ABSTRACT
Non-coding regulatory RNAs fine-tune gene expression post-transcriptionally. In the streptomycetes, rpfA – encoding a muralytic enzyme required for establishing and exiting dormancy – is flanked by non-coding regulatory RNA elements both upstream (riboswitch) and downstream [antisense small RNA (sRNA)]. In Streptomyces coelicolor, the upstream riboswitch decreases rpfA transcript abundance in response to the second messenger cyclic di-AMP, itself involved in cell wall metabolism and dormancy. There is, however, no obvious expression platform associated with this riboswitch and consequently, its mechanism of action is entirely unknown. Using in vitro transcription assays, we discovered that the rpfA riboswitch promoted premature transcription termination in response to cyclic di-AMP. Through an extensive mutational analysis, we determined that attenuation required ligand binding and involved an unusual extended stem-loop region unique to a subset of rpfA riboswitches in the actinobacteria. At the other end of the rpfA gene, an antisense sRNA, termed Scr3097, is expressed opposite the predicted rpfA terminator. Using northern blotting, we found that Scr3097 accumulation mirrored that of the rpfA mRNA. In liquid culture, we detected Scr3097 exclusively in exponential-phase cells, and in plate-grown culture, we observed the sRNA primarily in differentiating cultures. Using mutational analyses, we found that the sRNA increased rpfA mRNA abundance in cells. Taken together, our work revealed multiple regulatory RNAs controlling rpfA expression in the streptomycetes.
KEYWORDS: Attenuation, cyclic di-AMP, dormancy, resuscitation-promoting factor, riboswitch, small RNA, termination
Introduction
Non-coding RNAs, like riboswitches and small RNAs (sRNAs), provide an elegant means of fine-tuning gene expression after transcription has initiated. Riboswitches are cis-encoded regulatory RNA elements generally embedded in the 5′ untranslated region (UTR) of an mRNA. Like their protein counterparts, these regulators monitor the metabolic state of a cell through direct interaction with metabolites, and these metabolites are often the product of the pathway they regulate. Riboswitches exert their regulatory effects by switching between two mutually exclusive structures in response to ligand binding. Within the riboswitch itself, the sensor domain binds the ligand, whereas the downstream expression platform translates the biologic signal into a regulatory response by modulating transcript elongation, transcript stability or translation initiation.1
The Bacillus subtilis ydaO riboswitch, for example, prematurely terminates transcription of its cognate gene in response to high levels of cyclic di-AMP,2 a second messenger known to regulate sporulation in Bacillus3 and cell wall metabolism in a range of other bacteria.4-6 In the B. subtilis system, a canonical intrinsic terminator was predicted downstream of the sensor domain,2,7 and mutational analyses confirmed its involvement in ligand-dependent attenuation.2 Though it is clear that terminator formation is essential for riboswitch activity,2 a mutually exclusive antiterminator, which would enable read-through in the absence of cyclic di-AMP, has not been identified, and so the means by which the riboswitch toggles between its ‘ON’ and ‘OFF’ states remains unclear.
Like riboswitches, sRNAs provide another avenue to post-transcriptional regulation. These short non-coding transcripts are synthesized in response to specific conditions and in turn modulate the expression of their targets accordingly. sRNAs primarily impact transcript stability and translatability through their interaction with specific mRNA targets. They can also affect gene expression by titrating protein regulators away from their intended targets, often through target mimicry, though such protein sequestration has been observed less frequently than mRNA modulation.8
sRNAs are generally classified as either trans-encoded sRNAs or cis-encoded antisense sRNAs. Trans-encoded sRNAs are expressed from genes located elsewhere in the chromosome relative to their targets, and are only partially complementary to their target mRNAs. In contrast, antisense sRNAs are expressed opposite their target genes, and consequently share complete complementarity with their associated mRNA.8
In the streptomycetes, non-coding regulatory RNA elements flank either end of the rpfA gene: a cyclic di-AMP-responsive ydaO-like riboswitch motif is located within the 5′ UTR,2,7,9,10 and as described below, an antisense sRNA-encoding gene is found immediately downstream of the rpfA stop codon. In these bacteria, rpfA is required to both enter and exit dormancy.9,11 Bacterial dormancy allows survival under suboptimal growth conditions, and is characterized by low metabolism and an inability to replicate.12,13 Whereas some actinobacteria adopt ‘cyst-like’ dormant structures (e.g., Mycobacterium),14,15 the streptomycetes form spores. The Streptomyces developmental cycle begins with spore germination and the establishment of a vegetative mycelium, and culminates with the raising of aerial hyphae and metamorphosis of these hyphal compartments into spores.16 Although bacterial dormant states can vary, a shared feature is the need to remodel the cell wall not only at the onset of dormancy, but also during ‘resuscitation’, when growth resumes.14,15,17-20
The rpfA gene encodes a protein belonging to a family of secreted muralytic enzymes called the ‘resuscitation-promoting factors’ (Rpfs). In the actinobacteria, the Rpfs promote resuscitation through their peptidoglycan cleaving activities.21 Given the destructive potential of these enzymes, it is critical that their expression be tightly controlled to ensure the integrity of the cell wall throughout growth. In the streptomycetes, rpfA is regulated at multiple levels.10 Though sRNA-mediated control of rpfA expression has not been investigated, previous work has revealed post-transcriptional regulation by a ydaO-like riboswitch.10 This so-called rpfA riboswitch in Streptomyces coelicolor negatively impacts rpfA transcript abundance in response to cyclic di-AMP.10 Unlike its counterpart in B. subtilis,2,7 however, the rpfA riboswitch lacks a canonical intrinsic terminator, and riboswitch activity is Rho-independent.10
In this study, we set out to determine the mechanism by which the rpfA riboswitch regulates gene expression in S. coelicolor, and explore the contribution of the downstream sRNA-encoding gene to rpfA control. We found that the riboswitch prematurely terminated rpfA transcription in response to cyclic di-AMP binding. We further identified novel activity determinants that distinguished the S. coelicolor rpfA riboswitch from the B. subtilis model. Finally, our work uncovered a role for the sRNA Scr3097 in positively regulating rpfA mRNA abundance.
Results
rpfA riboswitch-dependent attenuation correlates with intracellular cyclic di-AMP levels
As a first step toward characterizing the regulatory mechanism used by the rpfA riboswitch, we monitored rpfA expression in plate-grown S. coelicolor throughout its developmental cycle. Using a transcriptional luciferase reporter system in which reporter genes were fused to the 3′ end of the rpfA riboswitch, we monitored reporter gene expression in wild-type S. coelicolor cells grown on solid medium. We found that expression levels were low throughout vegetative growth, but increased to maximal levels during aerial development and sporulation (Fig. 1A).
Figure 1.
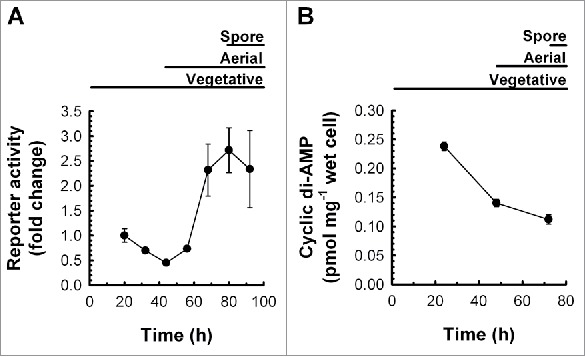
Riboswitch-mediated attenuation and cyclic di-AMP abundance in S. coelicolor throughout development. (A) Wild-type S. coelicolor, constitutively expressing the rpfA riboswitch transcriptionally fused to the luciferase reporter gene cluster (M145/pMC222), was grown on a sporulation-conducive solid medium. Reporter activity was quantified throughout growth and development. Background-subtracted luminescence was normalized to that of the positive control strain constitutively expressing the reporter without the rpfA riboswitch (M145/pFLUX-Pos). Normalized reporter activity was expressed as mean fold changes ± standard deviation (n = 3) relative to ‘20 h’. Figure is representative of 8 independent experimental replicates. (B) Wild-type S. coelicolor (M145) was grown at 30°C on a sporulation-conducive solid medium. At the indicated times, cyclic di-AMP was extracted from cells and quantified by LC-MS/MS. Cyclic di-AMP levels were normalized to wet cell weight. Data are presented as mean ± standard error (n = 4).
Because the rpfA riboswitch binds and responds to the second messenger cyclic di-AMP,10 we sought to connect post-transcriptional changes in rpfA expression with intracellular cyclic di-AMP levels. We followed ligand levels in plate-grown S. coelicolor using liquid chromatography-coupled tandem mass spectrometry (LC-MS/MS), and discovered that the second messenger was most abundant during vegetative growth (we were unable to monitor rpfA expression or cyclic di-AMP abundance during germination and early vegetative growth owing to insufficient biomass). Thereafter, cyclic di-AMP levels decreased, reaching minimal levels during sporulation (Fig. 1B). Our findings suggested that rpfA expression and cyclic di-AMP levels were inversely correlated, as would be expected for an ‘OFF’-acting riboswitch.
Cyclic di-AMP binds the rpfA riboswitch and alters the RNA structure
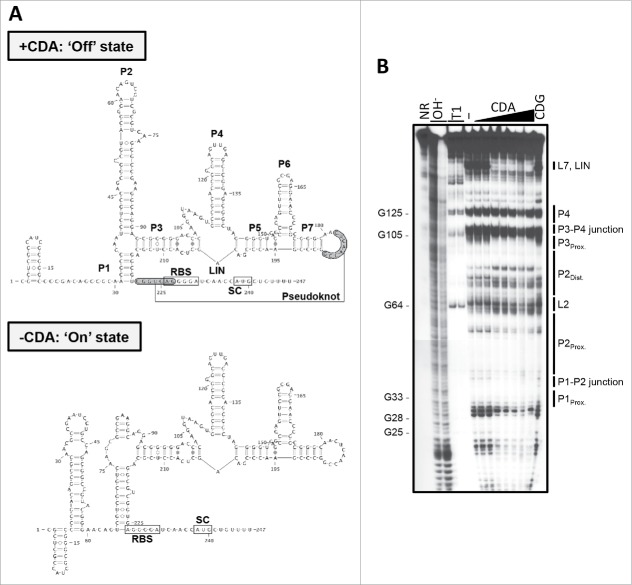
Though we had previously determined that the riboswitch negatively impacts rpfA transcript abundance,10 the mechanism by which it acts remained elusive. We therefore set out to characterize the ligand-binding domain and identify the expression platform within the rpfA 5′ UTR using a structural probing approach. Using in-line probing, we assessed the secondary structure of the rpfA riboswitch in both its unbound and cyclic di-AMP-associated states (Fig. 2). We determined that cyclic di-AMP binding protected against scission within the P1 stem, the P1-P2 junction, the proximal P2 bulge, the P3 stem, the single-stranded region bridging stems P3 and P5 (LIN), and the L7 loop, suggesting that these regions were stabilized upon ligand binding. Reduced cleavage in the presence of cyclic di-AMP was also noted in similar regions within the ydaO riboswitches from B. subtilis, Clostridium acetobutylicum, Nostoc punctiforme and Syntrophus aciditrophicus.2 These regions are highly conserved at the sequence and/or structure levels,2,7 and several of these regions harbor cyclic di-AMP-binding residues (e.g., P1, P1-P2 junction, P3, single-stranded region).22-24 In contrast, the P4 stem structure was consistently cleaved, irrespective of ligand status, suggesting a lack of structural rearrangement in this region. We also found that the L2 loop and the distal P2 stem were more prone to cleavage in the presence of cyclic di-AMP, suggesting enhanced structural flexibility upon ligand binding. This was unexpected, as in-line probing analyses of ydaO riboswitches from different species had indicated that the P2 stem-loop was not differentially cleaved,2 and co-crystallization of several ydaO-like riboswitch sensor domains with cyclic di-AMP suggested that this region lacked ligand-binding residues.22-24
Figure 2.
The secondary structure model of the rpfA riboswitch. (A) Predicted secondary structures of unbound and cyclic di-AMP (CDA)-associated rpfA riboswitch RNA. RBS, ribosome binding site; SC, start codon. (B) 32P-5′-end-labeled rpfA riboswitch RNA was incubated in the absence of ligand (−) or in the presence of either cyclic di-AMP (CDA) or cyclic di-GMP (CDG) for ∼40 h at room temperature. Cleaved products were size-fractionated on a denaturing urea-polyacrylamide gel and visualized by autoradiography. Image is representative of 2 independent replicates. NR, untreated RNA; OH−, alkali-treated RNA; T1, RNase T1-treated RNA.
Our structural probing data, coupled with our bioinformatic analyses, suggested that the rpfA 5′ UTR folded into two mutually exclusive structures (Fig. 2A). In its cyclic di-AMP-associated ‘OFF’ state, the rpfA riboswitch adopted a structure in which (i) nucleotides 31 to 35 base-paired with nucleotides 216 to 220 to form the P1 stem, (ii) nucleotides 55 to 60 base-paired with nucleotides 68 to 73 to form the upper (interrupted) P2 stem, and (iii) nucleotides 182 to 189 (L7 loop) base-paired with nucleotides 221 to 227 to form a pseudoknot (Fig. 2A, top panel). However, in the absence of cyclic di-AMP binding, the distal L2-P2 stem sequence (nucleotides 65 to 74) paired with the distal P1 stem sequence (nucleotides 216 to 226), disrupting the pseudoknot (Fig. 2A, bottom panel).
Cyclic di-AMP promotes general transcript degradation
Whereas ydaO-like riboswitches studied to date have obvious expression platforms (e.g., a typical intrinsic terminator in B. subtilis,2,7 and Thermoanaerobacter tengcongensis,24 and a ribosome binding site-sequestering stem in Thermoanaerobacter pseudethanolicus),23 the rpfA riboswitch appeared to consist solely of the sensor domain (Fig. 2A). Because the rpfA 5′ UTR lacked structural features commonly associated with expression platforms regulating transcript elongation, we extended our search into the upstream region of the coding sequence. We noted a string of U residues within the first dozen nucleotides of the rpfA coding region, and reasoned that it may comprise the poly(U) tail of an unconventional intrinsic terminator. However, substituting the poly(U) sequence with an arbitrary sequence (Fig. S1) increased – rather than alleviated – riboswitch-dependent attenuation and had little impact on ligand binding (Fig. S2).
Given the lack of an obvious intrinsic terminator associated with the rpfA riboswitch, and that riboswitch activity is Rho-independent,10 we hypothesized that the riboswitch was regulating transcript stability by recruiting specific RNases to the mRNA in response to cyclic di-AMP binding. To determine if the riboswitch promoted transcript degradation upon associating with the second messenger, we conducted in vitro stability assays. We incubated free and ligand-bound riboswitch RNA with exponential-phase S. coelicolor cell-free lysate as a source of RNases, and monitored transcript degradation. We found that riboswitch-associated transcripts were degraded more rapidly when cyclic di-AMP was present than when the closely related second messenger cyclic di-GMP was added (cyclic di-GMP is not recognized by the riboswitch)10 (Fig. S3 and S4). Boiling the cell lysate with 20 mM ethylenediaminetetraacetic acid (EDTA) before incubation with riboswitch RNA abrogated degradation, as would be expected if RNases in the lysate were responsible for transcript turnover (Fig. S3 and S4).
To determine whether transcript degradation was specific for the ligand-bound riboswitch, or if it represented a more general destabilization phenomenon, we repeated our assays using (i) a fragment of the human 18S rRNA that was predicted to be unresponsive to cyclic di-AMP, and (ii) a panel of riboswitch mutants, including variants able to bind cyclic di-AMP (P2Prox.) and ones unable to bind the ligand (LIN and P3–7) (described in detail below). We reproducibly observed greater transcript degradation in the presence of cyclic di-AMP compared with cyclic di-GMP, irrespective of the ligand-binding status of the RNA (Fig. S3 and S4). This suggested that the effect of cyclic di-AMP on rpfA transcript stability was riboswitch-independent, and that the second messenger served an as yet unexplored function in modulating general transcript stability.
Cyclic di-AMP promotes premature transcription termination
Given the riboswitch was not specifically modulating rpfA transcript stability in response to cyclic di-AMP, we postulated that the non-coding RNA could be mediating attenuation by stabilizing an atypical transcription terminator. To determine whether the riboswitch promoted premature transcription termination, we evaluated the impact of cyclic di-AMP on transcription of the rpfA riboswitch using an in vitro transcription termination assay with a bacterial RNA polymerase. We noted the accumulation of a truncated product in transcription reactions performed in the presence, but not in the absence, of the second messenger (Fig. 3 and S5), suggesting that the rpfA riboswitch promoted premature termination in response to cyclic di-AMP. Using an RNA ladder, we mapped the termination point to a site within the first 30 nt of the rpfA coding region, consistent with our RNA-seq data.10 To rule out any direct effect of cyclic di-AMP on RNA polymerase activity, we repeated our assay using a series of ligand-unresponsive mutant riboswitches (described below). We found that cyclic di-AMP did not impact the transcription of any of these mutant riboswitch sequences (Fig. 3 and S5), suggesting that the second messenger regulated rpfA expression through its interaction with the riboswitch, not the RNA polymerase.
Figure 3.
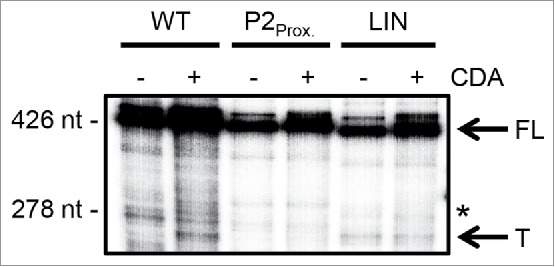
Cyclic di-AMP impact on rpfA transcript elongation by bacterial RNA polymerase. Wild-type (WT) and mutant (P2Prox., LIN) rpfA riboswitches were transcribed in vitro from an engineered B. subtilis lysC promoter using E. coli σ70-RNA polymerase. Reactions were supplemented with 0 µM (−) or 500 µM (+) cyclic di-AMP (CDA). Products were intrinsically labeled with 32P, size-fractionated on a denaturing urea-polyacrylamide gel and visualized by phosphorimaging. Image is representative of three biological replicates. The full-length gel image is presented in Fig. S5. FL, full-length transcripts; *, end of UTR(WT)-rpfA moiety (39 nt into the rpfA coding region); T, terminated transcripts.
Riboswitch-mediated attenuation requires cyclic di-AMP binding and the P2 stem
To further explore the mechanism underlying cyclic di-AMP-dependent regulation, we sought to identify the sequences and structures required for ligand-responsive riboswitch activity. We were particularly interested in identifying regions that contributed to gene regulation while being dispensable for cyclic di-AMP binding. To this end, we exploited our luciferase reporter assay to screen a panel of deletion and substitution riboswitch mutants (Fig. S1) for loss of attenuation during vegetative growth, when cyclic di-AMP levels were at their highest (Fig. 1B). To differentiate loss-of-function mutants that were unable to bind cyclic di-AMP, from ligand-binding mutants adopting a constitutively ‘ON’ state, we coupled these assays with in vitro ligand binding experiments.
We first modified regions of the rpfA riboswitch that in other ydaO-like riboswitch systems contribute to ligand binding and/or gene regulation, and confirmed the loss of ligand-responsive attenuation. Because stems P3 to P7 of other ydaO-like riboswitches contain approximately three-quarters of cyclic di-AMP-binding residues,22-24 we reasoned that deleting these regions from the rpfA riboswitch would relieve ligand-dependent attenuation. Consistent with our prediction, we noted a complete loss of ligand binding in vitro (Fig. 4) and a marked increase in rpfA expression in vivo (Fig. 5) upon deleting the majority of the sensor domain.
Figure 4.
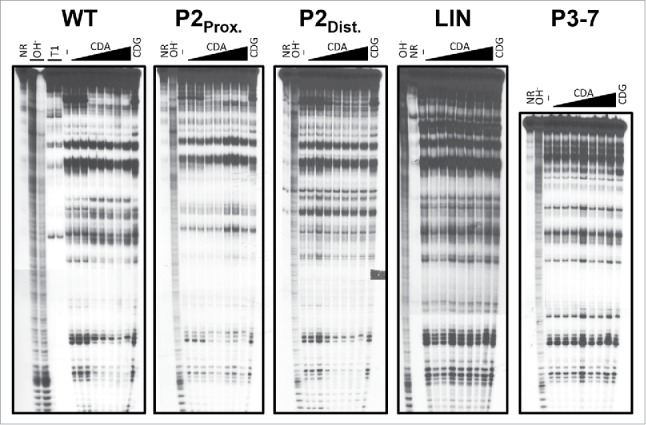
Cyclic di-AMP binding by wild-type and mutant rpfA riboswitches. 32P-5′-end-labeled wild-type and mutant rpfA riboswitch RNAs were incubated in the absence of ligand (−) or in the presence of either cyclic di-AMP (CDA) or cyclic di-GMP (CDG) for ∼40 h at room temperature. Cleaved products were size-fractionated on a denaturing urea-polyacrylamide gel and visualized by autoradiography. Each gel image is representative of two independent replicates. NR, untreated RNA; OH−, alkali-treated RNA; T1, RNase T1-treated RNA.
Figure 5.
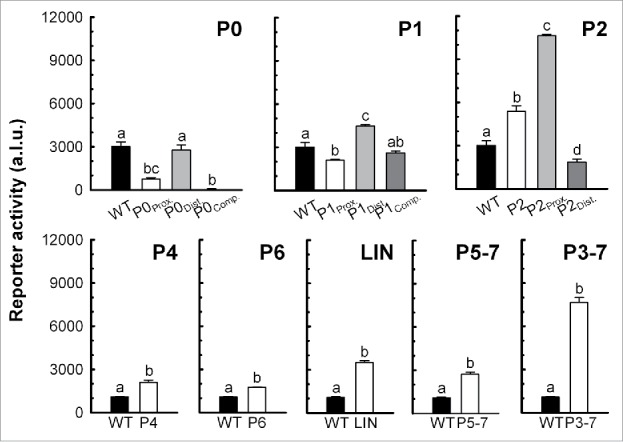
rpfA riboswitch activity during vegetative growth. S. coelicolor wild-type strains, constitutively expressing wild-type or mutant rpfA riboswitches transcriptionally fused to the luciferase reporter gene cluster, were grown on a sporulation-conducive solid medium. Background-subtracted reporter activity was measured after 44 h. Data are expressed as mean ± standard error (n = 3). Group means annotated with different letters are significantly different. P0: F(3,8) = 38.12, p < 0.001; P1: F(3,8) = 29.87, p < 0.001; P2: F(3,8) = 193.49, p < 0.001; P4: t(4) = −5.80, p = 0.004; P6: t(4) = −15.82, p < 0.001; LIN: t(4) = −17.42, p < 0.001; P5–7: t(4) = −10.15, p < 0.001; P3–7: t(4) = −18.01, p < 0.001. Figure is representative of at least 2 independent experimental replicates. a.l.u., arbitrary luminescence units; WT, M145/pMC222; P0Prox., M145/pMC232; P0Dist., M145/pMC233; P0Comp., M145/pMC234; P1Prox., M145/pMC235; P1Dist., M145/pMC236; P1Comp., M145/pMC237; P2, M145/pMC223; P2Prox., M145/pMC238; P2Dist., M145/pMC239; P4, M145/pMC224; P6, M145/pMC225; LIN, M145/pMC231; P5–7, M145/pMC226; P3–7, M145/pMC227.
We also examined the impact of substituting or deleting conserved stem-loop structures – P1, P4, P6, and P5 to P7 – on riboswitch activity. In Bacillus, mutating stem P4a residues impacts riboswitch activity,7 and destabilizing stem P6 abolishes cyclic di-AMP binding2 and increases expression.7 Furthermore, nucleotides within stems P1, P5 and P7, as well as residues within the P5-P6 junction, of different ydaO-like riboswitches interact directly with cyclic di-AMP.22-24 In B. subtilis, changes to the P7 stem sequence impede riboswitch-dependent attenuation,7 and substituting an unpaired adenine residue within the P5-P6 junction22 or altering the sequence of the P5 or P7 stem (with compensatory mutations maintaining structure)23 reduces or completely abolishes cyclic di-AMP binding. Consistent with these previous observations, we found that changing the P1 stem sequence impacted rpfA riboswitch activity, and deleting stem-loop P4, P6, or P5 to P7 significantly increased read-through in vivo (Fig. 5).
We also observed a loss of ligand binding (Fig. 4) and attenuation (Fig. 5) upon deleting LIN, consistent with previous works showing that cyclic di-AMP binding and gene regulation by the B. subtilis ydaO riboswitch rely on highly conserved nucleotides within the single-stranded region.2,22 These mutations also rendered the riboswitch unresponsive to cyclic di-AMP, as determined using our in vitro transcription termination system (Fig. 3 and S5). Our results suggested that, in contrast to our RNA stability experiments, cyclic di-AMP binding was essential for ligand-dependent inhibition of transcription.
Unlike other previously characterized ydaO-like riboswitches, the rpfA riboswitch has an elongated P2 stem-loop that undergoes substantial restructuring upon binding cyclic di-AMP (Fig. 2). As such, we hypothesized that the P2 stem-loop may be involved in ligand-responsive attenuation. To investigate this possibility, we deleted the entire P2 stem-loop and measured the attenuation activity of the resulting mutant riboswitch using our reporter assay. We noted a significant increase in gene expression (Fig. 5), suggesting that the P2 stem region was an important activity determinant.
To ascertain the means by which the P2 region impacted gene expression, we repeated our assays with a mutant riboswitch in which the proximal P2 stem region was substituted with its complementary sequence. We reasoned that these changes would result in constitutive read-through owing to the destabilization of the P2 stem and a corresponding increased likelihood of pairing between the distal P1 and P2 stem regions. Consistent with our prediction, we observed a dramatic increase in gene expression upon altering the proximal P2 stem sequence (Fig. 5). We confirmed our findings in vitro using our termination assay, and observed transcriptional read-through both in the presence and absence of cyclic di-AMP (Fig. 3 and S5). Interestingly, the substitution did not compromise ligand binding (Fig. 4), even though there was a substantial loss of structural modulation in the P2 stem region. This confirmed that the changes in expression were not owed to an inability to interact with the ligand, and instead suggested an altered expression platform.
We also evaluated the attenuation ability of a second mutant riboswitch in which we substituted the distal P2 stem region with its complementary sequence. We predicted that the substitution would prevent the stabilization of the ‘ON’ state in the absence of cyclic di-AMP owing to the loss of complementarity between the distal P1 and P2 stem regions. Consistent with this, we noted a significant decrease in gene expression upon mutating the distal P2 stem region (Fig. 5), though ligand binding was unaffected (Fig. 4). This further supported our proposal that the P2 stem was a key regulatory determinant in the rpfA riboswitch.
Folding of the rpfA riboswitch into its ‘ON’ state was predicted to involve pairing between nucleotides 56 to 59 (the proximal P2 stem region) and nucleotides 18 to 21 (the proximal P0 region; Fig. S1) (Fig. 2A). Therefore, we reasoned that any modification of the proximal P0 region would forcibly stabilize the P2 stem, locking the riboswitch in its ‘OFF’ state. We observed that substituting this region with its complementary sequence resulted in increased attenuation, and the impact of the proximal P0 region on riboswitch activity did not rely on a complementary sequence located downstream of the P1 stem (nucleotides 227 to 233; the distal P0 region; Fig. S1). Collectively, these data indicated that the P2 stem region was an integral component of the expression platform for the rpfA riboswitch.
An sRNA-encoding gene is located immediately downstream of the rpfA stop codon
In working to identify the transcription terminator associated with the riboswitch, we had taken advantage of RNA-seq data from S. coelicolor.25 In addition to a riboswitch-associated termination event that was apparent in the 5′ region of the rpfA transcriptional unit,10 there was evidence that an sRNA was expressed opposite the rpfA 3′ UTR. Given the proximity of this sRNA-encoding gene to rpfA (also known as sco3097), we termed this sRNA ‘Scr3097’. Scr3097 appeared to be an ∼80-nt transcript expressed from a gene located on the strand opposite rpfA, ending 10 bp downstream of the rpfA stop codon (Fig. 6A). Given the base-pairing potential between Scr3097 and the predicted rpfA terminator, we considered the possibility that Scr3097 affected rpfA expression.
Figure 6.
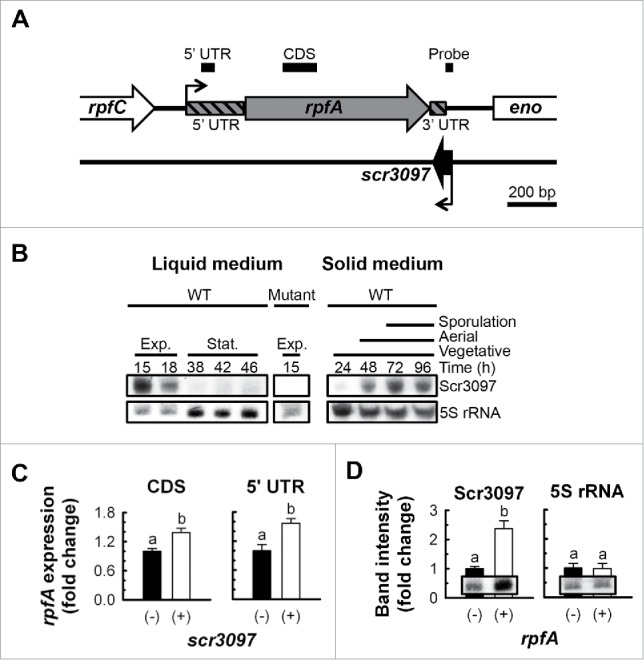
The antisense sRNA Scr3097. (A) Genetic context of scr3097 in S. coelicolor. Black elbow arrows denote the transcriptional start sites of rpfA and scr3097 as predicted using 5′ rapid amplification of cDNA ends9 and RNA-seq,25 respectively. Location of amplified PCR products (see panel C) and northern probes (see panel D) are shown as bold black lines above the block arrows. Genes, PCR products and northern probes were drawn to scale. (B) The wild-type S. coelicolor strain (WT, M145) and the rpfA-scr3097 deletion mutant (Mutant, E120/pIJ82; control) were grown at 30°C in a liquid rich medium or on a sporulation-conducive solid medium (MS agar). At the times indicated, total RNA was extracted from cells, and Scr3097 and 5S rRNA (control) transcripts were detected using northern blotting. Images are representative of 3 biological replicates. Exp., exponential phase; Stat., stationary phase. (C) scr3097− (E120/pMC241) and scr3097+ (E120/pMC243) strains of S. coelicolor were grown at 30°C in a rich liquid medium. Total RNA was extracted from exponential-phase cells, and rpfA transcripts were quantified by RT-qPCR using primer pairs targeting the coding sequence (CDS) or the 5′ UTR. Transcript levels were normalized to rpoB transcript levels and expressed as fold changes relative to normalized expression levels in scr3097− cells. Data are presented as mean ± standard error (n = 3). Group means annotated with different letters are significantly different. CDS: t(4) = −3.60, p = 0.023; UTR: t(4) = −3.56, p = 0.024. (D) rpfA− (E120/pMC242) and rpfA+ (E120/pMC243) strains were grown at 30°C in a rich liquid medium. Total RNA was extracted from exponential-phase cells, and Scr3097 and 5S rRNA (control) transcripts were detected using northern blotting. Transcripts were quantified using densitometry, and abundance was expressed relative to transcript levels in rpfA− cells. Data are presented as mean ± standard error (n = 3). Group means annotated with different letters are significantly different. Scr3097: t(4) = −2.84, p = 0.047; 5S rRNA: t(4) = 0.08, p = 0.941. Blot images are representative of 3 biological replicates.
We began our investigation by first determining how widespread scr3097 was among bacteria. We searched the nucleotide collection of the National Center for Biotechnology Information database for orthologous genes sharing sequence similarity using BLAST26 (January 2017), and discovered that the sRNA gene sequence was found in a range of streptomycetes in similar genetic contexts (Fig. S6). Interestingly, Streptomyces venezuelae lacked a sequence similar to that of scr3097 based on our sequence homology searches; however, a transcription start site was identified between rpfA and eno (encoding enolase) in the reverse orientation relative to both genes, and short ∼80-nt mature transcripts expressed from the rpfA-eno intergenic region accumulated in cells (Fig. S7), suggesting that the sRNA-encoding gene is likely more widely conserved than suggested by our in silico sequence similarity searches.
scr3097 is differentially expressed throughout growth
As a first step toward understanding the biologic function(s) of Scr3097, we monitored sRNA transcript levels in broth- and plate-grown wild-type S. coelicolor using northern blotting. We found that in liquid culture, Scr3097 transcripts were highly abundant in exponential-phase cells only; transcripts were not detectable once the culture transitioned into stationary phase (Fig. 6B). We observed a different abundance profile when S. coelicolor was grown on solid medium, with transcripts detected primarily during aerial development and sporulation (Fig. 6B). Similarly, in S. venezuelae, the sRNA-encoding gene promoter was most active during differentiation, with sRNA transcript levels peaking at this time (Fig. S7). Promoter activity and sRNA levels were comparatively low during vegetative growth (Fig. S7).
Scr3097 stimulates the expression of rpfA in exponential-phase cells
Given the base-pairing potential between Scr3097 and the rpfA 3′ UTR, we considered the likelihood that Scr3097 regulated rpfA expression. To address this possibility, we measured rpfA transcript abundance in scr3097− and scr3097+ strains by reverse transcription (RT)-coupled real-time PCR (qPCR) using a primer pair targeting the rpfA coding region. We found that rpfA transcripts were significantly more abundant in exponential-phase cells expressing the sRNA than in cells lacking scr3097 (Fig. 6C). In contrast, the presence of scr3097 had no significant impact on rpfA expression during stationary phase (data not shown), consistent with the fact that we did not detect Scr3097 during stationary phase (Fig. 6B). These results suggested that the sRNA positively affected rpfA transcript levels, and validated the proposal that Scr3097 activity contributed yet another level of regulation to rpfA expression and activity. To rule out any involvement of the rpfA riboswitch in sRNA-mediated regulation, we also quantified rpfA transcripts by RT-qPCR using a primer pair targeting the 5′ UTR. We noted a similar scr3097+-to-scr3097− rpfA expression ratio as when measured using the coding region-directed primer pair (Fig. 6C), confirming that the impact of Scr3097 on rpfA transcript levels did not involve the riboswitch.
To determine whether rpfA had any reciprocal effect on scr3097 expression, we measured Scr3097 transcript levels in both rpfA− and rpfA+ strains using northern blotting coupled with densitometric analyses. While sRNA expression was associated with increased rpfA transcript abundance, rpfA expression in turn resulted in increased Scr3097 transcript levels; 5S rRNA levels were unaffected by rpfA presence or absence (Fig. 6D).
Given the reciprocal regulation observed between Scr3097 and the rpfA 3′ UTR (Fig. 6), we were interested in probing the biological effect of Scr3097. Given that RpfA promotes S. coelicolor resuscitation,9,11 we hypothesized that Scr3097 accelerated spore germination and outgrowth by increasing rpfA expression. To examine the impact of scr3097 on spore germination kinetics, we monitored the optical density of spores suspended in a rich liquid medium, as the optical density of spores is known to decrease during the first stage of germination, characterized by the reactivation of endogenous metabolism, before increasing with germ tube emergence and outgrowth.27,28 We found that neither rpfA nor scr3097 dramatically impacted the initiation of spore germination (Fig. S8); however, efficient germ tube emergence and outgrowth, which typically began after 5–7 h, required both rpfA and scr3097 (Fig. S8). We found that spores of an rpfA-scr3097 deletion mutant germinated more slowly than those of the mutant complemented with rpfA and/or scr3097. Though complementing the mutant with rpfA alone accelerated outgrowth, maximum rates were noted only when the mutant was complemented with both rpfA and scr3097, suggesting Scr3097 promoted RpfA-dependent resuscitation. Interestingly, partial complementation was noted upon introducing scr3097 alone into the mutant, suggesting that Scr3097 may also regulate the expression of other genes to help compensate for the absence of RpfA.
We further examined the impact of scr3097 on sporulation and dormancy. Because RpfA promotes spore wall thickening,9,11 we reasoned that Scr3097-dependent stimulation of rpfA expression would increase spore wall thickness. We found that the spore wall of the rpfA-scr3097 deletion mutant was significantly thicker when complemented with an ectopic copy of rpfA (Fig. S8), consistent with the known biological function of this gene product.9,11 Unexpectedly, complementing the mutant with both rpfA and scr3097 had no noticeable effect on spore wall thickness (Fig. S8). This suggested that the activity of Scr3097 was epistatic to that of RpfA, and implied that Scr3097 may have other targets in the cell.
Discussion
In S. coelicolor, the rpfA gene is flanked by two non-coding regulatory RNA elements: (i) a cyclic di-AMP-responsive riboswitch motif within its 5′ UTR,2,7,9,10 and (ii) an sRNA-encoding gene immediately downstream of the rpfA coding region.25 Here, we have dissected the regulation of rpfA by each RNA element in the streptomycetes.
The rpfA riboswitch attenuates gene expression at a transcriptional level despite lacking a classical intrinsic terminator upstream of or near the rpfA start codon. Though the absence of such a staple of transcriptionally acting riboswitches could point to factor-dependent termination,29 our previous work suggested the rpfA riboswitch does not rely on Rho activity to attenuate gene expression.10 We nevertheless noted the production of a premature termination product in vitro, suggesting the rpfA riboswitch made use of an unusual terminator/antiterminator system to control gene expression. Transcription termination was not very strong, at least under our in vitro conditions, which suggested that the intrinsic terminator itself was not very strong, or that the Escherichia coli RNA polymerase used in our assays did not respond optimally to the S. coelicolor terminator. Terminator structures in the actinobacteria are poorly characterized. What little is known suggests a striking deviation from the canonical structure epitomized by the E. coli model [a GC-rich stem-loop followed by a U-rich tail]. In the mycobacteria30 and the streptomycetes,31,32 so-called ‘I-shaped’ intrinsic terminators (stable stem-loops lacking a U-rich tail), tandem stem-loops and multi-stem structures also trigger transcription termination. Given our evidence for transcriptional control, it is possible that the complex secondary structure of the cyclic di-AMP-bound rpfA riboswitch was sufficient to promote termination.
Though we were unable to identify the terminator structure proper, we discovered that a portion of the P2 stem-loop was crucial for attenuation activity. Indeed, cyclic di-AMP-mediated gene attenuation relied not only on the ability to interact with the ligand, but also on sequences within the P2 stem-loop. Though dispensable for binding, this region was crucial for ligand-dependent gene attenuation, both in vitro and in vivo. Our results suggested that at least a portion of the expression platform was found within the P2 stem-loop. Interestingly, the P2 stem region of the S. coelicolor rpfA riboswitch underwent prominent structural rearrangements in response to cyclic di-AMP, reflecting conformational changes within the expression platform.
When compared with other regions, particularly within the sensor domain, the P2 stem is poorly conserved, both in sequence and in length.2,7 It is interesting to note that the P2 stem is particularly elongated in the rpfA riboswitches from streptomycetes,10 being 4 to 9 times longer than the equivalent region in the characterized B. subtilis, C. acetobutylicum, S. aciditrophicus, T. pseudethanolicus and T. tengcongensis ydaO-like riboswitches.2,7,22-24 It seems that these systems compensate for the lack of an extended P2 stem with a separate expression platform, located downstream of the sensor domain.2,7,23,24 The P2 region has not, however, been subjected to mutational analyses in these other systems, and thus it remains possible that it also contributes to regulation in other ydaO-like riboswitches.
In addition to regulating transcript elongation, cyclic di-AMP may also influence rpfA transcript stability, albeit in a riboswitch-independent fashion. rpfA transcripts were more readily degraded in vitro by RNases in the presence of cyclic di-AMP than in the presence of cyclic di-GMP. Because (i) ligand binding was dispensable for degradation, and (ii) unrelated transcripts were also destabilized by the second messenger, cyclic di-AMP may impact general RNA stability in the streptomycetes, possibly via allosteric modulation of one or more RNases. With cyclic di-AMP levels decreasing throughout differentiation, and rpfA transcript levels concomitantly rising9,11 through both reduced riboswitch-mediated attenuation and increased transcript stability, the streptomycetes ultimately amass stores of rpfA mRNAs during sporulation. These transcripts may enable rapid protein synthesis during spore germination, when RpfA is required for resuscitation.9,11
Cyclic di-AMP was detected in both vegetative and differentiating cultures of S. coelicolor, suggesting that the riboswitch attenuated rpfA expression throughout growth, albeit to different extents. Because rpfA is required for proper spore germination,9,11 it is likely that cyclic di-AMP levels in germinating spores are very low to allow efficient rpfA transcription and mRNA accumulation. Indeed, rpfA promoter activity peaks during spore germination and outgrowth, and both rpfA transcripts and RpfA proteins are abundant in early exponential phase/vegetative growth.10,11 Though cyclic di-AMP levels were not measured at that time, owing to insufficient biomass for nucleotide extraction and quantification, others have noted a lack of cyclic di-AMP in S. venezuelae during the earliest stages of growth (N. Tschowri, personal communication).
In addition to negative regulation by the riboswitch, we have shown that rpfA expression was also positively affected by the antisense sRNA Scr3097. Given the base-pairing potential between Scr3097 and the 3′ end of the rpfA transcript, Scr3097 may have promoted increased rpfA transcript stability. Though less common than 5′ UTR-associated sRNAs, 3′ UTR-binding sRNAs are not unprecedented. In E. coli, the antisense sRNA GadY binds the 3′ UTR of gadX, and recruits the endoribonuclease RNase III. Subsequent cleavage of the resulting RNA duplex serves to stabilize the gadX transcript.33,34 While we do not have evidence of RNase recruitment by Scr3097, the sRNA did enhance rpfA transcript levels. Alternatively, given that Scr3097 was predicted to target the rpfA intrinsic terminator, it is also possible that the sRNA impacted transcription termination. In Vibrio anguillarum, the antisense sRNA RNAß promotes transcription termination downstream of fatA upon binding the 3′ end of the open reading frame and 3′ UTR.35
We had previously shown rpfA transcription to be controlled by the cyclic AMP-binding protein Crp, and RpfA protein levels to be modulated by a guanosine 5′-diphosphate-3′-diphosphate-dependent protease.10 The riboswitch and the antisense sRNA Scr3097 both regulate rpfA expression, adding yet another layer of control to the current model of rpf gene regulation (Fig. 7). During spore germination, when RpfA is required for resuscitation,9,11 levels of the dormancy-promoting second messenger cyclic di-AMP are predicted to be low. This would allow for efficient riboswitch read-through and rpfA expression. At the same time, the sRNA Scr3097 is expressed, enhancing rpfA mRNA levels. Once resuscitation is complete and normal vegetative growth has initiated, cyclic di-AMP accumulates within cells. This leads to the attenuation of rpfA expression. At the same time, Scr3097 levels drop, collectively contributing to decreased transcript and protein abundance. During sporulation, intracellular cyclic di-AMP levels decrease, and this is coupled with a modest increase in rpfA transcript levels.9,11 Increasing levels of Scr3097 further enhance rpfA mRNA accumulation.
Figure 7.
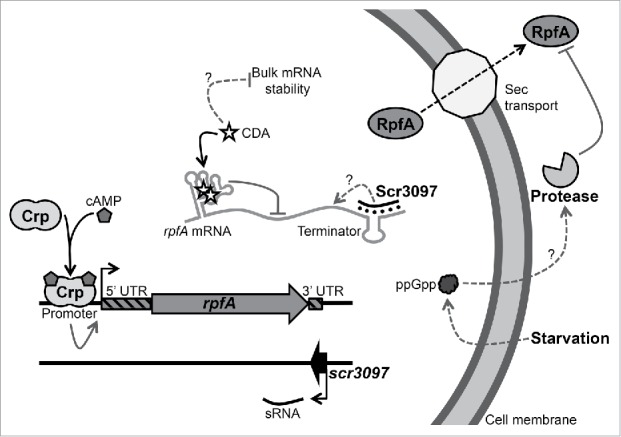
Model for rpfA regulation in the streptomycetes. rpfA is regulated at multiple levels by both protein and RNA regulators. The cyclic AMP (cAMP) receptor protein (Crp) activates rpfA transcription initiation in response to the second messenger cAMP during germination and outgrowth.10 A riboswitch prematurely terminates rpfA transcription in response to the second messenger cyclic di-AMP during vegetative growth. The sRNA Scr3097 increases rpfA transcript abundance during exponential phase (liquid growth) and sporulation (solid growth) through an as yet-to-be-determined mechanism. A secreted metalloprotease degrades RpfA proteins in the growth medium in response to the second messenger guanosine 5′-diphosphate-3′-diphosphate (ppGpp) and nutrient starvation.10 Arrows indicate positive regulation, whereas right-angle lines indicate negative regulation. Dotted lines indicate indirect regulation.
Here, we have shown that, in addition to proteins regulating rpfA promoter activity and RpfA protein stability,10 two non-coding regulatory RNAs – a cyclic di-AMP-responsive riboswitch and an antisense sRNA – influence rpfA expression post-transcriptionally. Such exquisite multi-level regulation of rpfA expression would enable the streptomycetes to enter dormancy, and initiate resuscitation, in response to specific environmental cues. These regulatory systems likely extend beyond the rpfs, to other muralytic enzyme-encoding genes, the expression of which must be tightly governed.
Materials and methods
In silico analyses
BLAST26 was used to search the genome collection of the National Center for Biotechnology Information database for sequences similar – and syntenous - to scr3097 (January 2017). Select sequences were aligned using Clustal Omega.36 sRNA secondary structures were predicted using Mfold,37 and the consensus structure was predicted using RNAalifold.38 Intrinsic terminators were predicted using WebGeSTer.39
Growth conditions
S. coelicolor strains (Table S1) were grown at 30°C on MYM agar40 or in tryptone soya broth (TSB) (Oxoid):YEME41 (50:50). When appropriate, agar plates were overlaid with sterile cellophane disks before inoculation to facilitate the recovery of Streptomyces biomass. E. coli strains (Table S1) were grown at 30°C (BW25113 derivatives) or 37°C (DH5α and ET12567/pUZ8002 derivatives) in SOB (without Mg2+ salts) and LB broth,42 and on nutrient (Difco) and LB agar (LB broth with 1.5%w/v agar). To select for mutations, plasmids and cosmids, media were supplemented with one or more of the following antibiotics: 100 µg mL−1 ampicillin, 50 µg mL−1 apramycin, 25 µg mL−1 chloramphenicol, 50 µg mL−1 hygromycin B, 50 µg mL−1 kanamycin, 25 µg mL−1 nalidixic acid.
Cyclic di-AMP extraction and quantification
S. coelicolor strain M145 (Table S1) was grown at 30°C on MYM agar overlaid with cellophane. After 24, 48 and 72 h, cyclic di-AMP was extracted from cells using a protocol adapted from Spangler, et al.43 Briefly, biomass (264 ± 42 mg) was recovered from the plate culture and homogenized in 3 mL ice-cold extraction solvent (methanol:acetonitrile:water, 40:40:20). The homogenate was incubated on ice for 15 min and then heated at ∼95°C for 10 min. After cooling on ice for 5 min, the homogenate was centrifuged at 1,900 × g for 10 min at 4°C, and the supernatant was collected. The pellet was extracted twice more using 2 mL ice-cold extraction solvent, as described above but omitting the heating and subsequent cooling steps. The recovered extract was placed at −20°C for at least 24 h, and centrifuged at 1,900 × g for 10 min at 4°C. The supernatant was retrieved and evaporated to dryness using a Genevac HT-4X (SP Scientific). The resulting residue was resuspended in 200 µL water, and cyclic di-AMP contained therein was quantified by reverse-phase LC-MS/MS, as described previously.43 Levels were normalized to wet cell weight.
Deletion of rpfA and scr3097
The rpfA-scr3097 gene locus of S. coelicolor (−277 to +864, numbering relative to the rpfA start codon) (Fig. S9) was deleted and replaced with the apramycin resistance cassette aac(3)IV using a PCR-directed gene replacement strategy optimized for use in the streptomycetes.44 Briefly, a 1,460-bp extended knockout cassette, comprising aac(3)IV bordered on either side by 39 bp of DNA flanking the region to be deleted, was PCR-amplified using Phusion High-Fidelity DNA polymerase (Thermo) with 5%v/v dimethylsulfoxide (DMSO), long primers SCO3097KOF and DBLKOR (Table S2), and aac(3)IV, excised from plasmid pIJ773 (Table S1) using restriction enzymes EcoRI and HindIII, as template. Optimized amplification conditions are detailed in Table S3. The PCR products were gel-purified using the MicroElute Gel Extraction kit (Omega BioTek).
The extended knockout cassette was electroporated into the recombinogenic E. coli strain BW25113/pIJ790 (Table S1) carrying the S. coelicolor cosmid StE41 (Table S1), in which a double crossover event replaced the wild-type locus with aac(3)IV by homologous recombination.44 Cosmid DNA was extracted from an apramycin-resistant transformant using standard methods.42 Isolated cosmid DNA was then electroporated into the methylation-deficient E. coli donor strain ET12567/pUZ8002 (Table S1) and mobilized into S. coelicolor strain M145 by conjugation, as described by Gust, et al.44 Apramycin-resistant, kanamycin-sensitive double crossover exconjugants were isolated, and their genotype was confirmed by PCR-amplifying the wild-type and mutant loci using Taq DNA polymerase (GeneDireX) with 5%v/v DMSO, 300 nM each appropriate primer (Tables S2 and S3) and crude cell lysate as template (Fig. S9).
Directed mutagenesis of the rpfA riboswitch
Nucleotide changes were introduced into the rpfA riboswitch, fused to the 3′ end of the ermE* promoter, using splicing by overlap extension (SOE)-PCR.45 The forward mutagenic primers were used with rpfAR2-K to amplify the downstream fragment of the target, whereas the reverse mutagenic primers were used with ermEF to amplify the corresponding upstream fragment (Tables S2 and S3). A 17-nt sequence – incorporating the modifications to be introduced into the riboswitch construct – was added to the 5′ end of each forward mutagenic primer. To promote the annealing of both the upstream and downstream products, the complementary mutations were also engineered into the 5′ end of each reverse mutagenic primer. Primer sequences are presented in Table S2.
Upstream and downstream overlapping products were PCR-amplified from plasmid pMC230 (Table S1) using Phusion High-Fidelity DNA polymerase (Thermo, NEB) with 5%v/v DMSO and the appropriate primer pairs (Table S3). Optimized amplification conditions and product lengths are indicated in Table S3. Products were column-purified using the PureLink PCR Purification kit (Invitrogen). The upstream and downstream products were then mixed, annealed together and extended by PCR using Phusion High-Fidelity DNA polymerase with external primers ermEF and rpfAR2-K, as described above (Table S3). The SOE-PCR products were gel-purified.
Compensatory mutant riboswitches P0Comp. and P1Comp. were engineered using a similar strategy, by introducing the P0Dist. and P1Prox. mutations into plasmids pMC245 and pMC236 (Table S1), respectively. P0Dist. and P1Prox. upstream and downstream fragments were thus PCR-amplified from plasmids pMC245 and pMC236, respectively.
Cloning of complementation and reporter constructs
DNA regions used for complementing the rpfA-scr3097 deletion mutant (rpfA, −504 to +816; scr3097, +457 to +1,046; the entire rpfA-scr3097 locus, −504 to +1,046; numbering relative to the rpfA start codon) (Fig. S10) were PCR-amplified using Phusion High-Fidelity DNA polymerase with 5%v/v DMSO, the appropriate primer pair (Table S3) and cosmid StE41 as template. Restriction sites were engineered into the 5′ end of each forward and reverse primer (Table S2). Primer pairs used, amplification conditions and product lengths are presented in Table S3. PCR products were column-purified.
rpfA, scr3097 and rpfA-scr3097 PCR products were digested with restriction enzymes BamHI and XbaI, while mutant rpfA riboswitch SOE-PCR products were digested with BglII and KpnI. Digested products were then column-purified and ligated to dephosphorylated, BamHI- and XbaI-digested plasmid pIJ82 (Table S1) (complementation) or dephosphorylated, BamHI- and KpnI-digested plasmid pFLUX (Table S1) (reporter) using T4 DNA ligase (Invitrogen). Recombinant plasmids were then electroporated into E. coli strain DH5α (Table S1), extracted, and sequenced to confirm insert sequences. Plasmids were electroporated into E. coli strain ET12567/pUZ8002 and mobilized into S. coelicolor strain E120 (Table S1) (complementation) or M145 (reporter) by conjugation. Successful plasmid integration was confirmed by colony PCR using Taq DNA polymerase with 5%v/v DMSO, 300 nM each appropriate primer (Table S3) and crude cell lysate as template (Fig. S10).
Total RNA extraction
Exponential-phase mycelia of S. coelicolor strain E120, complemented with or without an ectopic copy of rpfA and/or scr3097, were inoculated into TSB:YEME (50:50) at a concentration of 1.6 mg wet weight mL−1, and cultures were incubated for 15 and 47 h at 30°C. Cells were collected from exponential- and stationary-phase cultures by centrifugation (1,900 × g for 10 min at 4°C) and frozen at −80°C for ∼1 month. Total RNA was extracted from cells as described previously,25 and co-extracted DNA was completely digested using TURBO DNase (Ambion). Isolated RNA was quantified using an ND-1000 spectrophotometer (NanoDrop Technologies). RNA integrity was confirmed by size-fractionating 1.6 µg total RNA on a 2%w/v agarose gel at 100 V for 20 min in 1 × TBE buffer,42 and extract purity was verified by measuring the A260/A280 and A260/A230 ratios (1.86–2.03 and 2.14–2.34, respectively). Total RNA from wild-type S. coelicolor (M145) grown at 30°C in TSB:YEME (50:50) and on MS agar was previously isolated by Sexton, et al.11
Northern blotting
Scr3097- and 5S rRNA-specific probes were prepared by 32P-5′-end-labeling 0.1 µM ssDNA oligonucleotides (Table S2) with 0.5 U µL−1 T4 polynucleotide kinase (Fermentas, Invitrogen, NEB) using 250 nM [γ-32P]ATP (Perkin Elmer). After 30 min at 37°C, unincorporated nucleotides were removed using the PureLink PCR Purification kit or NucAway Spin Columns (Ambion).
DNase-treated total RNA (20–25 µg) was heated at ∼95°C for 5 min in RNA gel-loading buffer42 or Gel Loading Buffer II (Ambion), and size-fractionated at 200 V for 30–40 min on a continuous 6%w/v denaturing urea-polyacrylamide gel (BioShop, National Diagnostics) in 1 × TBE buffer using a Mini-PROTEAN Tetra Cell (Bio-Rad). RNA was then blotted onto a Zeta-Probe nylon blotting membrane (Bio-Rad) at 15 V for 40 min in 0.5 × TBE buffer using a Trans-Blot SD Semi-Dry Electrophoretic Transfer Cell (Bio-Rad). RNA was cross-linked to the membrane by baking the membrane at 55°C for 2 h with EDC cross-linking solution, as described previously.46 32P-5′-end-labeled probe complementary to the transcript of interest (0.4–2.3 pmol) was denatured at ∼95°C for 5 min, rapid-cooled on ice and then incubated with the membrane overnight at 42°C in ULTRAhyb-Oligo Hybridization Buffer (Ambion). Unbound probe was removed by washing the membrane 2–4 times for 2–5 min each at 42°C in 2 × SSC with 0.1%w/v sodium dodecyl sulfate (SDS).42 When appropriate, the membrane was also washed once at 42°C for 2–3 min in 0.2 × SSC with 0.1%w/v SDS.42 Bound transcripts were visualized by autoradiography or using a Typhoon FLA 9500 phosphorimager (GE Healthcare). Membranes were stripped of the probe by incubating the membrane for 40 min at 60–70°C in 0.2 × SSC with 0.1%w/v SDS. Successful removal of the probe was confirmed by autoradiography or phosphorimaging.
Reverse transcription and real-time PCR
rpfA and rpoB (RNA polymerase ß subunit) transcripts extracted from scr3097− and scr3097+ strains (E120/pMC241 and E120/pMC243, respectively; Table S1) were reverse transcribed using SuperScript III reverse transcriptase (Invitrogen) with 100 nM gene-specific reverse primers (Tables S2 and S3) and 100 ng µL−1 total RNA. ‘No RT’ negative controls were prepared as above, but without adding the enzyme. cDNA was used immediately, or stored at −20°C and used the next day.
cDNA was then amplified by qPCR using PerfeCTa SYBR Green SuperMix (Quanta Biosciences) with 300 nM each primer (Tables S2 and S3) and 12.5 ng µL−1 reverse transcribed total RNA. ‘No template’ controls, in which an equal volume of nuclease-free water replaced the cDNA template, were prepared in parallel and included in each run. Singleplex reactions were conducted, in triplicate, in clear 96-well PCR plates (Thermo) with a CFX96 Touch Real-Time PCR Detection System (Bio-Rad). The amplification conditions recommended by the manufacturer (Quanta Biosciences) were used with an optimized annealing/extension temperature (Table S3). A melt curve analysis (65–95°C with 5-sec fluorescence reads every 0.5°C increase) was conducted at the end of each run. Collected fluorescence data were baseline-corrected using the CFX Manager software (Bio-Rad), and transcript levels were calculated using the DART-PCR workbook.47 Levels of rpfA transcripts were normalized to those of the stably expressed endogenous reference gene rpoB.
Reporter assays
Fifteen microliters of S. coelicolor spore suspension [OD450 = 0.150, in 2 × YT broth]41 were spread, in triplicate, on 200 µL MYM agar in white flat-bottomed 96-well plates (Thermo). Plate cultures were incubated at 30°C, and the emitted luminescence was periodically measured for 2 sec using a TECAN UltraEvolution spectrometer. Luminescence emitted by the empty vector control (M145/pFLUX; Table S1) was subtracted from the raw luminescence of each sample. When appropriate, background-subtracted luminescence was normalized to that of the positive control strain constitutively expressing the reporter without the rpfA riboswitch (M145/pFLUX-Pos; Table S1).
In vitro transcription, RNA purification and 5′-end-labeling
In vitro transcription templates comprised phage T7 promoter-driven wild-type or mutant rpfA riboswitch fused to the first 39 bp of the rpfA coding region. Templates were PCR-amplified from recombinant pFLUX plasmids (Table S1) using Phusion High-Fidelity DNA polymerase with 5%v/v DMSO and primers IVTrpfAUTR5′ and IVTrpfAUTR3′ (Tables S2 and S3). The phage T7 RNA polymerase promoter sequence was engineered into the 5′ end of the forward primer IVTrpfAUTR5′, and 2 G residues were inserted between the promoter and the priming sequence to increase transcript yields. Products were subsequently column-purified. Plasmid pTRI-RNA 18S (Table S1) was used as template for 18S rRNA synthesis.
RNA was transcribed in vitro using the MEGAshortscript High Yield Transcription kit (Ambion). Briefly, a 40- or 80-µL reaction, comprising 1 × T7 Reaction Buffer, 7.5 mM each NTP, 50 nM template DNA and 4 or 8 µL T7 Enzyme Mix, was incubated at 37°C for 4 h. The template DNA was subsequently digested for 1 h at 37°C using 8 or 16 U TURBO DNase.
In vitro transcribed RNA was denatured at 95°C for 5 min in RNA gel-loading buffer or Gel Loading Buffer II, and immediately size-fractionated on a 50-cm-long continuous 6%w/v denaturing urea-polyacrylamide gel in 1 × TBE buffer for 70–90 min at 60 W using the Sequi-Gen GT Nucleic Acid Electrophoresis Cell (Bio-Rad). RNA was visualized by UV shadowing, excised from the gel and extracted 3 times at 37°C with 350–500 µL crush/soak buffer, as described previously.48 Gel-extracted RNA was then precipitated,42 resuspended in nuclease-free water and quantified using spectrophotometry.
Using the KinaseMax kit (Ambion), 20 pmol gel-purified RNA were subsequently dephosphorylated with calf intestine alkaline phosphatase and 5′-end-labeled with T4 polynucleotide kinase and [γ-32P]ATP. Unincorporated NTPs were removed from the reaction using NucAway Spin Columns.
In-line probing
32P-5′-end-labeled riboswitch RNA was re-folded by heating at 95°C for 2 min and cooling to room temperature for 10 min. Approximately 100 nM RNA was then incubated at room temperature for ∼40 h in 1 × in-line reaction buffer.48 When appropriate, reactions were supplemented with 1 nM to 1 mM cyclic di-AMP or cyclic di-GMP (Invivogen). RNA cleavage products were diluted in an equal volume of 2 × colorless gel-loading solution48 and size-fractionated on a denaturing urea-polyacrylamide gel, as for RNA gel purification. Undigested, alkaline-treated and RNase T1-treated precursor RNAs, prepared as described previously,48 were also size-fractionated alongside the digested RNA. The gel was dried at 80°C for 2 h using a Model 583 Gel Dryer (Bio-Rad), and RNA was visualized by autoradiography.
In vitro stability assays
Exponential-phase cells of S. coelicolor strain M145 (2.52 ± 0.83 g wet cells), grown at 30°C in TSB:YEME (50:50), were collected by centrifugation at 1,900 × g for 10 min. Cells were resuspended in 50 mL lysis buffer (10 mM Tris, 0.1 M NaCl, 5%v/v glycerol) and lysed at 30,000 psi using a Constant Cell Disruption System with a continuous-flow head (Constant Systems). Lysates were clarified by centrifugation at 15,000 × g for 10 min (4°C). When appropriate, RNases were inactivated by adding 20 mM EDTA to the lysate and heating the mixture at 95°C for 15 min.
32P-5′-end-labeled riboswitch RNA was re-folded by heating at 95°C for 2 min and cooling to room temperature for 10 min. To enable binding to the ligand, ∼0.1 µM labeled RNA was incubated at room temperature for 60–70 min with 751 µM cyclic di-AMP or cyclic di-GMP in 1 × in-line reaction buffer (final concentrations are indicated). RNA degradation was initiated immediately by adding 0.1 µL cell lysate µL−1 reaction or an equal volume of lysis buffer (‘0 min’ samples). The reaction was incubated at ∼30°C, and aliquots were taken periodically over the course of 90 min and flash-frozen in liquid nitrogen. RNA degradation products were diluted in an equal volume of 2 × colorless gel-loading solution and size-fractionated on a denaturing urea-polyacrylamide gel, as for RNA gel purification. The gel was dried as described above and visualized by phosphorimaging. RNA was quantified by densitometry using ImageQuant TL (GE Healthcare), and transcript abundance was expressed relative to untreated RNA (‘0 min’).
In vitro transcription termination assays
Templates for in vitro transcription termination assays comprised wild-type and mutant rpfA riboswitches fused to the first 63 bp of the rpfA coding sequence and 126 bp of pFLUX vector DNA, with sequences placed under the transcriptional control of the B. subtilis lysC promoter.2 DNA templates were PCR-amplified from recombinant pFLUX derivatives using Phusion High-Fidelity DNA polymerase with 5%v/v DMSO and the appropriate primer pair (Tables S2 and S3). Products were subsequently gel-purified.
Transcription from the lysC promoter was initiated by mixing the template with 1 × E. coli RNA Polymerase Reaction Buffer (NEB), 0.4 µCi µL−1 [α-32P]CTP (Perkin Elmer), 3 µM each GTP, CTP and UTP (NEB), 0 or 500 µM cyclic di-AMP, and 0.1 U µL−1 E. coli σ70-RNA polymerase (NEB) (final concentrations are indicated). After 10 min at 37°C, 50 U mL−1 heparin (prevents de novo transcription initiation) and 3 µM ATP (NEB) were added to the reaction. Transcript elongation continued at 37°C for 30 min, after which time the reaction was stopped by adding an equal volume of 2 × colorless gel-loading solution. Transcripts were then size-fractionated on a denaturing urea-polyacrylamide gel, as for RNA gel purification. The gel was dried and visualized by phosphorimaging, as described above.
Germination assays
Four hundred microliters of S. coelicolor spore suspension [OD450 ∼0.6, in TSB:YEME (50:50)] were deposited, in quadruplicate, in a clear flat-bottomed 48-well plate (Thermo). Cultures were incubated at 30°C with constant shaking. The OD450 was measured every 30 min for 24.5 h using a Cytation3 Cell Imaging Multi-Mode Reader (BioTek) (8 measurements per data point). The OD450 of uninoculated medium was subtracted from the OD450 of each sample at each time-point.
Transmission electron microscopy
Spore wall thickness was measured using ImageJ49 from transmission electron micrographs of sporulating S. coelicolor colonies, grown for 7 d at 30°C on MYM agar. Samples were prepared and viewed as described by Haiser, et al.9
Statistical analyses
Unpaired t-tests were performed to test for significant differences between (1) levels of reporter activity measured in strains expressing the luciferase reporter genes fused with wild-type or mutant rpfA riboswitch, (2) rpfA mRNA levels in the scr3097− and scr3097+ strains, and (3) Scr3097 and 5S rRNA levels in the rpfA− and rpfA+ strains. To investigate (1) the impact of rpfA riboswitch mutations on gene expression, and (2) the effect of strain genotype on spore wall thickness, a univariate one-way analysis of variance was performed with pairwise multiple comparisons conducted using the Holm-Šidák method. For all statistical tests, normality and homoscedasticity were confirmed using the Shapiro-Wilk test and the Levene Median test, respectively. To meet the assumptions of the parametric test, relative fold changes of Scr3097 levels in rpfA− and rpfA+ strains were first power-transformed (x3). p values ≤ 0.05 were deemed statistically significant. All analyses were performed using SigmaPlot v11.0 (Systat Software).
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors wish to thank Marcia Reid (McMaster University, Hamilton, Canada) and Dr. Volkhard Kaever (Hannover Medical School, Hannover, Germany) for their technical assistance, and Drs. Tina Henkin (Ohio State University, Columbus, USA), Daniel Lafontaine (Université de Sherbrooke, Sherbrooke, Canada) and Wade Winkler (University of Maryland, College Park, USA) for helpful discussions.
Funding
This work was supported by an NSERC Discovery Grant (No. 04681) and an NSERC Discovery Accelerator supplement to M.A.E. R.J.S. was supported by a Vanier Canada Graduate Scholarship.
References
- 1.Serganov A, Nudler E. A decade of riboswitches. Cell. 2013;152:17–24. doi: 10.1016/j.cell.2012.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nelson JW, Sudarsan N, Furukawa K, Weinberg Z, Wang JX, Breaker RR. Riboswitches in eubacteria sense the second messenger c-di-AMP. Nat Chem Biol. 2013;9:834–9. doi: 10.1038/nchembio.1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oppenheimer-Shaanan Y, Wexselblatt E, Katzhendler J, Yavin E, Ben-Yehuda S. c-di-AMP reports DNA integrity during sporulation in Bacillus subtilis. EMBO Rep. 2011;12:594–601. doi: 10.1038/embor.2011.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luo Y, Helmann JD. Analysis of the role of Bacillus subtilis σM in β-lactam resistance reveals an essential role for c-di-AMP in peptidoglycan homeostasis. Mol Microbiol. 2012;83:623–39. doi: 10.1111/j.1365-2958.2011.07953.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehne FMP, Gunka K, Eilers H, Herzberg C, Kaever V, Stülke J. Cyclic di-AMP homeostasis in Bacillus subtilis: Both lack and high level accumulation of the nucleotide are detrimental for cell growth. J Biol Chem. 2013;288:2004–17. doi: 10.1074/jbc.M112.395491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corrigan RM, Abbott JC, Burhenne H, Kaever V, Gründling A. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 2011;7:e1002217. doi: 10.1371/journal.ppat.1002217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Block KF, Hammond MC, Breaker RR. Evidence for widespread gene control function by the ydaO riboswitch candidate. J Bacteriol. 2010;192:3983–9. doi: 10.1128/JB.00450-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–28. doi: 10.1016/j.cell.2009.01.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haiser HJ, Yousef MR, Elliot MA. Cell wall hydrolases affect germination, vegetative growth, and sporulation in Streptomyces coelicolor. J Bacteriol. 2009;191:6501–12. doi: 10.1128/JB.00767-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.St-Onge RJ, Haiser HJ, Yousef MR, Sherwood E, Tschowri N, Al-Bassam M, Elliot MA. Nucleotide second messenger-mediated regulation of a muralytic enzyme in Streptomyces. Mol Microbiol. 2015;96:779–95. doi: 10.1111/mmi.12971 [DOI] [PubMed] [Google Scholar]
- 11.Sexton DL, St-Onge RJ, Haiser HJ, Yousef MR, Brady L, Gao C, Leonard J, Elliot MA. Resuscitation-promoting factors are cell wall-lytic enzymes with important roles in the germination and growth of Streptomyces coelicolor. J Bacteriol. 2015;197:848–60. doi: 10.1128/JB.02464-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dworkin J, Shah IM. Exit from dormancy in microbial organisms. Nat Rev Microbiol. 2010;8:890–6. doi: 10.1038/nrmicro2453 [DOI] [PubMed] [Google Scholar]
- 13.Rittershaus ESC, Baek S-H, Sassetti CM. The normalcy of dormancy: Common themes in microbial quiescence. Cell Host Microbe. 2013;13:643–51. doi: 10.1016/j.chom.2013.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anuchin AM, Mulyukin AL, Suzina NE, Duda VI, El-Registan GI, Kaprelyants AS. Dormant forms of Mycobacterium smegmatis with distinct morphology. Microbiology. 2009;155:1071–9. doi: 10.1099/mic.0.023028-0 [DOI] [PubMed] [Google Scholar]
- 15.Shleeva MO, Kudykina YK, Vostroknutova GN, Suzina NE, Mulyukin AL, Kaprelyants AS. Dormant ovoid cells of Mycobacterium tuberculosis are formed in response to gradual external acidification. Tuberculosis (Edinb). 2011;91:146–54. doi: 10.1016/j.tube.2010.12.006 [DOI] [PubMed] [Google Scholar]
- 16.Elliot MA, Flärdh K. Streptomycete spores. Encyclopedia of Life Sciences (eLS). 2012. doi: 10.1002/9780470015902.a0000308.pub2 [DOI] [Google Scholar]
- 17.Mukamolova GV, Yanopolskaya ND, Votyakova TV, Popov VI, Kaprelyants AS, Kell DB. Biochemical changes accompanying the long-term starvation of Micrococcus luteus cells in spent growth medium. Arch Microbiol. 1995;163:373–9. doi: 10.1007/BF00404211 [DOI] [Google Scholar]
- 18.Puspita ID, Uehara M, Katayama T, Kikuchi Y, Kitagawa W, Kamagata Y, Asano K, Nakatsu CH, Tanaka M. Resuscitation promoting factor (Rpf) from Tomitella biformata AHU 1821T promotes growth and resuscitates non-dividing cells. Microbes Environ. 2013;28:58–64. doi: 10.1264/jsme2.ME12122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradley SG, Ritzi D. Composition and ultrastructure of Streptomyces venezuelae. J Bacteriol. 1968;95:2358–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glauert AM, Hopwood DA. The fine structure of Streptomyces violaceoruber (S. coelicolor). III. The walls of the mycelium and spores. J Biophys Biochem Cytol. 1961;10:505–16. doi: 10.1083/jcb.10.4.505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kana BD, Mizrahi V. Resuscitation-promoting factors as lytic enzymes for bacterial growth and signaling. FEMS Immunol Med Microbiol. 2010;58:39–50. doi: 10.1111/j.1574-695X.2009.00606.x [DOI] [PubMed] [Google Scholar]
- 22.Jones CP, Ferré-D'Amaré AR. Crystal structure of a c-di-AMP riboswitch reveals an internally pseudo-dimeric RNA. EMBO J. 2014;33:2692–703. doi: 10.15252/embj.201489209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao A, Serganov A. Structural insights into recognition of c-di-AMP by the ydaO riboswitch. Nat Chem Biol. 2014;10:787–92. doi: 10.1038/nchembio.1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ren A, Patel DJ. c-di-AMP binds the ydaO riboswitch in two pseudo-symmetry-related pockets. Nat Chem Biol. 2014;10:780–6. doi: 10.1038/nchembio.1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moody MJ, Young RA, Jones SE, Elliot MA. Comparative analysis of non-coding RNAs in the antibiotic-producing Streptomyces bacteria. BMC Genomics. 2013;14:558. doi: 10.1186/1471-2164-14-558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 27.Hardisson C, Manzanal M-B, Salas J-A, Suárez J-E. Fine structure, physiology and biochemistry of arthrospore germination in Streptomyces antibioticus. J Gen Microbiol. 1978;105:203–14. doi: 10.1099/00221287-105-2-203 [DOI] [PubMed] [Google Scholar]
- 28.Hirsch CF, Ensign JC. Nutritionally defined conditions for germination of Streptomyces viridochromogenes spores. J Bacteriol. 1976;126:13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hollands K, Proshkin S, Sklyarova S, Epshtein V, Mironov A, Nudler E, Groisman EA. Riboswitch control of Rho-dependent transcription termination. Proc Natl Acad Sci U S A. 2012;109:5376–81. doi: 10.1073/pnas.1112211109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Unniraman S, Prakash R, Nagaraja V. Alternate paradigm for intrinsic transcription termination in eubacteria. J Biol Chem. 2001;276:41850–5. doi: 10.1074/jbc.M106252200 [DOI] [PubMed] [Google Scholar]
- 31.Pulido D, Jiménez A. Optimization of gene expression in Streptomyces lividans by a transcription terminator. Nucleic Acids Res. 1987;15:4227–40. doi: 10.1093/nar/15.10.4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ingham CJ, Hunter IS, Smith MCM. Rho-independent terminators without 3′ poly-U tails from the early region of actinophage øC31. Nucleic Acids Res. 1995;23:370–6. doi: 10.1093/nar/23.3.370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Opdyke JA, Fozo EM, Hemm MR, Storz G. RNase III participates in GadY-dependent cleavage of the gadX-gadW mRNA. J Mol Biol. 2011;406:29–43. doi: 10.1016/j.jmb.2010.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Opdyke JA, Kang J-G, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J Bacteriol. 2004;186:6698–705. doi: 10.1128/JB.186.20.6698-6705.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stork M, Di Lorenzo M, Welch TJ, Crosa JH. Transcription termination within the iron transport-biosynthesis operon of Vibrio anguillarum requires an antisense RNA. J Bacteriol. 2007;189:3479–88. doi: 10.1128/JB.00619-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al.. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. doi: 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bernhart SH, Hofacker IL, Will S, Gruber AR, Stadler PF. RNAalifold: Improved consensus structure prediction for RNA alignments. BMC Bioinformatics. 2008;9:474. doi: 10.1186/1471-2105-9-474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitra A, Kesarwani AK, Pal D, Nagaraja V. WebGeSTer DB—a transcription terminator database. Nucleic Acids Res. 2010;39:D129–35. doi: 10.1093/nar/gkq971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stuttard C. Temperate phages of Streptomyces venezuelae: Lysogeny and host specificity shown by phages SV1 and SV2. J Gen Microbiol. 1982;128:115–21. doi: 10.1099/00221287-128-1-115 [DOI] [Google Scholar]
- 41.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces genetics. Norwich (England): The John Innes Foundation. 2000. [Google Scholar]
- 42.Sambrook J, Russell DW. Molecular cloning - A laboratory manual. Cold Spring Harbor: (NY: ): Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 43.Spangler C, Böhm A, Jenal U, Seifert R, Kaever V. A liquid chromatography-coupled tandem mass spectrometry method for quantitation of cyclic di-guanosine monophosphate. J Microbiol Methods. 2010;81:226–31. doi: 10.1016/j.mimet.2010.03.020 [DOI] [PubMed] [Google Scholar]
- 44.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci U S A. 2003;100:1541–6. doi: 10.1073/pnas.0337542100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horton RM. PCR-mediated recombination and mutagenesis. Mol Biotechnol. 1995;3:93–99. doi: 10.1007/BF02789105 [DOI] [PubMed] [Google Scholar]
- 46.Pall GS, Hamilton AJ. Improved northern blot method for enhanced detection of small RNA. Nat Protoc. 2008;3:1077–84. doi: 10.1038/nprot.2008.67 [DOI] [PubMed] [Google Scholar]
- 47.Peirson SN, Butler JN, Foster RG. Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res. 2003;31:e73. doi: 10.1093/nar/gng073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Regulski EE, Breaker RR. In-line probing analysis of riboswitches. Methods Mol Biol. 2008;419:53–67. doi: 10.1007/978-1-59745-033-1_4 [DOI] [PubMed] [Google Scholar]
- 49.Abràmoff MD, Magalhães PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.