Abstract
Droplet single-cell RNA-sequencing (dscRNA-seq) has enabled rapid, massively parallel profiling of transcriptomes. However, assessing differential expression across multiple individuals has been hampered by inefficient sample processing and technical batch effects. Here we describe a computational tool, demuxlet, that harnesses natural genetic variation to determine the sample identity of each cell and detect droplets containing two cells. These capabilities enable multiplexed dscRNA-seq experiments in which cells from unrelated individuals are pooled and captured at higher throughput than in standard workflows. Using simulated data, we show that 50 SNPs per cell are sufficient to assign 97% of singlets and identify 92% of doublets in pools of up to 64 individuals. Given genotyping data for each of 8 pooled samples, demuxlet correctly recovers the sample identity of >99% of singlets and identifies doublets at rates consistent with previous estimates. We apply demuxlet to assess cell type-specific changes in gene expression in 8 pooled lupus patient samples treated with IFN-β and perform eQTL analysis on 23 pooled samples.
Droplet single cell RNA-sequencing (dscRNA-seq) has increased substantially the throughput of single cell capture and library preparation1, 10, enabling the simultaneous profiling of thousands of cells. Improvements in biochemistry11, 12 and microfluidics13, 14 continue to increase the number of cells and transcripts profiled per experiment. But for differential expression and population genetics studies, sequencing thousands of cells each from many individuals would better capture inter-individual variability than sequencing more cells from a few individuals. However, in standard workflows, dscRNA-seq of many samples in parallel remains challenging to implement. If the genetic identity of each cell could be determined, pooling cells from different individuals in one microfluidic run would result in lower per-sample library preparation cost and eliminate confounding effects. Furthermore, if droplets containing multiple cells from different individuals could be detected, pooled cells could be loaded at higher concentrations, enabling additional reduction in per-cell library preparation cost.
Here we develop an experimental protocol for multiplexed dscRNA-seq and a computational algorithm, demuxlet, that harnesses genetic variation to determine the genetic identity of each cell (demultiplex) and identify droplets containing two cells from different individuals (Fig. 1a). While strategies to demultiplex cells from different species1, 10, 17 or host and graft samples17 have been reported, simultaneously demultiplexing and detecting doublets from more than two individuals has not been possible. Inspired by models and algorithms developed for detecting contamination in DNA sequencing18, demuxlet is fast, accurate, scalable, and compatible with standard input formats17, 19, 20.
Figure 1. Demuxlet: demultiplexing and doublet identification from single cell data.
a) Pipeline for experimental multiplexing of unrelated individuals, loading onto droplet-based single-cell RNA-sequencing instrument, and computational demultiplexing (demux) and doublet removal using demuxlet. Assuming equal mixing of 8 individuals, b) 4 genetic variants can recover the sample identity of a cell, and c) 87.5% of doublets will contain cells from two different samples.
Demuxlet implements a statistical model for evaluating the likelihood of observing RNA-seq reads overlapping a set of single nucleotide polymorphisms (SNPs) from a single cell. Given a set of best-guess genotypes or genotype probabilities obtained from genotyping, imputation or sequencing, demuxlet uses maximum likelihood to determine the most likely donor for each cell using a mixture model. A small number of reads overlapping common SNPs is sufficient to accurately identify each cell. For a pool of 8 individuals and a set of uncorrelated SNPs each with 50% minor allele frequency (MAF), 4 reads overlapping SNPs are sufficient to uniquely assign a cell to the donor of origin (Fig. 1b) and 20 reads overlapping SNPs can distinguish every sample with >98% probability in simulation (Supplementary Fig. 1). We note that by multiplexing even a small number of individuals, the probability that a doublet contains cells from different individuals is very high (1 – 1/N, e.g., 87.5% for N=8 samples) (Fig. 1C). For example, if a 1,000-cell run without multiplexing results in 990 singlets with a 1% undetected doublet rate, multiplexing 1,570 cells each from 63 samples can theoretically achieve the same rate of undetected doublets, producing up to a 37-fold more singlets (36,600) if the sample identity of every droplet can be perfectly demultiplexed (Supplementary Fig. 2, see Methods for details). To minimize the effects of sequencing doublets, profiling 22,000 cells multiplexed from 26 individuals generates 23-fold more singlets at the same effective doublet rate (Supplementary Fig. 3).
We first assess the performance of multiplexed dscRNA-seq through simulation. The ability to demultiplex cells is a function of the number of individuals multiplexed, the depth of sequencing or number of read-overlapping SNPs, and relatedness of multiplexed individuals. We simulated 6,145 cells (5,837 singlets and 308 doublets) from 2 – 64 individuals from the 1000 Genomes Project21. We show that 50 SNPs per cell allows demultiplexing of 97% of singlets and identification of 92% of doublets in pools of up to 64 individuals (Supplementary Figs. 4–5, see Methods for details). Simulating a range of sequencing depths, we determined that 50 SNPs can be obtained with as few as 1,000 unique molecular identifiers (UMIs) per cell (Supplementary Fig. 6), and recommended sequencing depths of standard dscRNA-seq workflows would capture hundreds of SNPs. To assess dependence on the relatedness of multiplexed individuals, we simulated 6,145 cells from a set of 8 related individuals from 1000 Genomes21. In this simulation, 50 SNPs per cell would allow demuxlet to correctly assign over 98% of cells (Supplementary Fig. 7). These results suggest optimal multiplexed designs where cells from tens of unrelated individuals should be pooled, loaded at concentrations 2–10x higher than standard workflows, and sequenced to at least 1,000 UMIs per cell.
We evaluate the performance of demuxlet by analyzing a pool of peripheral blood mononuclear cells (PBMCs) from 8 lupus patients. By sequential pairwise pooling, three pools of equimolar concentrations of cells were generated (W1: patients S1-S4, W2: patients S5-S8 and W3: patients S1-S8) and each loaded in a well on a 10X Chromium instrument (Fig. 2a). 3,645 (W1), 4,254 (W2) and 6,205 (W3) cell-containing droplets were sequenced to an average depth of 51,000, 39,000 and 28,000 reads per droplet.
Figure 2. Performance of demuxlet.
a) Experimental design for equimolar pooling of cells from 8 unrelated samples (S1-S8) into three wells (W1-W3). W1 and W2 contain cells from two disjoint sets of 4 individuals. W3 contains cells from all 8 individuals. b) Demultiplexing single cells in each well recovers the expected individuals. c) Estimates of doublet rates versus previous estimates from mixed species experiments. d) Cell type identity determined by prediction to previously annotated PBMC data. e) t-SNE plot of two individuals (S1 and S5) from different wells are qualitatively concordant.
In wells W1, W2 and W3, demuxlet identified 91% (3332/3645), 91% (3864/4254), and 86% (5348/6205) of droplets as singlets (likelihood ratio test, L(singlet)/L(doublet) > 2), of which 25% (+/− 2.6%), 25% (+/− 4.6%) and 12.5% (+/− 1.4%) mapped to each donor, consistent with equal mixing of individuals in each well. From wells W1 and W2, each containing cells from two disjoint sets of 4 individuals, we estimated a demultiplexing error rate (number of cells assigned to individuals not in the pool) of less than 1% of singlets (W1: 2/3332, W2: 0/3864) (Fig. 2b).
We next assess the ability of demuxlet to detect doublets in both simulated and real data. 466/3645 (13%) droplets from W1 were simulated as synthetic doublets by setting the cellular barcodes of 466 cells each from individuals S1 and S2 to be the same. Applied to simulated data, demuxlet identified 91% (426/466) of synthetic doublets as doublets or ambiguous, correctly recovering the sample identity of both cells in 403/426 (95%) doublets (Supplementary Fig. 8). Applied to real data from W1, W2 and W3, demuxlet identified 138/3645, 165/4254, and 384/6205 doublets corresponding to doublet rates of 5.0%, 5.2% and 7.1%, consistent with the expected doublet rates estimated from mixed species experiments (Fig. 2c).
Demultiplexing of pooled samples allows for the statistical and visual comparisons of individual-specific dscRNA-seq profiles. Singlets identified by demuxlet in all three wells cluster into known immune cell types (Fig. 2d) and are correlated with bulk RNA-sequencing of sorted cell populations (R=0.76–0.92) (Supplementary Fig. 9). For the same individuals from different wells, t-distributed stochastic neighbor embedding (t-SNE) of dscRNA-seq data are qualitatively consistent, and estimates of cell type proportions are highly correlated (R = 0.99) (Fig. 2e and Supplementary Fig. 10). Further, t-SNE projections of the pool and each individual are not confounded by well-to-well effects (Supplementary Fig. 11a). While 6 genes were differentially expressed between wells W1 and W2 (DESeq2 on pseudobulk counts, FDR < 0.05), only 2 genes were differentially expressed between W1 and W2 individuals in well W3 (FDR < 0.05) (Supplementary Fig. 11b), suggesting multiplexing reduces technical effects due to separate sample processing22, 23.
We used multiplexed dscRNA-seq to characterize the cell type specificity and inter-individual variability of response to IFN-β, a potent cytokine that induces genome-scale changes in the transcriptional profiles of immune cells24, 25. From each of 8 lupus patients, PBMCs were activated with recombinant IFN-β or left untreated for 6 hours, a time point we previously found to maximize the expression of interferon-sensitive genes (ISGs) in dendritic cells (DCs) and T cells26, 27. Two pools, IFN-β-treated and control, were prepared with the same number of cells from each individual and loaded onto the 10X Chromium instrument.
We obtained 14,619 (control) and 14,446 (stimulated) cell-containing droplets, of which demuxlet identified 83% (12,138) and 84% (12,167) as singlets. The estimated doublet rate of 10.9% in each condition is consistent with predicted rates (Fig. 2C) and the observed and expected frequencies of doublets for each pair of individuals are highly correlated (R=0.98) (Supplementary Fig. 12). Detected doublets form distinct clusters near the periphery of other clusters defined by cell type (Supplementary Fig. 13).
Demultiplexing individuals enables the use of the 8 individuals within each pool as biological replicates to quantitatively assess cell type-specific IFN-β responses in PBMCs. Consistent with previous reports from bulk RNA-sequencing data, IFN-β stimulation induces widespread transcriptomic changes observed as a shift in the t-SNE projections of singlets24 (Fig. 3A). As expected, IFN-β did not affect cell type proportions between control and stimulated cells (Supplementary Fig. 14), and these were consistent with flow cytometry measurements (R=0.88) (Supplementary Fig. 15). Estimates of abundances for ~2000 homologous genes in each cell type and condition correlated with similar data from mice (Supplementary Fig. 16). We identified 3,055 differentially expressed genes (logFC > 2, FDR < 0.05) in at least one cell type (Supplementary Table 1). For 709 genes, estimates of fold change in response to IFN-β stimulation in myeloid and CD4+ cells are consistent with estimates in monocyte derived dendritic cells28 and CD4+ T cells27, respectively (Supplementary Fig. 17) and correlated with qPCR results of sorted CD4+ T cells (Supplementary Fig. 18). Differentially expressed genes cluster into modules of cell type-specific responses enriched for distinct gene regulatory programs (Fig. 3B, Supplementary Table 2). For example, genes upregulated in all leukocytes (Cluster III: 401 genes, logFC > 2, FDR < 0.05) or only in myeloid cells (Cluster I: 767 genes, logFC > 2, FDR < 0.05) are enriched for general antiviral response (e.g. KEGG Influenza A: Cluster III P < 1.6×10−5), chemokine signaling (Cluster I P < 7.6×10−3) and pathways active in systemic lupus erythematosus (Cluster I P < 4.4×10−3). The five clusters of downregulated genes are enriched for antibacterial response (KEGG Legionellosis: Cluster II monocyte down P < 5.5×10−3) and natural killer cell mediated toxicity (Cluster IV NK/Th cell down: P < 3.6×10−2). The analysis of multiplexed dscRNA-seq data recovers cell type-specific gene regulatory programs affected by interferon stimulation consistent with published IFN-β signatures in mouse and humans29.
Figure 3. Inter-individual variability in IFN-β response.
a) t-SNE plot of unstimulated (blue) and IFN-β-stimulated (red) PBMCs and the estimated cell types. b) Cell type-specific expression in stimulated (left) and unstimulated (right) cells. Differentially expressed genes shown (FDR < 0.05, |log(FC)| > 1). Each column represents cell type-specific expression for each individual from demuxlet. c) Observed variance (y-axis) in mean expression over all PBMCs from each of the 8 individuals versus expected variance (x-axis) over synthetic replicates sampled across all cells (light blue, pink) or replicates matched for cell type proportion (blue, red). d) Cell type proportions for each individual in unstimulated and stimulated cells. e) Correlation between sample replicates in control and stimulated cells. f) Number of significantly variable genes in each cell type and condition.
Over all PBMCs, the variance of mean expression across individuals is higher than the variance across synthetic replicates whose cells were randomly sampled (Lin’s concordance = 0.022, Pearson correlation= 0.69, Fig. 3C). The variance across synthetic replicates whose cells were sampled matching for cell type proportions is more concordant with the variance across individuals (Lin’s concordance = 0.54, Pearson correlation = 0.78, Fig. 3C–D), suggesting a contribution of cell type composition on expression variability. However, for each cell type, the variance across individuals22, 30 is also higher than the variance across synthetic replicates (Lin’s concordance = 0.007–0.20) suggesting additional inter-individual variability not due to cell type composition (Supplementary Fig. 19). In CD14+CD16− monocytes, the correlation of mean expression between pairs of synthetic replicates from the same individual (>99%) is greater than from different individuals (~97%), further indicating inter-individual variation beyond sampling (Fig. 3E). We found between 15 to 827 genes with statistically significant inter-individual variability in control cells and 7 to 613 in stimulated cells (Pearson correlation, FDR < 0.05), with most found in classical monocytes (cM) and CD4+ helper T (Th) cells. Inter-individual variable genes in stimulated cM and to a lesser extent in Th cells (P < 9.3×10−4 and 4.5×10−2, hypergeometric test, Fig. 3F) are enriched for differentially expressed genes, consistent with our previous discovery of more IFN-β response-eQTLs in monocyte-derived dendritic cells than CD4+ T cells26, 27. Comparing to 407 genes previously profiled in bulk monocyte-derived dendritic cells, the proportion of variance explained by inter-individual variability is more correlated in myeloid cells after stimulation (R = 0.26 – 0.3) than before (R = 0.05 – 0.19).
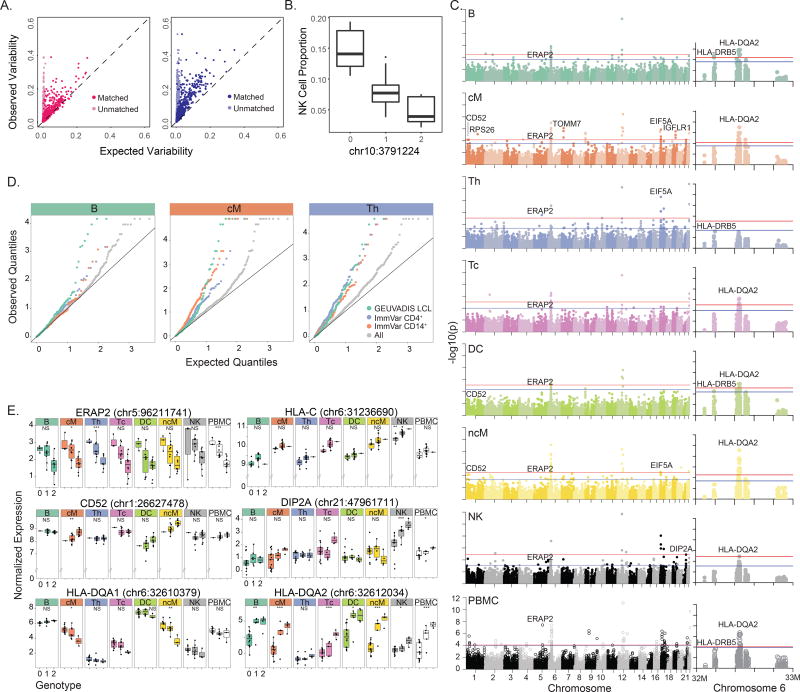
To map genetic variants associated with cell type proportions and cell type-specific expression using multiplexed dscRNA-seq, we sequenced an additional 15,250 (7 donors), 22,619 (8 donors) and 25,918 cells (15 donors; 8 lupus patients, 5 rheumatoid arthritis patients, and 2 healthy controls). Demuxlet identified 71% (10,766/15,250), 73% (16,618/22,619) and 60% (15,596/25,918) of droplets as singlets, correctly assigning 99% of singlets from the first two pools, W1 and W2 (10,740/10,766 and 16,616/16,618). The estimated doublet rates of 18%, 18% and 25% are consistent with the increased concentrations of loaded cells (Fig. 2C). Similar to the IFN-β stimulation experiment, we found that expression variability was determined by variability in cell type proportion (Fig. 4A) and reproducible between batches (Supplementary Fig. 20). Associating >150,000 genetic variants (MAF > 20%) with the proportion of 8 major immune cell populations, we identified a SNP (chr10:3791224) significantly associated (P = 1.03×10−5, FDR < 0.05) with the proportion of NK cells (Fig. 4B).
Figure 4. Genetic control over cell type proportion and gene expression (N=23).
a) Observed variance (y-axis) in mean expression over all PBMCs from each individual versus expected variance (x-axis) over synthetic replicates sampled across batch 1 (left, N=8) and batch 3 (right, N=15). b) Association of chr10:3791224 with NK cell type proportions. c) Genome-wide and chromosome 6 Manhattan plots across all major cell types. Horizontal lines correspond to FDR < 0.1 (blue) and FDR < 0.05 (red). d) Q-Q plots across all genes and subsets of previously published eQTLs in relevant cell types are shown for B, cM, and Th populations. e) Notable cis-eQTLs across all major immune cell types are marked with *(FDR < 0.25), ** (FDR < 0.1), and *** (FDR < 0.05). Lack of association is marked with NS (not significant).
Across 23 donors, we conducted an expression quantitative trait loci (eQTL) analysis to map genetic variants associated with expression variability in each major immune cell type. We found a total of 32 local eQTLs (+/− 100kb, FDR < 0.1), 22 of which were detected in only one cell type (Fig. 4C, Supplementary Table 3). Previously reported local eQTLs from bulk CD14+ monocytes, CD4+ T cells and lymphoblastoid cell lines are more significantly associated with gene expression in the most similar cell types (cM, Th and B cells, respectively) than other cell types (Fig. 4D). We used an inverse variance weighted meta-analysis to identify genes with pan-cell type eQTLs, including those in the major histocompatibility complex (MHC) class I antigen presentation pathway including ERAP2 (P < 3.57×10−32, meta-analysis), encoding an aminopeptidase known to cleave viral peptides34, and HLA-C (P < 1.74×10−29, meta-analysis), which encodes the MHC class I heavy chain (Fig. 4E). HLA-DQA1 has local eQTLs only in some cell types (P <2.11×10−15, Cochran’s Q) while HLA-DQA2 has local eQTLs in all antigen presentation cells (P < 1.02e10−43, Cochran’s Q). Among other cell type-specific local eQTLs are CD52, a gene ubiquitously expressed in leukocytes that only has eQTLs in monocyte populations, and DIP2A, a gene with an eQTL only in NK cells that is associated with immune response to vaccination in peripheral blood35. These results demonstrate the ability of multiplexed dscRNA-seq to characterize inter-individual variation in immune response and when integrated with genetic data, reveal cell type-specific genetic control of gene expression, which would be undetectable when bulk tissues are analyzed.
The capability to demultiplex and identify doublets using natural genetic variation reduces the per-sample and per-cell library preparation cost of single-cell RNA-sequencing, does not require synthetic barcodes or split-pool strategies36–40, and captures biological variability among individual samples while limiting unwanted technical variability. We find the optimal number of samples to multiplex is approximately 20, based on sample processing time and empirical doublet rates of current microfluidic devices and anticipate that number to increase with automated sample handling and lower doublet rates.
Compared to sorting known cell types followed by bulk RNA-seq, multiplexed dscRNA-seq is a more efficient and unbiased method for obtaining cell type-specific immune traits41. Demuxlet enables reliable estimation of cell type proportion, recovers cell type-specific transcriptional response to stimulation, and could facilitate further genetic and longitudinal analyses in relevant cell types and conditions across a range of sampled individuals, including between healthy controls and disease patients42–44. While demuxlet could in principle be applied to sequencing solid tissue, standardizing sample processing and preservation remain major challenges. Although we developed demuxlet specifically for RNA-sequencing, we anticipate that the computational framework could be easily extended to other single cell assays where synthetic barcodes or natural genetic variation are measured by sequencing.
Methods
Identifying the sample identity of each single cell
We first describe the method to infer the sample identity of each cell in the absence of doublets. Consider RNA-sequence reads from C barcoded droplets multiplexed across S different samples, where their genotypes are available across V exonic variants. Let dcv be the number of unique reads overlapping with the v-th variant from the c-th droplet. Let bcvi ∈ {R, A, O}, i ∈ {1, ⋯ , dcv} be the variant-overlapping base call from the i-th read, representing reference (R), alternate (A), and other (O) alleles respectively. Let ecvi ∈ {0,1} be a latent variable indicating whether the base call is correct (0) or not (1), then given ecvi = 0, bcvi ∈ {R = 0, A = 1} and ~ Binomial when g ∈ {0,1,2} is the true genotype of sample corresponding to c-th droplet at v-th variant. When ecvi = 1, we assume that Pr(bcvi|g, ecvi) follows Supplementary Table 4. ecvi is assumed to follow Bernoulli where qcvi is a phred-scale quality score of the observed base call. We use the standard 10X pipeline to process the raw reads which estimates the phred-scale quality score based on the alignment of each read to the reference human transcriptome using the STAR aligner49.
We allow uncertainty of observed genotypes at the v-th variant for the s-th sample using , the posterior probability of a possible genotype g given external DNA data Datasv (e.g. sequence reads, imputed genotypes, or array-based genotypes). If genotype likelihood Pr(Datasv|g) is provided (e.g. unphased sequence reads) instead, it can be converted to a posterior probability scale using where Pr(g) ~ Binomial(2, pv) and pv is the population allele frequency of the alternate allele. To allow errors ε in the posterior probability, we replace it with . The overall likelihood that the c-th droplet originated from the s-th sample is
| (1) |
In the absence of doublets, we use the maximum likelihood to determine the best-matching sample as argmaxs[Lc (s)].
Screening for droplets containing multiple samples
To identify doublets, we implement a mixture model to calculate the likelihood that the sequence reads originated from two individuals, and the likelihoods are compared to determine whether a droplet contains cells from one or two samples. If sequence reads from the c-th droplet originate from two different samples, s1, s2 with mixing proportions (1 − α) : α, then the likelihood in (1) can be represented as the following mixture distribution18,
To reduce the computational cost, we consider discrete values of α ∈ {α1, ⋯ , αM}, (e.g. 5 – 50% by 5%). We determine that it is a doublet between samples s1, s2 if and only if and the most likely mixing proportion is estimated to be argmaxαLc(s1, s2, α). We determine that the cell contains only a single individual s if , and less confident droplets are classified as ambiguous. While we consider only doublets for estimating doublet rates, we remove all doublets and ambiguous droplets to conservatively estimate singlets. Supplementary Fig. 8 illustrates the distribution of singlet, doublet likelihoods and the decision boundaries when t = 2 was used.
Theoretical expectation of deconvoluting singlets
The theoretical distribution of expected singlets with multiplexing (presented in Supplementary Fig. 2) is as follows. Let do (e.g. 0.01) be the proportion of true multiplets when xo (1,000) cells are loaded when multiplexing was not used. Then the expected multiplet rates when x cells are loaded can be modeled exponentially as . Let α be the fraction of true singlets incorrectly classified as non-singlets (i.e. doublet or ambiguous), and β be the fraction of multiplets correctly classified as non-singlets. When multiplexing x cells equally from n samples, the expected multiplet rates are d(x), and are expected to be undetectable doublets mixed between the cells from the same sample. Therefore, the overall effective multiplet rate is . Similarly, the expected number of correctly identified singlets becomes . Given α, β the expected number of singlets can be calculated by fixing the multiplet rate d(x) = d0. We used d0 = 0.01, x0 = 1000 for the simulation in Supplementary Fig. 2.
Dependence of demultiplexing performance on experimental design parameters
The demuxlet ‘plp’ option was used to generate a pileup format of 6,145 cells from one well of PBMC 10x data. The reads in the pileup were then modified to reflect the genotypes of individuals sampled from the 1000 Genomes Phase 3 cohort. The pileup was downsampled to obtain different numbers of read-overlapping exonic SNPs (ranging from 5,000 to 100,000) for the whole cohort. To create simulated doublets, we randomly sampled and merged pairs of barcodes within a dataset, resulting in a 5% doublet rate in the original data. For simulations with related individuals, we simulated transcriptomes from 8 individuals in 1000 Genomes with varying degrees of relatedness, ranging from unrelated to parent-child (HG00146, HG00147, HG00500, HG00501, HG00502, HG00512, HG00514, and HG00524).
Isolation and preparation of PBMC samples
Informed consent was obtained from all patients sequenced in this study. Peripheral blood mononuclear cells were isolated from patient donors, Ficoll separated, and cryopreserved by the UCSF Core Immunologic Laboratory (CIL). PBMCs were thawed in a 37°C water bath, and subsequently washed and resuspended in EasySep buffer (STEMCELL Technologies). Cells were treated with DNAseI and incubated for 15 min at RT before filtering through a 40um column. Finally, the cells were washed in EasySep and resuspended in 1x PBMS and 0.04% bovine serum albumin. Cells from 8 donors were then re-concentrated to 1M cells per mL and then serially pooled. At each pooling stage, 1M cells per mL were combined to result in a final sample pool with cells from all donors.
IFN-β stimulation and culture
Prior to pooling, samples from 8 individuals were separated into two aliquots each. One aliquot of PBMCs was activated by 100 U/mL of recombinant IFN-β (PBL Assay Science) for 6 hrs according to the published protocol26. The second aliquot was left untreated. After 6 hrs, the 8 samples for each condition were pooled together in two final pools (stimulated cells and control cells) as described above.
Fluorescence-activated cell sorting and analysis
1M PBMCs from each donor were stained using standard procedure (30 min, 4 C) with the following surface antibody panel (CD3-PerCP clone SK7 (BioLegend), CD4-APC clone OKT4 (BioLegend), CD8-BV570 clone RPA-T8 (BioLegend), CD14-FITC clone 63D3 (BioLegend), CD19-BV510 clone SJ25C1 (BD), and Ghost dye A710 viability stain (Tonbo)) (Life Sciences Reporting Summary). Samples were then analyzed and sorted using a BD FACSAria Fusion instrument at the UCSF flow cytometry core. To calculate cell type proportions, the number of events in each of CD3+ CD4+ CD8− (CD4+ T cells), CD3+ CD4− CD8+ (CD8+ T cells), CD3− CD19+ (B cells), and CD3− CD14+ (monocytes) were divided by the sum of events in these gates (Supplementary Fig. 21).
Quantitative polymerase chain reaction analysis
RNA was isolated from sorted CD4+ T cells following the RNeasy micro kit protocol (QIAGEN), and cDNA was prepared using MultiScribe Reverse Transcriptase (Applied Biosystems cat #4368814). The qPCR primers were chosen from the PrimerBank reference when available 50. Each sample was run in triplicate with the Luminaris HiGreen qPCR kit (Thermo Scientific #K0992) according to standard protocol using a Roche Light Cycler 96 instrument and fold change was calculated from ΔΔCT between control and stimulated samples with GAPDH as a reference gene.
Droplet-based capture and sequencing
Cellular suspensions were loaded onto the 10x Chromium instrument (10x Genomics) and sequenced as described in Zheng et al17. The cDNA libraries were sequenced using a custom program on 10 lanes of Illumina HiSeq2500 Rapid Mode, yielding 1.8B total reads and 25K reads per cell. At these depths, we recovered >90% of captured transcripts in each sequencing experiment.
Bulk isolation and sequencing
PBMCs from lupus patients were isolated and prepared as described above. Once resuspended in EasySep buffer, the EasyEights Magnet was used to sequentially isolate CD14+ (using the EasySep Human CD14 positive selection kit II, cat #17858), CD19+ (using the EasySep Human CD19 positive selection kit II, cat #17854), CD8+ (EasySep Human CD8 positive selection kitII, cat#17853), and CD4+ cells (EasySep Human CD4 T cell negative isolation kit (cat #17952) according to the kit protocol. RNA was extracted using the RNeasy Mini kit (#74104), and reverse transcription and tagmentation were conducted according to Picelli et al. using the SmartSeq2 protocol51, 52. After cDNA synthesis and tagmentation, the library was amplified with the Nextera XT DNA Sample Preparation Kit (#FC-131-1096) according to protocol, starting with 0.2ng of cDNA. Samples were then sequenced on one lane of the Illumina Hiseq4000 with paired end 100bp read length, yielding 350M total reads.
Alignment and initial processing of single cell sequencing data
We used the CellRanger v1.1 and v1.2 software with the default settings to process the raw FASTQ files, align the sequencing reads to the hg19 transcriptome, and generate a filtered UMI expression profile for each cell17. The raw UMI counts from all cells and genes with nonzero counts across the population of cells were used to generate t-SNE profiles.
Cell type classification and clustering
To identify known immune cell populations in PBMCs, we used the Seurat package to perform unbiased clustering on the 2.7k PBMCs from Zheng et al., following the publicly available Guided Clustering Tutorial17, 53. The FindAllMarkers function was then used to find the top 20 markers for each of the 8 identified cell types. Cluster averages were calculated by taking the average raw count across all cells of each cell type. For each cell, we calculated the Spearman correlation of the raw counts of the marker genes and the cluster averages, and assigned each cell to the cell type to which it had maximum correlation.
Differential expression analysis
Demultiplexed individuals were used as replicates for differential expression analysis. For each gene, raw counts were summed for each individual. We used the DESeq2 package to detect differentially expressed genes between control and stimulated conditions54. Genes with baseMean > 1 were filtered out from the DESeq2 output, and the qvalue package was used to calculate FDR < 0.05 55.
Estimation of inter-individual variability in PBMCs
For each individual, we found the mean expression of each gene with nonzero counts. The mean was calculated from the log2 single cell UMI counts normalized to the median count for each cell. To measure inter-individual variability, we then calculated the variance of the mean expression across all individuals. Lin’s concordance correlation coefficient was used to compare the agreement of observed data and synthetic replicates. Synthetic replicates were generated by sampling without replacement either from all cells or cells matched for cell type proportion. Cell type-specific variability estimated as the correlation between synthetic replicates was compared to variability estimates from 23 biological replicates of bulk IFN-stimulated monocyte-derived dendritic cells. Protein coding genes (407/414) originally measured using Nanostring (a hybridization based PCR-free quantification method) were assessed, and variability in the bulk dataset was estimated as repeatability using a linear mixed model56,26.
Estimation of inter-individual variability within cell types
For each cell type, we generated two bulk equivalent replicates for each individual by summing raw counts of cells sampled without replacement. We used DESeq2 to generate variance-stabilized counts across all replicates. To filter for expressed genes, we performed all subsequent analyses on genes with 5% of samples with > 0 counts. The correlation of replicates was performed on the log2 normalized counts. Pearson correlation of the two replicates from each of the 8 individuals was used to find genes with significant inter-individual variability.
Quantitative trait mapping in major immune cell types
Genotypes were imputed with EAGLE57 and filtered for MAF > 0.2, resulting in a total of 189,322 SNPs. Cell type proportions were calculated as number of cells for each cell type divided by the number of total cells for each person. Linear regression was used to test associations between each genetic variant and cell-type proportion with the Matrix eQTL software58. Cis-eQTL mapping was conducted in each cell type separately. All genes with at least 50 UMI counts in 20% of the individuals in all PBMCs were tested for each cell type, resulting in a total of 4,555 genes. Variance-stabilized and log-normalized gene expression was calculated using the ‘rlog’ function of the DESeq2 package54. All variants within a window of 100kbp of each gene were tested with linear regression using Matrix eQTL58. Batch information for each sample as well as the first 3 principal components of the expression matrix were used as covariates.
Supplementary Material
Acknowledgments
MS and CJY are supported by NIH R01AR071522 and R21AI133337. ST is supported by NIH F30DK115167. NZ is supported by NIH K25HL121295, R03DE025665, and Department of Defense W81WH-16-2-0018.
Footnotes
Contributions: HMK and CJY conceived the project. MS, ST, LM, RG, LB, EW, SW, and MN performed all experiments. HMK, MS, ST, EM, SM, and CJY analyzed the data. CL and LAC provided the patient samples. NZ and AM provided helpful comments and discussion. HMK, MS, ST, and CJY wrote the manuscript.
Competing financial interest: AM is a founder of Spotlight Therapeutics and serves as an advisor to Juno Therapeutics and PACT Pharma; the Marson lab has received sponsored research support from Juno Therapeutics and Epinomics.
Single cell and bulk RNA-sequencing data has been deposited in the Gene Expression Omnibus under the accession number GSE96583. Demuxlet software is freely available at https://github.com/statgen/demuxlet
References
- 1.Macosko EZ, et al. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pollen AA, et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat Biotech. 2014;32:1053–1058. doi: 10.1038/nbt.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buenrostro JD, et al. Nature. Vol. 523. Nature Research; 2015. pp. 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagano T, et al. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel AP, et al. Science. Vol. 344. American Association for the Advancement of Science; 2014. pp. 1396–1401. [Google Scholar]
- 6.Tirosh I, et al. Science. Vol. 352. American Association for the Advancement of Science; 2016. pp. 189–196. [Google Scholar]
- 7.Muraro MJ, et al. A Single-Cell Transcriptome Atlas of the Human Pancreas. Cell Syst. 2016;3:385–394. e383. doi: 10.1016/j.cels.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baron M, et al. A Single-Cell Transcriptomic Map of the Human and Mouse Pancreas Reveals Inter- and Intra-cell Population Structure. Cell Syst. 2016;3:346–360. e344. doi: 10.1016/j.cels.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shalek AK, et al. Nature. 2014. [Google Scholar]
- 10.Klein AM, et al. Droplet Barcoding for Single-Cell Transcriptomics Applied to Embryonic Stem Cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stegle O, Teichmann SA, Marioni JC. Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet. 2015;16:133–145. doi: 10.1038/nrg3833. [DOI] [PubMed] [Google Scholar]
- 12.Gawad C, Koh W, Quake SR. Single-cell genome sequencing: current state of the science. Nat Rev Genet. 2016;17:175–188. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]
- 13.Streets AM, et al. Microfluidic single-cell whole-transcriptome sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:7048–7053. doi: 10.1073/pnas.1402030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zilionis R, et al. Single-cell barcoding and sequencing using droplet microfluidics. Nat. Protocols. 2017;12:44–73. doi: 10.1038/nprot.2016.154. [DOI] [PubMed] [Google Scholar]
- 15.Ziegenhain C, et al. Comparative Analysis of Single-Cell RNA Sequencing Methods. Molecular Cell. 2017;65:631–643.e634. doi: 10.1016/j.molcel.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 16.Hicks SC, Teng M, Irizarry RA. bioRxiv. Cold Spring Harbor Labs Journals; 2015. p. 025528. [Google Scholar]
- 17.Zheng GXY, et al. Nature Communications. Vol. 8. Nature Publishing Group; 2017. p. 14049. [DOI] [Google Scholar]
- 18.Jun G, et al. The American Journal of Human Genetics. 2012;91:839–848. doi: 10.1016/j.ajhg.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The Genomes Project, C. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aguirre-Gamboa R, et al. Differential Effects of Environmental and Genetic Factors on T and B Cell Immune Traits. Cell Reports. 17:2474–2487. doi: 10.1016/j.celrep.2016.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, et al. A Functional Genomics Approach to Understand Variation in Cytokine Production in Humans. Cell. 2016;167:1099–1110.e1014. doi: 10.1016/j.cell.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 24.Mostafavi S, et al. Parsing the Interferon Transcriptional Network and Its Disease Associations. Cell. 164:564–578. doi: 10.1016/j.cell.2015.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stark GR, Kerr IM, Williams BRG, Silverman RH, Schreiber RD. in http://dx.doi.org/10.1146/annurev.biochem.67.1.227, Vol. 67 227–264 (Annual Reviews 4139 El Camino Way, P.O. Box 10139, Palo Alto, CA 94303-0139, USA, 2003).
- 26.Lee MN, et al. Science. 2014;343:1246980–1246980. doi: 10.1126/science.1246980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye CJ, et al. Science. 2014;345:1254665–1254665. doi: 10.1126/science.1254665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrés AM, et al. Balancing Selection Maintains a Form of ERAP2 that Undergoes Nonsense-Mediated Decay and Affects Antigen Presentation. PLOS Genetics. 2010;6:e1001157. doi: 10.1371/journal.pgen.1001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mostafavi S, et al. Parsing the Interferon Transcriptional Network and Its Disease Associations. Cell. 2016;164:564–578. doi: 10.1016/j.cell.2015.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palmer C, Diehn M, Alizadeh AA, Brown PO. Cell-type specific gene expression profiles of leukocytes in human peripheral blood. BMC Genomics. 2006;7:115. doi: 10.1186/1471-2164-7-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lappalainen T, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orrù V, et al. Genetic Variants Regulating Immune Cell Levels in Health and Disease. Cell. 2013;155:242–256. doi: 10.1016/j.cell.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brodin P, et al. Variation in the Human Immune System Is Largely Driven by Non-Heritable Influences. Cell. 2015;160:37–47. doi: 10.1016/j.cell.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saveanu L, et al. Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nat Immunol. 2005;6:689–697. doi: 10.1038/ni1208. [DOI] [PubMed] [Google Scholar]
- 35.Franco LM, et al. Integrative genomic analysis of the human immune response to influenza vaccination. eLife. 2013;2:e00299. doi: 10.7554/eLife.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao J, et al. Comprehensive single cell transcriptional profiling of a multicellular organism by combinatorial indexing. bioRxiv. 2017 doi: 10.1126/science.aam8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dixit A, et al. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell. 2016;167:1853–1866.e1817. doi: 10.1016/j.cell.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adamson B, et al. A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell. 2016;167:1867–1882.e1821. doi: 10.1016/j.cell.2016.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jaitin DA, et al. Dissecting Immune Circuits by Linking CRISPR-Pooled Screens with Single-Cell RNA-Seq. Cell. 2016;167:1883–1896.e1815. doi: 10.1016/j.cell.2016.11.039. [DOI] [PubMed] [Google Scholar]
- 40.Datlinger P, et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat Meth. 2017;14:297–301. doi: 10.1038/nmeth.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farh KK-H, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buettner F, et al. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotech. 2015;33:155–160. doi: 10.1038/nbt.3102. [DOI] [PubMed] [Google Scholar]
- 43.Tung P-Y, et al. Batch effects and the effective design of single-cell gene expression studies. Scientific Reports. 2017;7:39921. doi: 10.1038/srep39921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanay A, Regev A. Scaling single-cell genomics from phenomenology to mechanism. Nature. 2017;541:331–338. doi: 10.1038/nature21350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Habib N, et al. Div-Seq: Single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science. 2016;353:925. doi: 10.1126/science.aad7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lake BB, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science (New York, N.Y.) 2016;352:1586–1590. doi: 10.1126/science.aaf1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Habib N, et al. DroNc-Seq: Deciphering cell types in human archived brain tissues by massively-parallel single nucleus RNA-seq. bioRxiv. 2017 doi: 10.1038/nmeth.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wills QF, et al. Single-cell gene expression analysis reveals genetic associations masked in whole-tissue experiments. Nat Biotech. 2013;31:748–752. doi: 10.1038/nbt.2642. [DOI] [PubMed] [Google Scholar]
- 49.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, Spandidos A, Wang H, Seed B. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Research. 2012;40:D1144–D1149. doi: 10.1093/nar/gkr1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Picelli S, et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Meth. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 52.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protocols. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 53.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotech. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dabney A, Storey JD, Warnes GR. qvalue: Q-value estimation for false discovery rate control. R package version. 2010;1 [Google Scholar]
- 56.Falconer DS, Mackay TF, Frankham R. Introduction to quantitative genetics (4th edn) Trends in Genetics. 1996;12:280. [Google Scholar]
- 57.Loh PR, Palamara PF, Price AL. Fast and accurate long-range phasing in a UK Biobank cohort. Nat Genet. 2016;48:811–816. doi: 10.1038/ng.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.