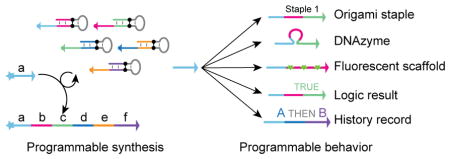
Abstract
DNA performs diverse functional roles in biology, nanotechnology, and biotechnology, but current methods for autonomously synthesizing arbitrary single-stranded DNA are limited. Here, we introduce the concept of Primer Exchange Reaction (PER) cascades, which grow nascent single-stranded DNA with user-specified sequences following prescribed reaction pathways. PER synthesis happens in a programmable, autonomous, in situ, and environmentally responsive fashion, providing a platform for engineering molecular circuits and devices with a wide range of sensing, monitoring, recording, signal processing, and actuation capabilities. We experimentally demonstrate a nanodevice that transduces the detection of a trigger RNA into the production of a DNAzyme that degrades an independent RNA substrate, a signal amplifier that conditionally synthesizes long fluorescent strands only in the presence of a particular RNA signal, molecular computing circuits that evaluate logic (AND, OR, NOT) combinations of RNA inputs, and a temporal molecular event recorder that records in the PER transcript the order in which distinct RNA inputs are sequentially detected.
Graphical Abstract
DNA is the information carrier for life and encodes the instructions for diverse molecular functions in the genome. DNA is also a foundational element in nanotechnology, and used to construct nanostructures,1–11 and dynamic circuits and devices12–20 with diverse technological applications21–32 (e.g. to spatially organize inorganic and organic materials for nanofabrication and biomedical applications,23,24,27 for super-resolution imaging,26 and as single molecule tools25,31,32). DNA is also widely used in biotechnology applications33–38 (e.g. biosensing, sequence amplification, genome editing, and even memory storage).
Despite the diverse applications of DNA in biology, nanotechnology, and biotechnology, current programmable methods for autonomously synthesizing single-stranded sequences are limited. DNA oligos are typically synthesized non-autonomously through a staged process that uses specific chemical conditions at each of several externally controlled steps.39 Enzyme-driven methods like the polymerase chain reaction (PCR),40 loop-mediated isothermal amplification (LAMP),35 strand displacement amplification (SDA),33 rolling circle amplification (RCA),34 and polymerase/exonuclease/nickase (PEN) reactions41,42 are typically limited to producing identical copies of fixed sequences. To overcome this limitation, we introduce Primer Exchange Reaction (PER) cascades, which isothermally produce single-stranded DNA with arbitrary user-prescribed sequences in a programmable, autonomous, stepwise fashion with the aid of a strand-displacing polymerase. The PER cascade starts with a prescribed DNA primer. Utilizing a catalytic DNA hairpin mediator, PER then appends to the existing primer a new primer with an independent, user-specified sequence (i.e. a Primer Exchange Reaction). The newly extended primer can then trigger the next step extension, thus forming a programmable PER cascade to autonomously grow a nascent DNA strand along a prescribed pathway to produce an arbitrary user-prescribed sequence (Fig. 1a). Although PEN reaction networks41,42 also utilize a strand displacing polymerase (together with a nicking enzyme) to cascade one primer sequence into another, the PER cascades here introduce the critical new capability of concatenating newly synthesized transcripts to grow a nascent DNA strand, which can serve diverse functional roles.
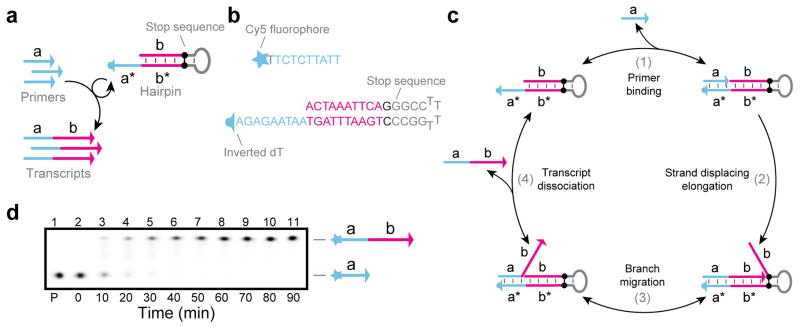
Figure 1. Primer Exchange Reactions (PER) overview.
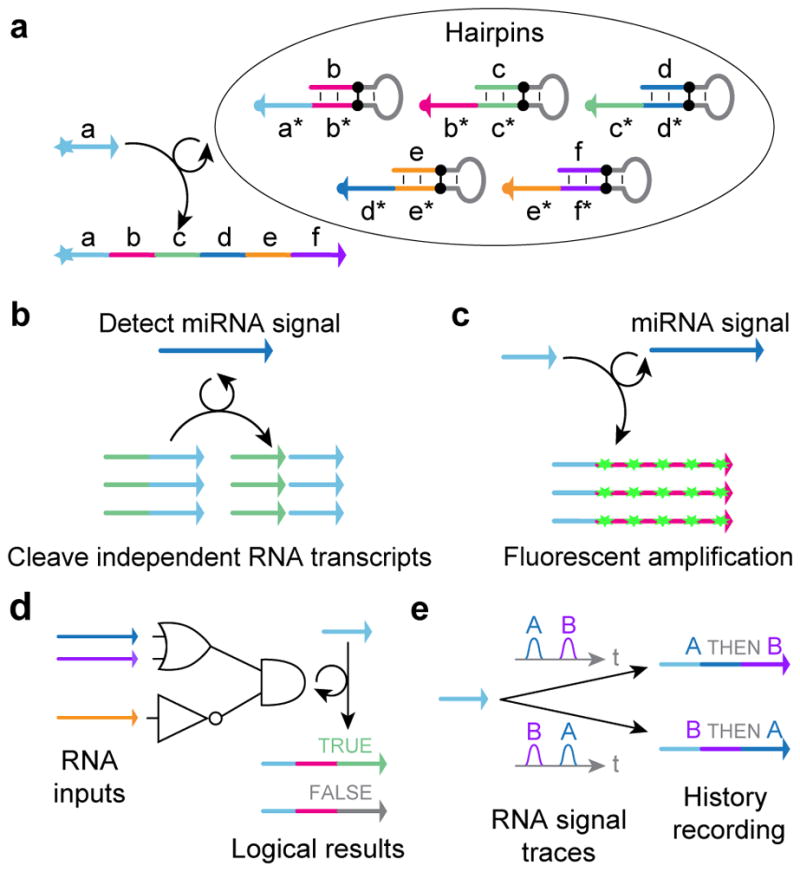
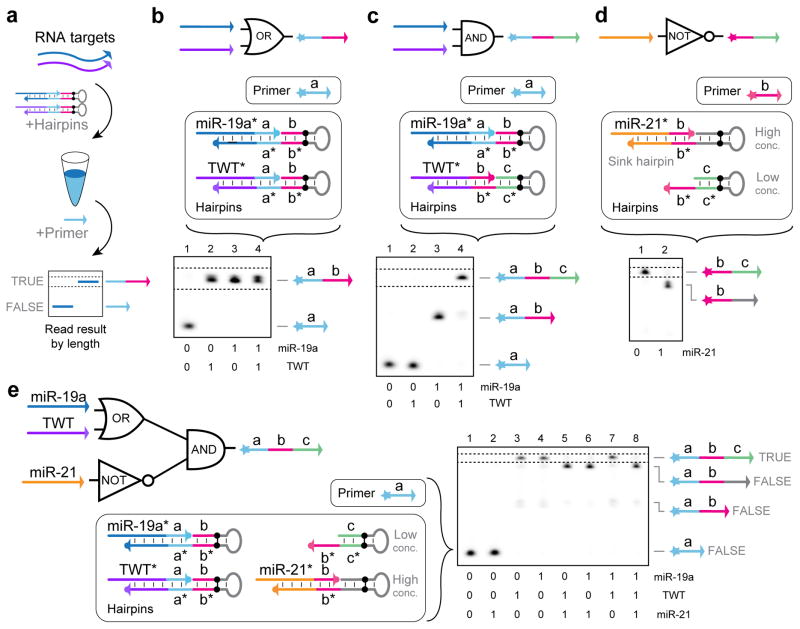
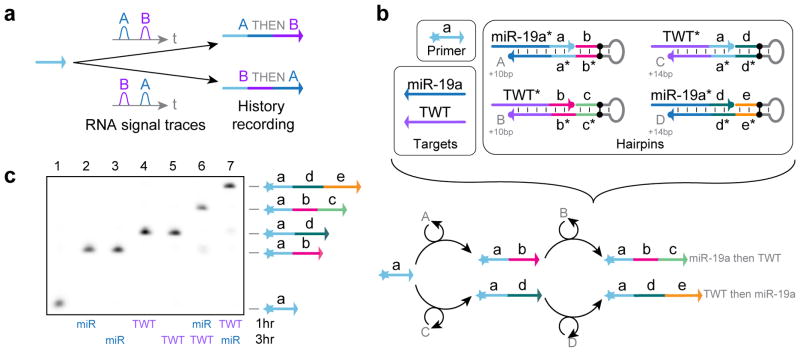
(a) PER cascades grow nascent strands of single-stranded DNA (ssDNA) with prescribed sequences. (b) A nanodevice which synthesizes a functional DNAzyme that cleaves an independent transcript in response to an input miRNA signal. (c) A PER signal amplifier that conditionally synthesizes fluorescent strands only in the presence of a particular RNA signal. (d) PER logic computation, which evaluates a logic expression based on which RNA inputs are present. (e) A PER molecular program which records the order in which two signals are encountered in a DNA transcript.
When and only when the right set of PER hairpins and the corresponding PER primer co-exist, a new strand is produced autonomously in situ. As the strand can be programmed to be of arbitrary user-specified sequence, it could serve many of the functional roles of DNA described above (e.g. structural elements for nanoconstructs, templates for organizing materials, chemically active species, etc), providing a way for programmable in situ control/delivery of such functions. Additionally, by interfacing the PER hairpins with external molecular elements, each of these functional responses could be programmed to respond to different environmental cues (e.g. the presence or absence of certain combinations of biomolecules, e.g. microRNAs) (Fig. 1b–e). Such responsive synthesis could be used to implement molecular behaviors that are tailored to particular environmental contexts, enabling programmable dynamic molecular circuits with a wide range of molecular sensing, monitoring, recording, signal processing, and actuation capabilities.
To explore these conceptual possibilities, in this paper, we experimentally demonstrated several distinct types of molecular behaviors that can be implemented with PER cascades. These include a nanodevice that transduces the detection of a trigger RNA into the production of a DNAzyme that degrades an independent RNA substrate, a signal amplifier that conditionally synthesizes long fluorescent strands only in the presence of a particular RNA signal, molecular computing circuits that evaluate logic (AND, OR, NOT) combinations of RNA inputs, and a temporal molecular event recorder that records in the PER transcript the order in which distinct RNA inputs are sequentially detected. Collectively, these results suggest that the PER cascades could provide the basis for a new generation of programmable molecular devices.
Results
Primer exchange reaction mechanism
A PER reaction is patterned by a single catalytic PER hairpin (Fig. 2a), which prescribes the sequence (domain b) that gets appended to primer strands with sequence domain a. Fig. 2c depicts the PER reaction cycle. In step 1, a primer with sequence domain a binds to its complement a* on the 3′ end of the primer. A strand displacing polymerase can subsequently extend the primer by copying the b domain (which is termed the copy region of the PER hairpin) before halting at the stop sequence (step 2). The copied b domain is then able to compete with the b domain on the hairpin through the random walk process of three-way branch migration43 (step 3). Finally, the extended primer can spontaneously dissociate from the hairpin once the copied domain b has been displaced (step 4). The hairpin is then free to interact with another primer in another cycle of primer exchange. In our experiments, we use 7–9 nt primer binding domains, which enable effective priming in step 2 and permit efficient spontaneous dissociation in step 4 at 37 °C. The kinetics of this reaction have been further characterized in Supplementary Note 1. Using a series of two minute PER reactions with different hairpin concentrations, we found that the reaction can be roughly approximated as a bimolecular reaction between primers and hairpins with a rate constant of 5.4 x 105 M−1s−1.
Figure 2. PER mechanism.
(a) A primer exchange reaction, which uses a catalytic hairpin to produce transcripts of the form a–b. (b) An example reaction implementation with a hairpin that uses a G-C pair as a stop sequence. See Supplementary Note 2 for other types of stop sequences that can be used. (c) The primer exchange reaction cycle. (d) PAGE denaturing gel depicting a reaction time series of a primer exchange reaction. Primer and hairpin concentrations were fixed at 100 nM and 1 nM respectively, and reactions were incubated at 37 °C with 10 μM of dATP, dTTP, and dCTP. See Methods section and Supplementary Note 3 for additional details.
Primer exchange hairpins were repurposed from the “Copy-and-Release Hairpin” that we recently described for the Autocyclic Proximity Recording mechanism.44 Each PER hairpin uses a stop sequence to halt polymerization.45–48 For example, this stop sequence can be a G-C pair (Fig. 2b) if the a (blue) and b (pink) domains comprise a three-letter code of A’s, T’s, and C’s, and dGTP is excluded from the mix. Other stop sequences include chemical modifications and synthetic base pairs, which permit all four letters to be synthesized (see Fig. 4b and Supplementary Note 2). Primers can be labeled with a dye on their 5′ ends for tracking in gel electrophoresis experiments (Fig. 2d). Hairpins also typically contain an inverted dT or other modification on their 3′ ends to prevent their extension on the 5′ unbound region of the primer (if any; not depicted), which could cause the primers to become irreversibly bound (data not shown).
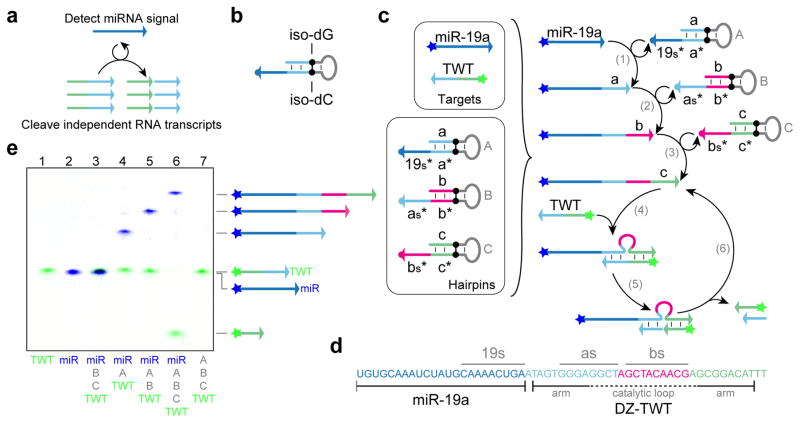
Figure 4. PER nanodevice for conditional RNA degradation.
(a) Schematic for nanodevice, which detects a miRNA target and cleaves an independent RNA transcript. (b) Synthetic nucleotides, iso-dG and iso-dC, which are used as a stop sequence for four-letter code synthesis. (c) System setup and reaction diagram for nanodevice. The oncogenic miR-19a sequence triggers the cleavage of a fragment of the Twist mRNA sequence (TWT) through the synthesis of a 10 – 23 DNAzyme sequence (DZ-TWT) by the A, B, and C hairpins. (d) Sequence breakdown of miRNA target (miR-19a) with DNAzyme (DZ-TWT) appended. Binding regions of the nascent primer strand are indicated by lines above the sequence, and the arms and catalytic domain of the DNAzyme are depicted below. (e) PAGE denaturing gel validating the miRNA and Twist mRNA fragment states given different hairpins in the incubation solution. Reactions were incubated for 4 hours at 37 °C with dNTPs at 10 μM each. The miRNA target and hairpins B and C were at 10 nM, and the TWT fragment and hairpin A were at 20 nM. See Methods section for full experimental details.
Primers and hairpins were incubated together with Bst Large Fragment polymerase to validate and characterize the basic single primer exchange reaction (Fig. 2d). The system comprises two components, the fluorophore-labeled primer and hairpin depicted in Fig. 2b, and denaturing gel electrophoresis was used to track the progression of the basic primer exchange reaction. Lane 1 shows the band corresponding to the primer with no hairpin, and lanes 2–11 depict the time progression of primer exchange reaction at 10 minute intervals with a 100:1 primer to hairpin ratio. These data confirm the catalytic recycling of the hairpin in solution. Bst Large Fragment polymerase, which has strong strand displacing and no exonuclease activity, was the first polymerase we tried. As it worked well initially, we used it for all other systems for consistency. However, other non-exonuclease polymerases (e.g. Klenow (exo-) and Bsm) were also observed to be compatible with PER.
PER cascades for synthesizing user-prescribed sequences
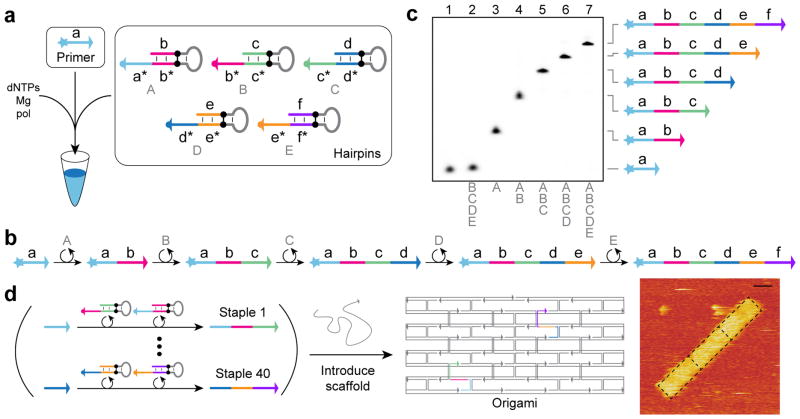
Primer exchange reactions can be chained together to form PER cascades that grow DNA strands of fixed length and prescribed sequence following a prescribed reaction pathway. The first PER pathway we implemented was a cascade of 5 elongation steps mediated by a set of catalytic hairpin species (Fig. 3a). With all 5 hairpins and the primer present in solution, the pathway proceeds through 5 elongation steps (Fig. 3b). Hairpin A catalyzes the extension of the primer domain a with domain b. Then, hairpin B catalyzes the extension of domain b with domain c. And hairpins C through E catalyze the extension of c with d, d with e, and e with f, respectively.
Figure 3. PER cascades.
(a) Schematic for autonomous stepwise growth of a primer sequence. (b) Reaction diagram for five elongation steps, patterned by hairpins A, B, C, D, and E. (c) Denaturing gel demonstrating differential extension with different subsets of the hairpin species present. Reactions were incubated for 4 hours at 37 °C, with primers at 100 nM, hairpins at 10 nM, and dATP, dTTP, and dCTP at 10 μM each. (d) Schematic for parallel synthesis of 40 staple strands for a DNA origami structure and AFM image of three of these structures aggregated with expected origami borders overlaid. PER synthesis reactions were incubated for 1 hour at 37 °C, with primers at 100 nM, hairpins at 10 nM, and dATP, dTTP, and dCTP at 100 μM each. After heat inactivation, scaffold was introduced directly to the reactions, and structures were annealed from 80 °C to 20 °C over 1 hour. See Methods section and Supplementary Note 4 for additional details.
Denaturing gel electrophoresis validates the ordered elongation of the primer strand when mixed with different subsets of the hairpins (Fig. 3c). Lane 1 shows a reaction where just the primer and no hairpins were incubated together. Lane 2 shows the primer incubated with hairpins B through E, which is all hairpins except the one that initiates the growth, hairpin A. This control shows no detectable leakage. Lane 3 shows the first elongation step with the primer and hairpin A incubated together. Lane 4 shows two elongations when the primer is incubated with the first two hairpins, A and B. Lanes 5, 6, and 7 show three, four, and five elongation steps of the primer when incubated with hairpins A through C, A through D, and A through E, respectively. Expected lengths of desired product bands were further verified by imaging gels after Sybr Gold staining. Catalytic turnover of all hairpins is verified through the near complete conversion of all primers to the last state, despite the hairpins being present at 1/10th the primer concentration.
To demonstrate how PER-synthesized strands can serve as elements for nanoconstruction, we sought to synthesize the staples for a DNA origami2 structure in a one-pot reaction. A structure comprising a 3-letter code scaffold held together by 40 staple strands was designed to fold into a compact rectangular shape that aggregates along its short end to produce chains of origami structures. A total of 80 hairpins were designed to synthesize the 40 staple strands from 40 primers (Fig. 3d), each in two reaction steps (see Supplementary Note 4 for additional details). The 40 reaction cascades were all run in parallel through incubation with polymerase for 1 hour at 37 °C. After heat inactivation, the scaffold strand was introduced directly into the PER mix and annealed for one hour. The designed morphology of the resultant origami structures were then verified by visualization with atomic force microscopy (see Methods and Supplementary Sections 10 and 11).
We call the primer exchange reaction a molecular primitive, due to its generality, modularity, and programmability (See Supplementary Note 5 for other examples of molecular primitives). Using the PER primitive, we can prescribe the number of steps as well as the sequence for each of these ordered steps in the reaction cascade. Additionally, the number of nucleotides added per step can be changed simply by including additional bases in the copy region of PER hairpin’s stem. Note that the copy region can be much longer than the primer binding region, which is typically kept at 7–9 nt under our typical reaction conditions to simultaneously allow (1) efficient primer binding and priming for polymerization and (2) dissociation of the elongated transcript from the hairpin.
These features enable the PER cascade to be programmed to produce arbitrary user prescribed sequences, as exemplified by the 5-step synthesis in Fig. 3a–c, the simultaneous operation of 40 orthogonal two-step reaction cascades in Fig. 3d, and the synthesis of a functional DNAzyme in next section.
Nanodevice for conditional RNA degradation
The DNA strand produced by PER cascades can serve as in situ synthesized, functional elements. In addition to serving as structural components for forming nanostructures (e.g. as staples for DNA origami in Fig. 3d), the strands can also function as chemically active species, as shown next. Additionally, the PER cascade can be programmed to initiate only when encountering specific molecular triggers. Utilizing these features, we next demonstrate the construction of a PER nanodevice for conditional RNA degradation. More specifically, the nanodevice detects the oncogenic miR-19a signal and subsequently synthesizes a functional DNAzyme that is programmed to cut an independent RNA substrate (Fig. 4a). For target detection, we designed a hairpin to bind the shared 3′ region of the oncogenic microRNAs (miRNAs) miR-19a and miR-19b.49 In response to detection, a PER pathway synthesizes a DNAzyme (DZ-TWT) that has been shown to cleave the full length mRNA of the Twist gene and promote apoptosis in vivo.50
For this application, a pair of synthetic nucleotides, iso-dG and iso-dC, were utilized as a stop sequence on all hairpins (Fig. 4b). This allowed us to utilize all four DNA bases in the synthesized sequences. As an example of an output construct for PER pathways, we chose to use the DZ-TWT DNAzyme, which has already been validated in vivo.50 DZ-TWT is part of the class of 10–23 DNAzymes that use a 15 nt catalytic loop domain to cut a purine-pyrimidine bond in a target RNA sequence.51 The catalytic loop is positioned at the cleavage site with the aid of two adjacent binding arms, whose sequences can be re-programmed to bind to and cleave arbitrary RNA targets.
To implement the system logic, three PER hairpins were designed to synthesize the DZ-TWT sequence only upon detection of the target microRNA (Fig. 4c). When the target is present, the A hairpin directs the synthesis of domain a onto the target strand (step 1). The B and C hairpins can subsequently pattern the appendage of domains b and c, respectively (steps 2–3). The completed a-b-c sequence forms the DZ-TWT sequence, which can bind to a 24 nt fragment of the Twist mRNA sequence (TWT) to form a loop that catalyzes the cleavage of the RNA at a particular base (steps 4–5). The DNAzyme can then be recycled after it dissociates from the cleaved fragments (step 6).
The sequences of the full DNAzyme appended to the target are shown in Fig. 4d. The three domains we split the DZ-TWT sequence into are shown in different colors. The lengths of the primer binding sites on the hairpins (19s*, as*, and bs* on hairpins A, B, and C respectively) were adjusted to bind 8–9 nt of the nascent strand to ensure the nascent strand would dissociate from hairpins after copying.
To evaluate the nanodevice function, we included different subsets of hairpins and the FAM-labeled TWT fragment with and without the Cy5-labeled miRNA target (Fig. 4e). Lanes 1 and 2 show the TWT fragment and miR-19a bands when incubated without any hairpins present. Lane 3 shows that there is no elongation of the miRNA in the presence of the last two hairpins, B and C, but not the first, A. Lane 4 shows the first step of extension when the miRNA is incubated with just the A hairpin and TWT fragment. Lane 5 shows incubation of the target with the first two hairpins, A and B, which results in two step elongation. When the target is mixed with all three hairpins (Lane 6), the full DZ-TWT sequence is appended to it and successfully cleaves the TWT fragment as expected. But when all the hairpins are mixed without target, no cleavage of the TWT fragment occurs (Lane 7). Catalytic turnover of the DNAzyme construct is indicated by essentially complete cleavage of 20 nM TWT RNA with only 10 nM of miRNA target.
Synthetic “telomerase” as an in situ signal amplifier
With the nanodevice, we were able to synthesize an arbitrary biologically relevant DNA sequence in response to an arbitrary input sequence. This programmability of PER pathways to transduce one sequence into another presents a modular framework for engineering environmentally responsive synthetic systems, which we explored further with several additional applications below. We next implemented a single-hairpin system that synthesizes long strands of repeated sequence domains (Fig. 5a), which we call a synthetic “telomerase”, and then used this construct as a signal amplifier which grows fluorescent concatemers upon detecting a particular miRNA input (Fig. 5c).
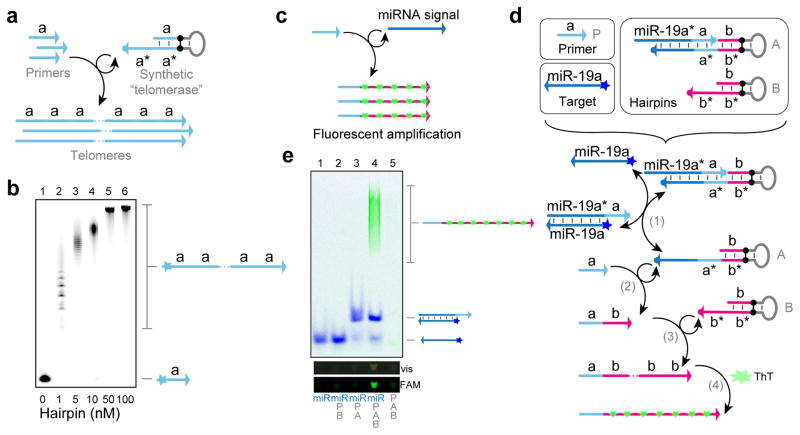
Figure 5. Signal amplifier with PER.
(a) Schematic for implementing a synthetic “telomerase” with a single PER hairpin. (b) A PAGE denaturing gel showing telomerization under different hairpin concentrations. 100 nM primers were incubated with the hairpins for 4 hours at 37 °C with dATP, dTTP, and dCTP at 100 μM. (c) Schematic for signal amplifier, where a miRNA target activates the synthesis of fluorescent telomere strands. (d) System components and reaction diagram for the signal amplifier. A gated hairpin (A) and telomerase hairpin (B) are designed to react to the detection of a miRNA signal and concatenate repeats of the human telomeric sequence TTAGGG, into which Thioflavin (ThT) intercalates. (e) A native PAGE gel showing conditional telomerization in the presence of 10 nM miRNA signal for 4 hours at 37 °C with dATP, dTTP, and dGTP at 100 μM. Primer P and hairpins A and B were held at 100 nM, 10 nM, and 1 μM, respectively. The protector for hairpin A was at 15 nM. Target detection can be visualized with a blue light transilluminator through the amber filter unit (vis) and on a Typhoon scanner (FAM). See Methods section for additional details.
The synthetic telomerase system comprises a single primer sequence (with domain a) and a hairpin that catalyzes the appendage of a repeated domain a onto the growing strand (Fig. 5a). For this demonstration, we chose the 10 nt (nucleotide) sequence ATCTCTTATT as the repeated domain, with the hairpin binding region corresponding to the last 9 nt of the sequence. As before, the primer was labeled with a Cy5 fluorophore for gel visualization and the hairpin was fitted with an inverted dT on its 3′ end to prevent its extension.
We validated the construct using a similar experimental setup as previous demonstrations, varying the hairpin concentration over two orders of magnitude to show how the reaction rate changes (Fig. 5b). Hairpins were incubated with the primer after 15 minutes of incubation with a specialized hairpin species designed to consume extraneous dGTP (see Supplementary Note 6 for details). Lane 1 shows the primer band with no hairpin added, and lanes 2 through 6 show “telomeres” grown with hairpin concentrations ranging from 1 to 100 nM. Increasing the hairpin concentrations resulted in longer telomeric sequences, which indicates the rate of telomerization can be tuned by adjusting hairpin concentration. We also discovered that the magnesium concentration can be used to modulate the telomerization rate, providing yet another method for adjusting reaction kinetics (see Supplementary Note 7).
We subsequently devised a strategy for implementing a signal amplifier that could conditionally grow this type of telomeric output only in response to the presence of a particular miRNA signal (Fig. 5c). By designing the sequences to bind a particular fluorescent dye, we were able to make these synthesized telomeres fluorescent, allowing for direct visualization of the result under a blue light. The fluorescence is achieved through the binding of the Thioflavin T (ThT) dye, which has been shown to become more fluorescent once intercalated into the quadruplex motifs formed by repeats of the human telomeric sequence TTAGGG.52 The target we chose was an oncogenic miRNA, miR-19a.49
Three components are required to implement the detector-telomerase system (Fig. 5d): a primer (P), a gated hairpin (A), and a telomerase hairpin (B). To transduce target detection into the synthesis of the human telomeric sequence, a toehold exchange reaction12,13 was designed such that the primer binding site of a PER hairpin (A) is only exposed in the presence of the cognate miR-19a signal. The miRNA target can bind onto a short exposed domain on a protector strand bound to the A hairpin and branch migrate through the remaining complementary sequence. Once the complementary miR-19a* domain has been fully displaced, the protector strand can spontaneously dissociate from the protected PER hairpin, exposing the primer binding site a* on the A hairpin (step 1). Once exposed, this PER hairpin facilitates the appendage of the b domain onto the a domain of a primer (step 2). The b domain corresponds to the human telomeric sequence TTAGGG, which is subsequently telomerized by a constitutively active telomerase hairpin B (step 3). Finally, these telomeres form a quadruplex structure, into which the ThT dye intercalates and becomes fluorescent (step 4).
The toehold exchange mechanism introduced here allows arbitrary input sequences to be connected to arbitrary primer exchange reactions using gated hairpins (see Supplementary Note 8). This is a more modular and scalable approach than using the input sequence directly as the primer as in Fig. 4 and expands the types of dynamic machinery that can be implemented with PER cascades, as each individual reaction can now be independently be programmed to be conditioned upon different environmental signals. This is exemplified in the two molecular systems below.
To evaluate the conditional telomerization reaction, we incubated reactions with different subsets of components and visualized the results on a native PAGE gel (see Methods section for full details) (Fig. 5e). Lane 1 shows a reaction with just the miRNA and none of the hairpins. With the primer P and telomerase hairpin B but no gated hairpin A, the miRNA target does not bind to any oligos, and there is no telomerization (Lane 2). With the primer P and gated hairpin A, but no telomerase hairpin B, the miRNA binds to the protector strand as indicated by the shift in its band (Lane 3). Telomerization only occurs when all three of P, A, and B are present with the miRNA target (Lane 4). In the control reaction, which contains none of the miRNA target, elements P, A, and B produce almost no background telomerization (Lane 5).
As a simpler readout method, the fluorescence of the reactions can be visualized using a Safe Imager 2.0 Transilluminator, through the amber filter unit (vis). This provides a safe, cost effective, and time efficient manner of reading out the signal. The tubes may also be visualized on a fluorescence scanner under the FAM channel (FAM) (see Methods section for full details).
Logic computation circuits
Signal processing of target sequences through logic expression evaluation has emerged as a valuable framework for programming complex dynamic molecular behaviors.12,16 Below, we show how PER can be used to implement AND, OR, and NOT logic for arbitrary sequences simply by programming which primer sequences get appended based on the presence (or absence) of target strands. The basic strategy is to equilibrate the RNA targets with the gated hairpins, introduce the primer, and read out the result by length on a gel after incubation (Fig. 6a).
Figure 6. Logic computation with PER.
(a) Operational schematic for evaluating logic expressions with RNA inputs. (b) miR-19a OR TWT gate reaction components and a PAGE denaturing gel depicting transcript production in response to the different RNA inputs. True outputs are read by looking for transcripts of a particular length, indicated by the dotted lines. Reaction setup and PAGE denaturing gel results are also shown for (c) miR-19a AND TWT, (d) NOT miR-21, and (e) (miR-19a OR TWT) AND (NOT miR-21). In all experiments, RNA targets and hairpin protectors were fixed at 250 nM final concentrations. Hairpins for part (b) were at 100 nM. Part (c) used 100 nM and 200 nM for the miR-19a and TWT hairpins, respectively. Part (d) held the miR-21 sink hairpin at a concentration of 150 nM, and the slow unprotected hairpin was held at 30 nM. Part (e) used 100 nM miR-19a and TWT hairpins, 200 nM miR-21 sink hairpin, and 40 nM slow unprotected hairpin. After 15 minutes of pre-incubation at 37 °C with the targets and protected hairpins, primers were introduced and incubated for 2 hours, 2 hours, 3 hours, and 5 hours for parts (b) through (e), respectively. All reactions used 10 μM each of dATP, dTTP, and dCTP. See Methods section for additional details.
Fig. 6b depicts the design for an OR gate for two RNA inputs, which is implemented with two of the gated hairpins introduced in previous section (Fig. 5d). Each target can activate one of the two hairpins, and when one or both of the two hairpins are activated, the b domain can be appended onto the primer’s a domain. Results are validated with a denaturing gel, which shows no extended products when neither target is present (lane 1). However, if one or both targets are present, the primer is extended by one domain (lanes 2–4).
To evaluate whether two targets are both present (AND logic), their presence is checked in a stepwise manner (Fig. 6c). The primer domain a does not get extended if neither target is present (lane 1) or if just the TWT target is present (lane 2). If just the miR-19a target is present, the primer is extended by one domain to a–b (lane 3). However, if both targets are present, the primer gets extended to the full a-b-c sequence, indicating successful evaluation of the AND expression (lane 4).
Implementing NOT logic requires a separation of timescales between target acceptance and rejection (Fig. 6d). This can be achieved by including a target-dependent sink reaction hairpin at a much higher concentration than the hairpin that evaluates to True (b–c). Thus, if the target, miR-21 in this case, is present, then the b primers get funneled into an inactive (gray) state. However, if no target is present, at a slow rate all of the b primers will get converted to b–c. The gate is validated by comparing incubation results without (lane 1) and with (lane 2) the target, respectively.
Finally, we demonstrated that several of these types of gates can be hooked together to compute the expression (miR-19a OR TWT) AN D (N OT miR-21) (Fig. 6e). This computation is achieved through the cascading of the result of the (miR-19a OR TWT) gate from Fig. 6b into the N OT miR-21 gate from Fig. 6d. The results were validated as before through gel electrophoresis. Primers remain predominantly in their unextended state when either no inputs or just the miR-21 target are present (lanes 1 and 2). When either just the TWT or just the miR-19a is present, the expression evaluates to True as indicated by the fully extended a-b-c product (lanes 3 and 4). However, when one of these is present in addition to miR-21, the primer is funneled into an inactive state (lanes 5 and 6). Having both miR-19a and TWT but no miR-21 again results in a True result (lane 7), but when all three targets are present the expression again evaluates to False (lane 8).
These circuit components demonstrate the modularity of the PER method. The results of one logical evaluation can be easily cascaded into each other by matching synthesized and binding primer domains, and this type of reconfiguration could in principle be extended to any primer domain given any single-stranded RNA or DNA inputs.
Temporal molecular event recorder
We next created a temporal molecular event recording system that encodes the sequential order in which two RNA targets are witnessed into dynamically synthesized transcripts (Fig. 7a).
Figure 7. PER temporal molecular event recorder.
(a) Schematic for event recorder, which produces PER transcripts in response to time-varying RNA signals. (b) System components and reaction diagram for recorder. Four gated hairpins (A, B, C, and D) are used to program the synthesis of different sequences in response to different orders of witnessing two RNA targets - miR-19a and a fragment of the Twist mRNA (TWT). (c) A PAGE denaturing gel shows transcripts of different lengths recorded for different 250 nM RNA signal spikes at 1 and 3 hours into a 5 hour reaction at 37 °C. Hairpins A and C were at 25 nM, protectors A and C were at 50 nM, hairpins B and D were at 75 nM, protectors B and D were at 100 nM, and the primer was at 100 nM. Reactions used 10 μM each of dATP, dTTP, and dCTP. All given concentrations are based on their final concentrations. See Methods section for additional details.
Each of the four hairpins used in this application utilize the same method of toehold exchange for target detection as the signal amplifier and logic circuits. Depending on the order in which the two RNA signals are introduced, a primer will undergo one of two elongation pathways (Fig. 7b). If miR-19a is introduced first, then the initial primer gets extended with b via hairpin A. Then, if the TWT target is introduced, hairpin B can append c to the primer. If, on the other hand, TWT is introduced first, then d gets added to the a primer sequence with the exposed C hairpin, and hairpin D can then append e if the miR-19a signal is encountered later on.
Because the hairpins are designed to copy different numbers of nucleotides per addition, differentiation of the sequence length is achieved based on the order in which the signals are introduced in solution. These results can be read out on a PAGE denaturing gel (Fig. 7c). By introducing signals at either 1 or 3 hours into a 5 hour incubation with the primer and hairpins A through D, the recording behavior was evaluated. Lane 1 shows the system with no targets introduced and shows no extension due to the continuous protection of all four hairpins. Lanes 2 and 3 show a single extension of 10 nt step patterned by hairpin A when miR-19a (miR) was introduced at 1 and 3 hours, respectively. Introducing TWT at 1 or 3 hours results in a single extension of 14 nt by the exposed C hairpin (lanes 4 and 5). Introducing first the miRNA and then TWT results in two 10 nt extensions to form the a-b-c sequence. Finally, introducing first the TWT target and then the miRNA results in two 14 nt extension steps to form a-d-e, thus validating our ability to read out the temporal relationship of the two RNA signals in the length of synthesized DNA.
Discussion
PER dynamically synthesizes arbitrary prescribed DNA sequences in an autonomous and stepwise fashion. PER cascades are programmed using catalytic PER hairpins, driven by polymerase, fueled by dNTPs, and happen in a homogenous solution under isothermal conditions. PER allows arbitrary sequences to be concatenated into a desired transcript, reactions can be simply cascaded together to form prescribed growth pathways, and the synthesis can be made responsive to current environmental conditions. Together, these features make PER a versatile programmable platform for implementing diverse environmentally responsive molecular behaviors (e.g. the conditional RNA degrader, the in situ signal amplifier, the logic RNA expression profiler, and the temporal recorder constructed in this paper).
PER provides a new platform for engineering synthetic DNA circuits and devices, expanding and complementing existing methods such as those based on toehold mediated branch migration,53,54 which has been the dominant mechanism for implementing dynamic DNA devices and circuits. Toehold based circuits and devices rely on dynamic conditional assembly and disassembly of presynthesized materials (i.e. DNA strands), and PER circuits primarily rely on conditional dynamic synthesis of new materials (i.e. DNA strands). Toehold circuits are typically engineered via controlled spatial concealing and revealing of cascading nucleation sites (i.e. toeholds), and are fueled by association (i.e. DNA hybridization driven) or dissociation (i.e. entropy-driven) of presynthesized units; PER circuits are engineered via controlled chemical synthesis of cascading nucleation sites (i.e. primers), and are fueled by polymerization of dNTP monomers.
Though both toehold and PER mechanisms can be utilized to engineer diverse dynamic behaviors, PER appears to be more compact in terms of its usage for construction materials (by avoiding using auxiliary strand fragments to effectively sequester toeholds from interacting with other meta-stably existing complements in the system) and fuels (by using dNTP monomers rather than DNA strands). PER’s compactness and simplicity, for example, enables facile design and construction of 40 orthogonal PER circuits for the in situ synthesis of DNA origami staples (Fig. 3d), and a single small hairpin based signal amplification circuit (synthetic “telomerase” in Fig. 5).
To construct dynamic circuits, both toehold and PER mechanisms need to engineer kinetic barriers which are essential for manipulating the reaction system’s free energy landscape to achieve desired component metastability. Toehold circuits implement such barriers by manipulating weak bonds and hence tend to suffer leakage, which is the unintended off pathway reaction in the absence of the trigger, typically caused by the sub-optimal spatial shielding of the pre-synthesized toeholds, e.g. due to “breathing” of the DNA duplex of toeholds’ incumbent and vicinity regions. PER circuits, by implementing such kinetic barriers via manipulation of covalent bonds, promise in principle to deliver steeper kinetic barriers, and thus to mitigate the leakage problem. While comprehensive and rigorous comparison of leakage in toehold v.s. PER systems are still underway, initial indication of PER’s resistance to leakage comes from the lack of detectable extension of a non-cognate primer in the presence of the highest hairpin concentration used for the synthetic “telomerase” system (e.g. in Supplementary Fig. 16).
PER differs from previous demonstrations of various polymerase based synthetic DNA circuits and molecular computing schemes in that it provides a general method for autonomous programmable synthesis of single-stranded DNA in an environmentally responsive fashion, and thus enables the construction of diverse synthetic dynamic devices beyond molecular information processing. Notable previous examples include the Polymerase/Exonuclease/Nickase Dynamic Network Assembly toolbox (or PEN DNA toolbox for short)41,42 and Whiplash PCR.55–59 The PEN DNA toolbox utilizes sophisticated coordination of polymerase, exonuclease, and nicking enzymes to achieve the controlled generation of primer cascades to implement various computing functions (e.g. oscillations41,60 and spatial patterns such as spirals61 and traveling waves62). Unlike PER, which utilizes the catalytic PER hairpin to append independent primers to existing primers, the PEN DNA toolbox typically generates a new cascading primer without concatenating it to the previous one, and hence is not suitable for the continuous growth of a nascent strand with arbitrary user-prescribed sequence. Additionally, the reliance on various nucleases put constraints on the sequences (e.g. enzyme recognition sites) that may be produced. Therefore, PEN reactions have similar cascading capabilities, but the concatenation of sequences together with PER cascades enables the synthesis of new functional materials. These outputs can serve as structural elements (e.g. origami staples, fluorescent scaffolds), functional elements (e.g. DNAzymes), and records (e.g. logic transcripts, history records). In contrast, the diffusible outputs of PEN cascades are still limited to being exact copies of existing sequences in solution. Whiplash PCR55–59 utilizes polymerase to append a new primer “state” domain to an existing one based on a set of state transition rules for performing a particular computing function, and as a consequence grows a DNA strand that encodes the state transition history. While suitable for implementing state transition based computing, Whiplash PCR typically involves staged external manipulation or thermal cycling to achieve successive transition and hence strand extension, with limited experimental demonstration. Theoretical schemes (e.g. in ref57,59) for isothermal, autonomous operation have been proposed, but not experimentally validated. Additionally, the strand extension is a by-product of implementing desired molecular computing, rather than the primary purpose and hence its general suitability as an autonomous programmable strand synthesis method remains to be explored.
In addition to RNA inputs, and the fluorescence and DNAzyme outputs demonstrated here, PER reactions could be potentially interfaced with other diverse environmental triggers and actuation functions, e.g. detecting protein input via aptamers63 or programing protein synthesis output via toehold switches.64 PER could also be directly interfaced with the rich set of DNA strand displacement circuits.54 The programmable sensing, processing, recording, and actuation capabilities of PER circuits could thus unleash a new generation of molecular programming devices and applications. One particularly interesting example would be molecular recording: as long polymers can be grown following different pathways depending on the current state of the environment, PER devices could be programmed to record environmental signals over extended periods of time, and the resultant transcripts could be sequenced to recover the molecular event reports.
Methods
DNA synthesis and purification
All oligos were ordered from IDT, either unpurified or HPLC-purified. Purified RNA molecules were ordered purified with RNase-free HPLC. Some unpurified oligos were purified in-house by running 100 μL of 100 μM unpurified oligo through a Qiagen MinElute PCR purification column and washed per kit instructions. Column-bound oligos were eluted to 15 μL and concentrations were measured using a Nanodrop and their extinction coefficients. Oligos were ordered pre-suspended in 1 x TE buffer at 100 μM, and these concentrations were assumed for all dilutions, with the exception of MinElute purified oligos. All oligos were diluted in 1 x TE to working concentrations of 10uM, with stock and working solutions of DNA stored at −20 °C and RNA stored at −80 °C. Oligo sequences for all experiments are listed in Supplementary Section 9.
PER incubation
All PER experiments were incubated at 37 °C for the indicated amount of time, usually with 1 x ThermoPol buffer with supplemented magnesium (20 mM Tris-HCl, 10 mM (NH4)2SO4, 10 mM KCl, 12 mM MgSO4, and 0.1% Triton®X-100), 0.8 units per μL of Bst Large Fragment polymerase, and 10–100 μM of the appropriate dNTPs. Typically, 20 μL reactions were quenched by heat inactivation of the enzyme at 80 °C for 20 minutes and loaded with 10 μL formamide. For RNA-sensitive samples, reactions were instead quenched with EDTA. For the signal amplifier, reactions were not quenched but loaded directly after incubation. Some experiments were pre-incubated for 15 minutes to allow the solution to equilibrate. See Supplementary Section 10 for reaction details for each experiment.
Gel electrophoresis
Most experiments used 15% TBE-Urea PAGE denaturing gels that were run at 200V for 35 minutes at 65 °C and scanned with the Cy5 and FAM channels. Gels were also stained with Sybr Gold for several minutes and subsequently imaged with the Sybr Gold channel (see Supplementary Figures 12 through 29 in Supplementary Section 11 for gel scans of all experiments). Some experiments used different gel conditions; see Supplementary Section 10 for full details.
AFM
AFM imaging was performed on a Nanoscope V machine.
Sequence design
Most sequences were designed using in-house Python optimization code paired with command line NUPACK executables.65 The NUPACK web application66,67 was also used to analyze constructs.
Data availability
The principal data supporting the findings of this work are available within the paper figures and the Supplementary Information. All other data are available from the corresponding author upon request.
Supplementary Material
Acknowledgments
The authors thank W. Shih, J. Kim, X. Chen, N. Hanikel, E. Winfree, B. Beliveau, and N. Liu for their discussions and helpful comments. This work was supported by the Office on Naval Research (under grants N000141310593, N000141410610, N000141612182, and N000141612410), the National Science Foundation (under grants CCF1317291, CMMI1334109, and 1540214), the National Institutes of Health (under grant 1R01EB01865901), and the Wyss Institute’s Molecular Robotics Initiative. J. Kishi was supported by an NSF graduate research fellowship, and T. Schaus was supported by the Jane Coffin Childs Postdoctoral Fellowship. Correspondence and requests for materials should be addressed to P.Y.
Footnotes
Author contributions: J. K. conceived of and designed the study, designed and performed the experiments, analyzed the data, and wrote the manuscript. T.S. designed and performed the experiments and analyzed the data. N.G. designed and performed the experiments and analyzed the data. F.X. designed and performed the experiments and analyzed the data. P.Y. conceived of and supervised the study, interpreted the data, and wrote the manuscript. All authors reviewed, edited, and approved the manuscript.
Competing financial interests: A provisional US patent has been filed based on this work. P.Y. is co-founder of Ultivue Inc. and NuProbe Global.
References
- 1.Winfree E, Liu F, Wenzler LA, Seeman NC. Design and self-assembly of two-dimensional DNA crystals. Nature. 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 2.Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 3.Douglas S, et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 2009;459:414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng JP, et al. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature. 2009;461:74–77. doi: 10.1038/nature08274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei B, Dai M, Yin P. Complex shapes self-assembled from single-stranded DNA tiles. Nature. 2012;485:623–626. doi: 10.1038/nature11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ke Y, Ong LL, Shih WM, Yin P. Three-dimensional structures self-assembled from DNA bricks. Science. 2012 doi: 10.1126/science.1227268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han D, et al. DNA gridiron nanostructures based on four-arm junctions. Science. 2013;339:1412–1415. doi: 10.1126/science.1232252. [DOI] [PubMed] [Google Scholar]
- 8.Gerling T, Wagenbauer KF, Neuner AM, Dietz H. Dynamic DNA devices and assemblies formed by shape-complementary, non–base pairing 3D components. Science. 2015;347:1446–1452. doi: 10.1126/science.aaa5372. [DOI] [PubMed] [Google Scholar]
- 9.Benson E, et al. DNA rendering of polyhedral meshes at the nanoscale. Nature. 2015;523:441–444. doi: 10.1038/nature14586. [DOI] [PubMed] [Google Scholar]
- 10.Dunn KE, et al. Guiding the folding pathway of DNA origami. Nature. 2015 doi: 10.1038/nature14860. [DOI] [PubMed]
- 11.Veneziano R, et al. Designer nanoscale DNA assemblies programmed from the top down. Science. 2016;352:1534–1534. doi: 10.1126/science.aaf4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seelig G, Soloveichik D, Zhang DY, Winfree E. Enzyme-free nucleic acid logic circuits. Science. 2006;314:1585–1588. doi: 10.1126/science.1132493. [DOI] [PubMed] [Google Scholar]
- 13.Zhang DY, Turberfield AJ, Yurke B, Winfree E. Engineering entropy-driven reactions and networks catalyzed by DNA. Science. 2007;318:1121–1125. doi: 10.1126/science.1148532. [DOI] [PubMed] [Google Scholar]
- 14.Yin P, Choi HMT, Calvert CR, Pierce NA. Programming biomolecular self-assembly pathways. Nature. 2008;451:318–322. doi: 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]
- 15.Omabegho T, Sha R, Seeman N. A bipedal DNA brownian motor with coordinated legs. Science. 2009 doi: 10.1126/science.1170336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qian L, Winfree E. Scaling up digital circuit computation with DNA strand displacement cascades. Science. 2011;332:1196–1201. doi: 10.1126/science.1200520. [DOI] [PubMed] [Google Scholar]
- 17.Chirieleison SM, Allen PB, Simpson ZB, Ellington AD, Chen X. Pattern transformation with DNA circuits. Nature Chemistry. 2013;5:1000–1005. doi: 10.1038/nchem.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weitz M, et al. Diversity in the dynamical behaviour of a compartmentalized programmable biochemical oscillator. Nature Chemistry. 2014;6:295–302. doi: 10.1038/nchem.1869. [DOI] [PubMed] [Google Scholar]
- 19.Karzbrun E, Tayar AM, Noireaux V, Bar-Ziv RH. Programmable on-chip DNA compartments as artificial cells. Science. 2014;345:829–832. doi: 10.1126/science.1255550. [DOI] [PubMed] [Google Scholar]
- 20.Mohammed AM, Šulc P, Zenk J, Schulman R. Self-assembling DNA nanotubes to connect molecular landmarks. Nature Nanotechnology. 2016 doi: 10.1038/nnano.2016.277. [DOI] [PubMed] [Google Scholar]
- 21.Choi HMT, et al. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nature Biotechnol. 2010;28:1208. doi: 10.1038/nbt.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang DY, Chen SX, Yin P. Optimizing the specificity of nucleic acid hybridization. Nature Chemistry. 2012;4:208–214. doi: 10.1038/nchem.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Douglas SM, Bachelet I, Church GM. A logic-gated nanorobot for targeted transport of molecular payloads. Science. 2012;335:831–834. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- 24.Kuzyk A, et al. DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature. 2012;483:311–314. doi: 10.1038/nature10889. [DOI] [PubMed] [Google Scholar]
- 25.Derr ND, et al. Tug-of-war in motor protein ensembles revealed with a programmable DNA origami scaffold. Science. 2012 doi: 10.1126/science.1226734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jungmann R, et al. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and exchange-PAINT. Nature Methods. 2014;11:313–318. doi: 10.1038/nmeth.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun W, et al. Casting inorganic structures with DNA molds. Science. 2014;346:1258361. doi: 10.1126/science.1258361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang JS, Zhang DY. Simulation-guided DNA probe design for consistently ultraspecific hybridization. Nature Chemistry. 2015;7:545–553. doi: 10.1038/nchem.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gopinath A, Miyazono E, Faraon A, Rothemund PWK. Engineering and mapping nanocavity emission via precision placement of DNA origami. Nature. 2016 doi: 10.1038/nature18287. [DOI] [PubMed] [Google Scholar]
- 30.Bhatia D, et al. Quantum dot-loaded monofunctionalized DNA icosahedra for single-particle tracking of endocytic pathways. Nature Nanotechnology. 2016;11:1112–1119. doi: 10.1038/nnano.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kilchherr F, et al. Single-molecule dissection of stacking forces in DNA. Science. 2016:353. doi: 10.1126/science.aaf5508. [DOI] [PubMed] [Google Scholar]
- 32.Nickels PC, et al. Molecular force spectroscopy with a DNA origami–based nanoscopic force clamp. Science. 2016;354:305–307. doi: 10.1126/science.aah5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker GT, et al. Strand displacement amplification - an isothermal, in vitro DNA amplification technique. Nucleic Acids Research. 1992;20:1691–1696. doi: 10.1093/nar/20.7.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lizardi PM, et al. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nature Genetics. 1998;19:225–232. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]
- 35.Notomi T, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Research. 2000;28:e63–e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang HH, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du Y, Dong S. Nucleic acid biosensors: Recent advances and perspectives. Analytical Chemistry. 2016 doi: 10.1021/acs.analchem.6b04190. [DOI] [PubMed] [Google Scholar]
- 38.Church GM, Gao Y, Kosuri S. Next-generation digital information storage in DNA. Science. 2012;337:1628–1628. doi: 10.1126/science.1226355. [DOI] [PubMed] [Google Scholar]
- 39.Kosuri S, Church GM. Large-scale de novo DNA synthesis: technologies and applications. Nature Methods. 2014;11:499–507. doi: 10.1038/nmeth.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saiki RK, et al. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 41.Montagne K, Plasson R, Sakai Y, Fujii T, Rondelez Y. Programming an in vitro DNA oscillator using a molecular networking strategy. Molecular Systems Biology. 2011:7. doi: 10.1038/msb.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baccouche A, Montagne K, Padirac A, Fujii T, Rondelez Y. Dynamic DNA-toolbox reaction circuits: A walkthrough. Methods. 2014;67:234–249. doi: 10.1016/j.ymeth.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 43.Lee CS, Davis RW, Davidson N. A physical study by electron microscopy of the terminally repetitious, circularly permuted DNA from the coliphage particles of Escherichia coli 15. Journal of Molecular Biology. 1970;48:1IN19–8IN322. doi: 10.1016/0022-2836(70)90215-9. [DOI] [PubMed] [Google Scholar]
- 44.Schaus TE, Woo S, Xuan F, Chen X, Yin P. A dna nanoscope via auto-cycling proximity recording. Nature Communications, accepted. 2017 doi: 10.1038/s41467-017-00542-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Newton CR, et al. The production of PCR products with 5′ single-stranded tails using primers that incorporate novel phosphoramidite intermediates. Nucleic Acids Research. 1993;21:1155–1162. doi: 10.1093/nar/21.5.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakamoto K, et al. State transitions by molecules. Biosystems. 1999;52:81–91. doi: 10.1016/s0303-2647(99)00035-0. [DOI] [PubMed] [Google Scholar]
- 47.Whitcombe D, Theaker J, Guy SP, Brown T, Little S. Detection of PCR products using self-probing amplicons and fluorescence. Nature Biotechnology. 1999;17:804–807. doi: 10.1038/11751. [DOI] [PubMed] [Google Scholar]
- 48.Aubert N, Rondelez Y, Fujii T, Hagiya M. Enforcing logical delays in DNA computing systems. Natural Computing. 2014;13:559–572. [Google Scholar]
- 49.Olive V, et al. miR-19 is a key oncogenic component of miR-17-92. Genes & development. 2009;23:2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hjiantoniou E, Iseki S, Uney JB, Phylactou LA. DNAzyme-mediated cleavage of twist transcripts and increase in cellular apoptosis. Biochemical and Biophysical Research Communications. 2003;300:178–181. doi: 10.1016/s0006-291x(02)02804-8. [DOI] [PubMed] [Google Scholar]
- 51.Santoro SW, Joyce GF. A general purpose RNA-cleaving DNA enzyme. Proceedings of the National Academy of Sciences. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mohanty J, et al. Thioflavin T as an efficient inducer and selective fluorescent sensor for the human telomeric G-quadruplex DNA. Journal of the American Chemical Society. 2012;135:367–376. doi: 10.1021/ja309588h. [DOI] [PubMed] [Google Scholar]
- 53.Yurke B, Turberfield AJ, Mills AP, Simmel FC, Neumann JL. A DNA-fuelled molecular machine made of DNA. Nature. 2000;406:605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- 54.Zhang DY, Seelig G. Dynamic DNA nanotechnology using strand-displacement reactions. Nature Chemistry. 2011;3:103–113. doi: 10.1038/nchem.957. [DOI] [PubMed] [Google Scholar]
- 55.Hagiya M, Arita M, Kiga D, Sakamoto K, Yokoyama S. Towards parallel evaluation and learning of boolean-formulas with molecules. DNA Based Computers III, DIMACS Series in Discrete Mathematics and Theoretical Computer Science. 1999;48:57–72. [Google Scholar]
- 56.Winfree E. Whiplash PCR for O(1) computing. Tech Rep, Caltech. 1998;1998:23. [Google Scholar]
- 57.Rose JA, Deaton RJ, Hagiya M, Suyama A. DNA Computing. Springer; 2002. PNA-mediated Whiplash PCR; pp. 104–116. [Google Scholar]
- 58.Komiya K, Yamamura M, Rose JA. International Workshop on DNA-Based Computers. Springer; 2008. Experimental validation of signal dependent operation in Whiplash PCR; pp. 1–10. [Google Scholar]
- 59.Reif JH, Majumder U. Isothermal reactivating Whiplash PCR for locally programmable molecular computation. Natural Computing. 2010;9:183–206. [Google Scholar]
- 60.Fujii T, Rondelez Y. Predator–prey molecular ecosystems. ACS Nano. 2012;7:27–34. doi: 10.1021/nn3043572. [DOI] [PubMed] [Google Scholar]
- 61.Padirac A, Fujii T, Estvez-Torres A, Rondelez Y. Spatial waves in synthetic biochemical networks. Journal of the American Chemical Society. 2013;135:14586–14592. doi: 10.1021/ja403584p. [DOI] [PubMed] [Google Scholar]
- 62.Zadorin AS, Rondelez Y, Galas J, Estevez-Torres A. Synthesis of programmable reaction-diffusion fronts using dna catalyzers. Physical Review Letters. 2015;114:068301. doi: 10.1103/PhysRevLett.114.068301. [DOI] [PubMed] [Google Scholar]
- 63.Dirks RM, Pierce NA. Triggered amplification by hybridization chain reaction. Proceedings of the National Academy of Sciences. 2004;101:15275–15278. doi: 10.1073/pnas.0407024101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Green AA, Silver PA, Collins JJ, Yin P. Toehold switches: de-novo-designed regulators of gene expression. Cell. 2014;159:925–939. doi: 10.1016/j.cell.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dirks RM, Bois JS, Schaeffer JM, Winfree E, Pierce NA. Thermodynamic analysis of interacting nucleic acid strands. Siam Review. 2007;49:65–88. [Google Scholar]
- 66.Zadeh JN, et al. Nupack: analysis and design of nucleic acid systems. Journal of Computational Chemistry. 2011;32:170–173. doi: 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- 67.Wolfe BR, Pierce NA. Sequence design for a test tube of interacting nucleic acid strands. ACS Synthetic Biology. 2014;4:1086–1100. doi: 10.1021/sb5002196. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The principal data supporting the findings of this work are available within the paper figures and the Supplementary Information. All other data are available from the corresponding author upon request.