Abstract
BACKGROUND
Human immunodeficiency virus–infected individuals (HIVIIs) have a higher incidence of head and neck squamous cell carcinoma (HNSCC), and clinical and histopathological differences have been observed in their tumors in comparison with those of HNSCC patients without a human immunodeficiency virus (HIV) infection. The reasons for these differences are not clear, and molecular differences between HIV-related HNSCC and non–HIV-related HNSCC may exist. This study compared the mutational patterns of HIV-related HNSCC and non–HIV-related HNSCC.
METHODS
The DNA of 20 samples of HIV-related HNSCCs and 32 samples of non–HIV-related HNSCCs was sequenced. DNA libraries covering exons of 18 genes frequently mutated in HNSCC (AJUBA, CASP8, CCND1, CDKN2A, EGFR, FAT1, FBXW7, HLA-A, HRAS, KEAP1, NFE2L2, NOTCH1, NOTCH2, NSD1, PIK3CA, TGFBR2, TP53, and TP63) were prepared and sequenced on an Ion Personal Genome Machine sequencer. DNA sequencing data were analyzed with Ion Reporter software. The human papillomavirus (HPV) status of the tumor samples was assessed with in situ hybridization, the MassARRAY HPV multiplex polymerase chain reaction assay, and p16 immunostaining. Mutation calls were compared among the studied groups.
RESULTS
HIV-related HNSCC revealed a distinct pattern of mutations in comparison with non–HIV-related HNSCC. TP53 mutation frequencies were significantly lower in HIV-related HNSCC. Mutations in HIV+ patients tended to be TpC>T nucleotide changes for all mutated genes but especially for TP53.
CONCLUSIONS
HNSCC in HIVIIs presents a distinct pattern of genetic mutations, particularly in the TP53 gene. HIV-related HNSCC may have a distinct biology, and an effect of the HIV virus on the pathogenesis of these tumors should not be ruled out.
Keywords: head and neck cancer, human immunodeficiency virus (HIV), human papillomavirus (HPV), mutation, TP53 gene
INTRODUCTION
Human immunodeficiency virus–infected individuals (HIVIIs) have a higher cancer incidence than the general population. Kaposi sarcoma and non-Hodgkin lymphoma are the most common neoplasms among HIVIIs and are AIDS-defining malignancies. However, since the advent of antiretroviral therapy, the incidence of other malignant tumors (non–AIDS-defining cancer) has increased, and they have become important causes of death for these patients.1–4
Three factors are considered important for the elevated cancer incidence among HIVIIs: high smoking rates, immunosuppression, and increased susceptibility to infection by oncoviruses. Smoking is the main preventable cause of cancer but is a common habit among HIVIIs. The prevalence of tobacco smoke exposure is 2 times higher in this group versus the overall population.5,6 Immunosuppression is the main characteristic of human immunodeficiency virus (HIV) infection, and it is considered to be an important cause of cancer development. Organ transplant patients on immunosuppressive regimens demonstrate an increased incidence of malignant tumors in comparison with the general population. Their cancer incidence is similar to the frequency observed among HIVIIs, and this suggests that immunodeficiency plays an important role in HIVII-related carcinogenesis.1 An important common effect of tobacco abuse and immunosuppression is an increased susceptibility to infection by oncoviruses with the subsequent development of virus-related carcinogenesis.7–10 Virus-related tumors are almost 10 times more frequent in HIVIIs versus the general US population.11 Human herpesviruses and human papillomaviruses (HPVs) are responsible for the majority of these cases, and they lead to the development of not only sarcomas and lymphomas but also squamous cell carcinomas in the anogenital and head and neck regions.11 Although human herpesviruses are related to AIDS-defining malignancies, HPVs are the major oncoviruses linked to non–AIDS-defining cancers.7
All these elements are also recognized as important risk factors for head and neck squamous cell carcinoma (HNSCC) development and may explain the higher incidence of these tumors among HIVIIs.1,2,4,12–14 HNSCC is diagnosed at an earlier age and at a more advanced stage in HIVIIs,15 and these tumors tend to be more aggressive16 and related to worse survival rates when they express high levels of TP53 in comparison with non-HIVIIs.17 The efficacy of current treatment approaches to HIVII-related HNSCC is still a matter of debate.18 Radiotherapy treatment has been found to be less effective for the control of tumor relapse and related to worse overall survival and more treatment-related toxicity in HIVIIs.18
Whether these differences in presentation and prognosis are related to the systemic effects of HIV-mediated immunosuppression or by particular biological characteristics of the primary tumor is still unclear.
Histopathological findings indicate that HIV-related HNSCC has unique features such as the enrichment of multinucleated giant tumor cells and the expression of HIV-related protein in some tumor cells.19 These observations indicate unique pathological processes that are associated with these tumors. To better understand the behavior of HIV-related HNSCC and possibly develop personalized treatment approaches, it is important to determine whether this represents a distinct molecular entity.
Recently, integrated genomic studies in HNSCC have dramatically expanded our knowledge about the pathogenesis, progression, and treatment of this tumor type. For instance, these studies have revealed the genes and pathways most frequently affected in HNSCC and have identified HPV-related HNSCC as a distinct molecular entity.20–23 In this way, we believe that a genomic analysis of HIV-related HNSCC might reveal whether this group of tumors has a distinct pattern of DNA alterations that could indicate differences in their pathogenesis and progression. To accomplish this, we compared the pattern of mutations between HIV-related HNSCC and non–HIV-related HNSCC by sequencing a panel of genes frequently mutated in head and neck cancer.
MATERIALS AND METHODS
Patient Selection
The cohort of patients used in this study was obtained from the Head and Neck Cancer Specialized Program of Research Excellence Human Immunodeficiency Virus Supplement Consortium. IRB approval or exemption to share deidentified data with the study data center was obtained from sample collection sites. A detailed description of the patient recruitment, sample collection, and clinicopathologic data collection has been provided in a previous publication.17 Briefly, formalin-fixed, paraffin-embedded (FFPE) tissue from patients diagnosed with HNSCC and an HIV infection were retrieved after a retrospective review of medical records. HNSCC patients not infected by HIV were also retrieved as an age-, subsite-, and stage-matched control group. DNA was extracted from the FFPE tissues, and only samples with sufficient DNA yields for sequencing were included in this study.
HPV Testing
For the HPV status classification, we considered p16 expression with immunohistochemistry and HPV detection with in situ hybridization (ISH) and the MassARRAY multiplex polymerase chain reaction (PCR) assay. A detailed description of these methods can be found in Walline et al’s work.17 Briefly, p16 expression (CINtec; mtm Laboratories) was evaluated semiquantitatively on the basis of the expression intensity ([1] no staining, [2] low intensity, [3] moderate intensity, or [4] high intensity) and the proportion of positive cells ([1] <5%, [2] 5%–20%, [3] 21%–50%, or [4] 51%–100%). The p16 expression score was determined as the product of the intensity and the proportion of p16+ cells. Scores of 1 to 4 were considered negative/low, scores of 5 to 11 were considered moderate, and scores of 12 to 16 were considered high.17
HPV detection based on ISH was performed with the Inform HPV III assay (Ventana Medical Systems). This assay allows the detection of 12 oncogenic HPV types (HPV-16, HPV-18, HPV-31, HPV-33, HPV-35, HPV-39, HPV-45, HPV-51, HPV-52, HPV-56, HPV-58, and HPV-66) but does not specify the HPV type.17
DNA extracted from tumors was used for PCR-based HPV detection with the MassARRAY HPV multiplex PCR assay, which was designed to detect 15 oncogenic HPV types (HPV-16, HPV-18, HPV-31, HPV-33, HPV-35, HPV-39, HPV-45, HPV-51, HPV-52, HPV-56, HPV-58, HPV-59, HPV-66, HPV-68, and HPV-73). Samples that had p16 expression scores higher than 5 and were positive according to ISH− or PCR-based HPV detection methods were considered HPV+. Tumors for which HPV was detected in the absence of p16 overexpression were considered HPV−.
Library Preparation and DNA Sequencing
Ten nanograms of DNA was used as the input for the target DNA library preparation with the Ion AmpliSeq Library Kit. For the amplification of the targeted DNA, we used a customized pool of primers designed for the amplification of all exon regions of the following genes: AJUBA, CASP8, CCND1, CDKN2A, EGFR, FAT1, FBXW7, HLA-A, HRAS, KEAP1, NFE2L2, NOTCH1, NOTCH2, NSD1, PIK3CA, TGFBR2, TP53, and TP63. The target DNA libraries were sequenced with the Ion Personal Genome Machine sequencer platform. Variant calls were made on the Ion Reporter server with the AmpliSeq tumor-normal pair comprehensive cancer panel pipeline with customized filters. Although HLA-A is included in the sequencing assay, it is currently excluded from the analysis because of the difficulty in accurately calling mutations in this highly polymorphic gene.
TP53 Mutation Classification
TP53 gene mutations were classified according to 2 functional-impact and risk-classification systems proposed by Poeta et al24 and Neskey et al.25 With Poeta et al’s system, the TP53 mutations were classified as disruptive and nondisruptive mutations. Disruptive mutations were those inducing a disruption of p53 protein production (nonsense, frameshift, in-frame, and splice site mutations) or any missense mutation occurring within L2 or L3 DNA binding domains (codons 163–195 and 236–251) and changing the original amino acid polarity or charge category. For Neskey et al’s classification, we used the online EAp53 server,26 which classifies TP53 mutations into high- and low-risk categories for HNSCC and determines a numeric risk score (Evolutionary Action score - EAscore). Only missense mutations are classifiable in this system.
Statistical Analysis
For the statistical analysis, we used IBM SPSS Statistics (version 22) and GraphPad Prism (version 6.07) for Windows. Associations between categorical variables were determined with Fisher’s exact test. Associations between categorical and quantitative variables were determined with the Mann-Whitney test when categories had 2 values and with the Kruskal-Wallis test when categories had 3 or more values. Significant associations were considered when the P value was lower than .05. The log-rank test was used to determine differences among survival curves.
RESULTS
To understand the mutation frequencies and patterns in HIV+ HNSCC, we sequenced tumor samples from 20 HIV+ HNSCC patients. We also sequenced 32 HIV−patients as controls (Table 1). Among HIV+ cases, 11 (55%) were HPV−, and 9 (45%) were HPV+; among HIV− cases, 6 (18.75%) were HPV+, and 26 (81.25%) were HPV−. Sex and tissue sites did not differ among these groups, but HIV+HPV+ patients were significantly younger than HIV−HPV+ patients. All HIV+ patients were smokers or former smokers, and all except one were alcohol users. No information about tobacco and alcohol consumption was available for HIV− cases.
TABLE 1.
Clinical Characteristics of the Studied Head and Neck Cancer Patients
| Characteristic | HIV+HPV+ (n = 9) | HIV+HPV− (n = 11) | HIV−HPV+ (n = 6) | HIV−HPV− (n = 26) | Pa | Pb | Pc |
|---|---|---|---|---|---|---|---|
| Sex, No. | |||||||
| Male | 8 | 9 | 6 | 25 | .309 | 1.000 | .205 |
| Female | 1 | 2 | 0 | 1 | |||
| Age, mean ± SD, y | 43.8 ± 10.5 | 52.3 ± 7.07 | 54.6 ± 7.03 | 54.3 ± 11.3 | .061 | .036 | .384 |
| Tumor site, No. | |||||||
| Oral cavity | 2 | 2 | 2 | 14 | .188 | .580 | .228 |
| Oropharynx | 5 | 3 | 3 | 5 | |||
| Larynx | 0 | 3 | 1 | 4 | |||
| Other head and neck sites | 2 | 3 | 0 | 3 | |||
Abbreviations: HIV, human immunodeficiency virus; HPV, human papillomavirus; SD, standard deviation.
Comparison among all 4 groups.
Comparison between HIV+ and HIV− patients with HPV+ tumors.
Comparison between HIV+ and HIV− patients with HPV− tumors.
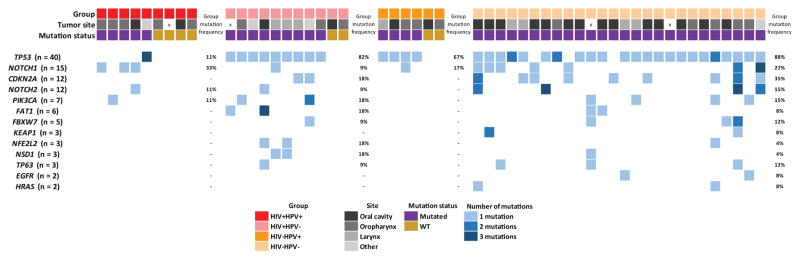
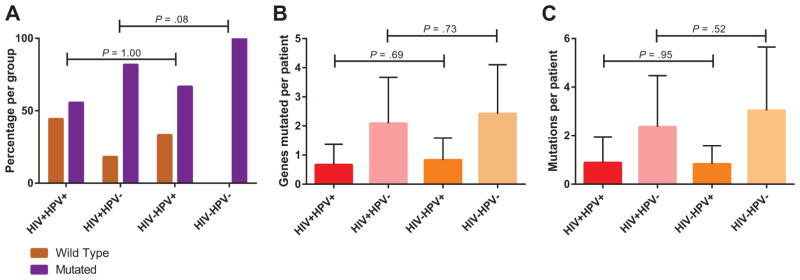
DNA was extracted from FFPE samples and sequenced on a custom Ion Torrent AmpliSeq panel containing 18 genes frequently altered in HNSCC. The identified mutations as well as the mutation frequencies described for HNSCC in The Cancer Genome Atlas (TCGA)22 are listed in Table 2, and summary oncoprints are shown in Figure 1. Because HPV is known to alter the mutational landscape of HNSCC, the cohort was divided into 4 groups for most analyses based on the HIV and HPV status. The associations were assessed through comparisons of all 4 groups together and also through comparisons of HIV+ and HIV− groups with the same HPV status. Overall, 84.6% of the patient tumors were found to have at least 1 mutation in the examined genes, and the number of patients with tumors harboring mutations was significantly different among the 4 groups (P = .002). Among the HPV+ cases, there was no difference in the number of tumors harboring mutations when HIV+ (55.6%) and HIV− patients (66.7%) were compared. However, among the HPV− cases, the number of mutations was higher in the HIV− group (100%) versus the HIV+ group (81.8%; Fig. 2A). The number of patients harboring mutations was significantly lower overall in the virus-related groups (HIV+ or/and HPV+) versus the HPV− cases from TCGA (P < .001) as well as the HIV−HPV− group (P = .004). The number of genes mutated per patient and the number of mutations per patient varied among the groups (Fig. 2B,C). Both variables showed higher values among HPV− patients, regardless of the HIV status.
TABLE 2.
Mutation Findings According to the HIV and HPV Status
| TCGA (279 Patients)a
|
HIV +HPV+ (9 Patients)
|
HIV +HPV− (11 Patients)
|
HIV−HPV+ (6 Patients)
|
HIV−HPV− (26 Patients)
|
Pb | Pc | Pd | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HPV+ Frequency (n = 36), % |
HPV− Frequency (n = 243), % |
No. of Cases |
Mutation Frequency, % |
No. of Mutations |
No. of Cases |
Mutation Frequency, % |
No. of Mutations |
No. of Cases |
Mutation Frequency, % |
No. of Mutations |
No. of Cases |
Mutation Frequency, % |
No. of Mutations |
|||||
| Mutation status | WT | 44 | 3 | 4 | 44.4 | — | 2 | 18.2 | — | 2 | 33.3 | — | 0 | 0 | — | .002 | 1.000 | .083 |
| MUT | 56 | 97 | 5 | 55.6 | 8 | 9 | 81.8 | 26 | 4 | 66.7 | 5 | 26 | 100 | 79 | ||||
| AJUBA | WT | 9 | — | 11 | — | 6 | — | 26 | — | — | — | — | ||||||
| MUT | 0 | 7 | 0 | 0 | — | 0 | 0 | — | 0 | 0 | — | 0 | 0 | — | ||||
| CASP8 | WT | 9 | — | 11 | — | 6 | — | 26 | — | — | — | — | ||||||
| MUT | 6 | 10 | 0 | 0 | — | 0 | 0 | — | 0 | 0 | — | 0 | 0 | — | ||||
| CCND1 | WT | 9 | — | 11 | — | 6 | — | 26 | — | — | — | — | ||||||
| MUT | 0 | 0.4 | 0 | 0 | — | 0 | 0 | — | 0 | 0 | — | 0 | 0 | — | ||||
| CDKN2A | WT | 9 | — | 9 | — | 6 | — | 17 | — | .090 | — | .445 | ||||||
| MUT | 0 | 26 | 0 | 0 | — | 2 | 18.2 | 2 | 0 | 0 | — | 9 | 34.6 | 11 | ||||
| EGFR | WT | 9 | — | 11 | — | 6 | — | 24 | — | 1.000 | — | 1.000 | ||||||
| MUT | 0 | 5 | 0 | 0 | — | 0 | 0 | — | 0 | 0 | — | 2 | 7.7 | 2 | ||||
| FAT1 | WT | 9 | — | 9 | — | 6 | — | 24 | — | .610 | — | .567 | ||||||
| MUT | 0 | 26 | 0 | 0 | — | 2 | 18.2 | 4 | 0 | 0 | — | 2 | 7.7 | 2 | ||||
| FBXW7 | WT | 9 | — | 10 | — | 6 | — | 23 | — | .881 | — | 1.000 | ||||||
| MUT | 2.8 | 5 | 0 | 0 | — | 1 | 9.1 | 1 | 0 | 0 | — | 3 | 11.5 | 4 | ||||
| HRAS | WT | 9 | — | 11 | — | 6 | — | 24 | — | 1.000 | — | 1.000 | ||||||
| MUT | 0 | 5 | 0 | 0 | — | 0 | 0 | — | 0 | 0 | — | 2 | 7.7 | 2 | ||||
| KEAP1 | WT | 9 | — | 11 | — | 6 | — | 24 | — | 1.000 | — | 1.000 | ||||||
| MUT | 0 | 5 | 0 | 0 | — | 0 | 0 | — | 0 | 0 | — | 2 | 7.7 | 3 | ||||
| NFE2L2 | WT | 9 | — | 9 | — | 6 | — | 25 | — | .306 | — | .205 | ||||||
| MUT | 0 | 7 | 0 | 0 | — | 2 | 18.2 | 2 | 0 | 0 | — | 1 | 3.8 | 1 | ||||
| NOTCH1 | WT | 6 | — | 10 | — | 5 | — | 19 | — | .583 | .604 | .391 | ||||||
| MUT | 8 | 20 | 3 | 33.3 | 3 | 1 | 9.1 | 1 | 1 | 16.7 | 1 | 7 | 26.9 | 10 | ||||
| NOTCH2 | WT | 8 | — | 10 | — | 6 | — | 22 | — | .924 | 1.000 | 1.000 | ||||||
| MUT | 0 | 5 | 1 | 11.1 | 1 | 1 | 9.1 | 1 | 0 | 0 | — | 4 | 15.4 | 10 | ||||
| NSD1 | WT | 9 | — | 9 | — | 6 | — | 25 | — | .306 | — | .205 | ||||||
| MUT | 8 | 11 | 0 | 0 | — | 2 | 18.2 | 2 | 0 | 0 | — | 1 | 3.8 | 1 | ||||
| PIK3CA | WT | 8 | — | 9 | — | 6 | — | 22 | — | .876 | 1.000 | 1.000 | ||||||
| MUT | 36 | 19 | 1 | 11.1 | 1 | 2 | 18.2 | 3 | 0 | 0 | — | 4 | 15.4 | 4 | ||||
| TGFBR2 | WT | 9 | — | 11 | — | 6 | — | 26 | — | — | — | — | ||||||
| MUT | 2.8 | 4 | 0 | 0 | — | 0 | 0 | — | 0 | 0 | — | 0 | 0 | — | ||||
| TP53 | WT | 8 | — | 2 | — | 2 | — | 3 | — | <.001 | .089 | .623 | ||||||
| MUT | 2.8 | 84 | 1 | 11.1 | 3 | 9 | 81.8 | 9 | 4 | 66.7 | 4 | 23 | 88.5 | 26 | ||||
| TP63 | WT | 9 | — | 10 | — | 6 | — | 23 | — | .881 | — | 1.000 | ||||||
| MUT | 0 | 2.5 | 0 | 0 | — | 1 | 9.1 | 1 | 0 | 0 | — | 3 | 11.5 | 3 | ||||
Abbreviations: HIV, human immunodeficiency virus; HPV, human papillomavirus; MUT, mutated; TCGA, The Cancer Genome Atlas; WT, wild type.
TCGA head and neck squamous cell carcinoma mutation frequencies per patient were taken from Lawrence et al.22
The P value was calculated on the basis of the HIV+HPV+, HIV+HPV−, HIV−HPV+, and HIV−HPV− groups.
The P value was calculated on the basis of the HIV+HPV+ and HIV−HPV+ groups.
The P value was calculated on the basis of the HIV+HPV− and HIV−HPV− groups.
Figure 1.
Oncoprint showing the frequencies of the mutations detected in each studied group. HIV indicates human immunodeficiency virus; HPV, human papillomavirus; WT, wild type.
Figure 2.
Overall mutation pattern according to each studied group. (A) Frequencies of mutated and wild-type cases for the studied genes (y-axis) according to each studied group. Groups with the same HPV status were compared, and a few HIV+ cases with mutations were found with HPV− tumors. (B) Number of genes mutated per patient in each studied group. No differences in the number of genes mutated per patient were observed between groups with the same HPV status. (C) Number of overall mutations per patient in each group. No differences in the number of mutations per patient were observed between groups with the same HPV status. HIV indicates human immunodeficiency virus; HPV, human papillomavirus.
No mutations were detected in 4 of the studied genes that are known to have low mutation frequencies in HNSCC (AJUBA, CASP8, CCND1, and TGFBR2). Mutations in the TP53 and NOTCH1 genes were detected in all groups. When we compared the frequencies of gene mutations among the groups, only TP53 mutations were significantly different (P < .001; Table 2). HIV−HPV− patients had the highest frequency of mutations in the TP53 gene (88.5%), whereas HIV+HPV+ patients had the least number of mutations in this gene (11.1%). A strikingly high frequency of TP53 mutations was observed in HPV+ patients (33.3% of all 15 HPV+ cases) in comparison with other HNSCC HPV+ sequencing studies. We do not currently have an explanation for this high rate. Mutations in the NFE2L2 gene were found in 2 of 20 HIV+ patients (10%); this was a higher frequency of detection in comparison with the frequency for non-HIV patients (3.1%) and that reported for the TCGA cohort (6.5%). NOTCH1 mutations were observed in 3 of 9 HIV+HPV+ patients (33.3%), and this was a higher mutation frequency in comparison with the frequency of our other groups and that reported in the TCGA data set. However, statistical significance was not reached for the NFE2L2 and NOTCH1 associations. NSD1 mutations were differentially distributed among tumor sites (P = .025): they were observed in the larynx (2 mutated cases) and other head and neck sites (1 mutated case) but were absent in oral cavity and oropharynx cases. This is consistent with the distribution of NSD1 mutations in TCGA. The frequency of mutations in any gene did not differ significantly between HIV+ and HIV− tumors with the same HPV status.
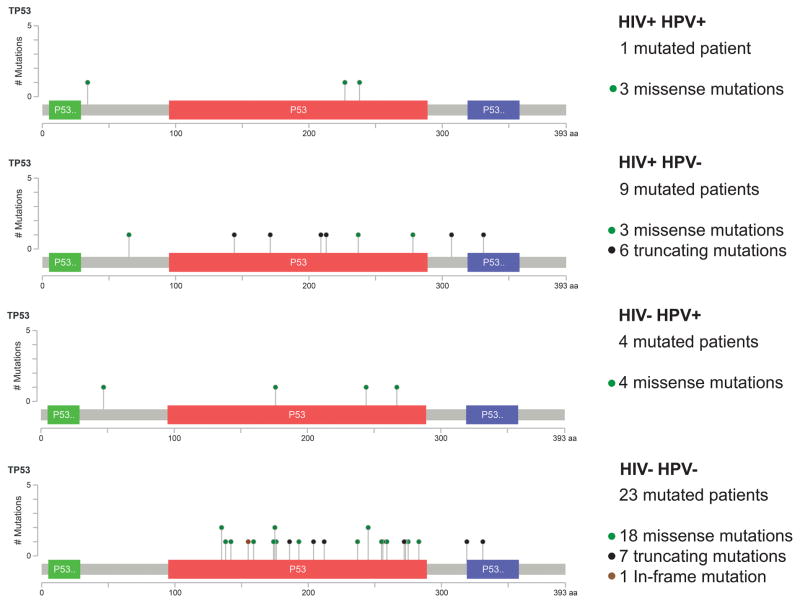
To assess qualitative differences in TP53 mutations, we compared the types of TP53 mutations among the groups (Fig. 3). Although HPV+ cases exhibited only missense TP53 mutations, HPV− tumors also contained truncating and in-frame indel mutations. Among HIV+HPV−cases, truncating mutations were the most common (66.7%), whereas these mutations represented just 26.9% of the HIV−HPV− events. Next, we used 2 TP53 functional-impact and risk-classification systems previously used for HNSCC (Table 3). With both systems, TP53 mutations related to aggressive tumors (high-risk and disruptive) were more prevalent in HIV− and HPV− cases. The mutations in virus-related cases were predominantly classified as wild-type or low-risk/nondisruptive mutations. Truncating and frameshift mutations are not classifiable in Neskey et al’s system25 and are, therefore, called other. These were more prevalent in the HIV+ group (45% of mutated cases) versus the HIV− group (30.8% of wild-type cases).
Figure 3.
Mutation maps of the TP53 gene in each studied group. Green dots represent missense mutations, red dots represent truncating mutations, and black dots represent in-frame indels; truncating and in-frame indels were observed only among HPV−cases. The frequency of truncating mutations was significantly higher among HIV+ cases versus HIV− patients. HIV indicates human immunodeficiency virus; HPV, human papillomavirus.
TABLE 3.
Association Between the TP53 Mutation Score and the Clinical Characteristics of Head and Neck Cancer Patients
| Characteristic |
TP53 Mutation Score: Neskey et al25
|
P | Pb |
TP53 Mutation Score: Poeta et al24
|
P | Pb | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | Low Risk | High Risk | Othera | WT | Nondisruptive | Disruptive | |||||
| Sex, No. | |||||||||||
| Male | 14 | 9 | 14 | 11 | 1.000 | — | 14 | 24 | 10 | 1.000 | — |
| Female | 1 | 0 | 1 | 2 | — | 1 | 2 | 1 | — | ||
| Age, mean ± SD, y | 47.2 ± 12.2 | 54.5 ± 8.36 | 53.8 ± 9.09 | 53.6 ± 11.1 | .419 | — | 47.2 ± 12.2 | 53.6 ± 10.2 | 54.7 ± 7.53 | .354 | — |
| Tumor site, No. | |||||||||||
| Oral cavity | 7 | 1 | 6 | 6 | .119 | — | 7 | 9 | 4 | .303 | — |
| Oropharynx | 7 | 3 | 5 | 1 | — | 7 | 7 | 2 | — | ||
| Larynx | 0 | 4 | 2 | 2 | — | 0 | 5 | 3 | — | ||
| Other head and neck sites | 1 | 1 | 2 | 4 | — | 1 | 5 | 2 | — | ||
| HIV status, No. | |||||||||||
| + | 10 | 3 | 2 | 5 | .011 | — | 10 | 6 | 4 | .020 | — |
| − | 5 | 6 | 13 | 8 | — | 5 | 20 | 7 | — | ||
| HPV status, No. | |||||||||||
| + | 10 | 2 | 3 | 0 | .001 | — | 10 | 3 | 2 | <.001 | — |
| − | 5 | 7 | 12 | 13 | — | 5 | 23 | 9 | — | ||
| Virus infection status, No. | |||||||||||
| Present | 12 | 5 | 4 | 5 | .022 | — | 12 | 8 | 6 | .010 | — |
| Absent | 3 | 4 | 11 | 8 | — | 3 | 18 | 5 | — | ||
| HIV/HPV status, No. | |||||||||||
| +/+ | 8 | 0 | 1 | 0 | .001 | .071 | 8 | 1 | 0 | .001 | .071 |
| −/+ | 2 | 2 | 2 | 0 | 2 | 2 | 2 | ||||
| +/− | 2 | 3 | 1 | 5 | .210 | 2 | 5 | 4 | .448 | ||
| −/− | 3 | 4 | 11 | 8 | 3 | 18 | 5 | ||||
Abbreviations: HIV, human immunodeficiency virus; HPV, human papillomavirus; SD, standard deviation; WT, wild type.
Other refers to mutations not classifiable by Neskey et al’s system.25
The P value was calculated through the comparison of groups with the same HPV status.
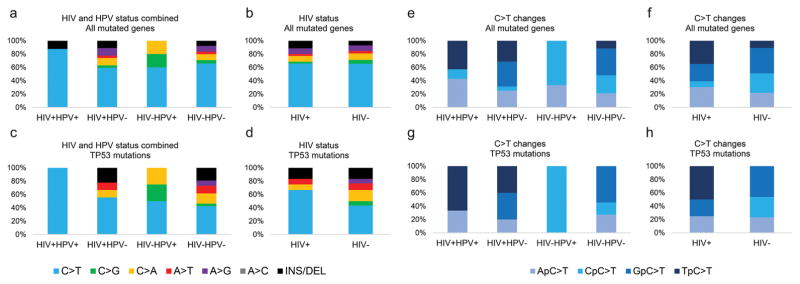
We also assessed the types of nucleotide changes in each studied group for all genes as well as the TP53 gene individually (Fig. 4). C>T mutations were the most prevalent nucleotide changes observed in all groups and for the majority of the genes. Only the genes NFE2L2, NSD1, and PIK3CA showed more C>G, C>A, and T>C nucleotide changes, respectively. No significant difference in the distribution of nucleotide changes was observed among the 4 groups (P = .93; Fig. 4A) or between HIV+ and HIV− groups with the same HPV status (P = .24 for HPV+ tumors and P = .98 for HPV−tumors). No difference in the pattern of nucleotide change distribution was observed when patients were grouped according to their HIV status (P = .95; Fig. 4B).
Figure 4.
Distribution of nucleotide changes among the studied groups. (A) Frequency of nucleotide changes in all mutated genes among the 4 studied groups. (B) Frequency of nucleotide changes in all mutated genes among tumors from HIV+ and HIV− patients, regardless of the HPV status. (C) Frequency of nucleotide changes in the TP53 gene among the 4 studied groups. (D) Frequency of nucleotide changes in the TP53 gene among tumors from HIV+ and HIV− patients, regardless of the HPV status. (E) Frequency of each 5′ nucleotide flanking C>T mutations in all mutated genes for the 4 studied groups. (F) Frequency of each 5′ nucleotide flanking C>T mutations in all mutated genes among tumors from HIV+ and HIV− patients, regardless of the HPV status. (G) Frequency of each 5′ nucleotide flanking C>T mutations in the TP53 gene for the 4 studied groups. (H) Frequency of each 5′ nucleotide flanking C>T mutations in the TP53 gene among tumors from HIV+ and HIV− patients, regardless of the HPV status. HIV indicates human immunodeficiency virus; HPV, human papillomavirus.
As for nucleotide changes in the TP53 gene, C>T changes were the most commonly observed, but no statistical difference was observed among the 4 groups (P = .80; Fig. 4C) or between HIV+ and HIV− samples with the same HPV status (P = .34 for HPV+ tumors and P = .92 for HPV− tumors). Although the frequency of C>T changes was higher among tumors with an HIV infection (66.7%) versus HIV− tumors (43.3%), the difference between them was not significant (P = .95; Fig. 4D).
Considering the high frequency of C>T mutations in these samples, we investigated the 5′ flanking nucleotide for each mutation (Fig. 4E). HIV+HPV− samples exhibited a higher frequency of TpC>T mutations (26.3%) than tumors of HIV−HPV− patients (10.2%) when all mutated genes were taken into account. As for HPV+ tumors, TpC>T changes were observed only among HIV+ patients (42.9% of all mutations) and were absent among HIV− ones (P = .02). When tumors were grouped according their HIV status, regardless of the HPV infection status, the frequency of TpC>T was significantly higher among HIV+ patients (34.8%) versus HIV− patients (10.9%; P = .02; Fig. 4F). The same trend was observed when only the TP53 gene was considered: only HIV+ tumors exhibited TpC>T nucleotide changes (P = .02; Fig. 4H).
Differences in 5-year survival rates were determined for patients with respect to their HIV/HPV group, TP53 status (wild-type vs mutated), and TP53 mutation type (insertion/deletion, transition, or transversion). No significant differences between survival curves were observed in any comparison (P = .45, P = .30, and P = .46, respectively). The presence of mutations in other genes (alone or in combination) was also not associated with survival in these patients. Because of the absence of TNM information for HIV− patients, the influence of the tumor clinical stage on these results was not able to be determined.
DISCUSSION
In this study, we observed an overall decreased frequency of mutations in HIV+ HNSCCs, and HIV-related HNSCCs had a distinct pattern of genetic mutations in the TP53 gene characterized by truncating mutations and TpC>T nucleotide changes.
HIVIIs are a unique group of patients in the context of HNSCC. They have greater exposure to tobacco and alcohol, have impaired immune function, and consequently are more prone to infection by high-risk HPV.4–7,14 In this way, these patients have multiple key risk factors for the development of cancer and particularly HNSCC. Not surprisingly, HIVIIs have higher rates of HNSCC than the general population. It has been reported that HIVIIs with HNSCC present at a more advanced tumor stage and have reduced survival rates.15,16,18,27 However, it is not clear whether these findings are caused by intrinsic features of the tumor cells or are related to host factors related to the HIV infection.22,23,28 We aimed to investigate this by comparing the patterns of mutations in HIV-related and non–HIV-related tumors in genes frequently mutated in HNSCC. Because an HPV infection causes a distinct subtype of HNSCC,22,23,28 we considered the HPV status as a key variable as well.
Our observations regarding genetic alterations in HNSCC from HIVIIs are unique in the literature. The only similar analysis that we could find was performed by Souza et al,29 who compared the patterns of TP53 mutations in DNA obtained from cervical swabs (normal and altered cytologies) from HIV-infected and uninfected patients (all HPV+). They found that the frequencies of TP53 mutations were similar in the 2 groups (approximately 19%).
Souza et al29 also found that the genomic position of TP53 mutations changed with the HIV status. HIV+ patients had more mutations located in exon 7, whereas HIV− patients had more mutations in exon 6. Our small sample size made it difficult to compare the genomic locations of TP53 mutations, but we did examine the types of mutations. HIV+HPV− patients presented with a higher frequency of truncating mutations, and the missense mutations were more frequently classified as low-risk and nondisruptive. Many TP53 mutations have been studied for their gain-of-function properties, which can provide novel characteristics to the tumors. Although missense mutations in the p53 DNA binding domain can promote a loss of DNA binding activity, this event may also change the protein structure and lead to new potential protein interactions, which in turn may result in p53 gain of function.30 The pattern of TP53 mutations in HIV+ cases suggests that there are fewer gain-of-function TP53 mutations, and it is intriguing to speculate that this could be related to the immune status of the individuals or HIV itself.
Another interesting qualitative observation is the different patterns of nucleotide changes in all mutated genes but especially in the TP53 gene among HIV+ and HIV− tumors. The presence of C>A changes in the TP53 gene is considered a hallmark of tobacco-related DNA mutations in lung cancer and also in some HNSCCs.21,22,31,32 In fact, only 1 HPV+ case in our cohort exhibited C>A transversions. Among the HIV+HPV− cases, the percentage of C>A changes was similar to that among the HIV−HPV− cases (12.5% vs 9.6%), and this might be expected because all the HIV+ patients were smokers. C>T transitions have been demonstrated as being more predominant in virally transformed tumors, including HNSCC tumors.22,33 However, we demonstrated that a specific subtype of C>T changes, the TpC>T mutations, were enriched in the HIV-infected patients, and they conferred a virus-related mutation fingerprint for these cases. Interestingly, the increased number of TpC>T changes among HIV+ tumors was independent of the presence of an HPV co-infection.
Cytosine deamination is a defense mechanism against retrovirus infections exerted by the APOBEC family of enzymes. Human APOBEC3G is induced by an HIV infection to impair virus infectivity by promoting mutations in its DNA.34–36 However, the mutagenic potential of APOBEC3G is not restricted to the viral genome, and cytosine deamination may also occur in human DNA. The APOBEC-related pattern of mutations, characterized by TpC>T transitions, can be observed in numerous human cancers, including HNSCC, and they are commonly described in the TP53 gene.37–41 Interestingly, oncogenic pathogens such as HPV and Helicobacter pylori have been proved to cause APOBEC3G-mediated mutations in oral and gastric epithelium.37,41 The enrichment of TpC>T changes in HIV-related tumors might suggest that HIV infection contributes to the TP53 mutation pattern observed in this cohort.
Some studies have demonstrated that HIV-encoded proteins may interact with the p53 protein and modulate its function in different ways.42–44 One of these HIV-encoded products, the Nef protein, is believed to interact with p53 and inhibit its function and thus promote HIV infection and replication. Greenway et al45 observed that Nef inhibits p53-dependent apoptosis, and McLemore et al19 found the expression of this protein in the cytoplasm of 7 HNSCC samples. These results might indicate that an HIV infection would have a direct role in HNSCC pathogenesis independent of an HPV infection.46 HIV DNA, RNA, and viral particles have been detected in the oral mucosa of HIVIIs,47 and it has been demonstrated that epithelial cells from the oral mucosa lining are susceptible to HIV infection.48 These findings might suggest a direct oncogenic effect of the virus on oral epithelial cells. More extensive sequencing would be necessary to confirm this.
Despite the genetic differences observed among these patients, no significant difference in survival was observed in our analysis with respect to the HIV and HPV status, TP53 mutation status, and TP53 mutation type. We believe that our small sample size might limit definitive conclusions on the impact of such variables on the survival of HNSCC patients. The impact of such variables should be further investigated in larger cohorts.
Considering these results, we conclude that HIV-related HNSCC likely represents a distinct genomic entity. The alterations observed in this study must be validated in larger cohorts, but they suggest that these tumors in HIVIIs are biologically distinct. More studies are needed to understand the unique etiology, pathogenesis, and biology of these tumors and to determine whether there are unique therapeutic modalities that would benefit these HNSCC patients.
Acknowledgments
FUNDING SUPPORT
This study was funded by the Head and Neck Cancer Specialized Program of Research Excellence Human Immunodeficiency Virus Supplement Consortium (the National Cancer Institute) and the American Recovery and Reinvestment Act through the following grants: P50 CA097248 to the University of Michigan; University of Michigan Cancer Center Core Grant P30 CA46592; 5P50 CA097007 to The University of Texas MD Anderson Cancer Center; P50 CA097190 to the University of Pittsburgh; P50 DE019032 and 3P50 DE019032-14S2 to Johns Hopkins University; and P50 CA128613 to Emory University. Sarah I. Pai is supported by grant 1R01 DE025340.
Footnotes
AUTHOR CONTRIBUTIONS
Frederico O. Gleber-Netto: Acquisition, analysis, or interpretation of the data; drafting of the article; and statistical analysis. Mei Zhao: Acquisition, analysis, or interpretation of the data. Sanchit Trivedi: Acquisition, analysis, or interpretation of the data. Jiping Wang: Acquisition, analysis, or interpretation of the data. Samar Jasser: Acquisition, analysis, or interpretation of the data. Christina McDowell: Acquisition, analysis, or interpretation of the data. Humam Kadara: Acquisition, analysis, or interpretation of the data. Jiexin Zhang: Acquisition, analysis, or interpretation of the data. Jing Wang: Acquisition, analysis, or interpretation of the data. William N. William, Jr: Acquisition, analysis, or interpretation of the data. J. Jack Lee: Acquisition, analysis, or interpretation of the data. Minh Ly Nguyen: Critical revision of the article for important intellectual content. Sara I. Pai: Acquisition, analysis, or interpretation of the data and critical revision of the article for important intellectual content. Heather M. Walline: Acquisition, analysis, or interpretation of the data. Dong M. Shin: Acquisition, analysis, or interpretation of the data and critical revision of the article for important intellectual content. Robert L. Ferris: Critical revision of the article for important intellectual content. Thomas E. Carey: Conception or design; acquisition, analysis, or interpretation of the data; and critical revision of the article for important intellectual content. Jeffrey N. Myers: Conception or design; acquisition, analysis, or interpretation of the data; and critical revision of the article for important intellectual content. Curtis R. Pickering: Conception or design; acquisition, analysis, or interpretation of the data; and drafting of the article.
CONFLICT OF INTEREST DISCLOSURES
Robert L. Ferris reports working on an advisory board and a clinical trial for Astra-Zeneca/MedImmune and receiving research funding from the company, working on an advisory board for Lilly, working on an advisory board and a clinical trial for Bristol-Myers Squibb and receiving research funding from the company, working on an advisory board and a clinical trial for Merck, working on an advisory board for Pfizer, and receiving research funding from VentiRx Pharmaceuticals outside the submitted work.
References
- 1.Grulich AE, Van Leeuwen MT, Falster MO, Vajdic CM. Incidence of cancers in people with HIV/AIDS compared with immunosuppressed transplant recipients: a meta-analysis. Lancet. 2007;370:59–67. doi: 10.1016/S0140-6736(07)61050-2. [DOI] [PubMed] [Google Scholar]
- 2.Patel P, Hanson DL, Sullivan PS, Novak RM, Moorman AC. Incidence of types of cancer among HIV-infected persons compared with the general population in the United States, 1992–2003. Ann Intern Med. 2008;148:728–736. doi: 10.7326/0003-4819-148-10-200805200-00005. [DOI] [PubMed] [Google Scholar]
- 3.Lin CS, Lin C, Weng SF, Lin SW, Lin YS. Cancer survival in patients with HIV/AIDS in the era of highly active antiretroviral therapy in Taiwan: a population-based cohort study. Cancer Epidemiol. 2013;37:719–724. doi: 10.1016/j.canep.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Brickman C, Palefsky JM. Cancer in the HIV-infected host: epidemiology and pathogenesis in the antiretroviral era. Curr HIV/AIDS Rep. 2015;12:388–396. doi: 10.1007/s11904-015-0283-7. [DOI] [PubMed] [Google Scholar]
- 5.Gritz ER, Vidrine DJ, Lazev AB, Amick BC, Arduino RC. Smoking behavior in a low-income multiethnic HIV/AIDS population. Nicotine Tob Res. 2004;6:71–77. doi: 10.1080/14622200310001656885. [DOI] [PubMed] [Google Scholar]
- 6.Mdodo R, Frazier EL, Dube SR, et al. Cigarette smoking prevalence among adults with HIV compared with the general adult population in the United States: cross-sectional surveys. Ann Intern Med. 2015;162:335–344. doi: 10.7326/M14-0954. [DOI] [PubMed] [Google Scholar]
- 7.Strickler HD, Burk RD, Fazzari M, et al. Natural history and possible reactivation of human papillomavirus in human immunodeficiency virus–positive women. J Natl Cancer Inst. 2005;97:577–586. doi: 10.1093/jnci/dji073. [DOI] [PubMed] [Google Scholar]
- 8.Xi LF, Koutsky LA, Castle PE, et al. Relationship between cigarette smoking and human papilloma virus types 16 and 18 DNA load. Cancer Epidemiol Biomarkers Prev. 2009;18:3490–3496. doi: 10.1158/1055-9965.EPI-09-0763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schabath MB, Villa LL, Lazcano-Ponce E, Salmeron J, Quiterio M, Giuliano AR. Smoking and human papillomavirus (HPV) infection in the HPV in Men (HIM) study. Cancer Epidemiol Biomarkers Prev. 2012;21:102–110. doi: 10.1158/1055-9965.EPI-11-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mesri EA, Feitelson MA, Munger K. Human viral oncogenesis: a cancer hallmarks analysis. Cell Host Microbe. 2014;15:266–282. doi: 10.1016/j.chom.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Martel C, Shiels MS, Franceschi S, et al. Cancers attributable to infections among adults with HIV in the United States. AIDS. 2015;29:2173–2181. doi: 10.1097/QAD.0000000000000808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engsig FN, Gerstoft J, Kronborg G, et al. Head and neck cancer in HIV patients and their parents: a Danish cohort study. Clin Epidemiol. 2011;3:217–227. doi: 10.2147/CLEP.S19875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D’Souza G, Carey TE, William WN, et al. Epidemiology of head and neck squamous cell cancer among HIV-infected patients. J Acquir Immune Defic Syndr. 2014;65:603–610. doi: 10.1097/QAI.0000000000000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helleberg M, Gerstoft J, Afzal S, et al. Risk of cancer among HIV-infected individuals compared to the background population: impact of smoking and HIV. AIDS. 2014;28:1499–1508. doi: 10.1097/QAD.0000000000000283. [DOI] [PubMed] [Google Scholar]
- 15.Singh B, Balwally AN, Shaha AR, Rosenfeld RM, Har-El G, Lucente FE. Upper aerodigestive tract squamous cell carcinoma. The human immunodeficiency virus connection. Arch Otolaryngol Head Neck Surg. 1996;122:639–643. doi: 10.1001/archotol.1996.01890180047012. [DOI] [PubMed] [Google Scholar]
- 16.Powles T, Powles J, Nelson M, et al. Head and neck cancer in patients with human immunodeficiency virus-1 infection: incidence, outcome and association with Epstein-Barr virus. J Laryngol Otol. 2004;118:207–212. doi: 10.1258/002221504322927982. [DOI] [PubMed] [Google Scholar]
- 17.Walline HM, Carey TE, Goudsmit CM, et al. High-risk HPV, bio-markers, and outcome in matched cohorts of head and neck cancer patients positive and negative for HIV. Mol Cancer Res. 2016;15:179–188. doi: 10.1158/1541-7786.MCR-16-0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mourad WF, Hu KS, Shasha D, et al. Long-term outcome of seropositive HIV patients with head and neck squamous cell carcinoma treated with radiation therapy and chemotherapy. Anticancer Res. 2013;33:5511–5516. [PubMed] [Google Scholar]
- 19.McLemore MS, Haigentz M, Smith RV, et al. Head and neck squamous cell carcinomas in HIV-positive patients: a preliminary investigation of viral associations. Head Neck Pathol. 2010;4:97–105. doi: 10.1007/s12105-010-0171-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agrawal N, Frederick MJ, Pickering CR, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawrence MS, Sougnez C, Lichtenstein L, et al. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D’Souza G, Kreimer AR, Viscidi R, et al. Case-control study of human papillomavirus and oropharyngeal cancer. N Engl J Med. 2007;356:1944–1956. doi: 10.1056/NEJMoa065497. [DOI] [PubMed] [Google Scholar]
- 24.Poeta ML, Manola J, Goldwasser MA, et al. TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N Engl J Med. 2007;357:2552–2561. doi: 10.1056/NEJMoa073770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neskey DM, Osman AA, Ow TJ, et al. Evolutionary action score of TP53 identifies high-risk mutations associated with decreased survival and increased distant metastases in head and neck cancer. Cancer Res. 2015;75:1527–1536. doi: 10.1158/0008-5472.CAN-14-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baylor College of Medicine. [Accessed June 1, 2016];EAp53 server. http://mammoth.bcm.tmc.edu/cgi-bin/panos/EAp53.cgi.
- 27.Picard A, Badoual C, Hourseau M, et al. HPV prevalence in HIV patients with head and neck squamous cell carcinoma. AIDS. 2016;30:1257–1266. doi: 10.1097/QAD.0000000000001030. [DOI] [PubMed] [Google Scholar]
- 28.Marur S, D’Souza G, Westra WH, Forastiere AA. HPV-associated head and neck cancer: a virus-related cancer epidemic. Lancet Oncol. 2010;11:781–789. doi: 10.1016/S1470-2045(10)70017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Souza RP, Gimenes F, De Abreu AL, et al. Differences in the mutation of the p53 gene in exons 6 and 7 in cervical samples from HIV− and HPV-infected women. Infect Agent Cancer. 2013;8:38. doi: 10.1186/1750-9378-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou G, Wang J, Zhao M, et al. Gain-of-function mutant p53 promotes cell growth and cancer cell metabolism via inhibition of AMPK activation. Mol Cell. 2014;54:960–974. doi: 10.1016/j.molcel.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfeifer GP, Denissenko MF, Olivier M, Tretyakova N, Hecht SS, Hainaut P. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene. 2002;21:7435–7451. doi: 10.1038/sj.onc.1205803. [DOI] [PubMed] [Google Scholar]
- 32.Pickering CR, Zhang J, Neskey DM, et al. Squamous cell carcinoma of the oral tongue in young non-smokers is genomically similar to tumors in older smokers. Clin Cancer Res. 2014;20:3842–3848. doi: 10.1158/1078-0432.CCR-14-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayes DN, Van Waes C, Seiwert TY. Genetic landscape of human papillomavirus–associated head and neck cancer and comparison to tobacco-related tumors. J Clin Oncol. 2015;33:3227–3234. doi: 10.1200/JCO.2015.62.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 35.Harris RS, Bishop KN, Sheehy AM, et al. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 36.Bishop KN, Holmes RK, Sheehy AM, Malim MH. APOBEC-mediated editing of viral RNA. Science. 2004;305:645. doi: 10.1126/science.1100658. [DOI] [PubMed] [Google Scholar]
- 37.Matsumoto Y, Marusawa H, Kinoshita K, et al. Helicobacter pylori infection triggers aberrant expression of activation-induced cytidine deaminase in gastric epithelium. Nat Med. 2007;13:470–476. doi: 10.1038/nm1566. [DOI] [PubMed] [Google Scholar]
- 38.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burns MB, Lackey L, Carpenter MA, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindley RA. The importance of codon context for understanding the Ig-like somatic hypermutation strand-biased patterns in TP53 mutations in breast cancer. Cancer Genet. 2013;206:222–226. doi: 10.1016/j.cancergen.2013.05.016. [DOI] [PubMed] [Google Scholar]
- 41.Henderson S, Chakravarthy A, Su X, Boshoff C, Fenton TR. APO-BEC-mediated cytosine deamination links PIK3CA helical domain mutations to human papillomavirus–driven tumor development. Cell Rep. 2014;7:1833–1841. doi: 10.1016/j.celrep.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 42.Izumi T, Io K, Matsui M, et al. HIV-1 viral infectivity factor interacts with TP53 to induce G2 cell cycle arrest and positively regulate viral replication. Proc Natl Acad Sci U S A. 2010;107:20798–20803. doi: 10.1073/pnas.1008076107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sato Y, Tsurumi T. Genome guardian p53 and viral infections. Rev Med Virol. 2013;23:213–220. doi: 10.1002/rmv.1738. [DOI] [PubMed] [Google Scholar]
- 44.Verma S, Ali A, Arora S, Banerjea AC. Inhibition of β-TrcP dependent ubiquitination of p53 by HIV-1 Vpu promotes p53 mediated apoptosis in human T cells. Blood. 2011;117:6600–6608. doi: 10.1182/blood-2011-01-333427. [DOI] [PubMed] [Google Scholar]
- 45.Greenway AL, McPhee DA, Allen K, et al. Human immunodeficiency virus type 1 Nef binds to tumor suppressor p53 and protects cells against p53-mediated apoptosis. J Virol. 2002;76:2692–2702. doi: 10.1128/JVI.76.6.2692-2702.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim RH, Yochim JM, Kang MK, Shin KH, Christensen R, Park NH. HIV-1 Tat enhances replicative potential of human oral keratinocytes harboring HPV-16 genome. Int J Oncol. 2008;33:777–782. [PubMed] [Google Scholar]
- 47.Qureshi MN, Barr CE, Hewlitt I, et al. Detection of HIV in oral mucosal cells. Oral Dis. 1997;3(suppl 1):S73–S78. doi: 10.1111/j.1601-0825.1997.tb00380.x. [DOI] [PubMed] [Google Scholar]
- 48.Moore JS, Rahemtulla F, Kent LW, et al. Oral epithelial cells are susceptible to cell-free and cell-associated HIV-1 infection in vitro. Virology. 2003;313:343–353. doi: 10.1016/s0042-6822(03)00283-6. [DOI] [PubMed] [Google Scholar]