Abstract
Pathogens use various mechanisms to manipulate host processes to promote infection. Decades of research on pathogens have revealed not only the molecular mechanisms that these microbes use to replicate and survive within host cells, but also seminal information on how host signaling machinery regulates cellular processes. Among these discoveries are mechanisms involving posttranslational modifications that alter the activity, localization, or interactions of the modified protein. Herein, we examine how pathogens have contributed to our basic understanding of three posttranslational modifications: phosphorylation, NMPylation, and ubiquitylation. Over the years, technologies, techniques and research tools have developed side by side with the study of pathogens, facilitating the discovery of protein modifications and furthering our understanding of how they contribute to both infection and cellular functions.
Keywords: Pathogens, Posttranslational modifications, Phosphorylation, AMPylation, Ubiquitylation
Introduction
Pathogens such as bacteria, viruses, fungi, and nematodes, use various mechanisms to alter cellular processes during their complex interactions with the host. Many of these interactions are mediated by proteins that are produced by the pathogen and act inside host cells to manipulate cellular processes and promote infection. These virulence proteins utilize different mechanisms to regulate host proteins, one of which is posttranslational modifications (PTMs).
As the host uses PTMs to regulate cellular processes, virulence proteins also use PTMs to manipulate and regulate host proteins. Some virulence proteins are enzymes that can directly mediate a PTM of a target, while others mediate the PTM of a host protein in an indirect manner, by recruiting host components. The virulence enzymes that directly mediate PTMs can do so by imitating enzymatic activities that are used by the host, or by using a unique activity. Interestingly, while some virulence enzymes are proposed to have been "taken" from a host and incorporated into the pathogen's virulence mechanisms, others are thought to have risen from convergent evolution to perform similar activities to those of host enzymes. Notably, virulence proteins can also take advantage of the host PTM machinery to be modified themselves, and thus achieve additional regulation of their activities, localization, or stability.
One of the first virulence enzymes that was reported to catalyze a direct PTM of a host protein was Diphtheria toxin produced by the pathogenic bacterium Corynebacterium diphtheriae. In the late 1960's, several groups described the mechanism by which Diphtheria toxin blocks host protein translation, leading to cell death. Two groups independently elucidated that the toxin hydrolyzes nicotinamide adenine dinucleotide (NAD) and then transfers the resulting ADP-ribose to aminoacyl transferase II (Elongation factor-2), resulting in its inactivation and thus a disruption in protein synthesis (Gill et al., 1969; Honjo et al., 1968). Since this discovery, many virulence proteins that mediate PTMs have been described, having a plethora of activities. The target proteins can be modified by small molecules (e.g. phosphorylation, acetylation, AMPylation) or small proteins (e.g. ubiquitin, SUMO, NEDD8) as substrates in a reversible manner (e.g. phosphorylation, AMPylation) or irreversible manner (e.g. deamidases, phosphothreonine-lyases, proteases) (Ribet and Cossart, 2010).
Here we present a historical discussion on how the study of host-pathogens interactions led to discoveries of PTM mechanisms. We focus our discussion on three pivotal mechanisms: phosphorylation, NMPylation, and ubiquitylation, while providing insights into the biochemical mechanisms and the use of developing technologies to discover new PTMs.
Phosphorylation
Phosphorylation, the transfer of a γ-phosphate of adenosine triphosphate (ATP) to a serine, threonine, or tyrosine residue of a protein (and also histidine and lysine), is a PTM used by all living organisms. Phosphorylation is a reversible modification, mediated by enzymes named kinases, that regulates protein activities and plays pivotal roles in pathogen infections (Ubersax and Ferrell, 2007). Kinases use the readily available ATP as a substrate and require the presence of a metal ion such as Mg2+ or Mn2+. Phosphorylation is often used in host immunity-related signal transduction cascades, such as mitogen activated protein kinase (MAPK) pathways, and is thus a lucrative target for manipulation by pathogens.
In 1954, Burnett and Kennedy provided direct evidence for the enzymatic phosphorylation of a protein when they described an enzyme from liver mitochondria that catalyzes the phosphorylation of casein, an artificial substrate, on a serine residue (Burnett and Kennedy, 1954). From this initial discovery until 1979, protein kinases were thought to phosphorylate proteins only on serine and threonine residues. However, the discovery of the Src kinase changed that perception.
The discovery of v-Src
In 1911, Peyton Rous showed that tumors (sarcomas) could be transmittable using liquid produced from other sarcomas (Rous, 1911). It was later shown that the causative agent in the liquid was Rous sarcoma virus (RSV). RSV was one of many viruses that were found to cause tumors in susceptible cells. In 1969, Huebner and Todaro coined the term "oncogenes" to describe viral genes that have the potential to cause cancer, and in the 1970's the RSV oncogene v-Src was found (Huebner and Todaro, 1969; Martin, 2001). The v-Src protein was isolated in 1977 by Brugge and Erikson (Brugge and Erikson, 1977). It was identified as a protein kinase a year later when the antibody heavy chain, as well as v-Src itself, were phosphorylated in immunoprecipitates incubated in kinase buffer with γ-32P-labeled ATP (Collett and Erikson, 1978). This protocol, which became known as the immune complex kinase assay, became widely used to study protein kinases.
Notably, in 1976, a "normal" cellular gene homologous to v-Src was discovered in avian genomes (Stehelin et al., 1976). This gene, named c-Src, was termed "proto-oncogene" to describe it as a cellular precursor of the viral oncogene. c-Src was also reported to have kinase activity, although lower than that of v-Src (Oppermann et al., 1979). Later studies revealed that Src is a member of a large family of non-receptor tyrosine kinases that are found in essentially all metazoan cells (Thomas and Brugge, 1997). c-Src is a modular protein containing SH2 and SH3 domains, which became prototypes of domains that mediate protein-protein interactions (Martin, 2001). The study of the Src modular domains also provided seminal lessons on protein autoinhibition (Boggon and Eck, 2004; Martin, 2001). Importantly, the discoveries of proto-oncogenes led to the realization that mutated versions of these genes can result in cancer.
Initially, Src was believed to phosphorylate substrates on threonine residues. However, work by Tony Hunter and colleagues on v-Src and the polyoma virus middle T antigen (also an oncogene) revealed a new mechanism of protein regulation - tyrosine phosphorylation (Eckhart et al., 1979; Hunter and Sefton, 1980). They discovered that the highly acidic buffer used to run hydrolyzed amino protein samples on thin layer plates to detect phosphorylated residues caused phosphothreonine and phosphotyrosine to co-migrate. Serendipitously, Hunter used an old buffer with a slightly higher pH, which caused phosphothreonine and phosphotyrosine to run separately and allowed him to identify tyrosine as the residue phosphorylated by Src (Hunter, 2008). And thus tyrosine phosphorylation, found later to be catalyzed by many cellular kinases, emerged as a central mechanism in cellular regulation.
Subsequent studies aimed at identifying Src phosphorylation substrates were successful using what was then ground breaking techniques and tools. For example, the multifunction cellular protein p36 (also known as calpactin I or annexin II) was identified in 1979 as a substrate of v-Src using high-resolution two-dimensional gel electrophoresis (Erikson and Erikson, 1980; Radke and Martin, 1979), a technique developed in 1975 that allowed for distinct separation of modified cellular proteins (O'Farrell, 1975). This technique was later improved by Cooper and Hunter to detect Src phosphotyrosine-containing substrates, exploiting the alkali stability of phosphotyrosine to reduce background signals (Cooper and Hunter, 1981). Another important tool used in the hunt for Src substrates, as well as for other kinases, came with the development of antibodies that recognized phosphorylated tyrosine residues (Frackelton et al., 1983).
Regulation of protein activities by kinases and phosphatases
After the discovery of tyrosine phosphorylation by Hunter and colleagues, many tyrosine kinases were identified in eukaryotes, and tyrosine phosphorylation was found to have an important role in various cellular processes, such as growth factor signaling and proliferation, cancer (as discussed above for viral oncogenes and cellular proto-oncogenes), and activation of MAPK pathways (Gschwind et al., 2004; Lemmon and Schlessinger, 2010). The importance of tyrosine phosphorylation for pathogenesis and immune responses was underscored by the discovery of a bacterial tyrosine phosphatase (PTP). In 1990, Guan and Dixon identified a bacterial PTP using database mining (Guan and Dixon, 1990). The gene encoded the Yersinia type III effector YopH, which was known to be required for the virulence of this bacteria (Bolin and Wolf-Watz, 1988). Interestingly, by aligning the catalytic residues of YopH with other known eukaryotic PTPs, the authors raised the possibility that YopH had a eukaryotic origin that was reminiscent of the eukaryotic origins for the viral tyrosine kinase v-Src (Guan and Dixon, 1990). It is worth noting that at the time YopH was discovered, tyrosine phosphorylation was not considered a non-metazoan signaling mechanism; however, the subsequent discovery of PTPs in various microorganisms refuted that paradigm (Heneberg, 2012). YopH is one of the most potent PTPs isolated to date and plays an essential role in virulence by targeting various signaling pathways required for host immunity (Bliska et al., 1991). Several phosphorylated tyrosine kinases and adaptors have been detected as YopH substrates (de la Puerta et al., 2009; Hamid et al., 1999; Persson et al., 1997). Notably, YopH was shown to inhibit the uptake of Yersinia by host cells by disrupting peripheral focal adhesion complexes, which contribute to cellular structure and spreading (Persson et al., 1997), and suppressing T and B lymphocyte activation (Yao et al., 1999).
A year after the report of the PTP YopH, Dixon and colleagues discovered another PTP-like protein in vaccinia virus, named VH1 (Guan et al., 1991). VH1 shares amino acid sequence identity with catalytic residues of PTPs, which are structurally and catalytically distinct from serine/threonine phosphatases (Barford, 1995). Surprisingly, however, VH1 was able to hydrolyze both substrates containing phosphotyrosine and substrates containing phosphoserine, identifying this viral protein as a dual specificity phosphatase (DSP) (Guan et al., 1991). VH1, which is highly conserved among poxviruses and is essential for vaccinia viability in tissue culture (Liu et al., 1995), dephosphorylates the transcription factor STAT1 to down-regulate the cellular antiviral response (Najarro et al., 2001).
Soon after the discovery of the DSP VH1, reports on eukaryotic PTPs began to appear (Guan et al., 1992). VH1 is now considered the prototype of a family of VH1-like DSPs; a sub-class of PTPs. Members of this family are found in viruses, yeast, plants, and higher eukaryotes, where they control many cellular aspects such as MAPK activation, immune responses, and embryogenesis (Camps et al., 2000; Fauman and Saper, 1996; Pulido and Hooft van Huijsduijnen, 2008).
In 1993, a direct role for serine/threonine phosphorylation was found in bacterial pathogenesis. Examination of amino acid sequences identified Yersinia YpkA (YopO), an effector delivered to target cells via a specialized type III secretion system, as a secreted bacterial protein kinase involved in pathogenicity (Galyov et al., 1993). Similar to other bacterial toxins, YpkA is a modular protein. Aside from its kinase domain, it also contains a domain that mimics host guanidine nucleotide dissociation inhibitors (GDIs), which prevent GTP exchange of small GTPases (Prehna et al., 2006). Remarkably, YpkA kinase activity is positively regulated by binding to host monomeric (G) actin (Juris et al., 2000). This form of positive regulation by binding a host co-factor prevents promiscuous activity of the effector before its translocation into the host. The kinase activity of YpkA is used to phosphorylate a serine residue on the conserved diphosphate binding loop of the active GTP-bound form of Gαq, involved in many cell pathways, which decreases its affinity for GTP and thereby inhibits Gαq signaling pathways in host cells (Navarro et al., 2007).
Following the discovery of YpkA, additional virulence determinants that are eukaryotic-like serine/threonine protein kinases have been described as substrates of various bacterial protein secretion systems (Hervet et al., 2011; Walburger et al., 2004). An interesting example is the Shigella type III effector, OspG, which was identified in 2005 as a kinase that exhibits similarities to eukaryotic serine/threonine kinases (Kim et al., 2005). However, OspG and its homologs NleH1/2 found in Escherichia coli are atypical kinases that lack sub-domains VIII–XI out of the eleven sub-domains found in typical serine/threonine kinases (Zhou et al., 2013). Therefore, OspG lacks the activation loop found in sub-domain VIII, which usually stimulates kinase activity upon phosphorylation. Nevertheless, OspG was recently found to use an activation mechanism similar to that of YpkA. Instead of binding monomeric (G) actin, OspG binds host ubiquitin to stimulate its kinase activity (Zhou et al., 2013). While the search for a target of OspG phosphorylation is ongoing, the kinase activity of OspG is required to prevent phospho-IκBα degradation and NFκB activation that are induced upon stimulation with the inflammatory cytokine TNF-α, thus dampening host innate immune responses (Kim et al., 2005; Zhou et al., 2013).
Antagonists of protein phosphorylation
Protein phosphorylation is a major cellular regulatory mechanism that is often used in host immunity-related signal transduction cascades such as the MAPK and NFκB pathways. While the "conventional" way of regulating the phosphorylation status of cellular proteins is through kinases and phosphatases, pathogens have evolved several additional unique mechanisms, both reversible and irreversible, to antagonize host protein phosphorylation.
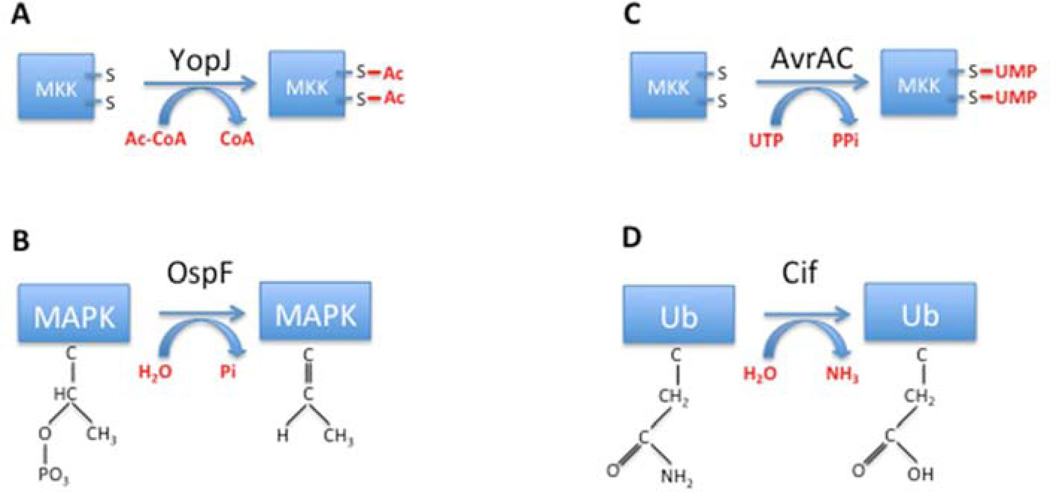
One efficient mechanism to antagonize the activation of a host protein by phosphorylation is competitive modification of the target residues. This mechanism was initially described in 2006 for the Yersinia type III effector YopJ. Based on similarities between its catalytic domain and predicted secondary structure of Clan CE cysteine proteases, YopJ was believed for many years to be a protease (Orth et al., 1999; Orth et al., 2000). It was known to inhibit the host inflammatory response and promote apoptosis of immune cells by preventing the activation of its interacting host proteins: MAPK kinases (MAPKKs) and the related kinase that activates the NFκB pathway, IκB kinase β (IKKβ) (Orth et al., 1999; Orth et al., 2000). However, there was no solid evidence indicating that inhibition of host inflammation was protease-mediated, and cleavage of the target host proteins, MAPKKs or IKKβ, was not observed (Mukherjee et al., 2006). The actual activity of YopJ was finally revealed when Mukherjee et al. developed a cellfree signaling system to recapitulate the inhibition of the MAPK and NFκB pathways in vitro (Mukherjee et al., 2006). They noticed that activation of these signaling pathways diminished in the presence of YopJ, and went on to show that YopJ acts directly on MAPKKs and IKKβ. The breakthrough in understanding YopJ’s inhibitory mechanism was made possible by emerging mass spectrometry techniques that can be used to study protein PTMs (Mann and Jensen, 2003). Mass spectrometry revealed that the total mass of a specific MAPKK, MAPKK6, increased in increments of 42 atomic mass units when co-expressed with YopJ. Subsequent liquid chromatography followed by tandem mass spectrometry identified these modifications as acetylation, or the addition of an acetyl group to a protein. Remarkably, the acetylated residues on MAPKK6 were a conserved serine and threonine in the kinase activation loop, which are the same residues that are phosphorylated by the MAPKK kinase to activate MAPKKs (Figure 1A). An in vitro transferase reaction using acetyl-coenzyme A (CoA) as a donor showed that YopJ directly functions as an acetyltransferase that prevents kinase phosphorylation and activation by adding a competitive acetyl group. These findings were later corroborated by Mittel and colleagues (Mittal et al., 2006; Mukherjee et al., 2006).
Figure 1. Pathogens antagonize host signaling.
(A) Yersinia YopJ acetylates serine/threonine residues on the activation lop of MAPK kinases (MKK) to prevent activation by phosphorylation. (B) Shigella OspF uses β-eliminylation to remove the phosphate from a phosphorylated MAPK, resulting in an irreversibly inactive kinase. (C) Xhanthomonas campestris AvrAc uses UMPylation to modify MAPK kinases on serine/threonine residues within the activation loop of MAPK kinases (MKK) to prevent phosphorylation-mediated activation. (D) The family of Cif-like effectors deamidate ubiquitin, resulting in substrates that cannot be used for ubiquitin elongation.
While lysine acetylation (N-acetylation) was a well-established regulatory mechanism, the studies on YopJ revealed a novel mechanism of serine and threonine acetylation that is used to directly compete with and prevent phosphorylation and activation of a kinase. Surprisingly, although YopJ was predicted to function as a protease based on a conserved catalytic triad and predicted secondary structure, it was found to be a transferase. From a mechanistic standpoint, both proteases and transferases can use a ping-pong mechanism for catalysis; however, the former uses water as a substrate, whereas the latter excludes water from its active site. It is worth noting that members of the YopJ family of acetyltransferases have been identified in many animal and plant pathogens, and that their targets are not limited to protein kinases.
The discovery of YopJ’s acetyltransferase activity was the first of many that helped define a strategy to study and discover novel PTMs: (1) Finding a target protein; (2) Identifying the molecular change using the new and evolving fields of biochemistry and mass spectrometry; and (3) Elucidating the catalytic mechanism used to modify a protein. This strategy has led to many surprising catalytic discoveries for virulence proteins and the identification of several novel PTMs.
Almost as proof of principle, Shao and colleagues independently discovered a unique PTM that antagonizes phosphorylation-mediated activation of kinases using a similar strategy. This PTM, named eliminylation, involves the irreversible removal of phosphate groups and is mediated by the OspF family of type III effectors, including the Shigella OspF, Salmonella SpvC, Chromobacterium VirA, and the Pseudomonas HopAI1 effectors (Li et al., 2007) (Figure 1B). Using a cell-free signaling system similar to that described by Mukherjee et al., Li et al. discovered that OspF targets the MAPK Erk and attenuates its phosphorylation status. These results led Li et al. to hypothesize that OspF harbors a phosphatase activity to reverse Erk phosphorylation. Indeed, OspF could dephosphorylate threonine, but not tyrosine, in the activation loop of MAPKs in vitro. Next, Li et al. subjected phosphorylated Erk2 that had been treated with OspF to tandem mass spectrometry analysis. The results revealed that in addition to the loss of a phosphate group, the threonine residue also underwent a dehydration reaction. Therefore, OspF functions as a phosphothreonine lyase, rather than a phosphatase, that carries out a β-eliminylation reaction to remove the phosphate moiety from the phosphothreonine in the pT-X-pY motif of phosphorylated MAPKs, resulting in an irreversibly inactive kinase (Li et al., 2007) (Figure 1B).
Interestingly, a similar mechanism of catalysis was identified recently in members of the bacterial LanL family of lanthionine synthetases that share sequence homology with OspF family members (Goto et al., 2011). This enzyme family is responsible for biosynthesis of lantibiotics, a class of peptide antibiotics, through a process that includes phosphorylation followed by dehydration of phosphothreonine and phosphoserine (Willey and van der Donk, 2007). While eliminylation can be found in bacteria, an interesting question is whether such activity exists in eukaryotes as a regulatory mechanism to irreversibly "dephosphorylate" proteins. Notably, dehydrated (unsaturated) serine and threonine were found in human proteins, but it is yet unclear whether these are enzyme-mediated PTMs (Brennan and Barford, 2009).
NMPylation
In 1967, Stadtman and colleagues discovered the enzyme glutamine synthetase-adenylyltransferase (GS-ATase) in E. coli. GS-ATase uses ATP and Mg2+ to covalently modify GS, an enzyme involved in nitrogen synthesis, by adding nucleotide adenosine monophosphate (AMP) to a tyrosine residue (Kingdon et al., 1967). This modification, termed adenylylation, regulates the activity and properties of GS. The following year, Stadtman and colleagues reported that GS-ATase can also catalyze the reverse reaction of deadenylylation and remove the AMP from GS (Shapiro and Stadtman, 1968). These two findings revealed a reversible PTM that uses AMP to modify protein characteristics. Remarkably, they went on to discover that the ATase itself undergoes PTM-mediated regulation by another nucleotidyltransferase through cycles of uridylylation and deuridylylation, in which uridine monophosphate (UMP) is reversibly added and removed from target residues. They showed that the uridylylated form catalyzes the deadenylylation of GS (Brown et al., 1971). Hence, nucleotidyltransferases can use various nucleotides as substrates to modify and regulate protein activities. Interestingly, in 1978, a non-regulatory use of uridylylation was discovered for a viral RNA-dependent RNA polymerase (RdRp) (Paul et al., 1998; Rothberg et al., 1978). However, the field of protein activity regulation by nucleoside monophosphate (NMP) modification remained relatively dormant for over 40 years until the discovery of the bacterial effector VopS.
VopS and the Fic domain
The field of NMP modifications re-emerged as an important regulatory PTM following the discovery of the catalytic activity of the Vibrio parahaemolyticus type III effector VopS in 2009. VopS was found to covalently modify host Rho GTPases with an AMP molecule on a threonine residue, leading to the disruption of Rho GTPase binding to downstream signaling machinery and subsequent collapse of the actin cytoskeleton (Yarbrough et al., 2009). This discovery relied on the strategy that was used to identify PTMs mediated by YopJ, and was ultimately confirmed by mass spectrometry analysis of the VopS-modified target proteins. The region within VopS that mediated the catalytic activity was the "filamentation induced by cyclic AMP" (Fic) domain, and the name AMPylation was coined for this PTM activity (Yarbrough et al., 2009).
Fic domains are found in a variety of bacterial species, and the role of AMPylation in signaling has accelerated since the discovery of VopS (Woolery et al., 2010). Dixon and colleagues reported that IbpA, which is a toxin secreted from the bacteria Histophilus somni and contains two Fic domains, can AMPylate Rho GTPases to induce cytotoxicity in mammalian cells (Worby et al., 2009). However, unlike VopS, the IbpA Fic domain AMPylated a tyrosine residue rather than a threonine residue (Worby et al., 2009). Notably, Dixon and colleagues also demonstrated that IbpA-mediated AMPylation could be reversed with a promiscuous phosphodiesterase from snake venom (Worby et al., 2009), further supporting Stadtman's previous observations on the reversibility of protein AMPylation (Shapiro and Stadtman, 1968) and UMPylation (uridylylation) (Brown et al., 1971).
Importantly, Fic domain-containing proteins are also found in eukaryotes, including humans (Kinch et al., 2009; Worby et al., 2009). This finding supports the hypothesis that the addition of a NMP or NMPylation, and specifically Fic-mediated NMPylation, is a universal PTM that is used to regulate protein activity. More recently, Rahman and colleagues demonstrated that the Drosophila Fic protein is required for neurotransmission in fly eyes, and that the deletion of this protein causes blindness. The wild-type form, but not a catalytically inactive mutant, of Drosophila Fic rescued the defects in fly vision (Rahman et al., 2012).
The nucleotidyltransferase DrrA
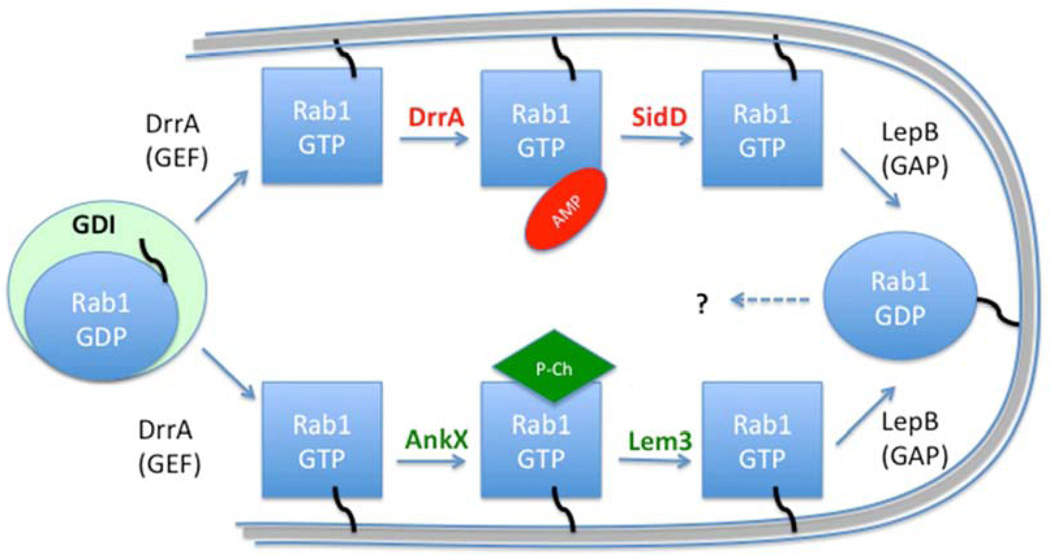
An exquisite example of the use of AMPylation as a reversible regulatory PTM came from the study of the Legionella type IV secretion system effector DrrA (SidM) (Figure 2). In 2010, Muller et al. determined the crystal structure of DrrA, which was known to play a major role in recruiting the small host GTPase Rab1 to the Legionella-containing vacuole (LCV), a specialized bacteria-generated compartment. They observed that in addition to the known guanine nucleotide exchange factor (GEF) domain and the phosphatidylinositol-4-phosphate binding domain, the N-terminal region of DrrA was structurally similar to nucleotidyltransferases and shared the same catalytic motif as the one found in GS-ATase (Muller et al., 2010). They went on to show that like GS-ATase, the DrrA nucleotidyltransferase domain mediated AMPylation of Rab1 on a tyrosine residue (Muller et al., 2010), implicating this domain in virulent activities involving the PTM of a host protein, similar to the Fic domain. However, subsequent studies revealed that AMPylation of Rab1 by DrrA is only part of this complex PTM regulatory mechanism. Legionella uses DrrA to recruit Rab1 to LCV membranes where it acquires endoplasmic reticulum (ER)-derived vesicles that are required to maintain the LCV. DrrA uses its GEF activity to activate Rab1 and maintain it on the LCV membrane. Another effector, LepB, is a GTPase-activating protein (GAP) that can facilitate Rab1 GTP hydrolysis and thus remove Rab1 from the LCV membrane (Ingmundson et al., 2007). However, there is a 4-hour time lapse between DrrA-mediated activation of Rab1 and its subsequent inactivation and release by LepB and host GAPs (Ingmundson et al., 2007). This delay is the result of DrrA-mediated AMPylation of Rab1, which renders Rab1 inaccessible to LepB (Muller et al., 2010). These observations led to the hypothesis that there is a de-AMPylating enzyme that removes the AMP from Rab1 to allow LepB to de-activate it after 4 hours. Indeed, within a year, two groups identified SidD, a de-AMPylating Legionella effector that de-AMPylates Rab1 (Neunuebel et al., 2011; Tan and Luo, 2011). Legionella achieves this complex reversible regulatory mechanism to modulate a host membrane trafficking pathway by using temporal regulation of effector translocation into the host cell (Neunuebel et al., 2011) (Figure 2). These discoveries underlined the regulatory and reversible role of AMPylation as a PTM.
Figure 2. Legionella uses AMPylation and phosphocholination to regulate the Rab1 GTPase. Rab1 is sequestered in the cytoplasm by a guanine dissociation inhibitor (GDI).
Upon infection, Rab1 is recruited to the Legionella-containing vacuole (LCV) membrane followed by activation by the Legionella GEF DrrA. It is then modified by either AMPylation or phosphocholination by the bacterial effectors DrrA or AnkX, respectively. The modified form of Rab1 cannot interact with host GEFs and GAPs. The PTMs on activated GTP-bound Rab1 can be reversed by deAMPylation or dephosphocholination by the effectors SidD or Lem3, respectively. The GTP-bound Rab1 can then be inactivated by the Legionella GAP LepB.
Regulation and substrate specificity for Fic domains
Interestingly, several subsequent studies demonstrated that like nucleotidyltransferases, Fic domains can use various substrates to catalyze different PTMs (Figure 2). The first non-AMPylating Fic domain to be described was the Legionella type IV effector AnkX. In 2011, Roy and colleagues used mass spectrometry to study the AMPylation of Rab1 by DrrA. Surprisingly, they found that Rab1 was also modified with another PTM, and that this modification was dependent on the Legionella type IV secretion system but independent of DrrA (Mukherjee et al., 2011). Mass spectrometry analysis identified the modification as phosphocholination in which a phosphocholine group is added to the serine residue directly preceding the tyrosine that is AMPylated by DrrA. This modification is mediated by the Fic domain of AnkX using CDP-choline as a substrate (Mukherjee et al., 2011). As shown for DrrA, the PTM mediated by the AnkX Fic domain was reversible. Two groups identified the Legionella effector Lem3 (lpg0609) as a de-phosphocholinase that can remove the AnkX-mediated phosphocholine from Rab proteins (Figure 2) (Goody et al., 2012; Tan et al., 2011). Interestingly, AMPylation and phosphocholination of Rab1 seem to be mutually exclusive (Goody et al., 2012).
Fic domains, like kinases, modify various residues (namely serine, threonine, and tyrosine). Moreover, Fic domain-containing proteins are not strictly AMPylators, but rather phosphotransferases with different substrate specificities. Crystal structures of several Fic domains and their substrate-bound states revealed that Fic domains cleave diphosphate-containing substrates and use a resulting phosphoryl-containing moiety to modify an accepting hydroxyl group. Dehio and colleagues suggested that most Fic domains are controlled by a conserved mechanism in which an inhibitory α-helix obstructs the ATP-binding site (Engel et al., 2012). This inhibition can occur in either intramolecular of intermolecular manner, and mutations in the conserved motif found in the inhibitory helix result in considerable increase in AMPylation activity of the enzymes (Engel et al., 2012). Cherfils and colleagues demonstrated that the diphosphate-containing substrates are specifically recognized by variable regions that are found outside of the Fic catalytic motif, at both ends of the active site (Campanacci et al., 2013). Interestingly, based on these observations, Cherfils and colleagues recently proposed that several structural elements in the Fic domain can determine the nature of the physiological substrates targeted by the enzyme. Furthermore, based on this model, they proposed that Fic domains that were previously suggested to have low AMPylation activity levels due to the presence of an inhibitory α-helix may actually have a physiological substrate other than ATP (Campanacci et al., 2013). Recently a number of molecular tools have been developed to study this PTM, including antibodies specific for modified residues, characteristic ions and fragmentation patterns of AMPylated peptides and nucleotide analogues (Grammel et al., 2011; Hao et al., 2011; Lewallen et al., 2012; Li et al., 2011).
Using NMPylation to antagonize host protein signaling
AvrAC, another Fic-domain containing type III effector from the phytopathogenic bacterium Xhanthomonas campestris, was recently shown to modify its substrates with UMP (Feng et al., 2012). AvrAC was shown to inhibit the plant immune response by UMPylating at least two receptor-like cytoplasmic kinases, BIK1 and RIPK, of the flowering plant Arabidopsis (Feng et al., 2012). Remarkably, the UMPylation sites were the conserved serine and threonine residues in the kinase activation loop that must be phosphorylated for kinase activation. Therefore, both AvrAC and YopJ act as antagonists and use similar mechanisms to competitively inhibit protein kinase activation. While YopJ competes with serine and threonine phosphorylation by modifying the residues with an acetyl group, AvrAC accomplishes this inhibition by modifying these residues with UMP. This example of phosphorylation-competitive modification further supports the hypothesis that similar PTM strategies can also be used in eukaryotes as a regulatory mechanism to fine-tune kinase activities (Figure 1C).
Ubiquitylation
The history of signaling mediated by small protein modifications goes back decades, long before our understanding of how proteins were degraded in a cell, and their significance is intertwined with the discovery of protein and membrane metabolism. The initial biochemical mechanism proposed for protein degradation was perplexing because cells were known to contain enzymes that efficiently hydrolyzed proteins, but these enzymes were separated from cytoplasmic proteins by a membrane (Turk and Turk, 2009). Many theories were proposed on how these enzymes might access their substrates, including the idea that this “suicide bag” of membrane bound enzymes could rupture, release proteases and degrade intracellular proteins. However, this compartment, the lysosome, is extremely acidic with a pH that is optimal for activities of the enzymes contained within it. Release of these enzymes into the cytoplasm exposes them to a neutral pH, resulting in irreversible inactivation of the majority of the lysosomal proteases (Turk and Turk, 2009).
The solution to the question of how cytoplasmic proteins were metabolized came with the discovery of the 26S proteasome and ubiquitin-mediated degradation of proteins, a discovery for which Hershko, Ciechanover, and Rose received the Nobel prize in chemistry in 2004 (Goldberg, 2005). This very large enzyme complex efficiently degrades cytosolic proteins, as well as nuclear and membranal proteins. Additional discoveries were made on the mechanisms that regulate protein degradation including a process involving the ATP-dependent modification of proteins with the small protein ubiquitin that directs the specific and efficient degradation of proteins by the proteasome. Seminal findings included the deciphering of the E1/E2/E3 enzyme cascades that are required to modify a target protein with ubiquitin. As discussed below, these mechanisms provide a plethora of targets that can be hijacked, mimicked, and manipulated by pathogens.
While further discoveries were being made on the regulation and biochemical mechanism of the proteasome, another pathway called autophagy emerged as a mechanism for the turnover of larger cytoplasmic complexes (Choi et al., 2013). This degradative pathway uses membranes to engulf components of the cytoplasm into autophagic vesicles and recycle their contents. The process involves fusion of autophagosomes with aforementioned “suicide bag” for the degradation of autophagosome components, resulting in the production of metabolites that can be reused by the cell. This pathway utilizes cellular machinery that mediates the movement and fusion of membranes, and these autophagic vacuoles are tagged with a protein that is modified using a set of enzymes similar to the E1/E2/E3 enzymes used for ubiquitylation. The autophagic pathway is an ideal target for pathogens because of the need to manipulate membranes during infection, as was exemplified above with the intricate manipulation of vesicle proteins by Legionella.
Modifying a protein with ubiquitin can result in a variety of outcomes other than degradation. These include activation of signaling proteins, regulation of protein-protein interactions, and relocalization of proteins within a cell (Jiang and Chen, 2012). Herein, we focus on the degradation of ubiquitylated proteins. This process involves an enzymatic cascade where the first enzyme E1 (ubiquitin-activating enzyme) is charged in an ATP-dependent manner with ubiquitin. E1 then transfers ubiquitin to an E2 (ubiquitin-conjugating enzyme), which can either transfer the ubiquitin to an E3 (ubiquitin-protein ligase) or be coordinated to transfer the ubiquitin directly to a target protein by an E3. All of the E3s contain binding sites for an E2 and the target protein(s) that are to be ubiquitylated. This recognition site on the target protein that is destined to be poly-ubiquitylated and degraded may consist of a protein sequence that in some cases must be modified by a PTM to be recognized (Pickart, 2001). All of these systems require tight regulation so that a cell can properly signal and maintain homeostasis. We discuss how the field of protein ubiquitylation, in particular E3-ubiquitin ligases, advanced due to the discoveries made by elucidating the function of virulence factors produced by pathogens.
HECT and RING E3 ubiquitin-protein ligases
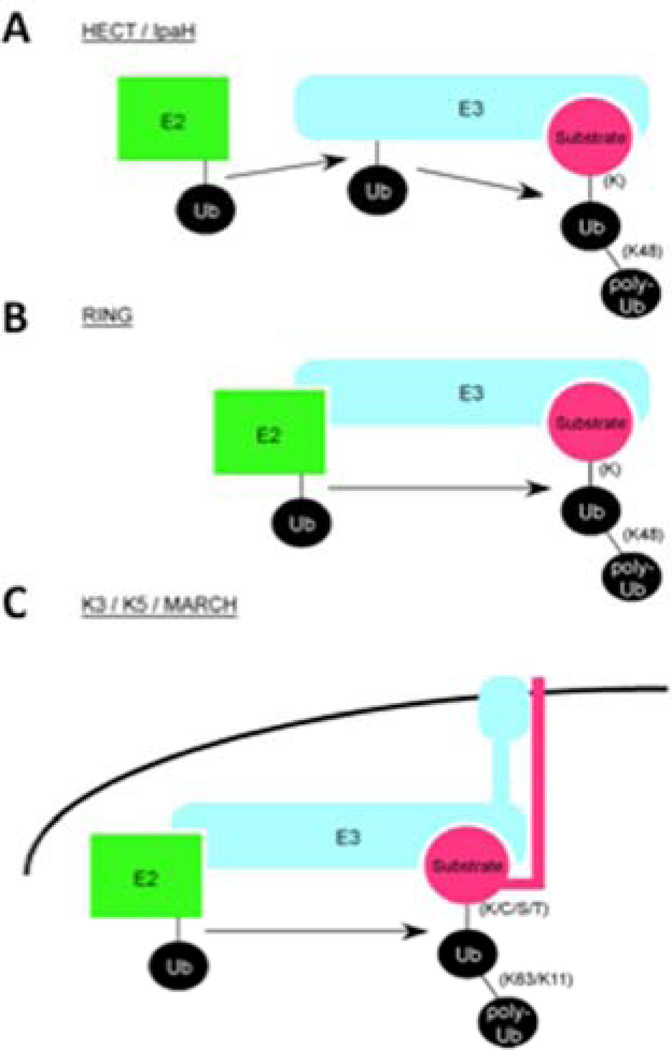
In 1995, Howley and colleagues made an important discovery through their studies on human papilloma virus (HPV) and one of its oncogenic proteins, E6 (Huibregtse et al., 1995). These investigators discovered that a complex of E6 together with the cellular protein E6-AP (E6-associated protein), an E3 ubiquitin ligase, recognized the tumor suppressor p53 and targeted it for degradation by ubiquitin-mediated proteolysis. Analysis of the E6-AP sequence revealed a number of cellular proteins that contain a domain homologous to the C-terminus of E6-AP, termed the HECT (Homologous to E6-AP Carboxy-Terminus) domain. E3 ubiquitin ligases containing HECT were shown to share the ability to accept ubiquitin from an E2 in the form of a thioester and directly transfer it to the target substrate. This discovery thus defined the class of HECT E3 ubiquitin ligases (Rotin and Kumar, 2009). Bacterial pathogens, such as Salmonella and E. coli, also use effector proteins containing HECT E3s to promote infection (Lin et al., 2011; Zhang et al., 2006). (Figure 3A).
Figure 3. Different mechanisms and substrates used by E3 ubiquitin ligase families.
(A) The HECT and IpaH families of E3 ligases accept ubiquitin from an E2 and then transfer it to a lysine residue on the substrate protein. (B) The RING E3 ligases do not accept ubiquitin from the E2, but rather facilitate the direct transfer of ubiquitin from the E2 to a lysine residue on the substrate. (C) The viral K3/K5 and the eukaryotic MARCH family of E3 ligases are membranal proteins that mediate the ubiquitylation of non-lysine residues on substrates, such as cysteine, serine, and threonine. They can also target membranal proteins by mediating non-canonical Lys63 and Lys11 linkages in polyubiquitin chains, as opposed to the canonical Lys48 linkages that are usually mediated by HECT and RING E3s.
Proteins containing a RING (Really Interesting New Gene) domain represent a second type of E3 ubiquitin-protein ligases (Figure 3B). These E3s act as scaffolds and coordinate the transfer of ubiquitin from the E2 to the substrate. Since their discovery is attributed to studies involving eukaryotic signaling rather than pathogens, we refer the readers to an in depth E3 review (Deshaies and Joazeiro, 2009). Interestingly, pathogens mimic this type of E3 to manipulate pathways in the host cell (Spallek et al., 2009). For example, the XopL effector from the plant pathogen Xanthomonas campestris pv. Vesicatoria contains a novel fold that mimics a scaffolding E3-ubiquitin ligase and is implicated in disrupting plant cell host defense (Singer et al., 2013).
Pathogen-encoded unique E3 ligases
Studies by Parsot and colleagues on bacterial T3SS effectors from Shigella flexineri revealed a third type of E3s that are encoded exclusively by bacterial pathogens called IpaH (Rohde et al., 2007). Structural analysis showed that IpaH proteins are distinct from the HECT and RING E3s, supporting the theory that these molecules evolved by a convergent mechanism, possibly through a “thioesterase” ancestor (Singer et al., 2013; Zhu et al., 2008). These bacterial effectors contain a leucine rich repeat (LRR) domain and a unique ubiquitin-protein ligase domain that transfers ubiquitin, using a catalytic cysteine residue, directly to its target substrate. The LRR is proposed to provide substrate specificity for the target proteins that will be tagged with poly-ubiquitin and to autoinhibit the E3 domain in the absence of substrate (Ashida et al., 2010; Boname and Lehner, 2011; Chou et al., 2012; Singer et al., 2013; Wang et al., 2013). IpaH E3s have essentially highjacked the entire ubiquitin/proteasome signaling machinery to direct the destruction of host proteins, enabling the pathogen to control the innate immune response. As this system is so successful, it is used by a large number of bacterial proteins from both pathogens and symbionts (Figure 3A).
Finally, a forth type of ubiquitin-protein ligase was discovered through studies on viral proteins called K3 and K5 (or MIR-1 and MIR-2, respectively) from Kaposi’s Sarcoma Associated Herpes virus (KSHV) that has been extensively reviewed by Boname and Lehner (Boname and Lehner, 2011). In brief, these E3s structurally resemble the RING E3 domains, but have unique functional and structural attributes that separate them from classic RING domain E3s. The K3 ubiquitin ligases are membrane bound enzymes that target cell surface receptors for destruction. Analysis of these E3s resulted in a number of discoveries about protein ubiquitylation. An initial surprise that emerged from the study of K3 viral ubiquitin ligases was that the enzymes could use non-conventional amino acids, including cysteine, serine and threonine residues, to mono-ubiquitylate target proteins with the help of a specific E2, UbcH5 (Cadwell and Coscoy, 2005; Wang et al., 2007). Next, it was discovered that this K3 E3 could use another E2, Ubc13, to add poly-ubiquitin chains to the mono-ubiquitylated target proteins. This E2 mediates addition of chains using ubiquitin-lysine 63 linkages rather than the canonical lysine 48 linkages, and is correlated with internalization and lysosomal degradation of cell surface receptors (Duncan et al., 2006). Another surprising discovery was made during further analysis of K5-mediated ubiquitylation showing that several cell surface receptors were targeted for destruction via mixed chain linkages (Lys11, Lys63) (Boname et al., 2010). These observations revealed multiple mechanisms that can be used to modify proteins with ubiquitin chains. Using bioinformatics, the viral E3s where found to be similar to a group of host proteins subsequently called Membrane Associated RING-CH family of E3 ligases (MARCH) that are predicted to play a role in the turnover of cell surface receptors (Lehner et al., 2005) (Figure 3C).
Antagonists of ubiquitin elongation
As observed with phosphorylation, ubiquitylation is also a target of pathogens, and a family of bacterial proteins called Cif-like effectors eloquently exemplifies a manipulation that antagonizes this PTM (Cui et al., 2010). These proteins contain a papain-like catalytic domain that is used to facilitate a deamidation reaction whereby a glutamine side chain is converted to a glutamate side chain (Figure 1D). One of these effectors, CHBP from Burkholderia pseudomallei, was shown to deamidate a specific glutamine residue within ubiquitin and an ubiquitin-like protein NEDD8 that is similarly conjugated to target proteins. This one Dalton change in molecular mass results in a ubiquitin molecule that can no longer be used in ubiquitin elongation reactions, and NEDD8 that can no longer modify the functions of its substrate proteins. This PTM causes a dramatic change in host signaling, and has thus far been shown to be irreversible.
Conclusions and future directions
The study of the functional mechanisms of pathogenic enzymes leads to a better understanding of virulence strategies as well as to discoveries of new cellular enzymatic activities and regulatory mechanisms. In this review, we have presented examples of how the study of host-pathogen interactions and PTMs that are mediated by pathogens provided valuable insights into eukaryotic cell biology. Pathogens use PTMs, both directly and indirectly, to manipulate the host cell and regulate host proteins activities, as well as the activities of their own proteins. Notably, while pathogens use various PTMs and different mechanisms, there are common host processes and pathways that are targeted and manipulated. These processes are mainly those required for immunity and signal transduction (such as MAPK and NFκB pathways), entry of the pathogen into the host cell (the cytoskeleton), and maintaining a replicative niche (vesicle trafficking). A significant point not discussed in detail is the use of the host PTM machinery to directly regulate the function of bacterial proteins within a host. For example, pathogenic proteins can be ubiquitylated to control their stability or subcellular localization. Some proteins have also been reported to undergo modification with a lipid moiety to ensure their membrane localization (Hicks and Galan, 2013; Ribet and Cossart, 2010).
It is interesting to look back and see how technological advancements led to seminal discoveries of novel PTMs. From the use of radioactivity and antibodies in the early years of PTM studies, we have recently entered the era of mass spectrometry. The ability to accurately identify minor changes (sometime only a one Dalton change) in a protein’s molecular weight presented a huge stepping stone for discoveries of PTMs. Based on the recently identified PTMs, it is apparent that a strategy to study virulence factors has emerged. This strategy follows three central steps: finding a target protein, using mass spectrometry techniques to identify how the target is modified by the enzyme of interest, and finally following appropriate biochemical strategies to elucidate/validate the mechanism.
Discoveries in the field of pathogen-mediated PTM now allow us to exploit the unique nature of these enzymes and use them as tools to control and fine-tune cellular processes. This concept was recently demonstrated by Lim and colleagues who used bacterial type III effectors with phosphothreonine lyase (OspF) and phosphatase (YopH) activities to rewire and fine tune kinase signaling pathways in T cells (Wei et al., 2012). Research on pathogen-mediated PTM is progressing and evolving rapidly as new PTMs and new targets are discovered. We will undoubtedly witness the discovery of more unique PTMs in the next few years that will shed light on new regulatory mechanisms in eukaryotic cells.
Acknowledgements
We thank members of the Orth lab for editing and helpful discussions. K.O. and D.S. are supported by grants from NIH-Allergy and Infectious Disease (R21-AI096133 and R01-AI087808) and the Welch Foundation (I-1561). K.O. is a Burroughs Wellcome Investigator in Pathogenesis of Infectious Disease and a W.W. Caruth Jr. Biomedical Scholar and has an Earl A. Forsythe Chair in Biomedical Science.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashida H, Kim M, Schmidt-Supprian M, Ma A, Ogawa M, Sasakawa C. A bacterial E3 ubiquitin ligase IpaH9.8 targets NEMO/IKKgamma to dampen the host NF-kappaB-mediated inflammatory response. Nat Cell Biol. 2010;12:66–73. doi: 10.1038/ncb2006. sup pp 61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barford D. Protein phosphatases. Curr Opin Struct Biol. 1995;5:728–734. doi: 10.1016/0959-440x(95)80004-2. [DOI] [PubMed] [Google Scholar]
- Bliska JB, Guan KL, Dixon JE, Falkow S. Tyrosine phosphate hydrolysis of host proteins by an essential Yersinia virulence determinant. Proc Natl Acad Sci U S A. 1991;88:1187–1191. doi: 10.1073/pnas.88.4.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggon TJ, Eck MJ. Structure and regulation of Src family kinases. Oncogene. 2004;23:7918–7927. doi: 10.1038/sj.onc.1208081. [DOI] [PubMed] [Google Scholar]
- Bolin I, Wolf-Watz H. The plasmid-encoded Yop2b protein of Yersinia pseudotuberculosis is a virulence determinant regulated by calcium and temperature at the level of transcription. Mol Microbiol. 1988;2:237–245. doi: 10.1111/j.1365-2958.1988.tb00025.x. [DOI] [PubMed] [Google Scholar]
- Boname JM, Lehner PJ. What has the study of the K3 and K5 viral ubiquitin E3 ligases taught us about ubiquitin-mediated receptor regulation? Viruses. 2011;3:118–131. doi: 10.3390/v3020118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boname JM, Thomas M, Stagg HR, Xu P, Peng J, Lehner PJ. Efficient internalization of MHC I requires lysine-11 and lysine-63 mixed linkage polyubiquitin chains. Traffic. 2010;11:210–220. doi: 10.1111/j.1600-0854.2009.01011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan DF, Barford D. Eliminylation: a post-translational modification catalyzed by phosphothreonine lyases. Trends Biochem Sci. 2009;34:108–114. doi: 10.1016/j.tibs.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Brown MS, Segal A, Stadtman ER. Modulation of glutamine synthetase adenylylation and deadenylylation is mediated by metabolic transformation of the P II -regulatory protein. Proc Natl Acad Sci U S A. 1971;68:2949–2953. doi: 10.1073/pnas.68.12.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugge JS, Erikson RL. Identification of a transformation-specific antigen induced by an avian sarcoma virus. Nature. 1977;269:346–348. doi: 10.1038/269346a0. [DOI] [PubMed] [Google Scholar]
- Burnett G, Kennedy EP. The enzymatic phosphorylation of proteins. J Biol Chem. 1954;211:969–980. [PubMed] [Google Scholar]
- Cadwell K, Coscoy L. Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science. 2005;309:127–130. doi: 10.1126/science.1110340. [DOI] [PubMed] [Google Scholar]
- Campanacci V, Mukherjee S, Roy CR, Cherfils J. Structure of the Legionella effector AnkX reveals the mechanism of phosphocholine transfer by the FIC domain. EMBO J. 2013 doi: 10.1038/emboj.2013.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps M, Nichols A, Arkinstall S. Dual specificity phosphatases: a gene family for control of MAP kinase function. FASEB J. 2000;14:6–16. [PubMed] [Google Scholar]
- Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. The New England journal of medicine. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- Chou YC, Keszei AF, Rohde JR, Tyers M, Sicheri F. Conserved structural mechanisms for autoinhibition in IpaH ubiquitin ligases. The Journal of biological chemistry. 2012;287:268–275. doi: 10.1074/jbc.M111.316265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collett MS, Erikson RL. Protein kinase activity associated with the avian sarcoma virus src gene product. Proc Natl Acad Sci U S A. 1978;75:2021–2024. doi: 10.1073/pnas.75.4.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JA, Hunter T. Changes in protein phosphorylation in Rous sarcoma virus-transformed chicken embryo cells. Mol Cell Biol. 1981;1:165–178. doi: 10.1128/mcb.1.2.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Yao Q, Li S, Ding X, Lu Q, Mao H, Liu L, Zheng N, Chen S, Shao F. Glutamine deamidation and dysfunction of ubiquitin/NEDD8 induced by a bacterial effector family. Science. 2010;329:1215–1218. doi: 10.1126/science.1193844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Puerta ML, Trinidad AG, del Carmen Rodriguez M, Bogetz J, Sanchez Crespo M, Mustelin T, Alonso A, Bayon Y. Characterization of new substrates targeted by Yersinia tyrosine phosphatase YopH. PLoS One. 2009;4:e4431. doi: 10.1371/journal.pone.0004431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Duncan LM, Piper S, Dodd RB, Saville MK, Sanderson CM, Luzio JP, Lehner PJ. Lysine-63-linked ubiquitination is required for endolysosomal degradation of class I molecules. The EMBO journal. 2006;25:1635–1645. doi: 10.1038/sj.emboj.7601056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhart W, Hutchinson MA, Hunter T. An activity phosphorylating tyrosine in polyoma T antigen immunoprecipitates. Cell. 1979;18:925–933. doi: 10.1016/0092-8674(79)90205-8. [DOI] [PubMed] [Google Scholar]
- Engel P, Goepfert A, Stanger FV, Harms A, Schmidt A, Schirmer T, Dehio C. Adenylylation control by intra- or intermolecular active-site obstruction in Fic proteins. Nature. 2012;482:107–110. doi: 10.1038/nature10729. [DOI] [PubMed] [Google Scholar]
- Erikson E, Erikson RL. Identification of a cellular protein substrate phosphorylated by the avian sarcoma virus-transforming gene product. Cell. 1980;21:829–836. doi: 10.1016/0092-8674(80)90446-8. [DOI] [PubMed] [Google Scholar]
- Fauman EB, Saper MA. Structure and function of the protein tyrosine phosphatases. Trends Biochem Sci. 1996;21:413–417. doi: 10.1016/s0968-0004(96)10059-1. [DOI] [PubMed] [Google Scholar]
- Feng F, Yang F, Rong W, Wu X, Zhang J, Chen S, He C, Zhou JM. A Xanthomonas uridine 5'-monophosphate transferase inhibits plant immune kinases. Nature. 2012;485:114–118. doi: 10.1038/nature10962. [DOI] [PubMed] [Google Scholar]
- Frackelton AR, Jr, Ross AH, Eisen HN. Characterization and use of monoclonal antibodies for isolation of phosphotyrosyl proteins from retrovirus-transformed cells and growth factor-stimulated cells. Mol Cell Biol. 1983;3:1343–1352. doi: 10.1128/mcb.3.8.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galyov EE, Hakansson S, Forsberg A, Wolf-Watz H. A secreted protein kinase of Yersinia pseudotuberculosis is an indispensable virulence determinant. Nature. 1993;361:730–732. doi: 10.1038/361730a0. [DOI] [PubMed] [Google Scholar]
- Gill DM, Pappenheimer AM, Jr, Brown R, Kurnick JT. Studies on the mode of action of diphtheria toxin. VII. Toxin-stimulated hydrolysis of nicotinamide adenine dinucleotide in mammalian cell extracts. J Exp Med. 1969;129:1–21. doi: 10.1084/jem.129.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AL. Nobel committee tags ubiquitin for distinction. Neuron. 2005;45:339–344. doi: 10.1016/j.neuron.2005.01.019. [DOI] [PubMed] [Google Scholar]
- Goody PR, Heller K, Oesterlin LK, Muller MP, Itzen A, Goody RS. Reversible phosphocholination of Rab proteins by Legionella pneumophila effector proteins. EMBO J. 2012;31:1774–1784. doi: 10.1038/emboj.2012.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Okesli A, van der Donk WA. Mechanistic studies of Ser/Thr dehydration catalyzed by a member of the LanL lanthionine synthetase family. Biochemistry. 2011;50:891–898. doi: 10.1021/bi101750r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grammel M, Luong P, Orth K, Hang HC. A chemical reporter for protein AMPylation. J Am Chem Soc. 2011;133:17103–17105. doi: 10.1021/ja205137d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gschwind A, Fischer OM, Ullrich A. The discovery of receptor tyrosine kinases: targets for cancer therapy. Nat Rev Cancer. 2004;4:361–370. doi: 10.1038/nrc1360. [DOI] [PubMed] [Google Scholar]
- Guan K, Hakes DJ, Wang Y, Park HD, Cooper TG, Dixon JE. A yeast protein phosphatase related to the vaccinia virus VH1 phosphatase is induced by nitrogen starvation. Proc Natl Acad Sci U S A. 1992;89:12175–12179. doi: 10.1073/pnas.89.24.12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan KL, Broyles SS, Dixon JE. A Tyr/Ser protein phosphatase encoded by vaccinia virus. Nature. 1991;350:359–362. doi: 10.1038/350359a0. [DOI] [PubMed] [Google Scholar]
- Guan KL, Dixon JE. Protein tyrosine phosphatase activity of an essential virulence determinant in Yersinia. Science. 1990;249:553–556. doi: 10.1126/science.2166336. [DOI] [PubMed] [Google Scholar]
- Hamid N, Gustavsson A, Andersson K, McGee K, Persson C, Rudd CE, Fallman M. YopH dephosphorylates Cas and Fyn-binding protein in macrophages. Microb Pathog. 1999;27:231–242. doi: 10.1006/mpat.1999.0301. [DOI] [PubMed] [Google Scholar]
- Hao YH, Chuang T, Ball HL, Luong P, Li Y, Flores-Saaib RD, Orth K. Characterization of a rabbit polyclonal antibody against threonine-AMPylation. J Biotechnol. 2011;151:251–254. doi: 10.1016/j.jbiotec.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneberg P. Finding the smoking gun: protein tyrosine phosphatases as tools and targets of unicellular microorganisms and viruses. Curr Med Chem. 2012;19:1530–1566. doi: 10.2174/092986712799828274. [DOI] [PubMed] [Google Scholar]
- Hervet E, Charpentier X, Vianney A, Lazzaroni JC, Gilbert C, Atlan D, Doublet P. Protein kinase LegK2 is a type IV secretion system effector involved in endoplasmic reticulum recruitment and intracellular replication of Legionella pneumophila. Infect Immun. 2011;79:1936–1950. doi: 10.1128/IAI.00805-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks SW, Galan JE. Exploitation of eukaryotic subcellular targeting mechanisms by bacterial effectors. Nat Rev Microbiol. 2013;11:316–326. doi: 10.1038/nrmicro3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjo T, Nishizuka Y, Hayaishi O. Diphtheria toxin-dependent adenosine diphosphate ribosylation of aminoacyl transferase II and inhibition of protein synthesis. J Biol Chem. 1968;243:3553–3555. [PubMed] [Google Scholar]
- Huebner RJ, Todaro GJ. Oncogenes of RNA tumor viruses as determinants of cancer. Proc Natl Acad Sci U S A. 1969;64:1087–1094. doi: 10.1073/pnas.64.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Beaudenon S, Howley PM. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:5249. doi: 10.1073/pnas.92.11.5249-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. Tony Hunter: kinase king. Interview by Ruth Williams. J Cell Biol. 2008;181:572–573. doi: 10.1083/jcb.1814pi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T, Sefton BM. Transforming gene product of Rous sarcoma virus phosphorylates tyrosine. Proc Natl Acad Sci U S A. 1980;77:1311–1315. doi: 10.1073/pnas.77.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingmundson A, Delprato A, Lambright DG, Roy CR. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature. 2007;450:365–369. doi: 10.1038/nature06336. [DOI] [PubMed] [Google Scholar]
- Jiang X, Chen ZJ. The role of ubiquitylation in immune defence and pathogen evasion. Nature reviews Immunology. 2012;12:35–48. doi: 10.1038/nri3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juris SJ, Rudolph AE, Huddler D, Orth K, Dixon JE. A distinctive role for the Yersinia protein kinase: actin binding, kinase activation, and cytoskeleton disruption. Proc Natl Acad Sci U S A. 2000;97:9431–9436. doi: 10.1073/pnas.170281997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DW, Lenzen G, Page AL, Legrain P, Sansonetti PJ, Parsot C. The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc Natl Acad Sci U S A. 2005;102:14046–14051. doi: 10.1073/pnas.0504466102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinch LN, Yarbrough ML, Orth K, Grishin NV. Fido, a novel AMPylation domain common to fic, doc, and AvrB. PLoS One. 2009;4:e5818. doi: 10.1371/journal.pone.0005818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingdon HS, Shapiro BM, Stadtman ER. Regulation of glutamine synthetase. 8. ATP: glutamine synthetase adenylyltransferase, an enzyme that catalyzes alterations in the regulatory properties of glutamine synthetase. Proc Natl Acad Sci U S A. 1967;58:1703–1710. doi: 10.1073/pnas.58.4.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner PJ, Hoer S, Dodd R, Duncan LM. Downregulation of cell surface receptors by the K3 family of viral and cellular ubiquitin E3 ligases. Immunological reviews. 2005;207:112–125. doi: 10.1111/j.0105-2896.2005.00314.x. [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewallen DM, Steckler CJ, Knuckley B, Chalmers MJ, Thompson PR. Probing adenylation: using a fluorescently labelled ATP probe to directly label and immunoprecipitate VopS substrates. Mol Biosyst. 2012;8:1701–1706. doi: 10.1039/c2mb25053e. [DOI] [PubMed] [Google Scholar]
- Li H, Xu H, Zhou Y, Zhang J, Long C, Li S, Chen S, Zhou JM, Shao F. The phosphothreonine lyase activity of a bacterial type III effector family. Science. 2007;315:1000–1003. doi: 10.1126/science.1138960. [DOI] [PubMed] [Google Scholar]
- Li Y, Al-Eryani R, Yarbrough ML, Orth K, Ball HL. Characterization of AMPylation on threonine, serine, and tyrosine using mass spectrometry. J Am Soc Mass Spectrom. 2011;22:752–761. doi: 10.1007/s13361-011-0084-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DY, Diao J, Zhou D, Chen J. Biochemical and structural studies of a HECT-like ubiquitin ligase from Escherichia coli O157:H7. The Journal of biological chemistry. 2011;286:441–449. doi: 10.1074/jbc.M110.167643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Lemon B, Traktman P. The dual-specificity phosphatase encoded by vaccinia virus, VH1, is essential for viral transcription in vivo and in vitro. J Virol. 1995;69:7823–7834. doi: 10.1128/jvi.69.12.7823-7834.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nat Biotechnol. 2003;21:255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- Martin GS. The hunting of the Src. Nat Rev Mol Cell Biol. 2001;2:467–475. doi: 10.1038/35073094. [DOI] [PubMed] [Google Scholar]
- Mittal R, Peak-Chew SY, McMahon HT. Acetylation of MEK2 and I kappa B kinase (IKK) activation loop residues by YopJ inhibits signaling. Proc Natl Acad Sci U S A. 2006;103:18574–18579. doi: 10.1073/pnas.0608995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Keitany G, Li Y, Wang Y, Ball HL, Goldsmith EJ, Orth K. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science. 2006;312:1211–1214. doi: 10.1126/science.1126867. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Liu X, Arasaki K, McDonough J, Galan JE, Roy CR. Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature. 2011;477:103–106. doi: 10.1038/nature10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller MP, Peters H, Blumer J, Blankenfeldt W, Goody RS, Itzen A. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science. 2010;329:946–949. doi: 10.1126/science.1192276. [DOI] [PubMed] [Google Scholar]
- Najarro P, Traktman P, Lewis JA. Vaccinia virus blocks gamma interferon signal transduction: viral VH1 phosphatase reverses Stat1 activation. J Virol. 2001;75:3185–3196. doi: 10.1128/JVI.75.7.3185-3196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Koller A, Nordfelth R, Wolf-Watz H, Taylor S, Dixon JE. Identification of a molecular target for the Yersinia protein kinase A. Mol Cell. 2007;26:465–477. doi: 10.1016/j.molcel.2007.04.025. [DOI] [PubMed] [Google Scholar]
- Neunuebel MR, Chen Y, Gaspar AH, Backlund PS, Jr, Yergey A, Machner MP. De-AMPylation of the small GTPase Rab1 by the pathogen Legionella pneumophila. Science. 2011;333:453–456. doi: 10.1126/science.1207193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Oppermann H, Levinson AD, Varmus HE, Levintow L, Bishop JM. Uninfected vertebrate cells contain a protein that is closely related to the product of the avian sarcoma virus transforming gene (src) Proc Natl Acad Sci U S A. 1979;76:1804–1808. doi: 10.1073/pnas.76.4.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth K, Palmer LE, Bao ZQ, Stewart S, Rudolph AE, Bliska JB, Dixon JE. Inhibition of the mitogen-activated protein kinase kinase superfamily by a Yersinia effector. Science. 1999;285:1920–1923. doi: 10.1126/science.285.5435.1920. [DOI] [PubMed] [Google Scholar]
- Orth K, Xu Z, Mudgett MB, Bao ZQ, Palmer LE, Bliska JB, Mangel WF, Staskawicz B, Dixon JE. Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science. 2000;290:1594–1597. doi: 10.1126/science.290.5496.1594. [DOI] [PubMed] [Google Scholar]
- Paul AV, van Boom JH, Filippov D, Wimmer E. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature. 1998;393:280–284. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- Persson C, Carballeira N, Wolf-Watz H, Fallman M. The PTPase YopH inhibits uptake of Yersinia, tyrosine phosphorylation of p130Cas and FAK, and the associated accumulation of these proteins in peripheral focal adhesions. EMBO J. 1997;16:2307–2318. doi: 10.1093/emboj/16.9.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Prehna G, Ivanov MI, Bliska JB, Stebbins CE. Yersinia virulence depends on mimicry of host Rho-family nucleotide dissociation inhibitors. Cell. 2006;126:869–880. doi: 10.1016/j.cell.2006.06.056. [DOI] [PubMed] [Google Scholar]
- Pulido R, Hooft van Huijsduijnen R. Protein tyrosine phosphatases: dual-specificity phosphatases in health and disease. FEBS J. 2008;275:848–866. doi: 10.1111/j.1742-4658.2008.06250.x. [DOI] [PubMed] [Google Scholar]
- Radke K, Martin GS. Transformation by Rous sarcoma virus: effects of src gene expression on the synthesis and phosphorylation of cellular polypeptides. Proc Natl Acad Sci U S A. 1979;76:5212–5216. doi: 10.1073/pnas.76.10.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M, Ham H, Liu X, Sugiura Y, Orth K, Kramer H. Visual neurotransmission in Drosophila requires expression of Fic in glial capitate projections. Nat Neurosci. 2012;15:871–875. doi: 10.1038/nn.3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribet D, Cossart P. Pathogen-mediated posttranslational modifications: A re-emerging field. Cell. 2010;143:694–702. doi: 10.1016/j.cell.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde JR, Breitkreutz A, Chenal A, Sansonetti PJ, Parsot C. Type III secretion effectors of the IpaH family are E3 ubiquitin ligases. Cell Host Microbe. 2007;1:77–83. doi: 10.1016/j.chom.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Rothberg PG, Harris TJ, Nomoto A, Wimmer E. O4-(5'-uridylyl)tyrosine is the bond between the genome-linked protein and the RNA of poliovirus. Proc Natl Acad Sci U S A. 1978;75:4868–4872. doi: 10.1073/pnas.75.10.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nature reviews Molecular cell biology. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- Rous P. A Sarcoma of the Fowl Transmissible by an Agent Separable from the Tumor Cells. J Exp Med. 1911;13:397–411. doi: 10.1084/jem.13.4.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro BM, Stadtman ER. Glutamine synthetase deadenylylating enzyme. Biochem Biophys Res Commun. 1968;30:32–37. doi: 10.1016/0006-291x(68)90708-0. [DOI] [PubMed] [Google Scholar]
- Singer AU, Schulze S, Skarina T, Xu X, Cui H, Eschen-Lippold L, Egler M, Srikumar T, Raught B, Lee J, et al. A pathogen type III effector with a novel E3 ubiquitin ligase architecture. PLoS pathogens. 2013;9:e1003121. doi: 10.1371/journal.ppat.1003121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spallek T, Robatzek S, Gohre V. How microbes utilize host ubiquitination. Cellular microbiology. 2009;11:1425–1434. doi: 10.1111/j.1462-5822.2009.01346.x. [DOI] [PubMed] [Google Scholar]
- Stehelin D, Varmus HE, Bishop JM, Vogt PK. DNA related to the transforming gene(s) of avian sarcoma viruses is present in normal avian DNA. Nature. 1976;260:170–173. doi: 10.1038/260170a0. [DOI] [PubMed] [Google Scholar]
- Tan Y, Arnold RJ, Luo ZQ. Legionella pneumophila regulates the small GTPase Rab1 activity by reversible phosphorylcholination. Proc Natl Acad Sci U S A. 2011;108:21212–21217. doi: 10.1073/pnas.1114023109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, Luo ZQ. Legionella pneumophila SidD is a deAMPylase that modifies Rab1. Nature. 2011;475:506–509. doi: 10.1038/nature10307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SM, Brugge JS. Cellular functions regulated by Src family kinases. Annu Rev Cell Dev Biol. 1997;13:513–609. doi: 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- Turk B, Turk V. Lysosomes as "suicide bags" in cell death: myth or reality? The Journal of biological chemistry. 2009;284:21783–21787. doi: 10.1074/jbc.R109.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubersax JA, Ferrell JE., Jr Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol. 2007;8:530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- Walburger A, Koul A, Ferrari G, Nguyen L, Prescianotto-Baschong C, Huygen K, Klebl B, Thompson C, Bacher G, Pieters J. Protein kinase G from pathogenic mycobacteria promotes survival within macrophages. Science. 2004;304:1800–1804. doi: 10.1126/science.1099384. [DOI] [PubMed] [Google Scholar]
- Wang F, Jiang Z, Li Y, He X, Zhao J, Yang X, Zhu L, Yin Z, Li X, Wang X, et al. Shigella flexneri T3SS effector IpaH4.5 modulates the host inflammatory response via interaction with NF-kappaB p65 protein. Cellular microbiology. 2013;15:474–485. doi: 10.1111/cmi.12052. [DOI] [PubMed] [Google Scholar]
- Wang X, Herr RA, Chua WJ, Lybarger L, Wiertz EJ, Hansen TH. Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. The Journal of cell biology. 2007;177:613–624. doi: 10.1083/jcb.200611063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P, Wong WW, Park JS, Corcoran EE, Peisajovich SG, Onuffer JJ, Weiss A, Lim WA. Bacterial virulence proteins as tools to rewire kinase pathways in yeast and immune cells. Nature. 2012 doi: 10.1038/nature11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willey JM, van der Donk WA. Lantibiotics: peptides of diverse structure and function. Annu Rev Microbiol. 2007;61:477–501. doi: 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]
- Woolery AR, Luong P, Broberg CA, Orth K. AMPylation: Something Old is New Again. Front Microbiol. 2010;1:113. doi: 10.3389/fmicb.2010.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worby CA, Mattoo S, Kruger RP, Corbeil LB, Koller A, Mendez JC, Zekarias B, Lazar C, Dixon JE. The fic domain: regulation of cell signaling by adenylylation. Mol Cell. 2009;34:93–103. doi: 10.1016/j.molcel.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T, Mecsas J, Healy JI, Falkow S, Chien Y. Suppression of T and B lymphocyte activation by a Yersinia pseudotuberculosis virulence factor, yopH. J Exp Med. 1999;190:1343–1350. doi: 10.1084/jem.190.9.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarbrough ML, Li Y, Kinch LN, Grishin NV, Ball HL, Orth K. AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science. 2009;323:269–272. doi: 10.1126/science.1166382. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Higashide WM, McCormick BA, Chen J, Zhou D. The inflammation-associated Salmonella SopA is a HECT-like E3 ubiquitin ligase. Mol Microbiol. 2006;62:786–793. doi: 10.1111/j.1365-2958.2006.05407.x. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Dong N, Hu L, Shao F. The Shigella Type Three Secretion System Effector OspG Directly and Specifically Binds to Host Ubiquitin for Activation. PLoS One. 2013;8:e57558. doi: 10.1371/journal.pone.0057558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Li H, Hu L, Wang J, Zhou Y, Pang Z, Liu L, Shao F. Structure of a Shigella effector reveals a new class of ubiquitin ligases. Nat Struct Mol Biol. 2008;15:1302–1308. doi: 10.1038/nsmb.1517. [DOI] [PubMed] [Google Scholar]