Abstract
Chondroitin sulfate (CS)/dermatan sulfate (DS) proteoglycans are abundant on the cell surface and in the extracellular matrix and have important functions in matrix structure, cell-matrix interaction and signaling. The DS epimerases 1 and 2, encoded by Dse and Dsel, respectively, convert CS to a CS/DS hybrid chain, which is structurally and conformationally richer than CS, favouring interaction with matrix proteins and growth factors. We recently showed that Xenopus Dse is essential for the migration of neural crest cells by allowing cell surface CS/DS proteoglycans to adhere to fibronectin. Here we investigate the expression of Dse and Dsel in Xenopus embryos. We show that both genes are maternally expressed and exhibit partially overlapping activity in the eyes, brain, trigeminal ganglia, neural crest, adenohypophysis, sclerotome, and dorsal endoderm. Dse is specifically expressed in the epidermis, anterior surface ectoderm, spinal nerves, notochord and dermatome, whereas Dsel mRNA alone is transcribed in the spinal cord, epibranchial ganglia, prechordal mesendoderm and myotome. The expression of the two genes coincides with sites of cell differentiation in the epidermis and neural tissue. Several expression domains can be linked to previously reported phenotypes of knockout mice and clinical manifestations, such as the Musculocontractural Ehlers-Danlos syndrome and psychiatric disorders.
Introduction
Chondroitin sulfate (CS)/dermatan sulfate (DS) is a linear polysaccharide that is covalently attached to core proteins of proteoglycans and widely distributed both at the cell surface and in the extracellular matrix [1, 2]. CS/DS occurs in many invertebrates and vertebrates [3] and is involved in a range of biological functions, including the upbuilding of the extracellular matrix, cell signaling, wound healing, regeneration, and anti-coagulation [4, 5]. CS/DS is composed of alternating units of a hexuronic acid, i.e. either D-glucuronic acid (GlcA) or L-iduronic acid (IdoA), and the aminosugar N-acetyl-D-galactosamine. After synthesis of the chondroitin backbone, two DS-epimerases convert GlcA into IdoA by epimerization of the C5-carboxyl group of GlcA [6, 7], leading to the formation of hybrid CS/DS chains with a varying IdoA content. The IdoA-containing units can form long blocks or be interspersed as single moieties among unmodified GlcA-containing units. C5-epimerization and subsequent O-sulfation provide CS/DS chains with considerable structural variability and flexibility that allows interaction with matrix proteins and growth factors.
The formation of IdoA is catalyzed by DS-epi1 and DS-epi2, which are encoded by Dse and Dse-like (Dsel), respectively [6, 7]. Loss-of-function of these DS epimerases leads to severe consequences in vertebrates, indicating that IdoA residues in CS/DS play an important role in life. Homozygous missense mutations in DSE cause the musculocontractural type of Ehlers-Danlos syndrome (MC-EDS), a connective tissue disorder with congenital malformations and progressive fragility-related complications [8, 9]. Human DSEL (C18orf4) has been genetically linked to bipolar disorder [10] and early-onset major depressive disorder [11]. Dse knockout mice exhibit skin fragility that is caused by fewer IdoA residues in the CS/DS chains of the small leucine-rich proteoglycans decorin and biglycan, which leads to altered assembly of collagen fibrils in the hypodermis and dermis [12, 13]. On the other hand, no morphological and histological abnormalities have been observed in a mouse null mutant of Dsel [14]. Double knockout mice, entirely devoid of IdoA in their CS/DS, die around birth [15]. We recently showed that DS-epi1 has an important function in neural crest specification and cell migration in Xenopus embryos [16]. Single IdoA moieties in CS/DS polysaccharide chains are important for neural crest cells to adhere to fibronectin. The resemblance of the craniofacial defects in DS-epi1-deficient embryos and craniofacial anomalies in MC-EDS patients led us to suggest that MC-EDS might manifest developmental disturbances of the neural crest and therefore should be added to the list of neurocristopathies [17]. An open question is whether other congenital defects in MC-EDS might be associated with a role of DS-epi1 in the embryo. Another conundrum is how DS-epi2 malfunction might contribute to psychiatric disorders.
Although knockout mice point to an important function of CS/DS chains in early development [12, 15], the topological distribution of the chains in the embryo remains elusive. Despite that the expression of CS/DS-modifying enzymes has been presented in zebrafish [18] and of DS-biosynthetic enzymes in Xenopus neural crest cells [16], a comprehensive investigation of Dse and Dsel expression during embryonic development has not been performed yet. Here, we report that the two genes have similar but not identical expression patterns in the developing Xenopus embryo. The expression domains are consistent with locations of phenotypic alterations in mutant mice. They also coincide with clinical manifestations in human MC-EDS patients and might substantiate the genetic associations with psychiatric disorders.
Materials and methods
Constructs and RNA synthesis
A full-length cDNA sequence of Xenopus laevis Dsel.L with 86 nucleotides of the 5’ untranslated region (5’UTR), the open reading frame and 8 nucleotides of the 3’UTR was PCR-amplified from pCR-XL-TOPO-Dsel.L [16], subcloned into the pCS2 vector and completely sequenced. X. laevis cDNA fragments of Dse.S (nucleotides 828–1823 from GenBank accession number KU877109) and Dsel.L (nucleotides 2582–3608 from GenBank accession number KU877110) were PCR amplified from pCS105-Dse.S (Osada/Taira NBRP Xenopus ANE library, clone ID: XL487g09ex; [19]) and pCR-XL-TOPO-Dsel.L [16], subcloned into pCR4-TOPO (Thermo Fisher Scientific) and completely sequenced.
Sense mRNA for microinjection was synthesized using the mMessage Machine Kit (Ambion). Full-length cDNA plasmids were linearized with a restriction enzyme and transcribed with an RNA polymerase as follows: pCS105-Dse.S (BstXI, Sp6), pCS2-Dsel.L (NotI, Sp6) and pCS2-nlacZ (NotI, Sp6; a kind gift of Dr. Tomas Pieler, Univ. Göttingen, Germany).
For in vitro transcription, plasmids were linearized and antisense RNAs transcribed as follows: pCR4-TOPO-Dse.S (NotI, T3; also used in [16]), pCR4-TOPO-Dsel.L (SpeI, T7; also used in [16]), En2 (XhoI, T7; [20]), Foxg1 (XhoI, Sp6; [21],Pax3 (BglII, Sp6, [22]), Snai2 (ClaI, Sp6; [23]), Twist1 (EcoRI, T7; [24]), Xag1 (NcoI, Sp6, a kind gift of Dr. Tomas Pieler, Univ. Göttingen, Germany). Sense RNA of pCR4-TOPO-Dse.S (Sp6, T7) was generated for in situ hybridization.
Frog handling
All Xenopus experiments reported in this study have been approved by the Lund/Malmö regional ethical committee (M140-14). Adult sexually mature Xenopus laevis frogs were purchased from Nasco (Fort Atkinson, WI, USA). Female and male frogs were housed in separate racks with a recirculation system (Tecniplast, Italy). A total of 20 female and 5 male frogs were used for this study. Ovulation was stimulated by injection of 750 units of human chorionic gonadotropin (Sigma CG10) into the lymph sac of a female frog. Female frogs were re-used for egg collection after a recovery period of at least 3 months. For testis isolation and in vitro fertilization, male frogs were sacrificed by incubation for 10 min in 0.05% benzocaine (Sigma E1501) and decapitation.
Embryo manipulations, whole-mount in situ hybridization and vibratome sections
Embryos were prepared, microinjected, cultured and analyzed by Red-Gal staining and whole-mount in situ hybridization as described in a previous publication [25]. For whole-mount in situ hybridization, treatment with Proteinase K (Ambion AM2546) and a hybridization temperature of 65°C were used. Stained embryos were sectioned with a surgical blade (Ref. 0301; Swann-Morton, Sheffield, U.K.). For vibratome sections, stained embryos were equilibrated in PBS containing 4.44 mg/ml gelatin (Merck, 1.04078) and 0.27 g/ml bovine serum albumin (Sigma, A3912). After equilibration, embryos were mounted by solidifying the solution with 0.075 volume glutaraldehyde (25% stock, Merck, 1.04239) and cut as 50 μm sections with a Leica VT 1200S vibratome (Leica Microsystems AB, Sweden).
RNA isolation and gene expression quantification by RT-qPCR
Total RNA from 10 embryonic explants per sample was extracted using the RNeasy Mini Kit (Qiagen) and reverse transcribed to cDNA (Quantitect Reverse Transcription Kit, Qiagen). Real-time quantitative PCR was run using the FAST SYBR Green Master Mix (Ambion Life Technologies) and the Mx3005P qPCR system (Stratagene) with standard cycling parameters. Fold expression values were calculated using the 2-ΔCt formula, using the housekeeping gene eEF1A1 as control. The sequences of the primers (Invitrogen) have been described previously [16].
Results
Antisense RNA probes specifically detect Dse and Dsel mRNAs in Xenopus embryos
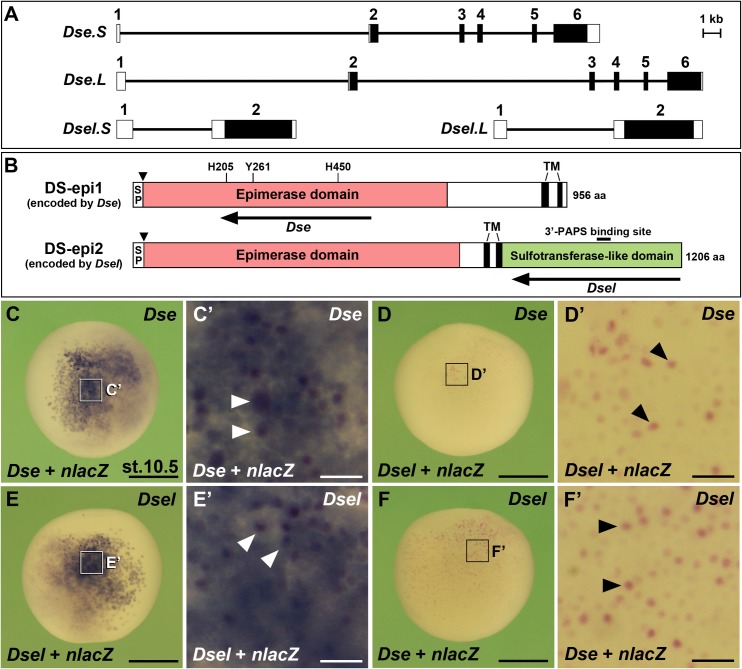
We recently isolated full-length cDNA clones of Dse and Dsel, for which each two homeologous copies exist in X. laevis [16]. Dse.S and Dse.L are located on chromosomes 5S and 5L, respectively, while Dsel.S and Dsel.L are on the 6S and 6L chromosomes (NCBI, International Xenopus Sequencing Consortium, UC Berkeley, USA). The genomic loci of Dse.S relative to Dse.L and of Dsel.S relative to Dsel.L are 18% and 14% smaller, respectively (Fig 1A), likely as a consequence of small-scale deletions due to intra-chromosomal rearrangements that are more common to the S (short) than the L (long) subgenome in X. laevis [26]. The Dse gene consists of six exons with an open reading frame that spans from exon 2 to exon 6, whereas the Dsel gene has two exons with a continuous open reading frame on the second exon. This genomic structure is conserved in human DSE and DSEL [7]. At the nucleotide level, the coding regions of the short and long homeologous X. laevis copies are 93% (Dse) and 95% (Dsel) identical. At the amino acid level, their identities are 94% (DS-epi1 encoded by Dse) and 96% (DS-epi2 encoded by Dsel) [16]. The amino acid similarity between the S and L copies of each protein is 97%. Xenopus DS-epi1 and DS-epi2 encompass 956 and 1206 amino acids, respectively (Fig 1B; [16]). Like their human counterparts [7], they share a cleavable signal peptide, a ~700 amino acid domain of an active epimerase domain and two transmembrane segments. A comparison of the X. laevis amino acid sequences reveals an identity of ~50% in the common epimerase domains of DS-epi1 and DS-epi2 (S1 Fig; [16]). Three amino acids in the epimerase domain that are required for its catalytic activity in human DS-epi1 (H205, Y261, H450; [27]) are conserved in each homeologous copy of the X. laevis DS-epi1 and DS-epi2 enzymes. In addition, DS-epi2 contains a carboxyterminal domain with homology to several carbohydrate sulfotransferases, including a short signature of the 3’-PAPS (3’-phosphoadenosine-5’-phosphosulfate)-binding site. This homology makes plausible a sulfotransferase activity of DS-epi2 although conclusive data are not available.
Fig 1. Genomic organization, protein structure and probe specificity of Dse/DS-epi1 and Dsel/DS-epi2 in X. laevis embryos.
(A) Genomic structures of the Dse and Dsel homeologs. Rectangles with numbers show exons and intervening lines demarcate introns. Filled boxes indicate open reading frames. Accession numbers of the Xenopus laevis genomic DNA / mRNA sequences are: DS-epi1.S, NC_030733 / KU877109; DS-epi1.L, NC_030732 / XM_018263281; DS-epi2.S, NC_030735 / XM_018223616; DS-epi2.L, NC_030734 / KU877110. (B) Overall protein structure of DS-epi1 and DS-epi2, which are encoded by Dse and Dsel, respectively. SP, cleavable signal peptide; TM, transmembrane domain. The arrows represent the antisense RNA probes of Dse and Dsel that were used for the in situ hybridization. (C-F’) Whole-mount in situ hybridization of early gastrula embryos in lateral view. The Dse probe detects injected Dse mRNA (C,C’) but not Dsel mRNA (D,D’). The Dsel probe specifically targets injected Dsel mRNA (E-F’). Arrowheads depict cells that received co-injected nlacZ mRNA as a lineage tracer (red nuclei). Each synthetic mRNA was injected at a dose of 100 pg into the animal pole of a single blastomere at the 4-cell stage. Scale bars are 500 μm (C-F) and 50 μm (C’-F’).
In order to analyze the gene expression of Dse and Dsel, we generated ~1 kb antisense RNA probes against the epimerase domain of DS-epi1 and the sulfotransferase-like domain of DS-epi2 (Fig 1B). Given the sequence homology of both genes in the epimerase domain, we first tested the specificity of the Dse probe in Xenopus embryos that were microinjected with either Dse or Dsel mRNA. Our previous RT-PCR analysis showed that endogenous mRNA levels of Dse and Dsel are relatively low at the early gastrula stage [16]. Whole-mount in situ hybridization at stage 10.5 showed that the Dse probe unambiguously detected cells derived from a Dse mRNA-injected blastomere (Fig 1C and 1C’). In contrast, the same probe failed to bind to exogeneous Dsel mRNA (Fig 1D and 1D’). As expected, the Dsel probe bound to injected Dsel mRNA (Fig 1E and 1E’) and not to exogeneous Dse mRNA (Fig 1F and 1F’). Thus the antisense RNA probes against Dse and Dsel specifically detect their respective target mRNAs.
Expression of the Dse gene
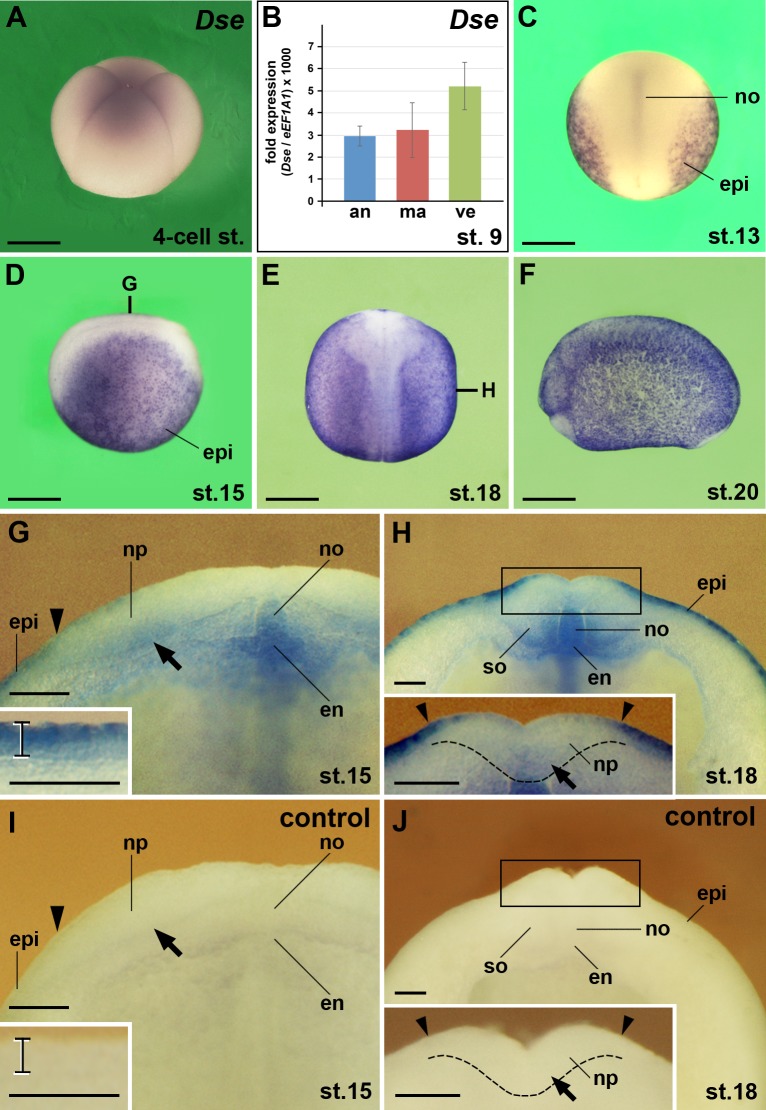
By whole-mount in situ hybridization, we detected low levels of Dse mRNA in 4-cell stage embryos, suggesting maternal Dse transcription (Fig 2A). Quantitative real time PCR (qPCR) showed Dse signals in animal, marginal and vegetal explants at stage 9 (Fig 2B). From (mid gastrula) stage 13 onwards, zygotic Dse expression was seen in the epidermis and notochord (Fig 2C–2H). A transversal hemi-section at stage 15 revealed robust Dse expression in the superficial layer and lower-level expression in the sensorial layer of the epidermis (inset in Fig 2G). The inner sensorial layer of the lateral neural plate showed weak Dse signals (Fig 2G). Additional signals were seen in the dorsal medial endoderm from which the hypochord will evolve. At (neurula) stage 18, low-level Dse expression shifted from the lateral to the medial (now ventral) sensorial layer of the neural groove at the level of the future mid- and hindbrain (inset in Fig 2H). Dse transcripts appeared in the ventro-medial edge of the somites (Fig 2H) that will give rise to the sclerotome [28]. In situ hybridization with a sense RNA probe did not show any signal in embryos at stage 15 and 18 (Fig 2I and 2J), confirming the specificity of Dse detection.
Fig 2. Dse mRNA is maternally deposited and expressed in the epidermis, neural ectoderm, notochord, somites and dorsal endoderm.
Xenopus embryos after whole-mount in situ hybridization in lateral view (A,D,F), dorsal view (C,E) and transversally sectioned through the anterior trunk (G-J). (A) Embryo at the 4-cell stage. Weak expression of Dse mRNA is visible. (B) qPCR analysis of embryonic explants at stage 9. Note Dse transcripts in the animal cap, marginal zone and vegetal region. (C) At stage 13, Dse is expressed in the epidermis and notochord. (D-F) Neurula embryos show ubiquitous Dse expression in the epidermis. The bold lines indicate the levels of sections in G and H. (G) Embryo at stage 15. The arrowhead indicates the border between epidermis and neural ectoderm. Note Dse transcripts in the sensorial layer of the lateral neural plate (arrow), notochord and dorsal endoderm. The bracket in the inset depicts the epidermis with robust Dse expression in the outer layer and lower mRNA levels in the inner layer. (H) Embryo at stage 18. Note Dse signals in the medio-ventral somites. The inset shows Dse expression in the sensorial layer of the ventral neural groove (arrow). The stippled line demarcates the border between the neural plate and the underlying dorsal mesoderm. (I,J) No signals appear in matching sections of sibling embryos that were probed with Dse sense RNA as control. an, animal cap; en, endoderm; epi, epidermis; ma, marginal zone; no, notochord; np, neural plate; so, somite; ve, vegetal region. Scale bars are 500 μm (A,C-F) and 50 μm (G-J).
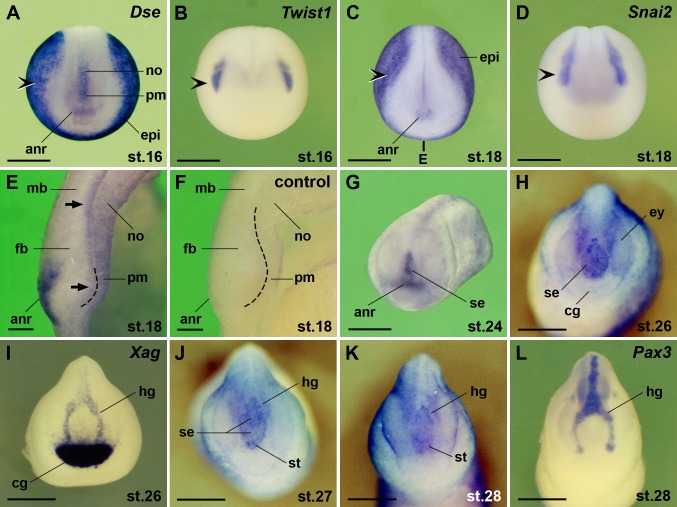
We previously showed that Dse is expressed in cranial neural crest cells [16]. Here we confirm its expression in the Twist1- and Snai2-positive pre-migratory neural crest at the border between epidermis and anterior neural plate (Fig 3A–3D). In early neurulae, Dse transcripts appeared in the superficial layer of the anterior neural ridge and in the prechordal mesendoderm (Fig 3A, 3C and 3E). At stage 18, weak Dse signals were also detected in the inner layer of the forebrain and midbrain anlage (Fig 3E). The absence of signals in a sibling embryo probed with a sense RNA underscore the specificity of the Dse signals (Fig 3F). At stage 24, Dse expression extended dorsally into the sense plate (Fig 3G). The sense plate arises from the anterior neural ridge and comprises the anlage of the stomodeum, adenohypophyseal placode and olfactory placodes [29–31]. At stage 26, the expression domain of Dse in the sense plate (Fig 3H) was flanked by Xag expression in the cement gland and hatching gland (Fig 3I). Dse expression in the sense plate then segregated into the anterior stomodeum-adenohypophysis anlage and a posterior bifurcated domain (Fig 3J and 3K) that abutted and partially overlapped with Pax3+ hatching gland cells (Fig 3L).
Fig 3. Dse is expressed in the pre-migratory neural crest, anterior surface ectoderm, brain, and prechordal mesendoderm.
Embryos are shown in anterior view (A-D, G-L) and midsagittally sectioned through the head (E,F). (A-D) Early neurulae. Note Dse expression in the anterior neural ridge and prechordal mesendoderm. Dse transcripts overlap with Twist1 and Snai2 expression in the cranial neural crest (indented arrowheads). The bold line indicates the level of section in E. (E) Embryo at stage 18. Dse is expressed in the superficial layer of the anterior neural ridge and the inner layer of the fore- and midbrain anlage (arrows). The stippled line demarcates the border between the anterior neural plate and the underlying prechordal mesendoderm. (F) Absence of signals in the matching section of a sibling embryo that was probed with Dse sense RNA as control. (G) Late neurula at stage 24. Dse is expressed in the anterior neural ridge and derived sense plate. (H) Tailbud embryo at stage 26. Dse is transcribed in the sense plate. (I) Xag expression demarcates the cement gland and hatching gland. (J-L) Tailbud embryos at stages 27 and 28. Note segregation of anterior Dse expression into the stomodeum-adenohypophysis anlage and posterior domain that partially overlaps with the Pax3+ hatching gland. anr, anterior neural ridge; cg, cement gland; epi, epidermis; ey, eye; fb, forebrain; hg, hatching gland; mb, midbrain; no, notochord; pm, prechordal mesendoderm; se, sense plate; st, stomodeum-adenohypophysis anlage. Scale bars are 500 μm (A-D,G-L) and 50 μm (E,F).
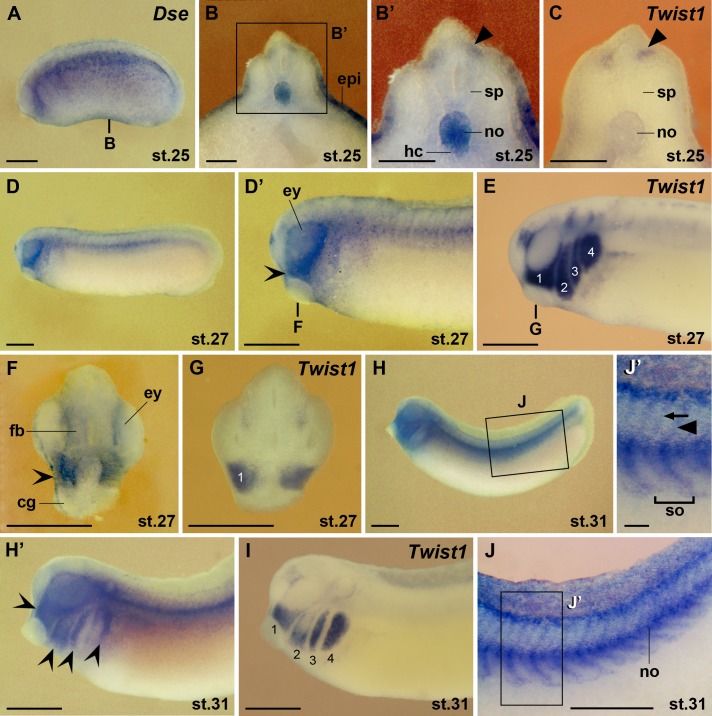
With the extension of the primary body axis, Dse expression faded in the epidermis and became more robust in the notochord and underlying hypochord (Fig 4A–4B’). At stage 25, Dse overlapped with the Twist1 marker in migrating trunk neural crest cells (Fig 4A–4C). At stage 27, Dse transcripts accumulated in the Twist1+ mandibular stream of the cranial neural crest cells and were also detected in the proximal portion of the eyes and the forebrain (Fig 4D–4G). Cranial neural crest cells arise rostral to the anterior limit of somite 4 and migrate along distinct routes as mandibular, hyoid, and branchial arch streams [32]. At stage 31, Dse followed Twist1 expression in hyoid and branchial cranial neural crest cells (Fig 4H and 4I; [16]). Dse mRNA was also localized in the trunk neural crest cells and neural crest-derived spinal nerves that migrate between and penetrate through the somites, respectively (Fig 4J and 4J’).
Fig 4. Dse is expressed in the eye, migratory neural crest and the hypochord.
Embryos are shown in lateral view (A,D-E,H-J’), and transversally sectioned through the trunk (B-C) and head (F,G). (A) Embryo at stage 25. The bold line indicates the level of the section in B. (B,B’) Dse is expressed in the epidermis, notochord and hypochord. The arrowhead shows Dse transcripts in trunk neural crest cells adjacent to the spinal cord. (C) Matching section of a sibling embryo showing Twist1 expression in trunk neural crest cells (arrowhead). (D-I) Tailbud embryos. The bold lines indicate the level of sections in F and G. Dse is expressed in the forebrain, proximal eye and Twist1+ cranial neural crest (indented arrowheads). The numbers indicate the mandibular (1), hyoid (2) and branchial neural crest (3,4). (J,J’) Embryo at stage 31 after removal of the epidermis. Dse is expressed in the spinal nerves (arrow) and migrating trunk neural crest cells (arrowhead). cg, cement gland; epi, epidermis; ey, eye; fb, forebrain; hc, hypochord; no, notochord; sp, spinal cord; so, somite. Scale bars are 500 μm (A,D-J) and 100 μm (B-C,J’).
At stage 33, distinct Dse expression was observed in the ectodermal adenohypophysis, forebrain, ventral midbrain, cranial neural crest and notochord (Fig 5A–5C). More posteriorly, Dse expression appeared in the neural crest-derived melanophores, in the somite-derived dermomyotome and sclerotome and in the endodermal hypochord (Fig 5D and 5E). At stage 40, Dse transcripts were maintained in post-migratory cranial neural crest cells and in the notochord (Fig 5F–5I).
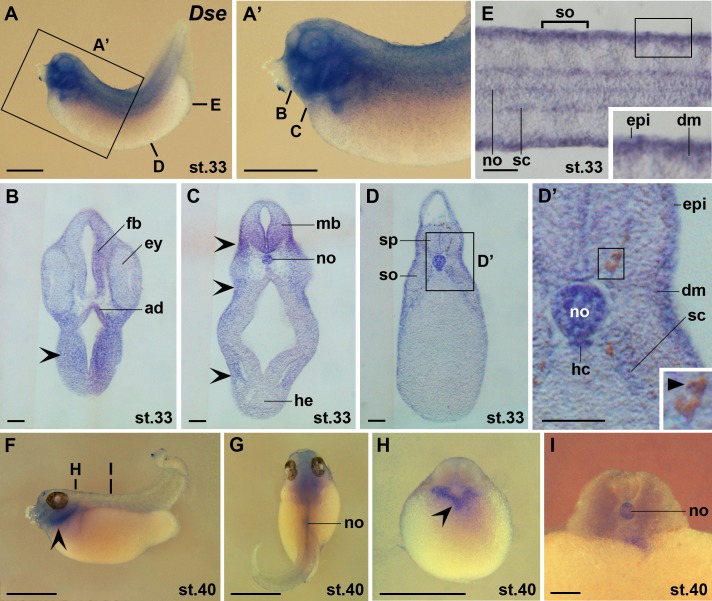
Fig 5. Dse is expressed in the adenohypophysis, melanophores, dermomyotome and sclerotome.
Embryos are shown in lateral view (A,A’,F), dorsal view (G), transversal sections (B-D’,H,I) and horizontal section (E). (A,A’) Embryo at stage 33. The bold lines indicate the level of sections in B-E. (B-E) Vibratome sections showing Dse expression in the adenohypophysis, forebrain, ventral midbrain, epidermis, notochord, hypochord, dermomyotome, and sclerotome. Note also Dse transcripts in cranial neural crest cells (indented arrowheads) and melanophores (filled arrowhead). (F-I) Embryo at stage 40. The bold lines indicate the level of sections in H and I. Dse is expressed in post-migratory cranial neural crest cells (indented arrowheads) and the notochord. ad, adenohypophysis; dm, dermomyotome; epi, epidermis; ey, eye; fb, forebrain; hc, hypochord; he, heart; mb, midbrain; no, notochord; sc, sclerotome; so, somite; sp, spinal cord. Scale bars are 1 mm (A,A’,F-H) and 100 μm (B-E,I).
In summary, Dse mRNA is maternally deposited and zygotically expressed in the epidermis, anterior surface ectoderm, neural ectoderm, neural crest, prechordal mesendoderm, notochord, somites and dorsal endoderm. Later, Dse transcripts are found in the adenohypophysis, eyes, brain, spinal nerves, melanophores, dermomyotome, sclerotome and hypochord.
Expression of the Dsel gene
Previous RT-PCR results showed that the maternal expression of Dsel is higher than that of Dse [16]. Our present analysis supports this conclusion. By whole-mount in situ hybridization, we found abundant maternal deposition of Dsel mRNA in 4-cell stage embryos (Fig 6A). At stage 9, Dsel transcripts were detected in the animal cap and marginal zone (Fig 6B and 6C). qPCR analysis showed additional Dsel mRNA in the vegetal region (Fig 5D), suggesting that the yolk quenches in situ hybridization signals in these cells. We previously demonstrated that low levels of Dsel transcripts are zygotically expressed in Twist1+ cranial neural crest cells [16]. Accordingly, we found weak Dsel expression in cranial neural crest cells of the mandibular, hyoid and branchial arch regions at stages 27 and 30 (Fig 6E–6H). Additional Dsel transcripts were observed in the eyes, distinct interneurons of the ventral midbrain and hindbrain, and in cranial neural crest-derived trigeminal (V) ganglia.
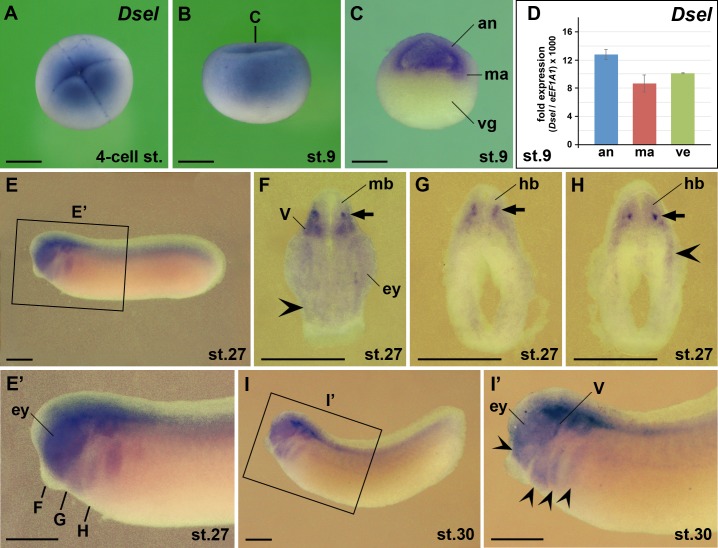
Fig 6. Dsel is expressed maternally and in the eye, brain and cranial neural crest.
Embryos after whole-mount in situ hybridization are shown in animal view (A), lateral view (B,E,E’,I,I’), hemi-sectioned (C) and transversally sectioned (F-H). (A) 4-cell stage embryo. (B,C) Blastula embryos. The bold line indicates the level of section in C. (D) qPCR analysis at stage 9. Note ubiquitous expression of Dsel in the animal cap, marginal zone and vegetal region. (E-I’) Tailbud embryos. The bold lines indicate the level of sections in F-H. Note Dsel expression in the eye, interneurons of the mid- and hindbrain (arrows), cranial neural crest (indented arrowheads) and bilateral trigeminal ganglia. an, animal cap; ey, eye; hb, hindbrain; ma, marginal zone; mb, midbrain; V, trigeminal ganglion, vg, vegetal region. Scale bars are 500 μm.
At stage 33, Dsel was maintained in cranial neural crest cells (Fig 7A and 7C–7G). Dsel overlapped with N-tubulin expression in the trigeminal (V) ganglia and three neural crest-derived epibranchial ganglia, including the geniculate (VII), petrosol (IX) and nodose (X) ganglia (Fig 7A–7B’ and 7E–7G). A more detailed analysis of histological sections showed weak Dsel expression in the adenohypophysis and ventrolateral forebrain (Fig 7D). More robust signals appeared bilaterally in the ventral marginal zone of the midbrain and hindbrain, where interneurons differentiate (Fig 7E, 7G and 7H). The lower expression levels in the isthmus (Fig 7F) suggest that the Dsel-positive interneurons in the adjacent midbrain (Fig 7E) and anterior hindbrain (Fig 7G) are non-continuous. Two distinct ventral columns of Dsel+ neurons were found on each side in the hindbrain (Fig 7H). Weaker bilateral Dsel expression domains appeared in the marginal zone of the posterior hindbrain and spinal cord (Fig 7I and 7J). At stage 34, more abundant Dsel transcripts were detected in post-migratory cranial neural crest cells of the head and branchial arches (Fig 7K and 7K’).
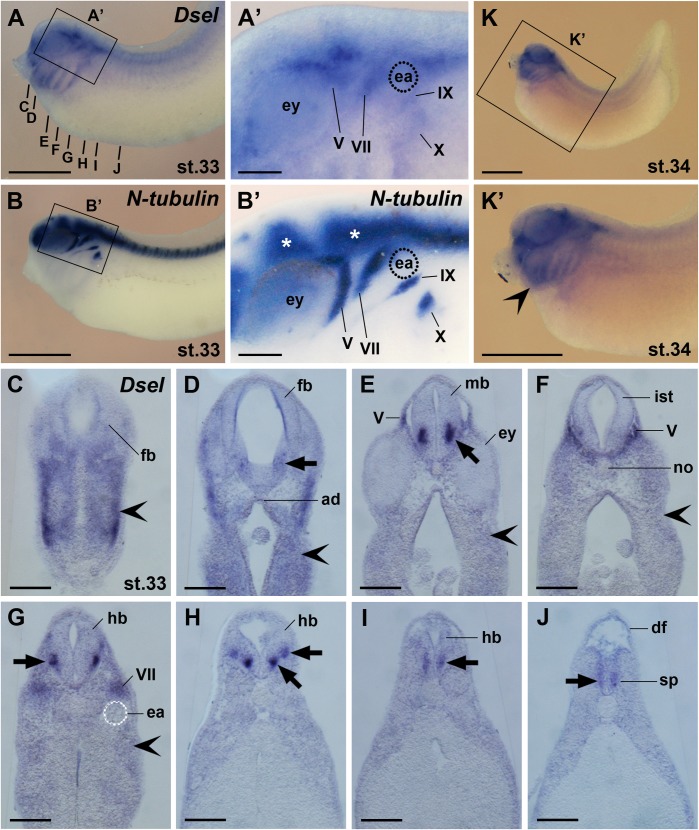
Fig 7. Dsel is expressed in differentiated neurons, cranial sensory ganglia and the spinal cord.
Embryos are shown in lateral view (A-B’,K,K’) and transversal vibratome sections (C-J). (A,A’) Embryo at stage 33. The bold lines indicate the level of sections in C-J. Magnification in A’ depicts Dsel mRNA in cranial sensory ganglia: V, trigeminal ganglion; VII, geniculate ganglion; IX, petrosal ganglion; X, nodose ganglion. (B,B’) Sibling embryo depicting N-tubulin expression in the central nervous system (stars) and cranial ganglia. (C-J) Dsel is expressed in the adenohypophysis and distinct neurons (arrows) of the forebrain, midbrain, hindbrain and spinal cord. The indented arrowhead labels signals in migrating cranial neural crest cells. (K,K’) Embryo at stage 34. The indented arrowhead labels robust Dsel transcripts in post-migratory cranial neural crest cells. ad, adenohypophysis; df, dorsal fin; ea, ear; ey, eye; fb, forebrain; hb, hindbrain; ist, isthmus; IX, petrosal ganglion; mb, midbrain; no, notochord; pm, sp, spinal cord; V, trigeminal ganglion; VII, geniculate ganglion; IX, petrosal ganglion; X, nodose ganglion. Scale bars are 1 mm (A,B,K,K’) and 200 μm (A’,B’,C-J).
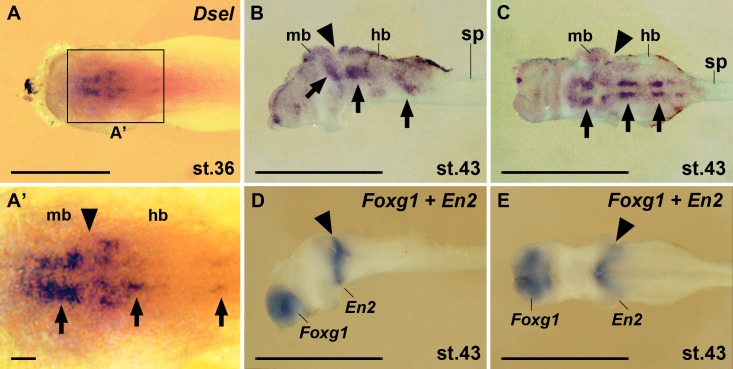
Swimming tadpole embryos at stages 36 and 43 showed distinct Dsel expression domains in the brainstem (Fig 8). Along the anteroposterior neuraxis, a single Dsel expression domain was seen in the ventral midbrain and at least two bilateral domains in the ventral hindbrain (Fig 8A–8C). Only weak Dsel expression appeared in the intervening En2+ isthmus region (Fig 8A’–8E). The Foxg1+ forebrain region and the spinal cord did not show detectable Dsel transcripts at these stages.
Fig 8. Dsel is expressed in the brainstem in tadpole embryos.
Specimen after whole-mount in situ hybridization are shown in dorsal view (A,A’), lateral view (B,D) and ventral view (C,E). (A,A’) Embryo at stage 36. Arrows label distinct Dsel expression domains in the midbrain and hindbrain. The arrowhead labels the position of the midbrain-hindbrain boundary. (B-E) Isolated central nervous system from stage 43 embryos after removing of surrounding tissues. Dsel mRNA accumulates in bilateral domains in the brainstem (arrows). Expression of Foxg1 demarcates the forebrain and En2 the midbrain-hindbrain isthmus (arrowhead). hb, hindbrain; mb, midbrain; sp, spinal cord. Scale bars are 1 mm (A-E) and 100 μm (A’).
Altogether, Dsel is a maternally enriched gene and exhibits zygotic expression from the tailbud stage onwards in the adenohypophysis, early eyes, differentiated neurons in the brain and spinal cord, cranial neural crest, cranial sensory ganglia, prechordal mesendoderm, and hypochord.
Dse and Dsel are co-expressed in the ventral brain, cranial neural crest cells and trigeminal ganglia
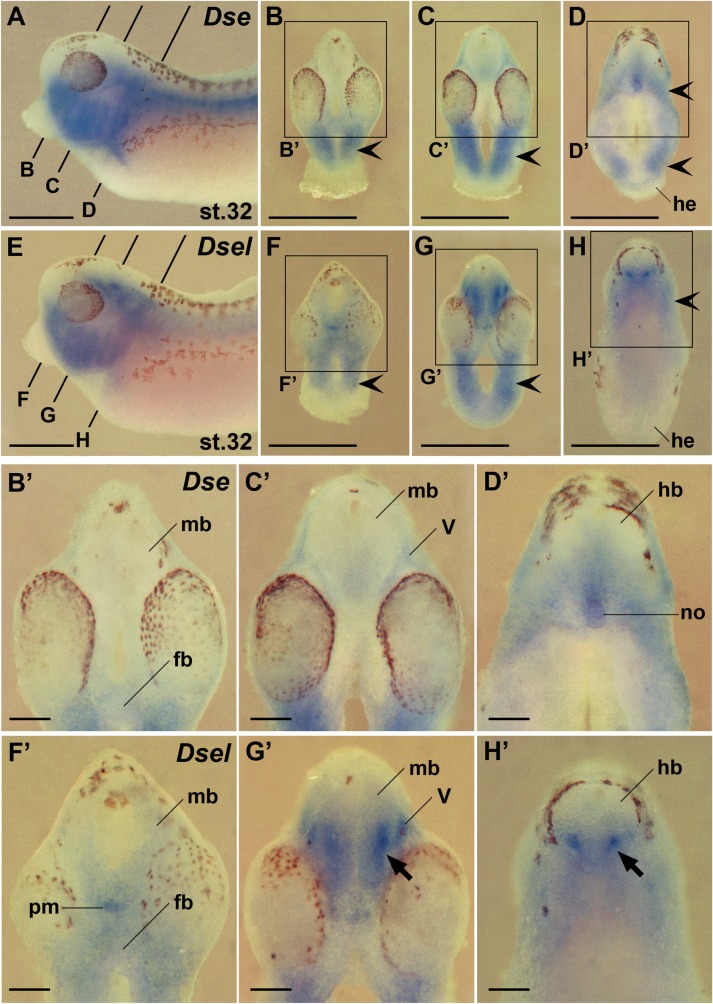
Next we compared the expression of Dse and Dsel in the head of transversally sectioned embryos after whole-mount in situ hybridization at stage 32 (Fig 9). The expression of both genes partially overlapped in the ventral forebrain, midbrain and hindbrain, with Dse mRNA (Fig 9A–9D’) being more restricted than Dsel mRNA (Fig 9E–9H’). Robust Dsel expression was observed in postmitotic ventral interneurons in the marginal zone of the mid- and hindbrain (Fig 9G’ and 9H’). Dse (Fig 9A–9D) and Dsel expression (Fig 9E–9H) largely overlapped in post-migratory cranial neural crest cells of the head mesenchyme and pharyngeal arches. Dse transcripts were more abundant than those of Dsel in posterior cranial neural crest cells adjacent to the heart (compare Fig 9A and 9D with Fig 9E and 9H). In the trigeminal (V) ganglia, Dsel was more robustly expressed than Dse (compare Fig 9A and 9C’ with Fig 9E and 9G’). Along the primary body axis, the Dsel gene was active in the prechordal mesendoderm (Fig 9F’), wherease Dse mRNA accumulated in the notochord (Fig 9D’).
Fig 9. Comparison of Dse and Dsel expression in the head.
Embryos at stage 32 are shown in lateral views (A,E) and transversal sections (B-D’, F-H’). (A) Dse expression. The bold lines indicate the level of sections in B-D. (B-D’) Dse expression is found in the cranial neural crest (indented arrowheads). Note ventral expression domains in the forebrain, posterior midbrain, hindbrain, and notochord. Weak expression is visible in the trigeminal ganglia. (E) Dsel expression. The bold lines indicate the level of sections in F-H. (F-H’) Dsel expression appears in the cranial neural crest (indented arrowheads) and trigeminal ganglia. Note expression domain in the prechordal mesendoderm. Apparent are low-level transcripts in the entire ventral brain and robust bilateral expression domains in the ventral marginal zone of the mid- and hindbrain (arrows). fb, forebrain; hb, hindbrain; he, heart; mb, midbrain; no, notochord; pm, prechordal mesendoderm; V, trigeminal ganglion. Scale bars are 1 mm (A-H) and 200 μm (B’-H’).
Overlapping and non-overlapping expression of Dse and Dsel in the trunk
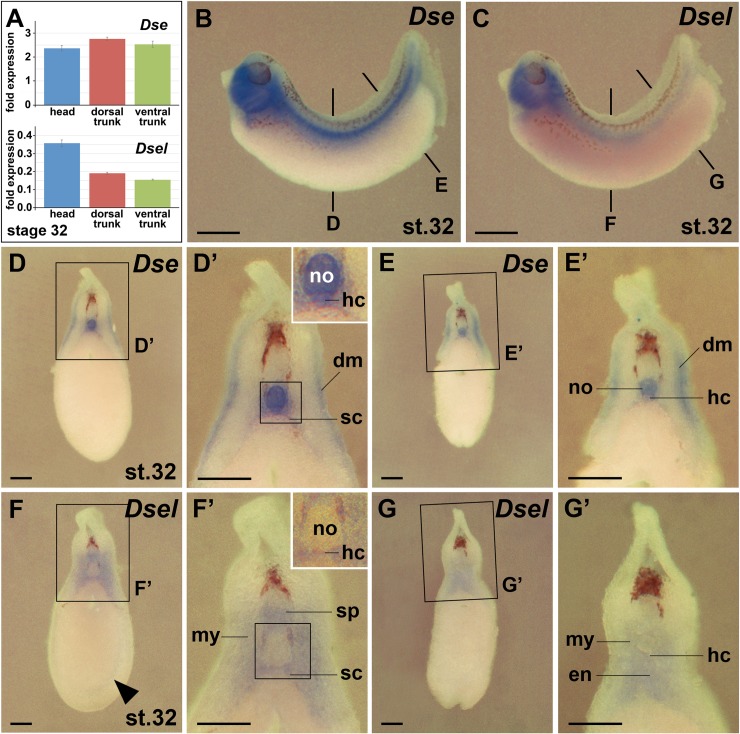
Using qPCR analysis, we quantified the Dse and Dsel mRNA levels in head, dorsal trunk and ventral trunk explants at stage 32 (Fig 10A). The Dse transcript levels were equal in these regions (Fig 10A top). The Dsel mRNA was 2-fold higher concentrated in the head than in the trunk, and comparable levels were found between the dorsal and ventral trunk region (Fig 10A bottom). Whole-mount in situ hybridization confirmed the relative distribution of both gene products in the head and dorsal trunk, but showed less intense staining in the ventral trunk (Fig 10B and 10C) likely as a consequence of quenching of in situ hybridization signals due to a high amount of yolk in the gut endoderm. A closer inspection of the trunk in transversally sectioned embryos (Fig 10D–10G’) revealed co-expression of Dse and Dsel in the hypochord and sclerotome cells that have migrated to the perineural and perinotochordal space [28]. Unique Dse expression was found in the notochord and dermatome (Fig 10D–10E’). Dsel expression alone appeared in the ventral hemisphere of the spinal cord, myotome and posteriorly in the dorsal hindgut endoderm (Fig 10F–10G’). Thus, Dse and Dsel share expression domains in the ventral brain, cranial neural crest, trigeminal (V) ganglia, sclerotome and hypochord.
Fig 10. Comparison of Dse and Dsel expression in the trunk.
Embryos at stage 32 are shown in lateral views (B,C) and transversal sections (D-G’) after whole-mount in situ hybridization. (A) qPCR analysis of embryonic explants at stage 32. The numbers on the y-axis indicate fold expression of (Dse / eEF1A1) x 1000 (top) and (Dsel / eEF1A1) x 1000 (bottom). Note equal distribution of Dse transcripts in the head, dorsal trunk and ventral trunk. Dsel mRNA is most abundant in the head and is expressed at comparable levels in the dorsal and ventral trunk. (B) Dse expression. The bold lines indicate the level of sections in D and E. (D-E’) Dse expression is detected in the notochord, dermatome, sclerotome and hypochord (inset). (C) Dsel expression. The bold lines indicate the level of sections in F and G. (F-G’) Dsel expression appears in the ventral spinal cord, migrating trunk neural crest cells (arrowhead), myotome, sclerotome, hypochord (inset) and posterior dorsal hindgut endoderm. dm, dermatome; en, endoderm; hc, hypochord; my, myotome; no, notochord; sc, sclerotome; sp, spinal cord. Scale bars are 1 mm (B,C) and 200 μm (D-G’).
Discussion
The frog Xenopus laevis arose from hybridization of two diploid Xenopus species, leading to allotetraploidy with two related subgenomes [26]. The gene loci of the two dermatan sulfate epimerases are conserved between human [7] and X. laevis (this study), with the open reading frame of Dse spanning over five exons and that of Dsel confined to a single exon. DS-epi1 and DS-epi2 have a common epimerase domain and their enzymatic activities have been experimentally validated in human and Xenopus [7, 16, 27]. Three catalytic amino acids that were previously shown to be indispensable for the epimerase activity in human DS-epi1 are also present in human DS-epi2 and are conserved in the four X. laevis sequences, suggesting that each homeolog might contribute to the production of IdoA structures in CS/DS hybrid chains.
Our comparative analysis of the two DS biosynthetic enzymes reveals both common and differential expression of Dse and Dsel (Table 1). Previous RT-PCR results revealed maternal deposits of mainly Dsel mRNA in Xenopus blastula embryos [16]. Based on whole-mount in situ hybridization and qPCR analysis, we now show maternal expression of both genes with Dsel exhibiting higher mRNA levels than Dse at the 4-cell stage and at stage 9. We previously reported distinct expression domains of Dse in the epidermis, pre-migratory cranial neural crest and the notochord at stage 17 and of both Dse and Dsel in migratory cranial neural crest cells at stage 30 [16]. Here, we introduce a series of new expression domains. Partially overlapping expression of the two genes was observed in the adenohypophysis, eyes, brain, trigeminal (V) ganglia, sclerotome, and the dorsal endoderm (hypochord). Additional distinct Dse transcripts were found in the anterior surface ectoderm, spinal nerves, and dermatome, while Dsel was expressed in the spinal cord, epibranchial (VII, IX, X,) ganglia, prechordal mesendoderm and myotome. The relative contribution of the two DS epimerases to early IdoA production in CS/DS chains might differ in other vertebrates, since e.g. in zebrafish both dse and dselb, but not the duplicated dsela ortholog, are maternally expressed [18]. Similarly as in Xenopus, zebrafish dse is expressed in the eye and notochord, dselb in the midbrain/hindbrain boundary region, and both dse and dselb in the somites.
Table 1. Summary of gene expression for Dse and Dsel in Xenopus laevis embryo.
| cleavage | blastula | gastrula | neurula | tailbud | tadpole | ||
|---|---|---|---|---|---|---|---|
| st. 1–6 | st. 7–9 | st. 10–13 | st. 14–22 | st. 23–40 | st. 41- | ||
| maternal | Dse | + | + | ||||
| Dsel | +++ | +++ | |||||
| eye | Dse | + | |||||
| Dsel | + | ||||||
| forebrain | Dse | + | + | - | |||
| Dsel | - | + | - | ||||
| midbrain | Dse | + | + | - | |||
| Dsel | - | +++ | ++ | ||||
| hindbrain | Dse | + | + | - | |||
| Dsel | - | +++ | ++ | ||||
| spinal cord | Dse | - | - | - | |||
| Dsel | - | ++ | - | ||||
| cranial sensory ganglia | Dse | + | |||||
| Dsel | ++ | ||||||
| spinal nerves | Dse | + | |||||
| Dsel | - | ||||||
| neural crest | Dse | ++ | +++ | ++ | |||
| Dsel | - | +++ | - | ||||
| anterior surface ectoderm | Dse | ++ | ++ | - | |||
| Dsel | - | - | - | ||||
| adenohypophysis | Dse | ++ | |||||
| Dsel | + | ||||||
| epidermis | Dse | +++ | +++ | ++ | - | ||
| Dsel | - | - | - | - | |||
| prechordal mesendoderm | Dse | + | - | ||||
| Dsel | - | ++ | |||||
| notochord | Dse | + | ++ | ++ | ++ | ||
| Dsel | - | - | - | - | |||
| dermatome | Dse | - | ++ | ||||
| Dsel | - | - | |||||
| myotome | Dse | - | - | ||||
| Dsel | - | ++ | |||||
| sclerotome | Dse | ++ | ++ | ||||
| Dsel | - | + | |||||
| dorsal endoderm | Dse | ++ | ++ | ||||
| Dsel | - | ++ |
Intensity levels based on whole-mount in situ hybridization are indicated as follows: -, no expression; +, weak; ++, intermediate; +++, strong. Note that not all stages were analyzed.
In Xenopus, Dse is dynamically expressed in distinct domains of the anterior surface ectoderm, including the anterior neural fold, sense plate and derived stomach anlage, adenohypophysis and hatching gland. Consistently, zebrafish dsela and dselb are expressed in mesoendodermal cells of the anterior polster [18]that gives rise to the hatching gland in teleosts. Hence the gene expression patterns of the DS-epimerases identifies the hatching gland as a possible site of CS/DS production and underscores its evolutionary origin from different germ layers in the frog and fish [33].
Dse shows abundant expression in the epidermis between stages 13 and 27 with low mRNA levels in the deep layer and higher levels in the superficial layer. The superficial epidermis is composed of polarized epithelial cells and abundantly expresses differentiation markers in contrast to the deep non-epithelial cells [34]. The bi-layered epidermis in Xenopus embryos is similar to the mammalian embryonic epidermis, which consists of an inner basal layer and a temporary outer periderm [35]. During mammalian skin development, basal stem cells divide and move to suprabasal locations, where they differentiate via a spinous cell intermediate to mature keratinocytes that eventually die and are shed from the skin surface [36]. Interestingly, newborn Dse-knockout mice have thickened basal and spinous layers of the epidermis with elevated levels of immature keratinocytes [13], supporting a positive role of DS-epi1 in epidermis differentiation.
Dse is dynamically expressed in the deep (sensorial) layer of the neural ectoderm. The site of expression changes from a lateral position in the open neural plate at stage 15 to a medial (then ventral) location in the invaginating neural groove at stage 18, where they stay until stage 32. Primary neurons develop in the sensorial layer and become post-mitotic in a wave, with Rohon-Béard sensory neurons differentiating first in lateral (then dorsal) positions followed by interneurons in intermediate and motoneurons in ventral positions of the closing neural tube [37, 38]. Hence, Dse expression appears to follow a spatio-temporal pattern in accordance with primary neuron differentiation. Dsel transcripts reach high levels between stages 27 and 43 in the inner mantle zone of the developing midbrain and hindbrain at sites where ventral interneurons differentiate. In tailbud embryos, Dsel and, to a lower extent, Dse are co-expressed in the trigeminal (V) ganglia. Dsel transcripts are also abundant in epibranchial ganglia, including the geniculate (VII), petrosol (IX) and nodose (X) ganglia, while Dse mRNA is transcribed in spinal nerves. Together, the gene expression of the two DS epimerases suggests that IdoA structures in CS/DS chains are produced at sites of neuronal cell differentiation in the developing nervous system.
Early in development, Dse is expressed in the epidermis and notochord from stage 13, in the neural plate, anterior neural ridge and prechordal mesendoderm from stage 15/16, and in the ventral brain primordia and somites from stage 18 onwards. These expression domains, together with Dsel gene activity in the ventral portion (alar plate) of the developing brain and spinal cord at tailbud stages are consistent with a possible contribution of CS/DS to neural tube closure. Knockout mice that lack either DS-epi1 alone (Dse-KO) or a combination of DS-epi1 and DS-epi2 (double-knockout, DKO) exhibit a low frequency (~5%) of exencephaly and spina bifida [13, 15]. Such neural tube abnormalities are common human birth defects and result from a failure to close the neural tube in the cranial region (exencephaly) or more caudally (spina bifida) due to misregulation of proliferation, differentiation and morphogenetic events in the neural plate as well as disturbed interaction with adjacent non-neural tissues [39].
In advanced tadpole embryos, Dsel expression was detected in ventral territories of the midbrain and anterior hindbrain, where dopaminergic and serotonergic neurons differentiate, respectively. Dopaminergic and serotonergic neurons are induced by morphogen signals that include FGF8 from the isthmic organizer and, in the case of serotonergic neurons, FGF4 from the notochord [40]. IdoA-containing CS/DS chains are known to bind and regulate FGF signals [41–43]. The expression of Dse and Dsel in the source (notochord) and/or receiving cells (mid- and hindbrain) of FGF signals might indicate a possible involvement of CS/DS in morphogen signaling during dopaminergic and serotonergic neuron differentiation. Our study supports previous studies in the postnatal mouse [44] and adult human [10] that Dsel is expressed at relatively high levels in the central nervous system. Two non-synonymous mutations in the coding region of DSEL (Y730C, I1113M) were reported in a heterozygous state in 3 individuals from a group of 113 bipolar disorder patients, but not in the control group [10]. Single nucleotide polymorphisms were also found upstream of Dsel in regions with possible regulatory function in early-onset major depressive disorder [11]. Since misregulation of dopamine and serotonine account for bipolar disorder and depression, a possible involvement of CS/DS in the differentiation or function of neuronal populations that express these monoamine neurotransmitters might help to explain the genetic link between human DSEL and these mood disorders.
While Dse is expressed in neural crest (NC) cells already at stage 15 and maintains expression therein at least until stage 40, Dsel is only transiently expressed in this migratory stem cell population between stages 27 and 34. Knockdown of Dse by injection of antisense oligonucleotides altered the expression of NC-specific genes and decreased the extent of NC cell migration, leading to reduction of craniofacial skeleton, lack of dorsal fin structures and reduced melanophores [16]. Indeed, transplantation experiments revealed a tissue-autonomous role of DS-epi1 in cranial NC cell migration and its indispensable role for cell adhesion, spreading and formation of polarized cell structures on fibronectin. These experiments suggested that the craniofacial abnormalities observed in MS-EDS patients might result from failure of cranial NC cell migration [17].
In the developing somites, Dse expression starts at stage 18 in the sclerotome and appears at stage 32 also in the dermatome, whereas Dsel expression at stage 32 is found in the sclerotome and myotome. The differential expression of DS epimerases in these paired blocks of paraxial mesoderm might correlate with previously reported phenotypic alterations in animal models and human conditions. First, the notochord induces the ventral portion of the somites to form the segmented sclerotome [45], which later gives rise to the axial skeleton (vertebrae, ribs). We suggest that reduced IdoA content in CS/DS chains in the notochord and sclerotome not only relates to the kinked tail phenotype in Dse-morphant Xenopus tadpoles [16] as well as Dse-KO and DKO mouse embryos and pups [12–15], but also explains the spinal and chest wall deformities in MS-EDS patients [46]. Second, decreased levels of DS-epi1 in the dermatome might contribute to lowered tensile strength of the skin and the abdominal wall closure defect in Dse-KO mice [12, 13] and tissue fragility involving the skin, joints and multiple organs in MS-EDS [46].
Thus, by mapping the spatio-temporal gene expression of two genes that are essential for IdoA biosynthesis on CS/DS proteoglycans, we show that clinical manifestations of MS-ESD and psychiatric disorders are correlated with sites where Dse and Dsel are expressed.
Conclusions
The expression of Dse and Dsel in the early Xenopus embryo coincides with sites of cell differentiation in the epidermis and neural tissue. Their expression in the ectoderm and underlying notochord and somites might account for the failed neural tube closure (exencephaly and spina bifida), which was reported in knockout mice. Robust expression of Dsel in the developing midbrain and anterior hindbrain, where dopaminergic and serotonergic neurons differentiate, might help to explain the genetic link of DSEL to bipolar disorder and depression. Several expression domains of Dse can be associated with congenital defects in the musculocontractural Ehlers-Danlos syndrome, such as the sclerotome to spinal/chest wall deformities and the dermatome to skin fragility.
Supporting information
The alignment was performed using ClustalW (EMBL-EBI) and BoxShade (ExPASy). The catalytic residues His205, Tyr261 and His450 in DS-epi1 are indicated with stars and are also conserved in DS-epi2. The total amino acid number and the percentage of amino acid identity to DS-epi1.S are indicated at the end of each sequence. Accession numbers of the Xenopus laevis protein sequences are: DS-epi1.S, KU877109; DS-epi1.L, XM_018263281; DS-epi2.S, XM_018223616; DS-epi2.L, KU877110.
(TIF)
Acknowledgments
We thank the Japanese National Bio-Resource Project (Xenopus), and Dr. T. Pieler for plasmids.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was financially supported by the Swedish Research Council (https://www.vr.se) to E.M.P. (2009-4951), Swedish Childhood Cancer Foundation (https://www.barncancerfonden.se) to E.M.P. (PROJ11/101) and to N.G. (NBCNSPDHEL12/012), Swedish Cancer Foundation (https://www.cancerfonden.se) to M.M. (140530), Director Albert Påhlsson’s Foundation (http://www.pahlssonsstiftelse.se) to E.M.P., and O.E. and Edla Johansson’s Scientific Foundation (http://www.oejvetstift.se) to E.M.P.. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Trowbridge JM, Gallo RL. Dermatan sulfate: new functions from an old glycosaminoglycan. Glycobiology. 2002;12(9): 117R–125R. [DOI] [PubMed] [Google Scholar]
- 2.Thelin MA, Bartolini B, Axelsson J, Gustafsson R, Tykesson E, Pera E, et al. Biological functions of iduronic acid in chondroitin/dermatan sulfate. FEBS J. 2013;280(10): 2431–2446. doi: 10.1111/febs.12214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamada S, Sugahara K, Ozbek S. Evolution of glycosaminoglycans: Comparative biochemical study. Commun Integr Biol. 2011;4(2): 150–158. doi: 10.4161/cib.4.2.14547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iozzo RV, Schaefer L. Proteoglycan form and function: A comprehensive nomenclature of proteoglycans. Matrix Biol. 2015;42: 11–55. doi: 10.1016/j.matbio.2015.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramachandra R, Namburi RB, Dupont ST, Ortega-Martinez O, van Kuppevelt TH, Lindahl U, et al. A potential role for chondroitin sulfate/dermatan sulfate in arm regeneration in Amphiura filiformis. Glycobiology. 2017;27(5): 438–449. doi: 10.1093/glycob/cwx010 [DOI] [PubMed] [Google Scholar]
- 6.Maccarana M, Olander B, Malmström J, Tiedemann K, Aebersold R, Lindahl U, et al. Biosynthesis of dermatan sulfate: chondroitin-glucuronate C5-epimerase is identical to SART2. J Biol Chem. 2006;281(17): 11560–11568. doi: 10.1074/jbc.M513373200 [DOI] [PubMed] [Google Scholar]
- 7.Pacheco B, Malmström A, Maccarana M. Two dermatan sulfate epimerases form iduronic acid domains in dermatan sulfate. J Biol Chem. 2009b;284(15): 9788–9795. doi: 10.1074/jbc.M809339200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Müller T, Mizumoto S, Suresh I, Komatsu Y, Vodopiutz J, Dundar M, et al. Loss of dermatan sulfate epimerase (DSE) function results in musculocontractural Ehlers–Danlos syndrome. Hum Mol Genet. 2013;22(18): 3761–3772. doi: 10.1093/hmg/ddt227 [DOI] [PubMed] [Google Scholar]
- 9.Syx D, Damme T, Symoens S, Maiburg MC, Laar I, Morton J, et al. Genetic Heterogeneity and Clinical Variability in Musculocontractural Ehlers–Danlos Syndrome Caused by Impaired Dermatan Sulfate Biosynthesis. Human Mut. 2015;36(5): 535–547. [DOI] [PubMed] [Google Scholar]
- 10.Goossens D, Van Gestel S, Claes S, De Rijk P, Souery D, Massat I, et al. A novel CpG-associated brain-expressed candidate gene for chromosome 18q-linked bipolar disorder. Mol Psychiatry. 2003;8(1): 83–89. doi: 10.1038/sj.mp.4001190 [DOI] [PubMed] [Google Scholar]
- 11.Shi J, Potash JB, Knowles JA, Weissman MM, Coryell W, Scheftner WA, et al. Genome-wide association study of recurrent early-onset major depressive disorder. Mol Psychiatry. 2011;16(2): 193–201. doi: 10.1038/mp.2009.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maccarana M, Kalamajski S, Kongsgaard M, Magnusson SP, Oldberg Å, Malmström A. Dermatan sulfate epimerase 1-deficient mice have reduced content and changed distribution of iduronic acids in dermatan sulfate and an altered collagen structure in skin. Mol Cell Biol. 2009;29(20): 5517–5528. doi: 10.1128/MCB.00430-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gustafsson R, Stachtea X, Maccarana M, Grottling E, Eklund E, Malmström A, et al. Dermatan sulfate epimerase 1 deficient mice as a model for human abdominal wall defects. Birth Defects Res A Clin Mol Teratol. 2014;100(9): 712–720. doi: 10.1002/bdra.23300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartolini B, Thelin MA, Rauch U, Feinstein R, Oldberg A, Malmström A, et al. Mouse development is not obviously affected by the absence of dermatan sulfate epimerase 2 in spite of a modified brain dermatan sulfate composition. Glycobiology. 2012;22(7): 1007–1016. doi: 10.1093/glycob/cws065 [DOI] [PubMed] [Google Scholar]
- 15.Stachtea XN, Tykesson E, van Kuppevelt TH, Feinstein R, Malmström A, Reijmers RM et al. Dermatan Sulfate-Free Mice Display Embryological Defects and Are Neonatal Lethal Despite Normal Lymphoid and Non-Lymphoid Organogenesis. PloS One. 2015;10(10): e0140279 doi: 10.1371/journal.pone.0140279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gouignard N, Maccarana M, Strate I, von Stedingk K, Malmström A, Pera EM. Musculocontractural Ehlers-Danlos syndrome and neurocristopathies: dermatan sulfate is required for Xenopus neural crest cells to migrate and adhere to fibronectin. Dis Model Mech. 2016;9(6): 607–620. doi: 10.1242/dmm.024661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pera EM, Gouignard N, Maccarana M. Aberrant neural crest development causes craniofacial and other malformations in an animal model of Musculocontractural Ehlers-Danlos syndrome. J Rare Dis Res & Treatment. 2016;1(3): 79–82. [Google Scholar]
- 18.Habicher J, Haitina T, Eriksson I, Holmborn K, Dierker T, Ahlberg PE, et al. Chondroitin / dermatan sulfate modification enzymes in zebrafish development. PLoS One. 2015;10(3): e0121957 doi: 10.1371/journal.pone.0121957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osada S-I, Ohmori S-y, Taira M. XMAN1, an inner nuclear membrane protein, antagonizes BMP signaling by interacting with Smad1 in Xenopus embryos. Development. 2003;130(9): 1783–1794. [DOI] [PubMed] [Google Scholar]
- 20.Hemmati-Brivanlou A, de la Torre JR, Holt C, Harland RM. Cephalic expression and molecular characterization of Xenopus En-2. Development. 1991;111(3): 715–724. [DOI] [PubMed] [Google Scholar]
- 21.Bourguignon C, Li J, Papalopulu N. XBF-1, a winged helix transcription factor with dual activity, has a role in positioning neurogenesis in Xenopus competent ectoderm. Development. 1998;125(24): 4889–4900. [DOI] [PubMed] [Google Scholar]
- 22.Espeseth A, Johnson E, Kintner C. Xenopus F-cadherin, a novel member of the cadherin family of cell adhesion molecules, is expressed at boundaries in the neural tube. Mol Cell Neurosci. 1995:6(3): 199–211. doi: 10.1006/mcne.1995.1017 [DOI] [PubMed] [Google Scholar]
- 23.Mayor R, Morgan R, Sargent MG. Induction of the prospective neural crest of Xenopus. Development. 1995;121(3): 767–777. [DOI] [PubMed] [Google Scholar]
- 24.Hopwood ND, Pluck A, Gurdon JB. A Xenopus mRNA related to Drosophila twist is expressed in response to induction in the mesoderm and the neural crest. Cell. 1998;59(5): 893–903. [DOI] [PubMed] [Google Scholar]
- 25.Pera EM, Acosta H, Gouignard N, Climent M. Whole-mount in situ hybridization and immunohistochemistry in Xenopus embryos In: Hauptmann G, editor. In Situ Hybridization Methods. Vol. 99 New York: Springer Science+Business Media; 2015. pp. 151–167. [Google Scholar]
- 26.Session AM, Uno Y, Kwon T, Chapman JA, Toyoda A, Takahashi S, et al. Genome evolution in the allotetraploid frog Xenopus laevis. Nature. 2016;538(7625): 336–343. doi: 10.1038/nature19840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pacheco B, Maccarana M, Goodlett DR, Malmström A, Malmström L. Identification of the active site of DS-epimerase 1 and requirement of N-glycosylation for enzyme function. J Biol Chem. 2009a;284(3): 1741–1747. doi: 10.1074/jbc.M805479200 [DOI] [PubMed] [Google Scholar]
- 28.Sánchez RS, Sánchez SS. Characterization of pax1, pax9, and uncx sclerotomal genes during Xenopus laevis embryogenesis. Dev Dyn. 2013;242(5): 572–579. doi: 10.1002/dvdy.23945 [DOI] [PubMed] [Google Scholar]
- 29.Drysdale TA, Elinson RP. Development of the Xenopus laevis hatching gland and its relationship to surface ectoderm patterning. Development. 1991;111(2): 469–478. [DOI] [PubMed] [Google Scholar]
- 30.Park BY, Saint-Jeannet JP. Induction and Segregation of the Vertebrate Cranial Placodes Developmental Biology. San Rafael (CA): Morgan & Claypool Life Sciences; 2010. [PubMed] [Google Scholar]
- 31.Schlosser G. Vertebrate cranial placodes as evolutionary innovations—the ancestor's tale. Curr Top Dev Biol. 2015;111: 235–300. doi: 10.1016/bs.ctdb.2014.11.008 [DOI] [PubMed] [Google Scholar]
- 32.Mayor R, Theveneau E. The neural crest. Development. 2013;140(11): 2247–2251. doi: 10.1242/dev.091751 [DOI] [PubMed] [Google Scholar]
- 33.Nagasawa T, Kawaguchi M, Yano T, Sano K, Okabe M, Yasumasu S. Evolutionary Changes in the Developmental Origin of Hatching Gland Cells in Basal Ray-Finned Fishes. Zoolog Sci. 2016;33(3): 272–281. doi: 10.2108/zs150183 [DOI] [PubMed] [Google Scholar]
- 34.Chalmers AD, Lachani K, Shin Y, Sherwood V, Cho KW, Papalopulu N. Grainyhead-like 3, a transcription factor identified in a microarray screen, promotes the specification of the superficial layer of the embryonic epidermis. Mech Dev. 2006;123(9): 702–718. doi: 10.1016/j.mod.2006.04.006 [DOI] [PubMed] [Google Scholar]
- 35.Gilbert SF, Barresi MJF. Developmental Biology. 11th ed. Sinaur Associates, Inc. Sunderland, Massachusetts U.S.A.; 2016. p. 528. [Google Scholar]
- 36.Hsu YC, Li L, Fuchs E. Emerging interactions between skin stem cells and their niches. Nat Med. 2014;20(8): 847–856. doi: 10.1038/nm.3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hartenstein V. Early neurogenesis in Xenopus: the spatio-temporal pattern of proliferation and cell lineages in the embryonic spinal cord. Neuron. 1989;3(4): 399–411. [DOI] [PubMed] [Google Scholar]
- 38.Hartenstein V. Early pattern of neuronal differentiation in the Xenopus embryonic brainstem and spinal cord. J Comp Neurol. 1993;328(2): 213–231. doi: 10.1002/cne.903280205 [DOI] [PubMed] [Google Scholar]
- 39.Wilde JJ, Petersen JR, Niswander L. Genetic, epigenetic, and environmental contributions to neural tube closure. Annu Rev Genet. 2014;48: 583–611. doi: 10.1146/annurev-genet-120213-092208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ye W, Shimamura K, Rubenstein JL, Hynes MA, Rosenthal A. FGF and Shh signals control dopaminergic and serotonergic cell fate in the anterior neural plate. Cell. 1998;93(5): 755–766. [DOI] [PubMed] [Google Scholar]
- 41.Penc SF, Pomahac B, Winkler T, Dorschner RA, Eriksson E, Herndon M, et al. Dermatan sulfate released after injury is a potent promoter of fibroblast growth factor-2 function. J Biol Chem. 1998;273(43): 28116–28121. [DOI] [PubMed] [Google Scholar]
- 42.Trowbridge JM, Rudisill JA, Ron D, Gallo RL. Dermatan sulfate binds and potentiates activity of keratinocyte growth factor (FGF-7). J Biol Chem. 2002;277(45): 42815–42820. doi: 10.1074/jbc.M204959200 [DOI] [PubMed] [Google Scholar]
- 43.Sun C, Marcello M, Li Y, Mason D, Lévy R, Fernig DG. Selectivity in glycosaminoglycan binding dictates the distribution and diffusion of fibroblast growth factors in the pericellular matrix. Open Biol. 2016;6(3): pii: 150277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akatsu C, Mizumoto S, Kaneiwa T, Maccarana M, Malmström A, Yamada S, et al. Dermatan sulfate epimerase 2 is the predominant isozyme in the formation of the chondroitin sulfate/dermatan sulfate hybrid structure in postnatal developing mouse brain. Glycobiology. 2011;21(5): 565–574. doi: 10.1093/glycob/cwq208 [DOI] [PubMed] [Google Scholar]
- 45.Scaal M. Early development of the vertebral column. Semin Cell Dev Biol. 2016;49: 83–91. doi: 10.1016/j.semcdb.2015.11.003 [DOI] [PubMed] [Google Scholar]
- 46.Kosho T. CHST14/D4ST1 deficiency: A new form of Ehlers‐Danlos syndrome. Pediatr Int. 2016;58(2): 88–99. doi: 10.1111/ped.12878 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The alignment was performed using ClustalW (EMBL-EBI) and BoxShade (ExPASy). The catalytic residues His205, Tyr261 and His450 in DS-epi1 are indicated with stars and are also conserved in DS-epi2. The total amino acid number and the percentage of amino acid identity to DS-epi1.S are indicated at the end of each sequence. Accession numbers of the Xenopus laevis protein sequences are: DS-epi1.S, KU877109; DS-epi1.L, XM_018263281; DS-epi2.S, XM_018223616; DS-epi2.L, KU877110.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.