3 and 4). By function annotation, two PsARF11-correlated genes (LEUNIG_HOMOLOG and LEUNIG) regulate mucilage extrusion, mainly composed of pectin, required for mucilage maturation during Arabidopsis fruits development [5]. Thus, PsARF11 may be involved in cell wall formation.
Here, the observed low transcript of PsARF10, 17 and 19 both in developing SAM and SAK (Fig. 3) indicated that these genes are not likely to play important roles in SAM and SAK development. As in the case of Arabidopsis, no phenotypic defects were reported for ARF10 and 19 single mutants [34]. Summary, lineage-specific expression of PsARFs may create a pattern enabling different developmental auxin responses required for normal SAM and SAK development.
Conclusions
As ARF transcriptional factor is known to be implicated in regulation of fruit and seed development, it is important to understand their structure, transcription and regulation. Based on our previous transcriptomic data, we identified 14 PsARF genes with a perfect ORF, and analyzed their phylogenetic relationship, gene structure and protein motifs. The current work has contributed to an increased knowledge of the ARF gene family in P. sibirica. The expression profiles and co-expression networks of PsARF genes will provide a fundamental basis for in-depth experimental studies of ARF function in Siberian apricot.
Additional files
Primer sequences used for real-time PCR. (DOCX 19 kb)
The electropherogram of 14 PsARF genes. (TIF 77 kb)
Amino acid sequence alignments of 14 conserved motifs. (TIF 219 kb)
Components analysis in the amino acid of middle regions of 14 PsARF proteins. (XLSX 14 kb)
Co-expression analysis of ARF genes. (XLSX 74 kb)
Function and GO annotation of co-expression genes correlated with ARFs. + means the correlation of co-expressed genes with corresponding ARF, and – means no correlation of co-expressed genes with corresponding ARF. (XLSX 198 kb)
Acknowledgements
Not applicable.
Funding
This study was supported by the Central Public Interest Scientific Institution Basal Research Fund (CAFYBB2016QA001), National Natural Science Foundation of China (31700586), and Hainan university research funded projects (KYQD(ZR)1701). These funding agencies had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The ARF sequences in P. sibirica are available from the Genbank database (accession nos. MF373590–603). The transcriptomic data of P. sibirica are available from NCBI Short Read Archive database under accession SRX339392, SRR1564517, SRR1568273, SRR1568275, SRR1568789 and SRR1568805. The transcriptomic data of E. guineensis are available from SRX059258–62, SRX059116–20, and SRX059798–802. The transcriptomic data of B. napus are available from SRX090767–9 and SRX090771. The transcriptomic data of R. communis are available from SRX007402–6. The transcriptomic data of E. alatus are available from SRX070806 and SRX029137–42.
Abbreviations
- AD
Activation domain
- ARF
Auxin response factor
- Aux/IAA
Auxin/indole-3-acetic acid
- AuxRE
Auxin response element
- CTD
Carboxy-terminal dimerization domain
- DBD
DNA-binding domain
- FPKM
Fragment Per Kilobase of exon model per Million mapped reads
- GO
Gene Ontology
- MD
Middle domain
- MEME
Multiple Expectation Maximization for Motif Elicitation
- N
Asparagine
- NLS
Nuclear localization signal
- ORF
Open reading frame
- P
Proline
- Q
Glutamine
- RD
Repression domain
- S
Serine
- SAK
Siberian apricot kernel
- SAM
Siberian apricot mesocarp
- T
threonine
Authors’ contributions
JN conceived and designed the experiments. QB wrote the manuscript and performed the bioinformatics analysis. SD, HC and HY conducted the expression analysis, LW and SL conducted the co-expression and GO analysis. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12870-017-1220-2) contains supplementary material, which is available to authorized users.
Contributor Information
Jun Niu, Phone: +86-898-66268107, Email: niujun5555@163.com.
Quanxin Bi, Email: biquanxin@126.com.
Shuya Deng, Email: dddssy1125@163.com.
Huiping Chen, Email: Chenhuiping@hainan.net.
Haiyan Yu, Email: 971320870@qq.com.
Libing Wang, Email: wlibing@caf.ac.cn.
Shanzhi Lin, Email: 735238859@qq.com.
References
- 1.Attia KA, Abdelkhalik AF, Ammar MH, Wei C, Yang J, Lightfoot DA, El-Sayed WM, El-Shemy HA. Antisense phenotypes reveal a functional expression of OsARF1, an auxin response factor, in transgenic rice. Curr Issues Mol Biol. 2009;11:129. [PubMed] [Google Scholar]
- 2.Bailey TL, Johnson J, Grant CE, Noble WS. The MEME suite. Nucleic Acids Res. 2015;43:W39. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanc G, Hokamp K, Wolfe KH. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 2003;13:137–144. doi: 10.1101/gr.751803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourgis F, Kilaru A, Cao X, Ngandoebongue GF, Drira N, Ohlrogge JB, Arondel V. Comparative transcriptome and metabolite analysis of oil palm and date palm mesocarp that differ dramatically in carbon partitioning. P Nat Acad Sci USA. 2011;108:12527–12532. doi: 10.1073/pnas.1106502108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bui M, Lim N, Sijacic P, Liu Z. LEUNIG_HOMOLOG and LEUNIG regulate seed mucilage extrusion in Arabidopsis. J Integr Plant Biol. 2011;53:399. doi: 10.1111/j.1744-7909.2011.01036.x. [DOI] [PubMed] [Google Scholar]
- 6.Chandler JW. Auxin response factors. Plant Cell Environ. 2016;39:1014–1028. doi: 10.1111/pce.12662. [DOI] [PubMed] [Google Scholar]
- 7.Di DW, Zhang C, Luo P, An CW, Guo GQ. The biosynthesis of auxin: how many paths truly lead to IAA? Plant Growth Regul. 2016;78:275–285. doi: 10.1007/s10725-015-0103-5. [DOI] [Google Scholar]
- 8.Ding ZJ, Friml J. Auxin regulates distal stem cell differentiation in Arabidopsis roots. P Nat Acad Sci USA. 2010;107:12046–12051. doi: 10.1073/pnas.1000672107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong SB, Liu YL, Niu J, Yu N, Lin SZ, Zhang ZX. De novo transcriptome analysis of the Siberian apricot ( Prunus sibirica L.) and search for potential SSR markers by 454 pyrosequencing. Gene. 2014;544:220–227. doi: 10.1016/j.gene.2014.04.031. [DOI] [PubMed] [Google Scholar]
- 10.Ellis CM, Nagpal P, Young JC, Hagen G, Guilfoyle TJ, Reed JW. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development. 2005;132:4563–4574. doi: 10.1242/dev.02012. [DOI] [PubMed] [Google Scholar]
- 11.Fan S, Liang T, Yu H, Bi Q, Li G, Wang L. Kernel characteristics, oil contents, fatty acid compositions and biodiesel properties in developing Siberian apricot ( Prunus sibirica L.) seeds. Ind Crop Prod. 2016;89:195–199. doi: 10.1016/j.indcrop.2016.05.012. [DOI] [Google Scholar]
- 12.Finet C, Berne-Dedieu A, Scutt CP, Marlétaz F. Evolution of the ARF gene family in land plants: old domains, new tricks. Mol Biol Evol. 2012;30:45–56. doi: 10.1093/molbev/mss220. [DOI] [PubMed] [Google Scholar]
- 13.Finet C, Fourquin C, Vinauger M, Berne-Dedieu A, Chambrier P, Paindavoine S, Scutt CP. Parallel structural evolution of auxin response factors in the angiosperms. Plant J. 2010;63:952–959. doi: 10.1111/j.1365-313X.2010.04292.x. [DOI] [PubMed] [Google Scholar]
- 14.Goetz M, Vivian-Smith A, Johnson SD, Koltunow AM. AUXIN RESPONSE FACTOR8 is a negative regulator of fruit initiation in Arabidopsis. Plant Cell. 2006;18:1873. doi: 10.1105/tpc.105.037192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldberg RB, De Paiva G, Yadegari R. Plant embryogenesis: zygote to seed. Science. 1994;266:605–614. doi: 10.1126/science.266.5185.605. [DOI] [PubMed] [Google Scholar]
- 16.Gray JA, Shalit-Kaneh A, Chu DN, Hsu PY, Harmer SL. The REVEILLE clock genes inhibit growth of juvenile and adult plants by control of cell size. Plant Physiol. 2017;173:2308. doi: 10.1104/pp.17.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guilfoyle TJ. The PB1 domain in auxin response factor and aux/IAA proteins: a versatile protein interaction module in the auxin response. Plant Cell. 2015;27:33–43. doi: 10.1105/tpc.114.132753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagen G, Guilfoyle T. Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol. 2002;49:373–385. doi: 10.1023/A:1015207114117. [DOI] [PubMed] [Google Scholar]
- 19.Hardtke CS, Berleth T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998;17:1405. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalluri UC, Difazio SP, Brunner AM, Tuskan GA. Genome-wide analysis of aux/IAA and ARF gene families in Populus trichocarpa. BMC Plant Biol. 2007;7:1–14. doi: 10.1186/1471-2229-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lau S, De SI, Kolb M, Meinhardt H, Jürgens G. Auxin triggers a genetic switch. Nat Cell Biol. 2011;13:611–615. doi: 10.1038/ncb2212. [DOI] [PubMed] [Google Scholar]
- 24.Li SB, Ouyang WZ, Hou XJ, Xie LL, Hu CG, Zhang JZ. Genome-wide identification, isolation and expression analysis of auxin response factor (ARF) gene family in sweet orange (Citrus sinensis) Front Plant Sci. 2015;6:119. doi: 10.3389/fpls.2015.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li SB, Xie ZZ, Hu CG, Zhang JZ. A review of auxin response factors (ARFs) in plants. Front Plant Sci. 2016;7:47. doi: 10.3389/fpls.2016.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li ZG, Chen HW, Li QT, Tao JJ, Bian XH, Ma B, Zhang WK, Chen SY, Zhang JS. Three SAUR proteins SAUR76, SAUR77 and SAUR78 promote plant growth in Arabidopsis. Sci Rep-UK. 2015;5:12477. doi: 10.1038/srep12477. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Liu SQ, Hu LF. Genome-wide analysis of the auxin response factor gene family in cucumber. Genet Mol Res. 2013;12:4317–4331. doi: 10.4238/2013.April.2.1. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Jiang HY, Chen W, Qian Y, Ma Q, Cheng B, Zhu S. Genome-wide analysis of the auxin response factor (ARF) gene family in maize (Zea mays) Plant Growth Regul. 2011;63:225–234. doi: 10.1007/s10725-010-9519-0. [DOI] [Google Scholar]
- 29.lv Z, Guo Z, Xiao J, Wang F. Karyotype analysis of Siberian apricot and apricot. North Fruits. 1986;1:18–20. [Google Scholar]
- 30.Mun JH, Yu HJ, Shin JY, Oh M, Hwang HJ, Chung H. Auxin response factor gene family in Brassica rapa: genomic organization, divergence, expression, and evolution. Mol Gen Genomics. 2012;287:765–784. doi: 10.1007/s00438-012-0718-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niu J, An JY, Wang LB, Fang CL, Ha DL, Fu CY, Qiu L, Yu HY, Zhao HY, Hou XY, Xiang Z, Zhou SF, Zhang ZX, Feng XY, Lin SZ. Transcriptomic analysis revealed the mechanism of oil dynamic accumulation during developing Siberian apricot (Prunus sibirica L.) seed kernels for the development of woody biodiesel. Biotechnol Biofuels. 2015;8:29–44. doi: 10.1186/s13068-015-0213-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niu J, Wang J, An J, Liu L, Lin Z, Wang R, Wang L, Ma C, Shi L, Lin S. Integrated mRNA and miRNA transcriptome reveal a cross-talk between developing response and hormone signaling for the seed kernels of Siberian apricot. Sci Rep-UK. 2016;6:35675. doi: 10.1038/srep35675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niu J, Zhu B, Cai J, Li P, Wang L, Dai H, Qiu L, Yu H, Ha D, Zhao H. Selection of reference genes for gene expression studies in Siberian apricot (Prunus sibirica L.) Germplasm using quantitative real-time PCR. PLoS One. 2014;9:e103900. doi: 10.1371/journal.pone.0103900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okushima Y, Overvoorde P, Arima K, Alonso J, Chan A, Chang C, Ecker J, Hughes B, Lui A, Nguyen D, Onodera C, Quach H, Smith A, Yu G, Theologis A. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell. 2005;17:444. doi: 10.1105/tpc.104.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rademacher Eike H, Lokerse Annemarie S, Schlereth A, LlavataPeris Cristina I, Bayer M, Kientz M, Rios AF, Borst JW, Lukowitz W, Jürgens G. Different auxin response machineries control distinct cell fates in the early plant embryo. Dev Cell. 2012;22:211–222. doi: 10.1016/j.devcel.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 36.Sagar M, Chervin C, Mila I, Hao Y, Roustan JP, Benichou M, Gibon Y, Biais B, Maury P, Latché A. SlARF4, an auxin response factor involved in the control of sugar metabolism during tomato fruit development. Plant Physiol. 2013;161:1362–1374. doi: 10.1104/pp.113.213843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sagar M, Chervin C, Roustan JP, Bouzayen M, Zouine M. Under-expression of the Auxin response factor Sl-ARF4 improves post-harvest behavior of tomato fruits. Plant Signal Behav. 2013;8:e25647. doi: 10.4161/psb.25647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schruff MC, Spielman M, Tiwari S, Adams S, Fenby N, Scott RJ. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development. 2006;133:251. doi: 10.1242/dev.02194. [DOI] [PubMed] [Google Scholar]
- 39.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen CJ, Wang SK, Bai YH, Wu YR, Zhang SN, Chen M, Guilfoyle TJ, Wu P, Qi YH. Functional analysis of the structural domain of ARF proteins in rice (Oryza sativaL.) J Exp Bot. 2010;61:3971–3981. doi: 10.1093/jxb/erq208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song J, Gao Z, Huo X, Sun H, Xu Y, Shi T, Ni Z. Genome-wide identification of the auxin response factor ( ARF ) gene family and expression analysis of its role associated with pistil development in Japanese apricot ( Prunus Mume Sieb. Et Zucc) Acta Physiol Plant. 2015;37:145. doi: 10.1007/s11738-015-1882-z. [DOI] [Google Scholar]
- 42.Sun R, Wang K, Guo T, Jones DC, Cobb J, Zhang B, Wang Q. Genome-wide identification of auxin response factor (ARF) genes and its tissue-specific prominent expression in Gossypium raimondii. Funct Integ Genom. 2015;15:1–13. doi: 10.1007/s10142-014-0393-0. [DOI] [PubMed] [Google Scholar]
- 43.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Troncoso-Ponce MA, Aruna K, Xia C, Durrett TP, Fan J, Jensen JK, Thrower NA, Markus P, Curtis W, Ohlrogge JB. Comparative deep transcriptional profiling of four developing oilseeds. Plant J. 2011;68:1014–1027. doi: 10.1111/j.1365-313X.2011.04751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ulmasov T, Hagen G, Guilfoyle TJ. Activation and repression of transcription by auxin-response factors. P Nat Acad Sci USA. 1999;96:5844–5849. doi: 10.1073/pnas.96.10.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang D, Pei K, Fu Y, Sun Z, Li S, Liu H, Tang K, Han B, Tao Y. Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa) Gene. 2007;394:13–24. doi: 10.1016/j.gene.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 48.Wang L. Resource investigation and distribution pattern of three Armeniaca species. Forest Resour Manage. 2011;5:65–70. [Google Scholar]
- 49.Wang L. Evaluation of Siberian apricot (Prunus sibirica L.) germplasm variability for biodiesel properties. J Am Oil Chem Soc. 2012;89:1743–1747. doi: 10.1007/s11746-012-2069-z. [DOI] [Google Scholar]
- 50.Wang Y, Zhang WZ, Song LF, Zou JJ, Su Z, Wu WH. Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol. 2008;148:1201. doi: 10.1104/pp.108.126375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Waterhouse A, Procter J, Martin D, Clamp M, Barton G. Jalview version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei H, Jiao Z, Hou X, Yan Y, Wei Y, Liu J, Li M, Xu B, Jin Z. The auxin response factor gene family in banana: genome-wide identification and expression analyses during development, ripening, and abiotic stress. Front Plant Sci. 2015;6:742. doi: 10.3389/fpls.2015.00742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao X, Ma J, Sun Y, Yao Y. A method for the further assembly of targeted unigenes in a transcriptome after assembly by trinity. Front Plant Sci. 2015;6:843. doi: 10.3389/fpls.2015.00843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J, Li S, Li R, Bolund L. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006;34:293–297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu H, Soler M, Mila I, Clemente HS, Savelli B, Dunand C, Paiva JAP, Myburg AA, Bouzayen M, Grimapettenati J. Genome-wide characterization and expression profiling of the AUXIN RESPONSE FACTOR (ARF) gene family in Eucalyptus grandis. PLoS One. 2014;9:e108906. doi: 10.1371/journal.pone.0108906. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer sequences used for real-time PCR. (DOCX 19 kb)
The electropherogram of 14 PsARF genes. (TIF 77 kb)
Amino acid sequence alignments of 14 conserved motifs. (TIF 219 kb)
Components analysis in the amino acid of middle regions of 14 PsARF proteins. (XLSX 14 kb)
Co-expression analysis of ARF genes. (XLSX 74 kb)
Function and GO annotation of co-expression genes correlated with ARFs. + means the correlation of co-expressed genes with corresponding ARF, and – means no correlation of co-expressed genes with corresponding ARF. (XLSX 198 kb)
Data Availability Statement
The ARF sequences in P. sibirica are available from the Genbank database (accession nos. MF373590–603). The transcriptomic data of P. sibirica are available from NCBI Short Read Archive database under accession SRX339392, SRR1564517, SRR1568273, SRR1568275, SRR1568789 and SRR1568805. The transcriptomic data of E. guineensis are available from SRX059258–62, SRX059116–20, and SRX059798–802. The transcriptomic data of B. napus are available from SRX090767–9 and SRX090771. The transcriptomic data of R. communis are available from SRX007402–6. The transcriptomic data of E. alatus are available from SRX070806 and SRX029137–42.


